Abstract
In Escherichia coli, 12 distinct RNA structures within the transcripts encoding ribosomal proteins interact with specific ribosomal proteins to allow autogenous regulation of expression from large multi-gene operons, thus coordinating ribosomal protein biosynthesis across multiple operons. However, these RNA structures are typically not represented in the RNA Families Database or annotated in genomic sequences databases, and their phylogenetic distribution is largely unknown. To investigate the extent to which these RNA structures are conserved across eubacterial phyla, we created multiple sequence alignments representing 10 of these messenger RNA (mRNA) structures in E. coli. We find that while three RNA structures are widely distributed across many phyla of bacteria, seven of the RNAs are narrowly distributed to a few orders of Gammaproteobacteria. To experimentally validate our computational predictions, we biochemically confirmed dual L1-binding sites identified in many Firmicute species. This work reveals that RNA-based regulation of ribosomal protein biosynthesis is used in nearly all eubacterial phyla, but the specific RNA structures that regulate ribosomal protein biosynthesis in E. coli are narrowly distributed. These results highlight the limits of our knowledge regarding ribosomal protein biosynthesis regulation outside of E. coli, and the potential for alternative RNA structures responsible for regulating ribosomal proteins in other eubacteria.
INTRODUCTION
In Escherichia coli, ribosomes and their associated cofactors comprise between 25 and 50% of cellular mass in actively growing cells (1), and the synthesis of ribosomal RNA and proteins is tightly coordinated to maintain stoichiometric levels of each component (2). Coordinating this synthesis is a complex task that is regulated at multiple levels (3). More than half of the ribosomal proteins (r-proteins) in E. coli are controlled by 12 distinct RNA autogenous regulatory elements (Figure 1) that occur within the mRNA transcripts, most frequently in the 5′-untranslated regions (UTRs) or leader sequences, of operons encoding r-proteins. Each regulatory element consists of a structured region of mRNA transcript that interacts with a specific r-protein to inhibit expression of an entire operon encoding multiple r-proteins (4). In many cases, the mRNA structure responsible for regulation is a mimic of the ribosomal RNA (rRNA)-binding site for the same protein (5). A variety of different mechanisms are used to inhibit gene expression including transcription termination (6), translation inhibition through ribosome-binding site occlusion (7) and ribosome entrapment (8,9). These regulatory RNAs provide direct feedback between the levels of ribosomal RNAs and the levels of r-proteins.
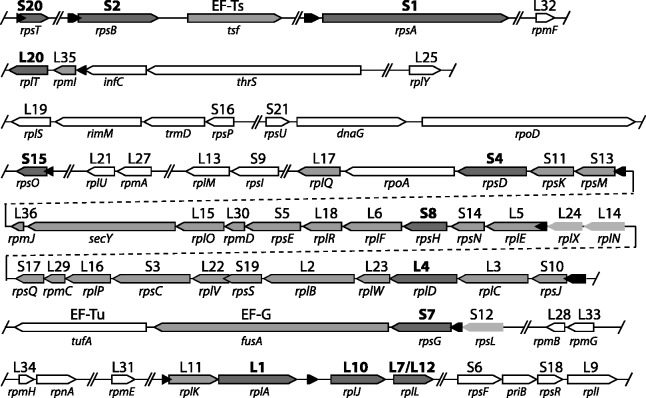
Figure 1.
Ribosomal protein gene organization in E. coli: Gene names are given below each arrow, and protein names for ribosomal proteins (r-proteins) are given above each arrow. Black arrows represent RNA autogenous regulatory structures. The two overlapping (due to pseudoknot) RNA structures interacting with L20 are represented by single black arrow. Gray arrows represent genes that are autogenously regulated; dark gray arrows (bolded protein names) encode proteins responsible for regulation, light gray arrows (no outline) are genes with reported retroregulation. Double slashes indicate breaks in the genomic sequence presented that correspond to long intervals between genes. White arrows represent genes with no known autogenous regulation. Gene organization is derived from the E. coli K12 substr. MG1655 genome [Refseq: NC_000913.2].
First identified in the late 1970s, r-protein autogenous regulators are some of the oldest known examples of effector-driven RNA-based gene regulation in bacteria, predating the discovery of riboswitches (10), T-boxes (11) and small RNAs (sRNAs) (12). Owing to the importance of ribosomal protein biosynthesis, the mechanism and structure for many of these regulators is well understood (4,13). However, most of the r-protein regulators described in E. coli are not annotated in standard genomic databases (14), and alignments for only 2 of the 12 RNAs are available in the RNA Families Database (Rfam) [Rfam: RF00140, RF00114] (15). While phylogenetic studies have been performed for several of these regulatory RNAs (16–20), such treatments were not systematic and the knowledge gained is not preserved in the databases used for genomic annotation. As a result, these important RNA molecules risk being overlooked as genome annotation moves further toward automation and online repositories. In addition, the extensive experimental data derived from E. coli sequences have not been put in context with homologous sequences identified through comparative genomic studies.
This work uses Infernal 1.0, an RNA-specific search tool (21), to identify homologues of the E. coli r-protein autogenous regulatory RNAs. Multiple-sequence alignments representing each of the structured mRNAs responsible for gene regulation were either expanded from existing alignments, or created based on the E. coli sequence and experimentally determined secondary structures in the literature. From these alignments, we integrated decades of experimental data with the RNA consensus secondary structures and characterized the phylogenetic distribution for each r-protein regulatory RNA. Additionally, for the L1-interacting RNA, we identified both a phyla-specific change in genomic locus and dual binding sites in a large proportion of sequenced Firmicute genomes. To verify the dual binding sites, we experimentally validated L1 interactions with the pair of RNA structures originating from Geobacillus kaustophilus.
MATERIALS AND METHODS
Alignment construction and homology searches
For the S4 and S15 binding RNAs, the seed multiple sequence alignments were downloaded from the Rfam database (Rfam families RF00140 and RF00114, respectively) (15). These alignments were manually examined to ensure that sequences were compatible with available experimental data and any alternative or pseudoknotted structures were represented by the structural annotation. The initial alignments for the L1-, L10(L12)4-, and S2-binding RNAs derived from non-coding RNA discovery efforts (22–25). To generate multiple sequence alignments for the remaining RNAs, BLAST matches outside of the Escherichia genus (exclude taxid: 0561) to the E. coli sequence corresponding to a minimal binding element were collected and aligned. To this multiple sequence alignment, experimentally determined secondary structure information was manually added, and the alignment was adjusted as necessary.
Once initial alignments were obtained, Infernal 1.0 (cmbuild) was used to create co-variance models corresponding to the RNA. These models were calibrated (cmcalibrate) and additional homologues identified for each RNA (cmsearch) (21). Both global and local searches were performed against a custom sequence database that contains genomic regions proximal to ribosomal proteins from all complete bacterial genomes in refseq46 (14). This database is ∼57 MB, which is an ∼100-fold reduction from the entire refseq46-microbial database. The use of this database significantly increased the speed and sensitivity of our search process. The complete database (refseq46-microbial) was used for a single search with the final alignments, and no additional homologues were identified. Prospective homologues were screened based on the appropriateness of the genomic context using a custom visualization tool, GenomeChart (26). Homologues were subsequently screened for their fit to the existing alignment and the consistency with experimental data. When necessary, alignments were manually adjusted as sequences were added, especially in cases with variable length helices. Searches that produced many potential sequences inconsistent with experimental data are discussed in the text (e.g. L4, S8 and S4 interacting autogenous regulatory RNAs). The search process was typically repeated two to three times to expand the sequence diversity present in the alignment. In cases with pseudoknotted or alternative structures, searches were conducted with each structure iteratively. The curated final alignments produced are available in Supplementary Datasets. Percentages of bacteria in each phyla containing each RNA were calculated based on the number of completed genomes within refseq46 and the alignments produced by this work. Consensus secondary structure diagrams were created from the alignments using GSC-weighting in R2R (27).
L1 RNA-protein binding assays
The gene encoding ribosomal protein L1 (rplA) was polymerase chain reaction (PCR) amplified from G. kaustophilus genomic DNA (28) using the following primers 5′-ggaattccatatgccgaaaagaggaaagaaatac-3′ and 5′-cgggatccttattgtgcaacagcaaccgtg-3′ and expressed in E. coli strain BL21 using T7 overexpression from pET-HT (29). The protein was purified from inclusion bodies using denaturing ion-exchange chromatography (30). The 37-nucleotide mRNA fragment preceding rplK and the 41-nucleotide mRNA fragment between rplK and rplA (including an appended T7 promoter) were purchased as synthetic DNA oligos and transcribed in vitro with T7 RNA polymerase and 5′-labeled with [α-32P] ATP. Mutants of these mRNA fragments (containing two G–A substitutions) were prepared in the same way. Filter-binding assays were performed similarly to established protocols (31). A fixed amount of RNA (500 cpm, ∼1 nM) was incubated with L1 protein in serial dilutions (0–500 nM) in a total volume of 50 μl for 15 min at 42°C (50 mM Tris–HCl pH 7.6 at 25°C, 20 mM MgCl2, 500 mM KCl, 1 mM beta-mercaptoethanol, 0.04% bovine serum albumin). The RNA–protein mixture was cooled to room temperature and the RNA–protein complexes were captured by vacuum suction through a nitrocellulose membrane (Optitran BA S-85 reinforced nitrocellulose, Whatman). RNA not interacting with L1 was captured by a positively charged membrane (N+ hybond, GE Healthcare). Membranes were washed once with 50 μl filter-binding buffer, and the radioactivity was quantified using a GE Healthcare STORM phosphoimager and ImageQuant. The fraction bound reflects the fraction of counts on the nitrocellulose membrane divided by the total counts on both membranes.
L1 5′-RACE
Bacillus subtilis total RNA was extracted from a log phase culture and 5′-RLM-RACE performed using the Invitrogen GeneRacer protocol with a homemade RNA-linker (32). First strand synthesis was conducted with gene-specific primers (rplK, 5′-AGCAATACTGCAGCAGGTGGAGTT, rplA 5′-TGCGAAAACGAGAACGCGCTGAGT). These reactions were used as template for PCR with an oligonucleotide corresponding to the 5′-linker (5′-GACTGGAGCACGAGGACACTGA) and a second set of gene-specific primers (rplK, 5′- CCAACTGGTGGTGCTGGGTTAGC; rplA, 5′- CGGTCTACAAGCTTAGCAGCTTCA). PCR products were cloned using a TOPO-cloning kit (Invitrogen) and the inserts sequenced to identify the 5′-end of the transcripts.
RESULTS
Overview of distributions
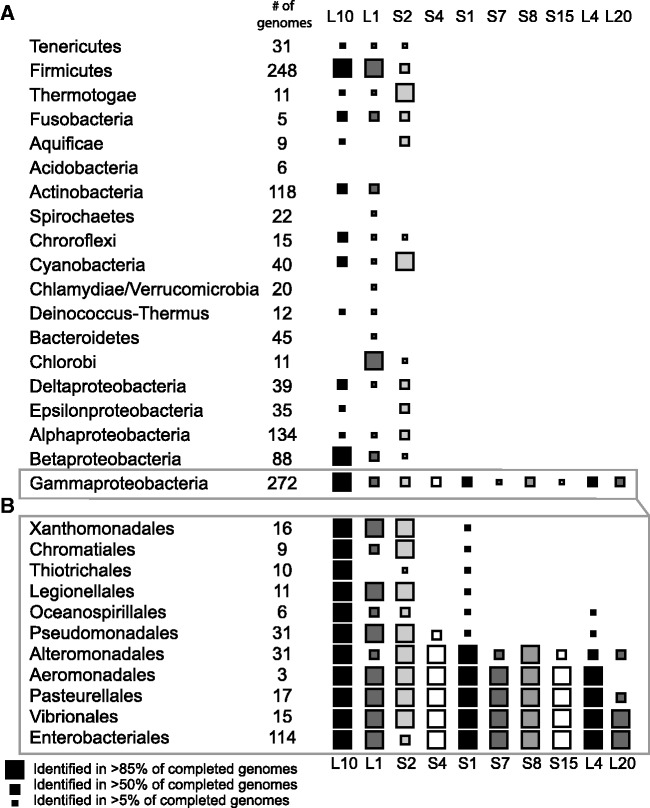
We compiled alignments for 10 of the 12 RNA structures controlling ribosomal protein biosynthesis in E. coli. Of the regulatory RNAs examined, three are widely distributed over many eubacterial phyla, and seven are narrowly distributed to a few orders of Gammaproteobacteria (Figure 2). The three widely distributed RNAs interact with ribosomal proteins L1, L10(L12)4 and S2, and each was identified in species from many bacterial phyla. All bacterial phyla, with the exception of Acidobacteria, harbor at least one of the RNAs. This sets a precedent for RNA-based regulation of ribosomal proteins in a wide range of bacterial diversity. The scattered distribution of the RNAs, and the low frequency of identification of some regulatory RNAs may point toward horizontal transfer, multiple inventions or a lack of sensitivity in our homology search methodologies.
Figure 2.
Phylogenetic distribution of E. coli r-protein regulatory RNAs. (A) Distribution of ribosomal protein autogenous regulatory RNAs in bacterial phyla. (B) Distribution of regulatory RNAs in orders of Gammaproteobacteria.
The seven narrowly distributed r-protein regulatory RNAs (interacting with S4, S1, S7, S8, S15, L20 and L4 proteins) were identified only in Gammaproteobacterial species, and in many cases, only within a subset of the Gammaproteobacterial orders (Figure 2). The S1-interacting RNA shows the widest apparent distribution appearing in species from each order. However, the remaining regulatory RNAs are present only in the Enterobacteriales, Vibrionales, Pasteurellales, Aeromonadales and Alteromonadales orders of Gammaproteobacteria. This pattern of conservation is consistent with vertical inheritance within Gammaproteobacteria (33). In addition, the loss of several RNA elements, including L1 and S2, is apparent in many genera of obligate intracellular species within Enterobacteria, such as Wiggesworthia, Blochmania and Buchnera. While Buchnera species typically retain the S1- and L4-binding RNAs, most enterobacterial endosymbionts appear to have lost these control mechanisms during their genome reduction process, although they typically retain the ribosomal proteins implicated in regulation (34). While the general gene order for most ribosomal proteins is typically conserved (35), there are significant changes to the operon structures of the spc, alpha and S10 operons (regulated in E. coli by S8, S4 and L4 respectively, see Figure 1). In B. subtilis, the majority of genes within these operons are co-transcribed as a single transcript (36), thus possibly removing the need to regulate these operons individually.
L20-interacting and S20-interacting RNAs
There are two RNAs previously described in E. coli for which we did not create alignments: a second L20 interaction site, and the reported S20-interacting mRNA. While the mRNA sequence preceding rpsT (encoding S20) has reported autogenous regulatory activity and apparent similarity to the ribosomal RNA, no secondary structure is reported for it in the literature (37,38). Given its small size, without a secondary structure it is impossible to create a convincing starting alignment for RNA homology searches that rely heavily on RNA secondary structure. This region is highly conserved in most Enterobacteria, but BLAST searches detect no similarity outside of Enterobacteriales (NCBI TAXID 91347).
A second L20-binding RNA contains a long-range pseudoknot between bases within the infC and rpmF coding regions. This large insertion overlaps significantly with the coding region of infC, making convincing identification of the RNA difficult with the tools used here. A previous phylogenetic study identified this RNA in Enterobacteriales, Vibrionales, Pasteurellales, Alteromonadales, Legionalles and Pseudomonadales (19).
L1-interacting RNA
L1-interacting mRNAs have been reported in both Enterobacteria (E. coli) (39) and in several archaea including Methanococcus vannielii (40,41). While the L1-binding sites in these organisms are similar, the genes regulated and the genomic loci of these sites are distinct in the two organisms. In E. coli, rplK and rplA are co-localized in the genome, and their translation is strongly coupled (Figure 1) (42). The L1-interacting structure precedes rplK, and L1 binding inhibits translation of both L1 and L11. In M. vannielii, rplK is not co-localized with rplA, instead rplA is co-transcribed with both rplJ and rplL, and the RNA structure directly precedes rplA. Consistent with the observed similarity between the binding sites, the E. coli L1 is able to regulate the M. vannielii production of L1 (40).
Our study identified L1-binding RNAs in the widest variety of different bacterial phyla. However, in many cases, only a few examples were identified in each phyla (Figure 2). Due to its small size (Figures 3 and 4) and minimal sequence conservation, the L1-binding site is somewhat difficult to track with computational tools, and it is likely that there are many individual occurrences that were not uncovered by our searches. The consensus figure produced from our alignment (Figure 4) for the L1-binding RNA is in good agreement with previous structural and biochemical studies. The consensus structure contains a 6–12-base-pair stem with an internal bulge that contains a potential non-canonical G–A pair followed by an unpaired adenosine. In structural studies, these nucleotides, and those in the base pairs immediately flanking the bulge, form a complex network of hydrogen bonds that is critical for protein binding (43).
Figure 3.
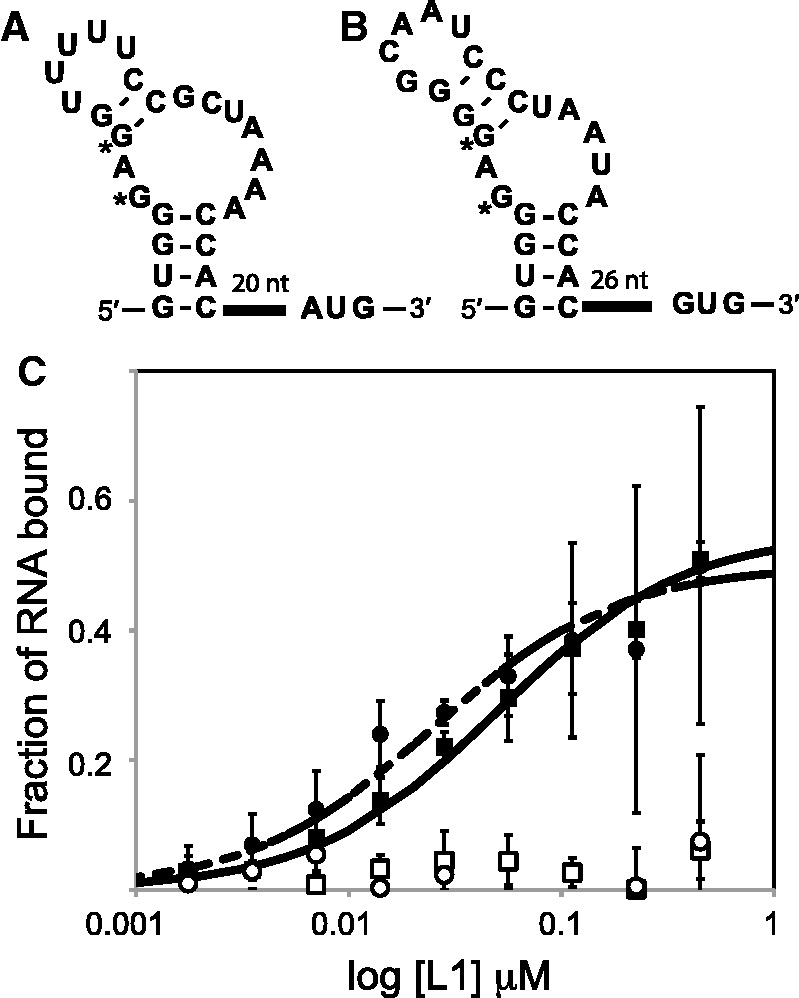
L1-binding RNA structures from G. kaustophilus. (A): G. kaustophilus RNA preceding rplK. (B): G. kaustophilus RNA preceding rplA. The guanosines marked by * were mutated to adenosine to create negative control RNAs. (C) Interaction between the RNAs and L1 protein as measured by filter-binding assays. RNAs pictured in parts A and B are represented by filled squares and circles, respectively. Their mutants are represented by open symbols. Error bars represent the standard error over three replicates. The maximum percentage bound was ∼50% for both RNAs, and the curves drawn correspond to dissociation constants of 50 and 25 nM. These values agree well with previously determined values for the L1 RNA–protein regulatory interaction (31).
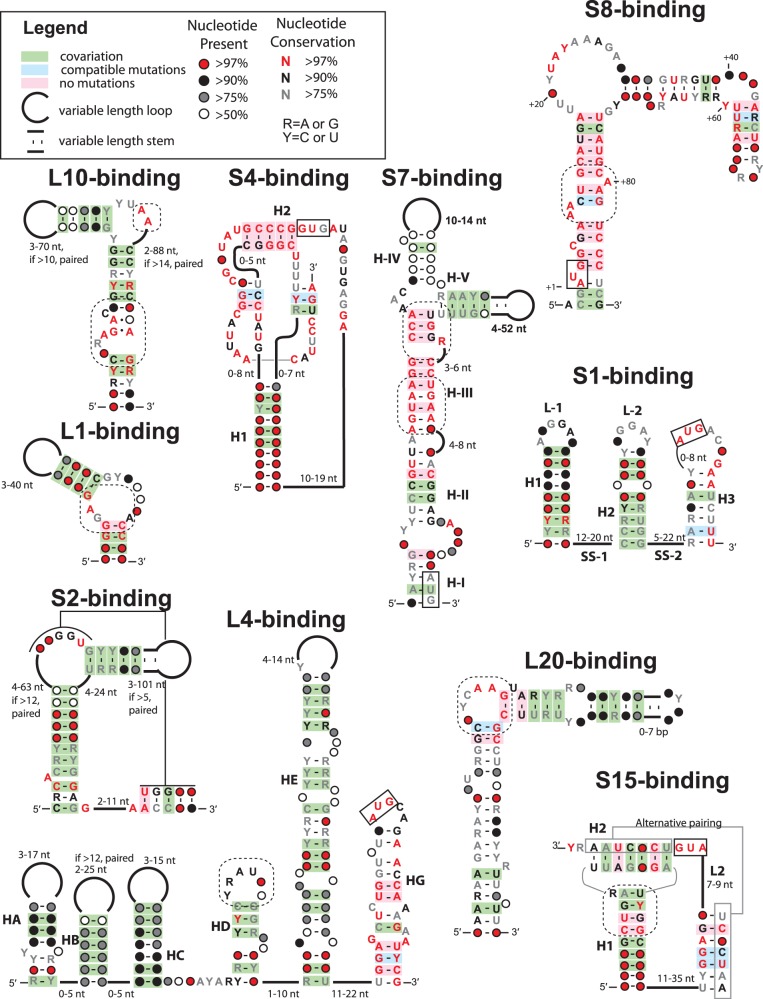
Figure 4.
Consensus sequence and secondary structures for E. coli r-protein regulatory RNAs. The alignments used to produce these figures are available as a Supplementary Dataset (Alignments.zip). Diagrams indicate the conserved secondary structure and sequence of the RNA structures. Base pairs supported by co-variation or compatible mutations are indicated by green or blue shading only if Watson–Crick pairing occurs in >95% of the sequences. Start codons (AUG or GUG) that occur within an RNA structure are boxed with solid lines, putative protein binding sites are indicated by dashed lines and helix numbering schemes are consistent with pre-existing literature for each RNA.
Despite its widespread distribution, the genomic locus of the L1-interacting RNA is not consistent within eubacteria. In Cyanobacteria, Actinobacteria and Chloroflexi, the L1-interacting RNA appears directly preceding rplA (typically between rplA and rplK), and in Proteobacteria, Spirochaetes, Thermotoga and Tenericutes, it appears preceding rplK. Surprisingly, we identified two binding sites in >40% of completed Firmicute genomes, one preceding rplK, and one preceding rplA.
To ensure that the dual binding sites were not false-positives resulting from the small size of the L1-binding site and the sensitivity of our searches, we experimentally validated the interaction between ribosomal protein L1 and the two RNA structures originating from G. kaustophilus using filter-binding assays (Figure 3A and B). These studies show that each of the RNA structures has nM affinity for the protein (Kd ∼25–50 nM) that is abolished by point mutations to the consensus-binding site (Figure 3C) (31). To verify that rplK and rplA are co-transcribed in Firmicutes (as they are in E. coli) we performed 5′-RACE on total RNA extracted from B. subtilis. We found that all the products isolated from gene-specific primer within rplA (encoding L1) contain the entirety of rplK, indicating that the two genes are co-transcribed (Supplementary Figure S1). In addition, ∼40% of the sequences isolated from gene specific primers within rplK extend to include the second RNA element that precedes rplK. The combination of these findings suggests that both sites are biologically relevant and play a role in regulating these genes in Firmicutes.
L10(L12)4-interacting RNA
The L10(L12)4-interacting autogenous regulatory RNA is also widely distributed, and the RNA has been described in both Proteobacteria and Firmicutes (20). Our study expands the phylogenetic spread of this RNA considerably, identifying it in more than half of sequenced Fusobacteria, Actinobacteria, Cyanobacteria and Chloroflexi. In addition, examples were identified in Tenericutes, Thermotogae, Aquificae and Deinococcus-Thermus. The RNA is responsible for regulating rplJ and rplL in response to the L10(L12)4 complex (44,45) (Figure 1). The minimal binding site identified using phylogenetic analyses and in vitro assays is characterized by a kink-turn motif followed by a bulged cytosine in the non-canonical stem (20). This motif is a direct mimic of the rRNA binding site for L10 (20). In the alignment produced by this study, the kink-turn is always present, and the bulged cytosine is present in >90% of examples (Figure 4). However, examples from Epsilonproteobacteira, Aquificae and Cyanobacteria are often missing this bulged cytosine. The stem is topped with a hairpin that contains an internal loop with two unpaired adenosines. These adenosines are present in all sequences examined, even in those where the bulge is extended into a second hairpin of varying length (Figure 4). Previous experimental work has shown that the two bulged adenosines and the kink-turn are both critical for L10(L12)4 binding (20).
The mechanism that the L10(L12)4-binding RNA leader from E. coli uses to control gene expression is dependent on complex folding patterns with other portions of the mRNA leader (46–49) and appears to act post-transcriptionally, but before translation (50). However, based on the current Rfam alignment (which includes a terminator), this RNA is followed by a rho-independent intrinsic terminator in Firmicutes, Cyanobacteria and Fusobacteria (25,51), suggesting that there may be different regulatory mechanisms in different bacterial species, similarly to many riboswitch aptamers, which may have distinct expression platforms depending on the species of bacteria (52).
S2-interacting RNA
Despite its wide distribution (Figure 2), the S2-interacting regulatory RNA in E. coli was discovered relatively recently (53), and no mechanistic studies have been performed to date. The RNA appears to control the synthesis of both ribosomal protein S2 and EF-Ts (Figure 1), and the RNA was independently discovered through two additional avenues. It was first described in a study of the E. coli transcriptome as sRNA t44 [Rfam: RF00127] (54), and subsequently was identified in a de novo search for non-coding RNAs in marine Alphaproteobacteria using comparative genomics (22). A good alignment of the S2-binding RNA is already available (22); this work re-evaluated the existing alignment and identified the RNA in a wider range of organisms by incorporating data from more recent sequencing projects.
Although the native transcript in E. coli is significantly longer than the conserved S2-binding site, deletion experiments have shown that only the conserved portion of the RNA represented by the alignment is necessary for regulation (53). In addition, deletion of the GGU internal loop (predicted to form a pseudoknot by this alignment) is sufficient to abolish activity (Figure 4). Subsequent experiments have also shown that instances of this RNA from other Gammaproteobacteria, including Pseudomonas, function as regulatory elements in response to the E. coli S2 protein (17). The S2 interacting regulatory RNA also stands in contrast to the other widely distributed RNA autogenous regulatory elements in that it is not an obvious mimic of the ribosomal RNA, and its binding partner is not a primary RNA binding protein (55).
L4-interacting RNA
The S10 operon in E. coli consists of 11 genes (rpsJ, rplC, rplD, rplW, rplB, rpsS, rplV, rpsC, rplP, rpmC, rpsQ) and is autogenously controlled by L4, encoded by rplD (Figure 1). A structured portion of mRNA preceding all of the coding genes allows both transcription termination (56) and translation inhibition (57) in response to L4. The L4-interacting RNA is perhaps the most comprehensively studied of the r-protein regulatory RNAs. Genetic and in vitro assays have shown that translation inhibition and transcription termination are conferred by distinct, although slightly overlapping, portions of the mRNA and together can account for a nearly 30-fold decrease in gene expression in the presence of excess L4 (58).
The L4-interacting RNA from E. coli consists of five hairpins, one of which overlaps the start codon of S10 (59) (Figure 4). The overall structure is largely conserved, and homologous sequences from Vibrio cholerae and Haemophilus influenzae have been experimentally demonstrated to regulate gene expression in response to E. coli L4 (16). The alignment of L4-interacting RNAs in this work indicates that helices HA-HC are highly variable in both sequence and in length. Without the annotation of transcription start sites, in many organisms, it is challenging to identify all of the hairpins based solely on genomic sequences. This is consistent with experimental results indicated that these helices are not necessary for regulation (60).
In contrast, helices HD-HG, which are required for transcription termination (61), show a much greater degree of sequence conservation allowing greater confidence in their alignment. In HD, the most conserved area encompasses the loop and the bases proximal to the loop, and this region has been implicated in L4 binding (62). While HE is present in every sequence in the alignment, there is a significant amount of sequence variation and many non-conserved single-nucleotide bulges. Portions of HE are required for transcription termination, and have been implicated in the NusA-dependent RNA polymerase pausing that precedes termination (61,63). While the string of uridines necessary for rho-independent transcription termination is not rigorously conserved in position, there are strings of uridines present in the lower part of HE or the linker between predicted HE and HG in all the sequences in the alignment. Helix HG is the most highly conserved region of the RNA, likely due in part to overlap with the S10 coding sequence (Figure 4). While we identified examples of the full-length RNA in the Pseudomonadas family Moraxellaceae, we also identified sequences with similarity to HG alone in several species of Xanthomonas and the Pseudomonas family Pseudomonadaceae (16). As these regions have not been demonstrated to control gene expression in the absence of the upstream regions of the RNA, sequences from these species were omitted from the alignment.
The L4-interacting RNA translation inhibition mechanism has received less attention than its transcription termination mechanism. Deletion of HA-HD or HE is known to have little effect on translation regulation, while point mutations in HE have significant effects on translational efficiency (58). The proposed mechanism involves formation of an alternative stem that incorporates bases within both HG and HE (59). However, this stem is not exceedingly stable in the E. coli sequence and there is no evidence from the alignment for a conserved stem in this area.
S8-interacting RNA
The spc operon consists of twelve genes (rplN, rplX, rplE, rpsN, rpsH, rplF, rplR, rpsE, rpmD, rplO, secY, rpmJ) and in E. coli is controlled by the binding of ribosomal protein S8 to an mRNA structure that overlaps the start of rplE (64,65) (Figure 1). The binding of S8 to this region inhibits translation of L5 and the proteins encoded by the subsequent genes (rpsN-rplO) that are translationally coupled to rplE (66,67). Regulation of rplN and rplX, which occur 5′ of the S8 binding site, has also been reported (68). However, the mechanism of this retroregulation is not known.
Like several r-protein autogenous regulatory RNAs, the S8 binding site bears a strong resemblance to the S8 binding site on the 16S rRNA (64). Consistent with previous mutagenesis results (64,69) and three-dimensional structure information (70), the alignment indicates that the distal portion of the stem can be variable despite the large overlap between the RNA structure and the L5 coding sequence. In contrast to variability of the distal stem, the core S8 binding site within the internal bulge is highly conserved with few or no mutations (Figure 4). Bases that comprise the core S8-binding site and directly contact S8 in a crystal structure of the mRNA–protein complex (70) are well-conserved in our alignment. The binding site is directly adjacent to the conserved pair of bulged adenosines (A8 and A9 in our diagram where numbering starts at the AUG) and extends through the conserved G12-C79 base pair. The bulged C5 and U2 have been implicated in reducing the S8 affinity for the mRNA over the rRNA (69). These bases did not contact the S8 protein in a crystal structure of the mRNA–S8 complex, and displayed high beta factors suggesting local flexibility (70). Our alignment shows that these bases are well conserved. However, it is not clear whether this conservation is due to functional constraints on the RNA or overlap with the L5-coding region.
The nearly 100% overlap of the S8 regulatory RNA with the L5 coding sequence introduces challenges for the computational homology searches. Purely sequence-based searches (e.g. BLAST) return spurious hits that lack a recognizable hairpin structure. The examples included in our alignment were stringently selected to preserve not only a highly conserved binding site, but also extended secondary structure in the variable region. In addition, there are several sequences originating from various species of Blochmania and Baumania that display a weak hairpin structure, but also contain mutations in the conserved binding site that are not likely to be compatible with binding. These sequences were also excluded from the alignment.
S7-interacting RNA
The str operon in E. coli consists of four genes, rpsL, rpsG, fusA and tufA, that encode for ribosomal proteins S12, S7 and elongation factors EF-G and EF-tu (Figure 1). The translation of S12 and S7 is coupled, with independent S7 translation accounting for only 10–20% of protein production (71). S7 binds to the mRNA region between rpsL and rpsG and inhibits only the coupled translation of S12 and S7 (72). Retroregulation of S12 has been observed (73). The exact mechanism of action is unknown, but the mRNA structure formed in the absence of S7 binding is proposed to facilitate the coupled translation of S12 and S7 (74). On S7 binding, the RNA structure and the translational coupling are disrupted.
Based on in vitro nuclease protection assays, there are two proposed binding sites for S7. Both of these sites resemble S7-binding sites on the rRNA (73,75). The first is composed of the 4–8 nucleotide bulge in H-III (Figure 4), and bases within the lower portion of H-III (5′-UGUAA and 5′-UGAAU in E. coli, 5′-AGUAA and 5′-UGAAN in our consensus diagram). However, the bulge is poorly conserved in the alignment compiled here. The second proposed binding site consists of the 5′-CCA and 5′-UUGGA sequences at the three-stem junction in the E. coli S7-binding mRNA (5′-CCA and 5′-UUGGR in our consensus diagram) (Figure 4). Bases implicated in S7-binding by UV-crosslinking (the UU from E. coli 5′-UUGGA) (74) are well-conserved. However, two of the three helices that form the three-stem junction (H-IV and H-V) are absent from the RNA in many Shewanella species, and there is a great deal of variability in both helices outside the regions proposed for S7-binding. In particular, H-IV is often not thermodynamically stable.
It has been reported that nearly one-third of all bacterial genomes contain an extended distance between rpsL and rpsG, and a sequence with the same genomic location and some similarity to the 16S ribosomal RNA has been described in Cyanobacteria (76). Our investigations of other bacterial phyla reveal that while extended distances between S12 and S7 exist in many species, the RNA structure observed in E. coli is not obviously conserved in these species.
S1-interacting RNA
The S1-binding RNA is somewhat unique because ribosomal protein S1 interactions with the ribosome are still a subject of debate (77). S1 is known to interact non-specifically with many RNAs and is required for translation of many transcripts (78), including its own (18). The mRNA regulatory structure is found in the 5′-UTR of rpsA (Figure 1) and regulates S1 synthesis at the translational level (79). Ternary complexes composed of the RNA, S1 protein and 30S particles have been reconstituted in vitro (18).
Our alignment is consistent with previous phylogenetic analysis (18), indicating that the S1-interacting regulatory RNA is narrowly distributed (Figure 2). The conserved GG sequences in loops L1 and L2 are important for regulation (Figure 4) (18). The sequences of H2 and H3 are somewhat conserved, but the linker between them (single-stranded region 2, ss-2) is variable in sequence and in length, but it is nearly always AU rich. These findings are consistent with mutagenesis studies showing that the lower portion of helix H2 is important for regulation, but that the sequence and length of the second single-stranded region (SS-2) is not important for regulation (18). Toe-printing assays show that the S1 protein largely interacts with the AU-rich single stranded regions that occur between the helices and in the loop region of helix H3.
There are no additional experimentally verified examples of S1-binding autogenous regulatory motifs in other organisms. However, a putative RNA structure identified through comparative genomics precedes rpsA in many species of Cyanobacteria (23). This RNA bears no resemblance to the RNA in E. coli, and these RNAs are not included in our alignment.
S4-interacting RNA
The alpha operon in E. coli consists of five genes: rpsM, rpsK, rpsD, rpoA and rplQ, encoding ribosomal proteins S13, S11, S4, the α component of RNA polymerase and ribosomal protein L17, respectively (Figure 1). Ribosomal protein S4 regulates the synthesis of rpsM, rpsK, rpsD and rplQ as a translational repressor, but does not affect expression of the intervening rpoA (80,81). The RNA structure responsible for this regulation occurs in the 5′-UTR, partially overlaps rpsM and is a complex double pseudoknot (Figure 4). The start codon and a significant stretch of the rpsM protein coding sequence are contained within the structure. The mRNA and rRNA structures bound by E. coli are different from one another (82), but the same domain of S4 protein appears to be responsible for binding both (83). The mechanism of action for this RNA involves the 30S particle and the RNA inhibits translation through ribosomal entrapment (9).
The consensus structure of the S4-interacting regulatory RNA produced by our alignment (Figure 4) is in good agreement with previous experimental studies. Helix H1 shows almost no sequence conservation, but does have extensive co-variation indicating the structure rather than the sequence is important (82). There is no co-variation apparent in the pseduoknotted structure, likely due to the overlap of these regions with the coding sequence. Mutagenesis studies have shown that compensatory mutations in H2 that maintain the stem do not restore S4-binding (82).
The existing Rfam seed alignment for this RNA [Rfam: RF00140] includes several examples that are not consistent with the available experimental evidence. These include examples from Bordetella pertussis, Bordetella bronchiseptica, Dechloromonas aromatica and Pseudomonas syringae. All of these sequences contain base changes inconsistent with one or more of the pseudoknots, and are missing two or more bases of the H2 stem, which is highly conserved and critical for S4 binding (82). After careful consideration, these sequences were removed and not included in the alignment. As a result of the large overlap between the RNA structure and the coding region of S4, purely sequenced-based search tools often return such spurious examples.
In many eubacteria, rpsD occurs in isolation in the genome, rather than as a portion of the alpha operon. As a result, studies to examine rpsD regulation in B. subtilus lead to the discovery of an mRNA structure that interacts with ribosomal protein S4 in this organism (84). However, the mRNA structure present in B. subtilis bears no obvious resemblance to the mRNA structure in E. coli.
S15-interacting RNA
The mRNA structure interacting with S15 only regulates rpsO, encoding S15 (Figure 1). The RNA structure partially overlaps the coding sequence for rpsO (85) and like the S4-interacting regulatory RNA, the S15-interacting RNA is pseudoknotted (86) and appears to act through a ribosomal entrapment mechanism (87). There are two possible structures (88), one of which is stabilized by the binding of S15 (Figure 4). The current Rfam alignment for this RNA [Rfam: RF00114] only reflects the alternative unbound structure for this RNA.
A great deal of mutagenesis as well as structural probing has been performed to elucidate the secondary and tertiary structure for the S15-interacting regulatory RNA (89–91), and our alignment is consistent with this data. Conserved structures corresponding to both possible conformations of the RNA are observed, although the pseudoknotted structure stabilized by S15 binding is better supported by the alignment with more co-varying positions and fewer non-conical pairs and insertions (Figure 4). The nucleotides in the lower portion of the H1 helix are variable, but base-pairing is largely maintained. Consistent with previous mutagenesis studies, the C-G/U•G base pairs in the upper portion of H1 and the purine between H1 and H2 are rigorously conserved (89) and likely play an important role in recognition. Deletion studies have shown that the L2 loop may be reduced to 7 nucleotides (including the AUG) (91), and our alignment shows that L2 is variable in length.
Two additional mRNA structures that interact with S15 to regulate rpsO in other organisms are known, one in Geobacillus stearothermophilus (92) and the other Thermus thermophilus (93). Neither of these RNAs bears any resemblance to the RNA structure in E. coli, and cross-species RNA–protein binding assays indicate that the S15 proteins in E. coli and G. stearothermophilus may have distinct determinants for binding their regulator RNAs (94).
L20-interacting RNA
There are two L20 binding sites in E. coli, and an alignment was generated for only one of these sites for reasons discussed above. L20 regulates the translation of itself and L35, and both the L20 regulatory sites are at the junction of infC and rpmI, encoding initiation factor IF3 and L35, respectively (95,96) (Figure 1). The two L20 binding sites are independent (7) and have similar in vitro affinity for L20 (19). We compiled an alignment for the L20 site encompassing the infC/rmpI intergenic region and some flanking sequence that overlaps the coding regions. While it is known that regulation occurs at the translational level, the exact mechanism of action is not currently known (7).
Like the binding sites for L1 and S8, the L20 binding site is proposed to act as a mimic of the ribosomal RNA (19). The bulge containing a rigorously conserved pair of adenosines (Figure 4) is similar to the L20 binding site on the 23S RNA (19). The pairing elements show little base-conservation, and the length of the helix is variable. All of these aspects are consistent with previous mutagenesis and truncation experiments (7,19).
Like S15, L20 interacts with a different mRNA structure in B. subtilis (97). In B. subtilis, the RNA structure occurs upstream of infC, and regulation occurs at the transcriptional level rather than the translational level. The RNA itself bears only a faint resemblance to the one described in E. coli, and thus is not included in our alignment. However, both RNAs are considered mimics of the ribosomal RNA and the C-terminal half of the E. coli L20 protein has the similar effects to L20 from B. subtilis during in vitro single-round transcription attenuation assays (97). These results indicate that the necessary binding determinates are replicated in each of the RNA structures despite their dissimilar structures.
DISCUSSION
Structured mRNA elements autogenously controlling ribosomal protein biosynthesis are common in E. coli, and their discovery and characterization represents some of the initial work performed on RNA-based gene regulation. Despite their importance for an essential process and the extensive mechanistic studies performed, the regulatory RNAs responsible for ribosomal protein regulation are not well annotated in genomic databases. Owing to the technological limitations associated with RNA annotation and homology searches (98), and the lack of a complete archive of known RNAs, it is often difficult to determine whether a putative RNA structure represents a new finding or is a homologue of one that has been extensively studied in the past.
This study examines important RNAs in the light of new genomic data, places the consensus sequences and structures in the context of experimental data collected in the past and documents them for modern databases. In addition to creating alignments for many autogenous regulatory RNAs controlling r-protein synthesis, we have made several corrections to existing alignments to ensure that they are consistent with experimental data. We also experimentally validated some of our most surprising findings by verifying the transcription of dual L1-binding sites in G. kaustophilus and the interaction of both RNAs with L1 protein.
The exceedingly narrow distribution for most of the RNAs is striking. The presence of individual RNA structures within a few orders of Gammaproteobacteria has been remarked on in the past (16,17). However, there is no clear explanation for the explosion of regulatory mechanisms in the more recently evolved branches of Gammaproteobacteria (33). We speculate that most other bacteria do not lack such mechanisms, but rather a large number of distinct RNA structures are yet to be identified. The presence of RNA-based regulation of ribosomal protein biosynthesis in the vast majority of bacterial phyla and the sporadic identification of distinct RNA control structures in other bacterial phyla (84,91,97) support this hypothesis. In addition, as new sequence data continue to fuel comparative genomic-based non-coding RNA discovery efforts, RNA structures associated with ribosomal proteins are frequently encountered. For example, structured RNAs have been reported preceding the coding region of S1 and between the coding regions of S7 and S12 in Cyanobacteria (23,76), and several structured RNAs associated with ribosomal protein genes have been reported in B. subtilis and other Firmicutes (24,25).
The growing diversity of distinct RNA regulators using various mechanisms to regulate ribosomal protein genes across different bacterial phyla when operon structure is largely preserved suggests that there is significant pressure to tightly regulate ribosomal protein synthesis. In addition, most r-proteins are RNA-binding proteins, potentially increasing the probability of developing RNA-based autogenous regulatory mechanisms. However, it also seems that many distinct RNAs may solve the same biological problem. Indeed, diversity rather than conservation is becoming the dominant theme in RNA-based regulation in bacteria. RNA-based regulation of processes such as methionine (99) and sugar metabolism (100) is extraordinarily diverse, and trans-acting RNAs such as sRNAs evolve rapidly (101). This study illustrates the limited extent of our knowledge regarding ribosomal protein regulation in bacteria and sets the stage to allow the discovery and characterization of diverse RNA structures and mechanisms of regulation for ribosomal proteins in other microorganisms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figure 1 and Supplementary Datasets.
FUNDING
Boston College; the Pharmaceutical Research and Manufacturers of America (PhRMA) Foundation and the Alfred P. Sloan Foundation. Funding for open access charge: Alfred P. Sloan Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Zasha Weinberg and Prof. Ming Hammond for the discussions that lead to this work and Prof. Ronald Breaker for the freedom to work on it. We also thank the Yale University Life Sciences High Performance Computing Center and the High Performance Computing Center at Boston College. Genomic DNA for G. kaustophilus was a kind gift from Hideo Takami at the Japan Agency of Marine-Earth Science and Technology, Microbial Genome Research Group.
REFERENCES
- 1.Hardy SJ, Kurland CG, Voynow P, Mora G. The ribosomal proteins of Escherichia coli. I. purification of the 30S ribsomal proteins. Biochemistry. 1969;8:2897–2905. doi: 10.1021/bi00835a031. [DOI] [PubMed] [Google Scholar]
- 2.Harvey RJ. Regulation of ribosomal protein synthesis in Escherichia coli. J. Bacteriol. 1970;101:574–583. doi: 10.1128/jb.101.2.574-583.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson DN, Nierhaus KH. The weird and wonderful world of bacterial ribosome regulation. Crit. Rev. Biochem. Mol. Biol. 2007;42:187–219. doi: 10.1080/10409230701360843. [DOI] [PubMed] [Google Scholar]
- 4.Zengel JM, Lindahl L. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Prog. Nucleic Acid Res. Mol. Biol. 1994;47:331–370. doi: 10.1016/s0079-6603(08)60256-1. [DOI] [PubMed] [Google Scholar]
- 5.Nomura M, Yates JL, Dean D, Post LE. Feedback regulation of ribosomal protein gene expression in Escherichia coli: structural homology of ribosomal RNA and ribosomal protein MRNA. Proc. Natl Acad. Sci. USA. 1980;77:7084–7088. doi: 10.1073/pnas.77.12.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zengel J, Lindahl L. Escherichia coli ribosomal protein L4 stimulates transcription termination at a specific site in the leader of the S10 operon independent of L4-mediated inhibition of translation. J. Mol. Biol. 1990;213:67–78. doi: 10.1016/S0022-2836(05)80122-6. [DOI] [PubMed] [Google Scholar]
- 7.Guillier M, Allemand F, Raibaud S, Dardel F, Springer M, Chairuttini C. Translational feedback regulation of the gene for L35 in Escherichia coli requires binding of ribosomal protein L20 to two sites in its leader mRNA: a possible case of ribosomal RNA-messenger RNA molecular mimicry. RNA. 2002;8:878–889. doi: 10.1017/s1355838202029084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marzi S, Myasnikov AG, Serganov A, Ehresmann C, Romby P, Yusupov M, Klaholz BP. Structured mRNAs regulate translation initiation by binding to the platform of the ribosome. Cell. 2007;130:1019–1031. doi: 10.1016/j.cell.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 9.Schlax PJ, Xavier KA, Gluick TC, Draper DE. Translational repression of the Escherichia coli alpha operon mRNA: importance of an mRNA conformational switch and a ternary entrapment complex. J. Biol. Chem. 2001;276:38494–38501. doi: 10.1074/jbc.M106934200. [DOI] [PubMed] [Google Scholar]
- 10.Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu. Rev. Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green NJ, Grundy FJ, Henkin TM. The T box mechanism: tRNA as a regulatory molecule. FEBS Lett. 2010;584:318–324. doi: 10.1016/j.febslet.2009.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aseev LV, Boni IV. Extraribosomal functions of bacterial ribosomal proteins. Mol. Biol. (Mosk) 2011;45:739–750. [PubMed] [Google Scholar]
- 14.Pruitt K, Tatusova T, Maglott D. NCBI reference sequence project: update and current status. Nucleic Acids Res. 2003;31:34. doi: 10.1093/nar/gkg111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allen T, Shen P, Samsel L, Liu R, Lindahl L, Zengel J. Phylogenetic analysis of L4-mediated autogenous control of the S10 ribosomal protein operon. J. Bacteriol. 1999;181:6124–6132. doi: 10.1128/jb.181.19.6124-6132.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aseev LV, Levandovskaya AA, Skaptsova NV, Boni IV. Conservation of regulatory elements controlling the expression of the rpsB-tsf operon in γ-proteobacteria. Mol. Biol. 2009;43:101–107. [PubMed] [Google Scholar]
- 18.Boni IV, Artamonova VS, Tzareva NV, Dreyfus M. Non-canonical mechanism for translational control in bacteria: synthesis of ribosomal protein S1. EMBO J. 2001;20:4222–4232. doi: 10.1093/emboj/20.15.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guillier M, Allemand F, Dardel F, Royer C, Springer M, Chiaruttini C. Double molecular mimicry in Escherichia coli: binding of ribosomal protein L20 to its two sites in mRNA is similar to its binding to 23S rRNA. Mol. Microbiol. 2005;56:1141–1456. doi: 10.1111/j.1365-2958.2005.04644.x. [DOI] [PubMed] [Google Scholar]
- 20.Iben J, Draper D. Specific interactions of the L10 (L12)4 ribosomal protein complex with mRNA, rRNA, and L11. Biochemistry. 2008;47:2721–2731. doi: 10.1021/bi701838y. [DOI] [PubMed] [Google Scholar]
- 21.Nawrocki E, Kolbe D, Eddy S. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer MM, Ames TD, Smith DP, Weinberg Z, Schwalbach MS, Giovannoni SJ, Breaker RR. Identification of candidate structured RNAs in the marine organism 'Candidatus Pelagibacter ubique'. BMC Genomics. 2009;10:268. doi: 10.1186/1471-2164-10-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, Wang JX, Lee ER, Block KF, Sudarsan N, et al. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 2007;35:4809–4819. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol. 2010;11:1–17. doi: 10.1186/gb-2010-11-3-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao Z, Barrick J, Weinberg Z, Neph S, Breaker R, Tompa M, Ruzzo WL. A computational pipeline for high- throughput discovery of cis-regulatory noncoding RNA in prokaryotes. PLoS Comput. Biol. 2007;3:e126. doi: 10.1371/journal.pcbi.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller C, Anthony J, Meyer MM, Marth G. Scribl: an HTML5 Canvas-based graphics library for visualizing complex genomic data over the web. Bioinformatics. 2012;29:381–383. doi: 10.1093/bioinformatics/bts677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weinberg Z, Breaker RR. R2R—software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinformatics. 2011;12:3. doi: 10.1186/1471-2105-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takami H, Nishi S, Lu J, Shimamura S, Takaki Y. Genomic characterization of thermophilic Geobacillus species isolated from the deepest sea mud of the Mariana Trench. Extremophiles. 2004;8:351–356. doi: 10.1007/s00792-004-0394-3. [DOI] [PubMed] [Google Scholar]
- 29.Block KF, Puerta-Fernandez E, Wallace JG, Breaker RR. Association of OLE RNA with bacterial membranes via an RNA-protein interaction. Mol. Microbiol. 2011;79:21–34. doi: 10.1111/j.1365-2958.2010.07439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Semrad K, Green R, Schroeder R. RNA chaperone activity of large ribosomal subunit proteins from Escherichia coli. RNA. 2004;10:1855–1860. doi: 10.1261/rna.7121704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohrer C, Mayer C, Neumair O, Grobner P, Piendl W. Interaction of ribosomal L1 proteins from mesophilic and thermophilic Archaea and Bacteria with specific L1-binding sites on 23S rRNA and mRNA. Eur. J. Biochem. 1998;256:97–105. doi: 10.1046/j.1432-1327.1998.2560097.x. [DOI] [PubMed] [Google Scholar]
- 32.Weinberg Z, Perreault J, Meyer MM, Breaker RR. Exceptional structured noncoding RNAs revealed by bacterial metagenome analysis. Nature. 2009;462:656–659. doi: 10.1038/nature08586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao B, Mohan R, Gupta RS. Phylogenomics and protein signatures elucidating the evolutionary relationships among the Gammaproteobacteria. Int. J. Syst. Evol. Microbiol. 2009;59:234–247. doi: 10.1099/ijs.0.002741-0. [DOI] [PubMed] [Google Scholar]
- 34.Gil R, Sabater-Munoz B, Latorre A, Silva FJ, Moya A. Extreme genome reduction in Buchnera spp.: toward the minimal genome needed for symbiotic life. Proc. Natl Acad. Sci. USA. 2002;99:4454–4458. doi: 10.1073/pnas.062067299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nikolaichik YA, Donachie WD. Conservation of gene order amongst cell wall and cell division genes in Eubacteria, and ribosomal genes in Eubacteria and Eukaryotic organelles. Genetica. 2000;108:1–7. doi: 10.1023/a:1004077806910. [DOI] [PubMed] [Google Scholar]
- 36.Li X, Lindahl L, Sha Y, Zengel JM. Analysis of the Bacillus subtilis S10 ribosomal protein gene cluster identifies two promoters that may be responsible for transcription of the entire 15-kilobase S10-spc-alpha cluster. J. Bacteriol. 1997;179:7046–7054. doi: 10.1128/jb.179.22.7046-7054.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parsons GD, Donly BC, Mackie GA. Mutations in the leader sequence and initiation codon of the gene for ribosomal protein S20 (rpsT) affect both translational efficiency and autoregulation. J. Bacteriol. 1988;170:2485–2992. doi: 10.1128/jb.170.6.2485-2492.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parsons GD, Mackie GA. Expression of the gene for ribosomal protein S20: effects of gene dosage. J. Bacteriol. 1983;154:152–160. doi: 10.1128/jb.154.1.152-160.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dean D, Nomura M. Feedback regulation of ribosomal protein gene expression in Escherichia coli. Proc. Natl Acad. Sci. USA. 1980;77:3590–3594. doi: 10.1073/pnas.77.6.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanner M, Mayer C, Kohrer C, Golderer G, Grobner P, Piendl W. Autogenous translational regulation of the ribosomal MvaL1 operon in the archaebacterium Methanococcus vannielii. J. Bacteriol. 1994;176:409–418. doi: 10.1128/jb.176.2.409-418.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kraft A, Lutz C, Lingenhel A, Grobner P, Piendl W. Control of ribosomal protein L1 synthesis in mesophilic and thermophilic archaea. Genetics. 1999;152:1363. doi: 10.1093/genetics/152.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baughman G, Nomura M. Localization of the target site for translational regulation of the L11 operon and direct evidence for translational coupling in Escherichia coli. Cell. 1983;34:979–988. doi: 10.1016/0092-8674(83)90555-x. [DOI] [PubMed] [Google Scholar]
- 43.Nevskaya N, Tishchenko S, Gabdoulkhakov A, Nikonova E, Nikonov O, Nikulin A, Platonova O, Garner M, Nikonov S, Piendl W. Ribosomal protein L1 recognizes the same specific structural motif in its target sites on the autoregulatory mRNA and 23S rRNA. Nucleic Acids Res. 2005;33:478–485. doi: 10.1093/nar/gki194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brot N, Caldwell P, Weissbach H. Autogenous control of Escherichia coli ribosomal protein L10 synthesis in vitro. Proc. Natl Acad. Sci. USA. 1980;77:2592. doi: 10.1073/pnas.77.5.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holowachuk E, Friesen J, Fiil NP. Bacteriophage lambda vehicle for the direct cloning of Escherichia coli promoter DNA sequences: feedback regulation of the rplJL-rpoBC operon. Proc. Natl Acad. Sci. USA. 1980;77:2124. doi: 10.1073/pnas.77.4.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnsen M, Christensen T, Dennis PP, Fiil NP. Autogenous control: ribosomal protein L10-L12 complex binds to the leader sequence of its mRNA. EMBO J. 1982;1:999–1004. doi: 10.1002/j.1460-2075.1982.tb01284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Christensen T, Johnsen M, Fiil NP, Friesen JD. RNA secondary structure and translation inhibition: analysis of mutants in the rplJ leader. EMBO J. 1984;3:1609–1612. doi: 10.1002/j.1460-2075.1984.tb02018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Climie SC, Friesen JD. Feedback regulation of the rplJL-rpoBC ribosomal protein operon of Escherichia coli requires a region of mRNA secondary structure. J. Mol. Biol. 1987;198:371–381. doi: 10.1016/0022-2836(87)90287-7. [DOI] [PubMed] [Google Scholar]
- 49.Climie SC, Friesen JD. In vivo and in vitro structural analysis of the rplJ mRNA leader of Escherichia coli. Protection by bound L10-L7/L12. J. Biol. Chem. 1988;263:15166–15175. [PubMed] [Google Scholar]
- 50.Robakis N, Meza-Basso L, Brot N, Weissbach H. Translational control of ribosomal protein L10 synthesis occurs prior to formation of first peptide bond. Proc. Natl Acad. Sci. USA. 1981;78:4261–4264. doi: 10.1073/pnas.78.7.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;37:D136–D140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aseev LV, Levandovskaya AA, Tchufistova LS, Scaptsova NV, Boni IV. A new regulatory circuit in ribosomal protein operons: S2-mediated control of the rpsB-tsf expression in vivo. RNA. 2008;14:1882–1894. doi: 10.1261/rna.1099108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tjaden B, Saxena RM, Stolyar S, Haynor DR, Kolker E, Rosenow C. Transcriptome analysis of Escherichia coli using high-density oligonucleotide probe arrays. Nucleic Acids Res. 2002;30:3732. doi: 10.1093/nar/gkf505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Held WA, Ballou B, Mizushima S, Nomura M. Assembly mapping of 30 S ribosomal proteins from Escherichia coli. J. Biol. Chem. 1974;249:3103. [PubMed] [Google Scholar]
- 56.Lindahl L, Archer R, Zengel JM. Transcription of the S10 ribosomal protein operon is regulated by an attenuator in the leader. Cell. 1983;33:241–248. doi: 10.1016/0092-8674(83)90353-7. [DOI] [PubMed] [Google Scholar]
- 57.Yates JL, Nomura M. E. coli ribosomal protein L4 is a feedback regulatory protein. Cell. 1980;21:517–522. doi: 10.1016/0092-8674(80)90489-4. [DOI] [PubMed] [Google Scholar]
- 58.Freedman L, Zengel J, Archer R, Lindahl L. Autogenous control of the S10 ribosomal protein operon of Escherichia coli: genetic dissection of transcriptional and posttranscriptional regulation. Proc. Natl Acad. Sci. USA. 1987;84:6516. doi: 10.1073/pnas.84.18.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen P, Zengel J, Lindahl L. Secondary structure of the leader transcript from the Escherichia coli S10 ribosomal protein operon. Nucleic Acids Res. 1988;16:8905. doi: 10.1093/nar/16.18.8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zengel J, Lindahl L. A hairpin structure upstream of the terminator hairpin required for ribosomal protein L4-mediated attenuation control of the S10 operon of Escherichia coli. J. Bacteriol. 1996;178:2383. doi: 10.1128/jb.178.8.2383-2387.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sha Y, Lindahl L, Zengel J. RNA determinants required for L4-mediated attenuation control of the S10 r-protein operon of Escherichia coli. J. Mol. Biol. 1995;245:486–498. doi: 10.1006/jmbi.1994.0040. [DOI] [PubMed] [Google Scholar]
- 62.Zengel J, Sha Y, Lindahl L. Surprising flexibility of leader RNA determinants for r-protein L4-mediated transcription termination in the Escherichia coli S10 operon. RNA. 2002;8:572–578. doi: 10.1017/s1355838202026237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zengel JM, Lindahl L. Ribosomal protein L4 and transcription factor NusA have separable roles in mediating terminating of transcription within the leader of the S10 operon of Escherichia coli. Genes Dev. 1992;6:2655–2662. doi: 10.1101/gad.6.12b.2655. [DOI] [PubMed] [Google Scholar]
- 64.Cerretti D, Mattheakis L, Kearney K, Vu L, Nomura M. Translational regulation of the spc operon in Escherichia coli: identification and structural analysis of the target site for S8 repressor protein. J. Mol. Biol. 1988;204:309–329. doi: 10.1016/0022-2836(88)90578-5. [DOI] [PubMed] [Google Scholar]
- 65.Gregory R, Cahill P, Thurlow D, Zimmermann RA. Interaction of Escherichia coli ribosomal protein S8 with its binding sites in ribosomal RNA and messenger RNA. J. Mol. Biol. 1988;204:295–307. doi: 10.1016/0022-2836(88)90577-3. [DOI] [PubMed] [Google Scholar]
- 66.Dean D, Yates J, Nomura M. Escherichia coli ribosomal protein S8 feedback regulates part of spc operon. Nature. 1981;289:89–91. doi: 10.1038/289089a0. [DOI] [PubMed] [Google Scholar]
- 67.Olins P, Nomura M. Translational regulation by ribosomal protein S8 in Escherichia coli: structural homology between rRNA binding site and feedback target on mRNA. Nucleic Acids Res. 1981;9:1757. doi: 10.1093/nar/9.7.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mattheakis L, Vu L, Sor F, Nomura M. Retroregulation of the synthesis of ribosomal proteins L14 and L24 by feedback repressor S8 in Escherichia coli. Proc. Natl Acad. Sci. USA. 1989;86:448–452. doi: 10.1073/pnas.86.2.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu H, Jiang L, Zimmermann RA. The binding site for ribosomal protein S8 in 16S rRNA and spc mRNA from Escherichia coli: minimum structural requirements and the effects of single bulged bases on S8-RNA interaction. Nucleic Acids Res. 1994;22:1687–1695. doi: 10.1093/nar/22.9.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Merianos HJ. The structure of a ribosomal protein S8/spc operon mRNA complex. RNA. 2004;10:954–964. doi: 10.1261/rna.7030704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saito K, Mattheakis LC, Nomura M. Post-transcriptional regulation of the str operon in Escherichia coli. Ribosomal protein S7 inhibits coupled translation of S7 but not its independent translation. J. Mol. Biol. 1994;235:111–124. doi: 10.1016/s0022-2836(05)80020-8. [DOI] [PubMed] [Google Scholar]
- 72.Dean D, Yates JL, Nomura M. Identification of ribosomal protein S7 as a repressor of translation within the str operon of E. coli. Cell. 1981;24:413–419. doi: 10.1016/0092-8674(81)90331-7. [DOI] [PubMed] [Google Scholar]
- 73.Saito K, Nomura M. Post-transcriptional regulation of the str operon in Escherichia coli. Structural and mutational analysis of the target site for translational repressor S7. J. Mol. Biol. 1994;235:125–139. doi: 10.1016/s0022-2836(05)80021-x. [DOI] [PubMed] [Google Scholar]
- 74.Golovin A, Spiridonova V, Kopylov A. Mapping contacts of the S12-S7 intercistronic region of str operon mRNA with ribosomal protein S7 of E. coli. FEBS Lett. 2006;580:5858–5862. doi: 10.1016/j.febslet.2006.09.050. [DOI] [PubMed] [Google Scholar]
- 75.Robert F, Brakier-Gingras L. Ribosomal protein S7 from Escherichia coli uses the same determinants to bind 16S ribosomal RNA and its messenger RNA. Nucleic Acids Res. 2001;29:677–682. doi: 10.1093/nar/29.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meng BY, Shinozaki K, Sugiura M. Genes for the ribosomal proteins S12 and S7 and elongation factors EF-G and EF-Tu of the cyanobacterium, Anacystis nidulans: structural homology between 16S rRNA and S7 mRNA. Mol. Gen. Genet. 1989;216:25–30. doi: 10.1007/BF00332226. [DOI] [PubMed] [Google Scholar]
- 77.Sengupta J, Agrawal RK, Frank J. Visualization of protein S1 within the 30S ribosomal subunit and its interaction with messenger RNA. Proc. Natl Acad. Sci. USA. 2001;98:11991–11996. doi: 10.1073/pnas.211266898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sørensen MA, Fricke J, Pedersen S. Ribosomal protein S1 is required for translation of most, if not all, natural mRNAs in Escherichia coli in vivo. J. Mol. Biol. 1998;280:561–569. doi: 10.1006/jmbi.1998.1909. [DOI] [PubMed] [Google Scholar]
- 79.Boni IV, Artamonova VS, Dreyfus M. The last RNA-binding repeat of the Escherichia coli ribosomal protein S1 is specifically involved in autogenous control. J. Bacteriol. 2000;182:5872–5879. doi: 10.1128/jb.182.20.5872-5879.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jinks-Robertson S, Nomura M. Ribosomal protein S4 acts in trans as a translational repressor to regulate expression of the alpha operon in Escherichia coli. J. Bacteriol. 1982;151:193–202. doi: 10.1128/jb.151.1.193-202.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thomas M, Dewell DM, Nomura M. Regulation of alpha operon gene expression in Escherichia coli: a novel form of translational coupling. J. Mol. Biol. 1987;196:333–345. doi: 10.1016/0022-2836(87)90694-2. [DOI] [PubMed] [Google Scholar]
- 82.Tang CK, Draper DE. Unusual mRNA pseudoknot structure is recognized by a protein translational repressor. Cell. 1989;57:531–536. doi: 10.1016/0092-8674(89)90123-2. [DOI] [PubMed] [Google Scholar]
- 83.Baker A, Draper D. Messenger RNA recognition by fragments of ribosomal protein S4. J. Biol. Chem. 1995;270:22939–22945. doi: 10.1074/jbc.270.39.22939. [DOI] [PubMed] [Google Scholar]
- 84.Grundy FJ, Henkin TM. The rpsD gene, encoding ribosomal protein S4, is autogenously regulated in Bacillus subtilis. J. Bacteriol. 1991;173:4595–4602. doi: 10.1128/jb.173.15.4595-4602.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Portier C, Dondon L, Grunberg-Manago M. Translational autocontrol of the Escherichia coli ribosomal protein S15. J. Mol. Biol. 1990;211:407. doi: 10.1016/0022-2836(90)90361-O. [DOI] [PubMed] [Google Scholar]
- 86.Philippe C, Benard L, Portier C, Westhof E, Ehresmann B, Ehresmann C. Molecular dissection of the pseudoknot governing the translational regulation of Escherichia coil ribosomal protein S15. Nucleic Acids Res. 1995;23:18–28. doi: 10.1093/nar/23.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Philippe C, Eyermann F, Benard L, Portier C, Ehresmann B, Ehresmann C. Ribosomal protein S15 from Escherichia coli modulates its own translation by trapping the ribosome on the mRNA initiation loading site. Proc. Natl Acad. Sci. USA. 1993;90:4394–4398. doi: 10.1073/pnas.90.10.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Philippe C, Portier C, Mougel M, Grunberg-Manago M, Ebel JP, Ehresmann B, Ehresmann C. Target site of Escherichia coli ribosomal protein S15 on its messenger RNA: conformation and interaction with the protein. J. Mol. Biol. 1990;211:415–426. doi: 10.1016/0022-2836(90)90362-P. [DOI] [PubMed] [Google Scholar]
- 89.Bénard L, Mathy N, Grunberg-Manago M, Ehresmann B, Ehresmann C, Portier C. Identification in a pseudoknot of a UG motif essential for the regulation of the expression of ribosomal protein S15. Proc. Natl Acad. Sci. USA. 1998;95:2564. doi: 10.1073/pnas.95.5.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bénard L, Philippe C, Dondon L, Grunberg-Manago M, Ehresmann B, Ehresmann C, Portier C. Mutational analysis of the pseudoknot structure of the S15 translational operator from Escherichia coli. Mol. Microbiol. 1994;14:31–40. doi: 10.1111/j.1365-2958.1994.tb01264.x. [DOI] [PubMed] [Google Scholar]
- 91.Serganov A, Ennifar E, Portier C, Ehresmann B, Ehresmann C. Do mRNA and rRNA binding sites of E. coli ribosomal protein S15 share common structural determinants? J. Mol. Biol. 2002;320:963–978. doi: 10.1016/s0022-2836(02)00553-3. [DOI] [PubMed] [Google Scholar]
- 92.Scott LG, Williamson JR. Interaction of the Bacillus stearothermophilus ribosomal protein S15 with its 5′-translational operator mRNA1. J. Mol. Biol. 2001;314:413–422. doi: 10.1006/jmbi.2001.5165. [DOI] [PubMed] [Google Scholar]
- 93.Serganov A, Polonskaia A, Ehresmann B, Ehresmann C, Patel DJ. Ribosomal protein S15 represses its own translation via adaptation of an rRNA-like fold within its mRNA. EMBO J. 2003;22:1898–1908. doi: 10.1093/emboj/cdg170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Scott L, Williamson J. The binding interface between Bacillus stearothermophilus ribosomal protein S15 and its 5′-translational operator mRNA. J. Mol. Biol. 2005;351:280–290. doi: 10.1016/j.jmb.2005.06.030. [DOI] [PubMed] [Google Scholar]
- 95.Lesage P, Chiaruttini C, Graffe M, Dondon J, Milet M, Springer M. Messenger RNA secondary structure and translational coupling in the Escherichia coli operon encoding translation initiation factor IF3 and the ribosomal proteins, L35 and L20. J. Mol. Biol. 1992;228:366–386. doi: 10.1016/0022-2836(92)90827-7. [DOI] [PubMed] [Google Scholar]
- 96.Lesage P, Truong HN, Graffe M, Dondon J, Springer M. Translated translational operator in Escherichia coli auto-regulation in the infC-rpmI-rplT operon. J. Mol. Biol. 1990;213:465–475. doi: 10.1016/S0022-2836(05)80208-6. [DOI] [PubMed] [Google Scholar]
- 97.Choonee N, Even S, Zig L, Putzer H. Ribosomal protein L20 controls expression of the Bacillus subtilis infC operon via a transcription attenuation mechanism. Nucleic Acids Res. 2007;35:1578–1588. doi: 10.1093/nar/gkm011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mosig A, Zhu L, Stadler PF. Customized strategies for discovering distant ncRNA homologs. Brief. Funct. Genomic Proteomic. 2009;8:451–460. doi: 10.1093/bfgp/elp035. [DOI] [PubMed] [Google Scholar]
- 99.Wang JX, Breaker RR. Riboswitches that sense S-adenosylmethionine and S-adenosylhomocysteine. Biochem. Cell Biol. 2008;86:157–168. doi: 10.1139/O08-008. [DOI] [PubMed] [Google Scholar]
- 100.Gorke B, Vogel J. Noncoding RNA control of the making and breaking of sugars. Genes Dev. 2008;22:2914–2925. doi: 10.1101/gad.1717808. [DOI] [PubMed] [Google Scholar]
- 101.Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 2011;3:pii:a003798. doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.