Abstract
The serotonin receptor 2C plays a central role in mood and appetite control. It undergoes pre-mRNA editing as well as alternative splicing. The RNA editing suggests that the pre-mRNA forms a stable secondary structure in vivo. To identify substances that promote alternative exons inclusion, we set up a high-throughput screen and identified pyrvinium pamoate as a drug-promoting exon inclusion without editing. Circular dichroism spectroscopy indicates that pyrvinium pamoate binds directly to the pre-mRNA and changes its structure. SHAPE (selective 2′-hydroxyl acylation analysed by primer extension) assays show that part of the regulated 5′-splice site forms intramolecular base pairs that are removed by this structural change, which likely allows splice site recognition and exon inclusion. Genome-wide analyses show that pyrvinium pamoate regulates >300 alternative exons that form secondary structures enriched in A–U base pairs. Our data demonstrate that alternative splicing of structured pre-mRNAs can be regulated by small molecules that directly bind to the RNA, which is reminiscent to an RNA riboswitch.
INTRODUCTION
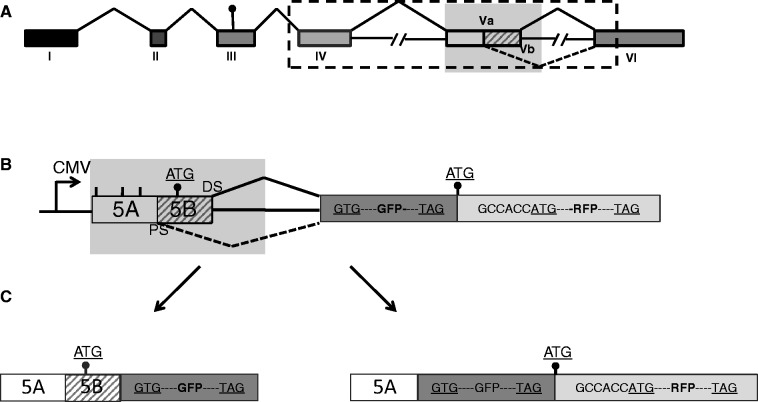
Obesity is a major and growing health problem. Mouse studies and the development of weight-loss drugs like fenfluramine–phentermine (fen–phen) validated the serotonin receptor 2C (HTR2c) protein as an anti-obesity drug target (1). HTR2c is a G-protein–coupled receptor located on the X chromosome. As shown in Figure 1, exon Vb is alternatively spliced. Only when this exon is included, a functional receptor can be made, as skipping of the exon generates a frame shift, which likely generates a mRNA, which is not translated into protein and is likely degraded.
Figure 1.
Screen to identify substances that promote exon Vb inclusion. (A) Schematic overview of the HTR2c gene. The dot indicates the start codon. The shaded area is used to construct the fluorescence-based screening reporter genes. The dotted area containing exon Va, Vb and IV is used to generate the RT–PCR-based reporter minigenes. (B) Reporter construct: the shaded area from the HTR2c gene in (A) was introduced into the reporter pFlare that can generate both GFP and RFP. Lines indicate methionine codons that were mutated. (C) Protein products generated by the reporter: inclusion of exon Vb creates GFP protein, and skipping of exon Vb destroys this open reading frame. In both cases, RFP is expressed and serves as a control for non–splicing-related effects. The start codons used are indicated as dots.
The receptor pre-mRNA undergoes A → I editing in at least five editing sites located in exon Vb, together with alternative splicing generating at least 25 isoforms.
The inclusion of exon Vb can be achieved in two known ways: (i) changing one or more of five adenosines in exon Vb to inosine using the deaminases ADAR (adenosine deaminase acting on RNA) (2); or (ii) by promoting exon Vb inclusion via processed small nucleolar RNAs (psnoRNAs) (3,4) without any change in mRNA sequence. The inosines generated by ADAR are interpreted as guanosines by the ribosome. Therefore, the edited versions of the HTR2c have a different protein sequence. This difference in composition has a functional effect: the edited versions of the receptor are less active than the non-edited versions, as the coupling efficiency towards the G-protein is reduced (5). Mice lacking HTR2c expression are hyperphagic and obese (6,7). Although re-introducing the non-edited receptor reverts this phenotype, introduction of the fully edited HTR2c does not revert the hyperphagic behaviour (8,9), suggesting that expression of the non-edited most active receptor is important to prevent hyperphagia.
The Prader–Willi syndrome is the most frequent genetic cause for obesity in humans. Genetic data indicate that the loss of regulatory RNAs generated from the HBII-52 and -85 clusters cause the disease (10,11). The HBII-52 and -85 loci encode RNAs that structurally resemble snoRNAs, but the protein complexes they form are biochemically distinct from canonical small nucleolar ribonucleoprotein (snoRNPs) (3,12). Furthermore, the HBII-52 and -85 loci express shorter RNAs, termed psnoRNAs (13). HBII-52–derived psnoRNAs change alternative splicing of the serotonin receptor 2C. This promotes the formation of an mRNA encoding a protein isoform that has the strongest response to serotonin (3,14). Collectively, these data show that the HTR2c pre-mRNA processing controls appetite in vertebrates.
The use of RNA-binding antibiotics, such as gentamycin, chloramphenicol and tetracycline, demonstrates that drugs can target RNA molecules (15–17). The target for these antibiotics is bacterial ribosomal RNA that is highly structured. Most pre-mRNAs have short rapidly changing secondary structures, if they have structures at all. In contrast, the HTR2c pre-mRNA is edited at several positions (Figure 6C). This is only possible when the pre-mRNA forms a transiently stable secondary structure in vivo, which is necessary for its editing by ADAR enzymes that need double-stranded RNA as substrates (18). This scenario is unique, as pre-mRNA processing likely occurs concomitant with transcription, and the structures of pre-mRNAs in vivo are generally not known (19,20). As the pre-mRNA is structured and at least transitionally stable in vivo, it presents defined molecular binding sites, making the HTR2c pre-mRNA a potential drug target similar to the structured ribosomal RNAs recognized by antibiotics.
Figure 6.
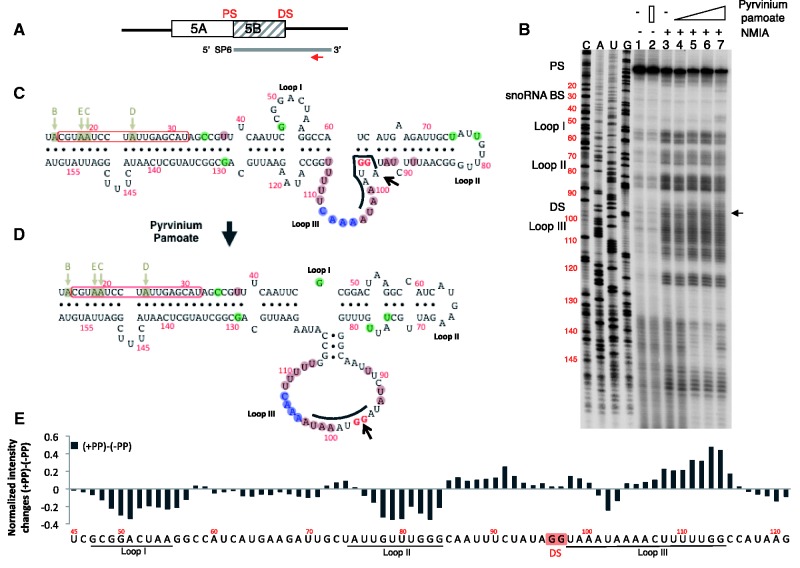
Structure probing of the exon Vb region. (A) Schematic representation of the probe used. 5B is the regulated exon and DS and PS are proximal and distal splice sites, respectively. The line indicates the SP6 polymerase-generated probe, and the arrow indicates the location of the reverse transcription primer. (B) SHAPE assay of the RNA treated with pyrvinium pamoate. RNA was analysed by SHAPE assay in the presence of an increasing amount of pyrvinium pamoate, 1, 2 and 5 µM (lanes 4–6). Lanes 1 and 2 have no NMIA, and lane 2 contains 5 µM pyrvinium pamoate. The primer used for reverse transcription and DNA sequencing is 5′-AATCCGAAAGTATTG-3′, located chrX: 114082824–114082809. The numbering of all gels refers to the structures shown in panel C that encompasses the reference sequence chrX: 114082671–114082860 in Hg19. The sequencing gel is labelled in sense orientation. The black arrow indicates the change at the 5′ splice site. (C) Secondary structures determined from the SHAPE assay. The program ‘RNAstructure’ predicts structure C to form in the absence of pyrvinium pamoate (lane 3). In the presence of pyrvinium pamoate, structure D is formed (land 6). The editing sites B–E are indicated by arrows. The boxed sequence corresponds to the binding site of the HBII-52 snoRNA. The evolutionary conservation is indicated by shaded circles around the nucleotides: pink: changes in evolution that would disrupt this structure; green: changes in evolution that will keep this structure; blue: nucleotides that are deleted in some organisms. (E) Difference in NMIA reactivity between the presence and absence of pyrvinium pamoate. The graph shows the difference in intensity between structure probing without pyrvinium pamoate (lane 3) and with pyrvinium pamoate (lane 7) for each nucleotide. Loops I, II and III are indicated; DS and PS are distal and proximal splice sites, respectively. A positive difference indicates that the nucleotide is more reactive; a negative difference indicates a closer conformation.
Most substances that change alternative splicing work indirectly. They influence mainly post-translational modifications, such as phosphorylation (21), or cause a change in regulatory factors (22) [reviewed in (23)]. However, pre-mRNA riboswitches have been identified in bacteria, and a vitamin B1 riboswitch regulating alternative splicing has been found in fungi (24). This demonstrates that pre-mRNA splicing can be directly regulated by interaction of pre-mRNA with small molecules.
Given the importance of proper alternative splicing regulation for human health, screening systems for substances that change alternative splicing have been developed (25).
Here, we report the identification of pyrvinium pamoate as a drug that promotes inclusion of the alternative exon of the serotonin receptor 2C through direct binding to the pre-mRNA. Pre-mRNA binding causes a conformational change, which makes the regulated splice site more accessible to the splicing machinery, promoting exon inclusion.
MATERIALS AND METHODS
Screening
The screen was performed as described previously (26). The screening reporter construct described in Figure 1B was stably integrated into HEK293 cells, and several cell lines with single integrations were selected. The screening reporter contained the human sequence chrX: 114082566–114083052 in Hg19. The first three in frame ATG were changed into GTG.
We validated the screen using the HBII-52 snoRNA expression construct (4) as a positive control and, subsequently, selected two cell lines. These cell lines were used to screen 1692 compounds of the UCLA chemical library in a 96-well format. The drugs were dissolved in DMSO. Their final concentration in the screen was 6 µM. Fluorescence data were acquired using the typhoon phosphor imager. The GFP/RFP ratio was measured at three different time points (1, 2 and 3 days) after LOWESS smoothing of the RFP to GFP ratios that removed reading artifacts. We then identified the active compounds by the slope of the RFP to GFP ratio change over the time course of the experiment. Compared with using a single data point, this procedure reduces the chance of a false call because of data variability.
Next, we removed all hits that contained known transcriptional activators and fluorescent compounds and concentrated on the hits common to both cell lines.
Circular dichroism spectroscopy
Circular dichroism (CD) spectra were measured with a JASCO J-810 Spectropolarimeter. Briefly, the RNA corresponding to chrX: 114082666–114082860, Hg19 (exon Vb and intronic region) was gel purified from the in vitro transcription reaction and resuspended in RNA structure buffer (10 mM Tris, pH 7, 100 mM KCl and 10 mM MgCl2) at a concentration of 20 µg/ml. Poly U RNA (Sigma) was used as a control. The RNA fragment was denatured in the presence of pyrvinium pamoate or DMSO at 75°C for 5 min and then incubated at room temperature for 15 min. The CD signal was recorded from 250 to 300 nm at 2-nm intervals.
Cell culture
HEK293T cells (ATCC) were cultured in Dulbecco’s modified Eagle’s medium containing 10% (v/v) fetal bovine serum (Invitrogen). DNA plasmids were transfected into the cells with calcium phosphate as described previously (3). Serotonin receptor 2c gene splicing analysis was performed as described previously (4). Cycloheximide (Sigma) was added to the cells at 0.25 pM. Stable cell lines were made from HEK293 cells transfected with pFlareHT construct trough clonal selection.
DNA constructs
pFlareHT reporter construct was derived from pFlare by cloning chrX: 114082566–114083052, Hg19 into pFLare-5G (25) using BglII/XhoI, having the first three in frame start codons mutated from ATG to GTG. The reporter minigene pRSR-5HTcons for the splicing analysis of serotonin receptor 2c was described previously (4). To prepare the RNA probe, SE6 construct was made as follow: DNA fragment, including 3′-end of exon 5 (100 nt) and 5′-end of intron 5 (97 nt) from HTR2c gene, was polymerase chain reaction (PCR) amplified and cloned into pCRII vector (Invitrogen) with SP6 promoter sequence introduced in the forward primer.
Array experiments
RNA was isolated using Quiagen RNeasy kit. Its quality was determined by RNA integrity (RIN) number analysis, and samples with a RIN > 9.5 were used following the Affymetrix labelling procedure.
For the analysis, the signal from Affymetrix human junction arrays was normalized using the ‘probe scaling’ method. The background was corrected with ProbeEffect from GeneBase (27). The gene expression index was computed from probes that were selected using ProbeSelect from GeneBase (27). The gene expression signals were computed using these probes. Genes were considered expressed if the mean intensity was ≥500. Genes were considered regulated if (i) they were expressed in at least one condition (i.e. pyrvinium pamoate and/or control); (ii) the fold-change was ≥1.5; and (iii) the unpaired t-test P-value between gene intensities was ≤0.05. For each probe, a splicing-index was computed. Unpaired t-tests were performed to determine the difference in probe expression between the two samples as described previously (28). Probe P-values in each probeset were then summarized using Fisher’s method. Using annotation files, splicing patterns (cassette exons, 5′/3′ alternative splice sites and mutually exclusive exons) were tested for a difference between isoforms, selecting the ones with a minimum number of regulated probeset (with a P ≤ 0.01) in each competing isoform (at least one-third of ‘exclusion’ probesets have to be significant; at least one-third of ‘inclusion’ probesets have to be significant and show an opposite regulation for the splicing-index compared with the ‘exclusion’ probesets). For example, for a single cassette exon, the exclusion junction and at least one of the three inclusion probesets (one exon probeset and two inclusion junction probesets) have to be significant and have to show an opposite regulation for the splicing-index.
SHAPE assay
SHAPE assay was performed as previously described (29) using in vitro transcribed RNA corresponding to coordinates chrX: 114082666–114082860 in Hg19. The reactions were separated on a 10% acrylamide/8 M urea sequencing gel. The gel was dried with a vacuum gel dryer and exposed into a phosphor screen. The screen was then scanned with Typhoon 9410 phosphorimager, and the scanned bands were quantified with ImageQuant 5.2. Primers sequence: reverse transcription primer 5′-AATCCGAAAGTATTG-3′.
Analysis of SHAPE data
The background band volumes in absence of N-methylisatoic anhydride (NMIA) (lane 1 in Figure 6B) were subtracted from the band volumes in presence of NMIA (lanes 3 or 7 in Figure 6B) to determine the extent of SHAPE reactivity with and without pyrvinium pamoate. The resulting SHAPE reactivities were then normalized using the boxplot analysis method, where the number of outliers was capped at 5%, as explained by Deigan et al. (30). Normalized SHAPE data for nucleotides U45 to A121 were used to refine the prediction of secondary structure by free-energy minimization using RNAstructure version 5.4 and the default slope and intercept parameter values to determine pseudo free-energy changes.
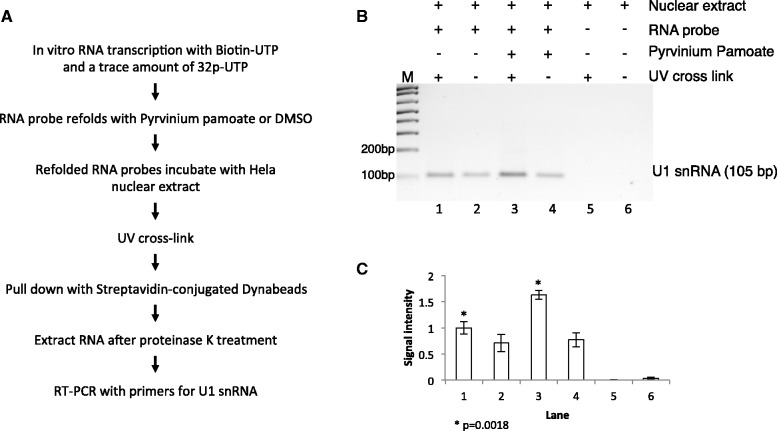
U1 binding assay
Biotin-11-UTP (Ambion) and a trace amount of γ-32P-UTP (Ambion) were used to in vitro transcribe the RNA fragment used in SHAPE assay. The biotinylated RNA probe was gel purified and refolded with 100 nM pyrvinium pamoate or DMSO. Ten nanomolars of folded biotinylated RNA probe was incubated in a 50 µl reaction with 44% (vol/vol) HeLa nuclear extract (Dundee Cell Products), 2 mM adenosine triphosphate, 20 mM creatine phosphate, 3 mM MgCl2 and 1 mM DTT at 30°C for 20 min. One reaction without RNA probe was used as a control. Twenty-five microlitres of the reaction was set aside as non–cross-linking control, and the other half of the reaction was ultraviolet (UV) cross-linked using the Stratalinker (Stratagene) at 4°C for 10 min. The biotinylated RNA was then isolated with streptavidin-conjugated Dynabeads (Invitrogen) and was used as the template for reverse transcription reaction and 18-cycle PCR with U1 snRNA-specific primers.
Isolation of supraspliceosomes
Nuclear supernatants enriched in supraspliceosomes were prepared as described previously (31). Briefly, nuclear supernatants enriched for supraspliceosomes were prepared from purified nuclei of HEK293 cells by microsonication of the nuclei and precipitation of the chromatin. The nuclear supernatants were fractionated in 10–45% glycerol gradients. Aliquots from gradient fractions were analysed by western blot. RNA was extracted from gradient fractions as described previously (31) and analysed by reverse transcriptase–PCR (RT–PCR) as described previously (32).
Fluorometric assay
All the RNA oligonucleotides were synthesized and purified by IDT (Integrated DNA Technologies Inc.). Fluorescence emission spectra in the wavelength range 560–680 nm for pyrvinium pamoate were obtained using an ISS K2 Multifrequency Phase Fluorometer (ISS Inc., Champaign, IL, USA) at room temperature using 460 nm as the excitation wavelength. The intensities at 620 nm were used to calculate the fluorescent signal changes.
Primers used
HTR2c reporter gene for splicing assay:
Forward: GCAATCCTTTATGATTATGTC
Reverse: GGTTCATTGGTATGCCGA
pFlare HT reporter gene:
Forward: GGCCACTACCTAGATATTTGTG
Reverse: CGTCGCCGTCCAGCTCGACCAG
pFRHT reporter gene:
Forward: GGCCACTACCTAGATATTTGTG
Reverse: CAGCGGTTCCATCTTCCAGCGG
SE6 construct for RNA probe:
Forward: GCCCTTATTTAGGTGACACTATAGAA GGATCGGTATGTAGCAATAC
Reverse: CTAGAAAGCTTGTTACCAGTCGACGTCTGTACG
U1 snRNA:
Forward: GGGGAGATACCATGATCACG
Reverse: GTCGAGTTTCCCACATTTGG
RESULTS
Screen for substances that change alternative splicing of the HTR2c
To identify substances that change the splicing of the HTR2c, we generated a reporter construct that expresses green fluorescent protein when exon Vb is included in the pre-mRNA (Figure 1A and B). We synthesized an exon V fragment that had all but the last ATG mutated into GTG, which allows protein expression from the last ATG in exon Vb. This construct was cloned into pFLare-5G (25). Exon Vb inclusion generates a short peptide KIAIVWAISI corresponding to amino acids 76–85 of the serotonin receptor 2C. The resulting pFlare-HT construct expresses GFP with the attached short peptide when exon Vb is included. Conversely, skipping of exon Vb prevents GFP expression, as it destroys the open reading frame. The GFP cassette is flanked downstream by RFP, which is expressed in the absence of GFP, as RFP contains the only start codon. RFP is, therefore, always expressed and serves as a control for non–splicing-related effects, such as RNA stability, transcriptional activity and an increase in translation. We used the change in the ratio of GFP to RFP as a measure of splicing-dependent exon Vb inclusion.
The reporter construct was stably integrated into HEK293 cells, and several cell lines with single integration of the reporter construct were selected. We validated the screen using an HBII-52 psnoRNA expression construct as a positive control (14), and subsequently selected two cell lines that showed an increase of the GFP to RFP ratio in response to HBII-52 overexpression. These cell lines were used to screen 1692 compounds of the UCLA chemical library in a 384-well format (25).
The final concentration of the compounds dissolved in DMSO was 6 µM in the screen. Fluorescence data were acquired using the typhoon phosphoimager. The GFP/RFP ratio was measured at four different time points (0, 1, 2 and 3 days) (Supplementary Figure S1A). We then performed locally weighted scatterplot smoothing (LOWESS) (33) of the RFP to GFP ratios that removed reading artifacts (Supplementary Figure S1B).
Compounds that changed the slope of the RFP to GFP ratio during the time course were considered active (Supplementary Figure S1C). Compared with using a single data point, this procedure reduces the chance of a false call because of data variability. We considered points outside the 3× standard deviation of the data significant. Our five plates showed similar background distribution (Figure 3C). The overall Z factor (34), the ratio of the sum of standard deviations and differences in means was 0.61.
Figure 3.
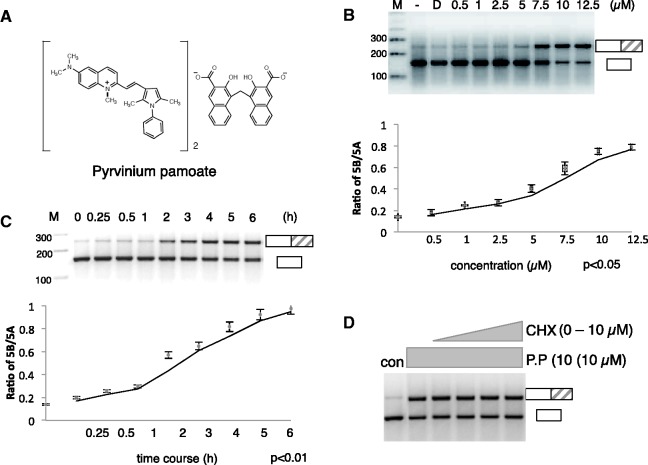
Characterization of pyrvinium pamoate action. (A) Chemical structure of pyrvinium pamoate. (B) Titration curve of pyrvinium pamoate using cells transiently transfected with the HTR2c reporter construct. The changes in exon Vb usage were determined using the primers schematically indicated in Figure 1. The graph underneath shows the statistical evaluation of three independent experiments. (C) Time course of pyrvinium pamoate using an assay similar to the one in (B). (D) Effect of pyrvinium pamoate on exon Vb inclusion in the presence of cycloheximide. Cells were transiently transfected with the HTR2c gene and treated with 10 µM cyclohexamide overnight. The cells were then treated with pyrvinium pamoate for 2 h.
Next, we removed all hits that contained known transcriptional activators and fluorescent compounds and concentrated on the hits common for both cell lines. Using this primary screen, we identified 22 compounds that promote exon Vb inclusion (Supplementary Figure S2).
In summary, we successfully set-up a screen that identified chemical compounds that promote exon Vb inclusion.
A secondary screen validates pyrvinium pamoate as a substance that promotes exon Vb inclusion
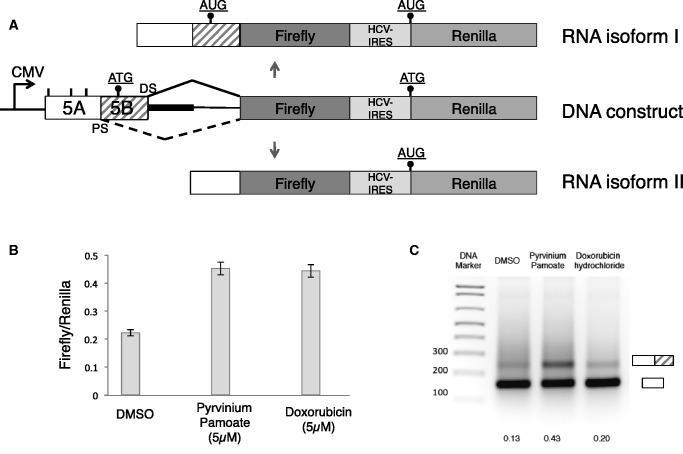
We next validated the hits observed in the primary screen in secondary screens. First, we used a Renilla luciferase construct, which is similar to pFlare-HT, but it uses luciferase and renilla as reporters, as shown in Figure 2A. Two compounds, pyrvinium pamoate and doxorubicin from the primary screen had an effect with this protein-based reporter after 12 h of treatment (Figure 2B). We next used RT–PCR for further validation using a reporter minigene that contains all coding exons and their introns (Figure 1). Intron 5 has been shortened from 50 kb to 3000 nt, leaving 1500 nt from the 5′ and 3′ intronic region, respectively (Figure 1A). Pyrvinium pamoate showed the highest efficacy in this RNA-based secondary screen and was further characterized. Pyrvinium pamoate (Figure 3A) has been previously used as an FDA approved pinworm drug and is known to bind DNA (35,36). The compound has been recently described as an activator of the Wnt signalling pathway acting through casein kinase I, which is inhibited at a concentration of 10 nM (37). Interestingly, doxorubicin, an anti-tumour drug also showed a small increase in exon Vb inclusion when tested by RT–PCR (Figure 2C). Doxorubicin consists of an aromatic ring structure that intercalates with DNA and an amino sugar that binds to AT base pairs (38). The other primary hits shown in Supplementary Figure S2 could not be confirmed in both secondary screens.
Figure 2.
Confirmation of pyrvinium pamoate in secondary screens. (A) Structure of the firefly/renilla construct. The start codons are indicated as dots, similar to Figure 1. (B) Validation of pyrvinium pamoate and doxorubicin as drugs that change alternative splicing of the reporter using the firefly/renilla chemoluminescence as readout. (C) RT–PCR-based screen of the compounds, using a reporter minigene, shown in Figure 1A, dotted area. Treatment was overnight using 5 µM of each drug. The numbers indicate the ratio between exon inclusion and exon skipping.
Thus, our screen identified two compounds with binding affinity to double-stranded nucleic acids. We characterized the compound with the strongest effect, pyrvinium pamoate, in more detail.
Characterization of pyrvinium pamoate action
We next determined the concentration of pyrvinium pamoate (Figure 3A) needed to promote exon Vb inclusion. We analysed the effect of increasing pyrvinium pamoate concentrations on HEK293 cells transfected with the HTR2c reporter gene. Cells were transfected with the reporter gene overnight, and pyrvinium pamoate was then added at the concentrations and time indicated. As shown in Figure 3B, we observe a switch from exon skipping to predominant inclusion with an IC50 of ∼6 µM. Next, we determined the time course of pyrvinium pamoate action and analysed its effect on exon Vb usage between 15 min and 6 h, again using cells transfected with a reporter gene. As shown in Figure 3C, exon inclusion starts around 2 h. This time scale is much shorter than the effects seen with other substances that change splice site selection, which typically occurs over night (23).
The rapid onset of the pyrvinium pamoate effect suggests that it might be independent of protein synthesis (Figure 3C). We tested this idea by treating cells transfected with the HTR2c reporter minigene with cycloheximide and pyrvinium pamoate overnight, which blocks protein synthesis. As shown in Figure 3D, even without protein synthesis, there is a promotion of exon Vb inclusion by pyrvinium pamoate.
We conclude that pyrvinium pamoate acts rapidly without the need of protein synthesis in the low micromolar range on the HTR2c pre-mRNA.
Pyrvinium pamoate does not change the editing of the HTR2c pre-mRNA
The alternative exon Vb of the HTR2c pre-mRNA can be included via two major mechanisms. The first mechanism is A–I editing at one or more of five editing sites in exon Vb. As the inosine is interpreted as a guanosine by the ribosome, editing changes the amino acid composition of the encoded receptor and leads to receptors with less efficacy (2,9,39). The second pathway is the inclusion of the non-edited exon, which is achieved in neurons by processed snoRNAs derived from HBII-52 (SNORD115) (psnoRNAs) that promote exon Vb inclusion through an unknown mechanism (3,4). To determine which of these pathways is promoted by pyrvinium pamoate, we directly sequenced the RT–PCR-derived band that contained exon Vb. As there is no endogenous expression of HTR2c in HEK293 cells, we used reporter genes for the analysis. As shown in Figure 4, the sequence shows adenosine at the five editing positions, indicating that pyrvinium pamoate does not cause editing of the HTR2c pre-mRNA and could potentially act similar to the psnoRNA HBII-52, which also promotes exon Vb inclusion without changing the editing status of the receptor pre-mRNA (4).
Figure 4.
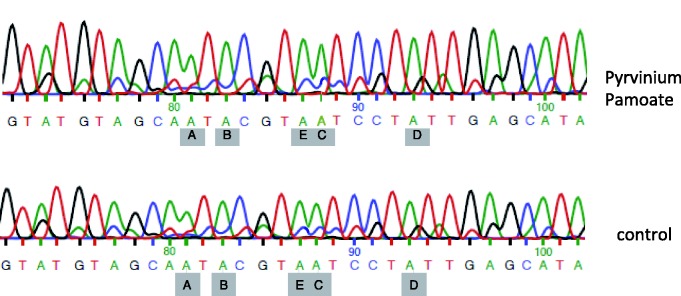
Direct sequencing of exon Vb after pyrvinium pamoate treatment. The direct sequencing result from the PCR product corresponding to the exon inclusion band after pyrvinium pamoate treatment is shown. The five major editing sites A–E are indicated and point to the edited adenosines. The control was DMSO treated. Note that all editing sites read as adenosine, demonstrating the absence of RNA editing.
Pyrvinium pamoate binds directly to HTR2c RNA
Previously, it has been shown that pyrvinium pamoate intercalates with DNA and binds with an affinity of 0.1 mM (35). As the serotonin receptor pre-mRNA forms a secondary structure in vivo, as indicated by the presence of functional RNA editing sites, we investigated whether pyrvinium pamoate directly binds to the HTR2c RNA. We used a 200-nt-long RNA fragment corresponding to the double-stranded region shown in Figure 6C.
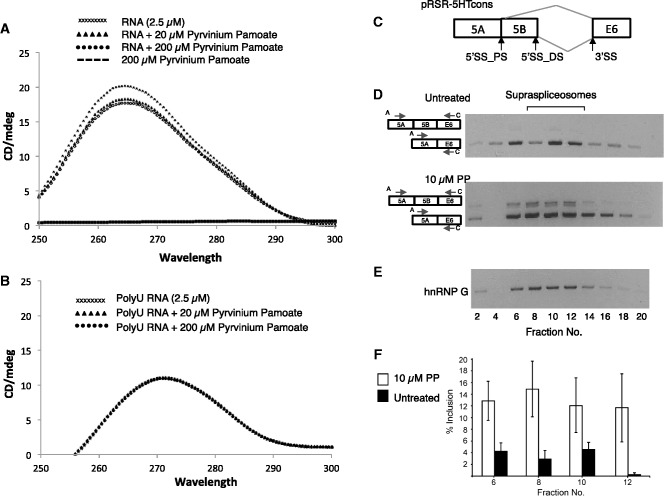
The 2.5 µM RNA was incubated with 20 and 200 µM pyrvinium pamoate, and CD was measured. As shown in Figure 5A, pyrvinium pamoate changed the CD spectrum of the RNA, with the maximum change ∼265 nm.
Figure 5.
Pyrvinium pamoate changes the CD of the structured part of the HTR2c pre-RNA and affects alternative splicing in the supraspliceosome. (A) CD spectrum of 2.5 µM HTR2c RNA incubated with an increasing amount of pyrvinium pamoate (0, 20 and 200 µM), as well as pyrvinium pamoate without RNA. (B) CD spectrum of 2.5 µM poly U RNA incubated with an increasing amount of pyrvinium pamoate (0, 20 and 200 µM). Note that there is no change in the CD spectrum, and all the spectra are superimposed. (C–F) Human 293 cells transfected with a serotonin receptor expression construct pRSR-5HTcons were treated with 10 µM pyrivinium pamoate for 6 h. Next, nuclear supernatants enriched in supraspliceosomes were prepared from treated and untreated cells and were fractionated in 10–45% glycerol gradients. Aliquots from even gradient fractions were analysed by RT–PCR and western blotting. (C) A schematic drawing of the HTR2c pre-mRNA showing the alternative 5′ splice sites (5′SS_PS, 5′ss_DS) that give rise to the alternatively spliced 5a and 5b isoforms, respectively. (D) RT–PCR analyses of aliquots of RNA extracted from even gradient fractions, untreated (upper panel) or treated with 10 µM pyrvinium pamoate (lower panel) using primer pairs that flank the respective alternative exons as indicated. The alternatively spliced isoforms of the HTR2c transcript are schematically drawn on the left, and the respective PCR primer pairs are indicated by arrowheads. Supraspliceosomes peak in fractions 8–12. The gradients were calibrated with 200S TMV (tobacco mosaic virus) particles, which sedimented in fractions 9 and 10 of a parallel gradient. (E) Western blot analyses using anti-hnRNP G antibodies. (F) Percentage of inclusion of exon 5b with- and without treatment with pyrvinium pamoate (from D, average of three independent experiments).
As a control, 2.5 µM poly U RNA (homopolymer 300–800 nt) was incubated with 20 and 200 µM pyrvinium pamoate, and we did not observe a change in CD (Figure 5B).
This suggests that pyrvinium pamoate binds directly to the structured HTR2c RNA and likely changes its tertiary conformation.
Pyrvinium pamoate acts on HTR2c RNA within the spliceosome
We next asked whether pyrvinium pamoate acts through the spliceosome. It has been shown that pre-mRNAs are assembled within supraspliceosomes during their entire lifetime in the nucleus (40). Furthermore, several regulatory splicing factors and alternative isoforms of a number of tested transcripts were found predominantly in supraspliceosomes (32,40,41). As HTR2c exon 5 b is edited by the A->I editing enzymes, it is pertinent to note that supraspliceosomes harbour the A->I editing enzymes ADAR1 and ADAR2 (42). To search for pyrvinium pamoate function in spliceosomes, HEK293 cells were transfected for 24 h with serotonin receptor expression construct pRSR-5HTcons, and they were then treated with 10 µM pyrvinium pamoate. After 6 h, the cells were harvested, and nuclear supernatants enriched in supraspliceosomes were prepared and fractionated in 10–45% glycerol gradients as described previously (31). As shown in Figure 5C and D, we found that the serotonin receptor mRNA peaks with supraspliceosomes together with the regulatory splicing factor hnRNP G (Figure 5E), which was previously shown to be associated with supraspliceosomes (43). Furthermore, we see a change in pre-mRNA splicing within the fractions containing splicing components on addition of pyrvinium pamoate (Figure 5D and F). It is, therefore, likely that pyrvinium pamoate changes alternative pre-mRNA splicing and not the stability or transcription of one isoform.
Pyrvinium Pamoate changes the conformation of the HTR2c RNA in vitro
Based on the CD spectrum analysis, we hypothesized that pyrvinium pamoate changes the secondary structure of the HTR2c pre-mRNA.
To determine the exact changes in RNA structure caused by pyrvinium pamoate, we performed SHAPE (selective 2′ hydroxyl acylation analysed by primer extension) analysis. A 200-nt-long fragment of HTR2c pre-mRNA was analysed by SHAPE, as described previously (29). Briefly, RNA was denatured, renatured in the presence or absence of pyrvinium pamoate and incubated with NMIA, which reacts with accessible 2′-OH groups. After a further denaturing step, reverse transcription with an end-labelled primer is performed. The reverse transcriptase stops at each modified nucleotide, generating a band that indicates a single-stranded conformation.
We compared RNA without pyrvinium pamoate addition with RNA, where an increasing amount of pyrvinium pamoate (1–5 µM) was added after the initial denaturation. Changes in the band intensities of each individual nucleotide are shown in Figure 6B and are quantified in Figure 6E.
We then used the program ‘RNAstructure’ to determine the secondary structure of the HTR2c RNA pre-mRNA fragment, both in the presence and absence of pyrvinium pamoate (44). In the absence of pyrvinium pamoate, the SHAPE assay indicates the structure shown in Figure 6C, which is also predicted by the centroid and minimum free-energy RNA structure prediction (45). The structure consists of a stem and three loops. The 45-nt-long stem contains the five editing sites and, therefore, likely forms in vivo, as the editing enzymes require double-stranded RNA. These stem structures flank a region of ∼90 nt that contains the alternative 5′ splice site and undergoes structural changes. Importantly, the exonic parts and the first intronic G of the regulated splice site are localized in a stem, which could explain why the splice site is blocked (Figure 6C, arrow). Interestingly, mutation in the splice site that leads to its activation is predicted to destroy this stem formation (4).
The presence of pyrvinium pamoate causes a structural change and generates the structure shown in Figure 6D. Through an interaction between loop I and II, a longer stem is formed, and loop III is largely extended. Importantly, the regulated distal splice site is now no longer present in a stem structure (Figure 6D, arrow).
The two structures are energetically similar (ΔG° = −56.8 and −52.6 kcal/mol for structures in Figure 6C and D). This is consistent with a small molecule or protein being able to change the conformation, which likely aids in biological regulation (46).
The structure probing data indicate that pyrvinium pamoate promotes a rearrangement of the RNA structure that moves the regulated 5′ splice site from a partially bound to a completely free conformation.
Pyrvinium pamoate promotes U1 snRNP binding to exon Vb
The structure probing data suggest that pyrvinium pamoate makes exon Vb more accessible to U1 snRNP binding. We next tested whether pyrvinium pamoate actually increases U1 binding and performed cross-linking experiments. A biotinylated RNA corresponding to the RNA used in the SHAPE experiments was refolded using pyrvinium pamoate and DMSO control, followed by incubation with nuclear extract and cross-linking. RNA bound to the beads was released by proteinase K digestion of the attached proteins, followed by RNA purification. We then detected U1 snRNA by RT–PCR. As shown in Figure 7, U1 snRNA occupancy increased ∼50% in the presence of pyrvinium pamoate.
Figure 7.
Pyrvinium pamoate increases snRNA binding to HTR2c exon Vb. (A) Schematic outline of the method used to detect U1snRNA and exon 5B binding. The biotinylated probe corresponds to the RNA used in Figure 6. (B) RT–PCR analysis of RNA extracted from streptavidin-conjugated Dynabeads. Lane 1 and 2: RNA probe folded in DMSO; lane 3 and 4, RNA probe folded in 10 µM pyrvinium pamoate; lane 5 and 6: nuclear extract only; lane 2, 4 and 6: no UV cross-linking. (C) Quantification and statistic analysis of the RT–PCR results.
The data indicate that pyrvinium pamoate causes a change in secondary structure that increases U1 binding to the regulated splice site.
Phylogenetic analysis indicates evolutionary conservation of the structures
The HTR2c gene is present in placental mammals. To identify evolutionary conserved sequences, we performed a CLUSTAL W alignment of the regulated region (Supplementary Figure S3). The changes throughout evolution are indicated in Figure 6C and D. The alignment indicates that the stem structures are highly conserved. Mutations that keep the structure forming without pyrvinium pamoate are present in five stem locations, whereas only three locations in the stems are affected by non-compensatory mutations. In contrast, 13 positions in loops are altered by mutations. The last 65 nt of the sequences shown in Figure 6C and D are encoded by an intron that does not have a coding requirement, but especially the stem remains highly conserved. Therefore, the data indicate a high-evolutionary pressure to maintain the double-stranded RNA structure, suggesting a functional relevance.
Genome-wide effects of pyrvinium pamoate on alternative splicing indicate a preference for double-stranded RNA regions rich in A–U base pairs
We next determined whether pyrvinium pamoate is selective for the HTR2c pre-mRNA and performed genome wide exon junction array analysis. We found (Figure 3) that pyrvinium pamoate acts within 6 h on the serotonin receptor 2C reporter gene; therefore, we incubated HEK293 cells for 6 h with 10 µM pyrvinium pamoate.
The array analysis indicated 376 changes in alternative splice site selection and 176 changes in overall gene expression after 6 h of treatment. RT–PCR analysis showed a validation rate of >73% (11/15) for splicing events (Supplementary Figure S4). To identify possible long-term effects, we repeated this array analysis by treating HEK293 cells with pyrvinium pamoate for 16 h. The longer treatment resulted in 5284 changes in overall gene expression and 1120 changes in splicing of cassette exons. Changes in alternative splicing had a similar validation rate of >87% (14/16). The changes after 6 h affected mostly mitogen-activated protein kinase (MAPK) signalling pathways and toll-like receptor pathways. Changes after 16 h affected multiple biological pathways but were most predominant in the p53 pathway and in the spliceosome (Supplementary Figures S5–S7). These data indicate that pyrvinium pamoate acts in two stages: first, it rapidly changes alternative splice site usage, with only a minor influence on overall gene expression. On longer exposure, pyrvinium pamoate affects mainly overall gene expression, possibly through the activation of MAPK pathways.
We next analysed the pyrvinium pamoate regulated exons for common features. The 6 and 16 h treatment gave non-overlapping sets of changed RNAs. Given the activation of MAPK pathways after 6 h, we assumed that most of the later changes are indirect and concentrated on the exons changed after 6 h.
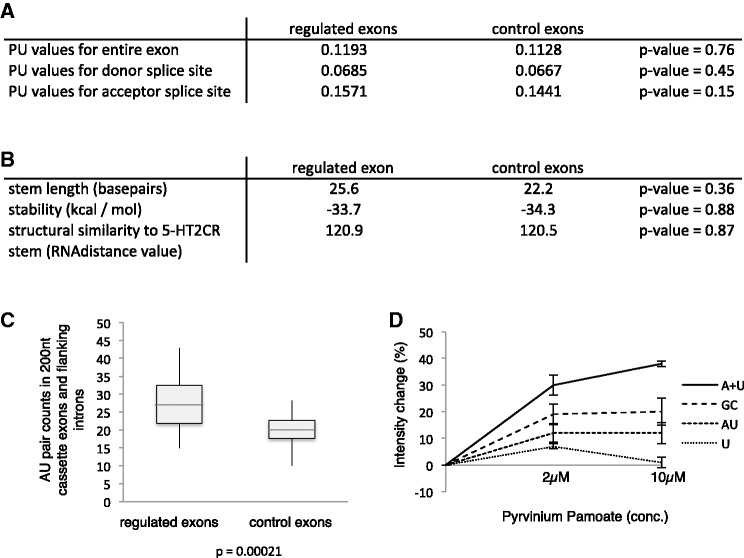
Exons that were regulated by pyrvinium pamoate within 6 h showed similar overall base composition, enhancer/silencer presence and splice site strength. We, therefore, asked whether pyrvinium pamoate targets preferential double-stranded pre-mRNA regions and compared pyrvinium pamoate regulated alternative exons with randomly selected non-regulated alternative exons (control exons). We found no difference between double strandedness of the exons or their splice sites between the two groups (Figure 8A and B). The double strandedness was calculated as the likelihood of these bases to be unpaired, expressed as a PU value (possibility that all base pairs in a substring are unpaired). The PU value is an average of possible conformations (20).
Figure 8.
General features of pyrvinium pamoate regulated exons. (A) Probability of being unpaired (PU) values for alternative exons and their splice sites regulated by pyrvinium pamoate after 6 h and random control alternative exons. (B) Stem length, stability and structural similarity to the serotonin receptor pre-mRNA of the regulated and control exons. (C) Number of AU base pairs in stem structures forming around regulated and control exons. (D) Change in fluorescence at 620 nm after adding pyrvinium pamoate to 10 µM of the indicated oligo nucleotides. A + U: double-stranded RNA of A18 and U18; GC: double-stranded RNA of CGCGCGCCCGGGCGCGCG; AU: double-stranded RNA of AUAUAUAAAUUUAUAUAU; and U: single-stranded U18.
It has been suggested that pyrvinium pamoate interacts with A–T rich base pairs in the minor groove of DNA (36). The DNA complex of the second drug, doxorubicin, which we identified, has been determined by crystallization and nuclear magnetic resonance. The structure of the doxorubicin–DNA complex indicates that in addition to the intercalation of the anthracycline structure, the amino sugar interacts with A–T base pairs in the minor groove of DNA (38). It is thus possible that both substances promoting exon Vb usage could have a preference for A–U rich base pairs on double-stranded RNA. We, therefore, tested the occurrence of AU base pairs in double-stranded regions of pyrvinium pamoate regulated exons. We looked at a 200-nt-long segment where the regulated exon was placed in the middle and determined the structure with minimum energy using RNA fold (45). We then counted the occurrence of all A–U, G–C and G–U base pairs in this region. As shown in Figure 8C, we found a significant enrichment of A–U base pairs in pyrvinium pamoate regulated exons that was statistical significant (P = 0.00021).
We next tested biochemically whether pyrvinium pamoate can discriminate between RNA sequences. Pyrvinium pamoate shows an increase in fluorescence when added to DNA that reflects its binding to DNA (36). We found a similar increase in fluorescence at 620 nM, when we added pyrvinium pamoate to dsRNA. We next tested the change in fluorescence using 18mer RNA oligos. As shown in Figure 8D, the change in pyrvinium pamoate fluorescence was strongest when it was added to A18:U18 double-stranded RNAs. The change was about half as strong for a G:C rich dsRNA or a double-stranded RNA composed of mainly alternating A:U sequences. This indicates that pyrvinium pamoate discriminates between double-stranded RNAs with different composition.
Together, these data indicate that pyrvinium pamoate selects alternative exons that show a higher abundance of A–U base pairs in their vicinity.
DISCUSSION
Successful identification of a substance that changes alternative splicing of the serotonin receptor 2C
Defects in pre-mRNA processing are increasingly recognized as a cause or consequence of human disease. Therefore, several screening systems have been developed that identified substances modifying alternative splicing. These efforts have uncovered substances influencing the alternative splicing of SMN2 exon 7, as well as compounds that bind to tau exon 10 pre-mRNA. These genes are involved in spinal muscular atrophy and tauopathies, respectively.
Compounds changing alternative splicing of SMN2 exon 7 work by modulating trans-acting factors regulating the alternative exon through post-translational modifications. Aminoglycosides have been identified that bind the structured 5′ splice sites in tau exon 10 (47), but they do not influence alternative splicing of the exon. In contrast, aminoglycoside antibiotics, such as neomycin B, tobramycin and gentamycin C1 inhibit bacterial protein synthesis after binding to ribosomal RNA (17). It is likely that the lack of defined stable structures in pre-mRNAs makes it difficult to target pre-mRNA directly, as no stable binding sites for interacting drugs are present.
We, therefore, concentrated on the serotonin receptor pre-mRNA that forms a secondary structure in vivo. This pre-mRNA is edited at five sites, which occurs only in double-stranded RNA regions that are formed long enough to undergo modifications by ADAR. Previously, a synthetic helix-threading peptide was shown to bind to the double-stranded region of the serotonin pre-mRNA in vitro, which further suggests a stable structure (48). Screening a chemical library, we identified pyrvinium pamoate as a substance that strongly promotes exon Vb usage. Pyrvinium pamoate likely acts through the spliceosome, as we show that treatment with pyrvinium pamoate lead to increase in exon 5b inclusion within splicing complexes. CD and fluorescent spectra, as well as shape analysis, indicate that pyrvinium pamoate binds directly to the RNA and changes its conformation.
The data show the principle that pre-mRNAs with a strong secondary structure can be drug targets and potential alternatives to proteins. The identification of fen–phen validated the HTR2c as an anti-appetite drug target, but it also illustrated the need for alternative treatment options, as the drug cross-reacted with the 5HT2A receptor protein, causing valvular heart disease (49). The cross-reactivity is caused by the similarity of the proteins, which is not present on the pre-mRNA level, as the intronic regions are different. Therefore, ligands binding to the RNA could be valuable tools to manipulate the HTR2 system.
As pyrvinium pamoate promotes the inclusion of the non-edited form of the receptor that has the strongest anorexic effect, we tested the effect of pyrvinium pamoate on feeding behaviour through injection in the brain. We observed seizures of the animals as well as a reduction in food intake (data not shown). The inert toxicity of the drug makes it, however, impossible to identify an effect on food uptake via the receptor.
Model for pyrvinium pamoate action
Pyrvinium pamoate has previously been shown to bind to DNA through both intercalation and electrostatic interactions (35). The staining pattern in nuclei suggests that pyrvinium pamoate binds to A–T rich DNA (36). Binding to DNA leads to its unwinding (35). Using CD spectroscopy, we found that pyrvinium pamoate binds also to double-stranded RNA, which is similar to other nucleic acid stains. The concentration of pyrvinium pamoate needed in vitro to see a change in CD spectroscopy is two times larger than the concentration needed in SHAPE assays and in vivo. This likely reflects differences between the techniques.
Using short RNA oligonucleotides, we found that A18:U18 double-stranded RNA showed the highest change in fluorescence of pyrvinium pamoate, suggesting that the drug preferentially binds to double-stranded RNAs having complementary A and U stretches. The stem formed by the serotonin pre-mRNAs contains five AA:UU dinucleotides that are evenly distributed. It is possible that pyrvinium pamoate binds to these regions.
The detailed SHAPE analysis indicates that in the absence of pyrvinium pamoate, the regulated 5′ splice site is partially in a double-stranded conformation, which likely limits the access of U1 snRNP to the splice site and inhibits its recognition. In vitro, we found less binding of U1 snRNP to the HTR2c pre-mRNA, which supports this model and in agreement with skipping of exon Vb being the default mode (4). Pyrvinium pamoate binds to the RNA and causes a conformational change, which brings the regulated 5′ splice site into an unbound conformation that is likely recognized. The mechanism of this conformational change is not clear. Pyrvinium pamoate addition changes the number of nucleotides that are not bound in the tested RNA from 44 to 52, suggesting that the drug causes a partial unwinding of double-stranded RNA, which has also been observed in DNA (35).
In summary, all the data support a model where pyrvinium pamoate binds to A–U rich regions in the dsRNA of the serotonin receptor, which leads to a relaxation of the RNA structure making the 5′ splice site more accessible.
Screen shows that a small ligand can discriminate a related set of exons
To determine the genome-wide effect of pyrvinium pamoate, we performed array analysis and found that the drug changes numerous alternative exons after a 6-h treatment time. Analysis of these exons showed they contain a significantly higher number of A–U base pairs in double-stranded regions than control exons. This indicates that the composition of transiently forming double-stranded RNA structures can contribute to splice site selection. The results also show that a coordinated change in splicing can be triggered by small molecules that interact with these structures. However, natural compounds, possibly tissue-specific metabolites that have properties similar to pyrvinium pamoate remain to be determined.
The action of pyrvinium pamoate on the HTR2c pre-mRNA resembles a riboswitch
HTR2c pre-mRNA reacts to the direct binding of pyrvinium within minutes with a conformational change that causes a change in splice site selection. This rapid structural change to a ligand is reminiscent to a riboswitch, RNA structures mainly found in bacteria. The only known eukaryotic riboswitch is the thiamine pyrophosphate riboswitch found in Neurospora (24) and Arabidopsis (50). In both systems, the riboswitch changes alternative splicing. The HTR2c pre-mRNA interacting with pyrvinium pamoate resembles this scenario, but the change is triggered by a synthetic drug.
Given the phylogenetic conservation of the HTR2c system, it is possible that endogenous small ligands exist having the same effect.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–7.
FUNDING
NIH [RO1 GM083187, P20RR020171; R01 GM076485 to D.H.M.]; Foundation for Prader–Willi Research (to S.S.); Binational Science Foundation (BSF), USA-Israel, Transformative Grant [2010508 to S.S. and R.S.]. Funding for open access charge: NIH [RO1 GM083187].
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Miller KJ. Serotonin 5-ht2c receptor agonists: potential for the treatment of obesity. Mol. Interv. 2005;5:282–291. doi: 10.1124/mi.5.5.8. [DOI] [PubMed] [Google Scholar]
- 2.Flomen R, Knight J, Sham P, Kerwin R, Makoff A. Evidence that RNA editing modulates splice site selection in the 5-HT2C receptor gene. Nucleic Acids Res. 2004;32:2113–2122. doi: 10.1093/nar/gkh536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kishore S, Khanna A, Zhang Z, Hui J, Balwierz PJ, Stefan M, Beach C, Nicholls RD, Zavolan M, Stamm S. The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum. Mol. Genet. 2010;19:1153–1164. doi: 10.1093/hmg/ddp585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 5.Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 6.Tecott LH, Sun LM, Akana SF, Strack AM, Lowenstein DH, Dallman MF, Julius D. Eating disorder and epilepsy in mice lacking 5-HT2c serotonin receptors. Nature. 1995;374:542–546. doi: 10.1038/374542a0. [DOI] [PubMed] [Google Scholar]
- 7.Nonogaki K, Strack AM, Dallman MF, Tecott LH. Leptin-independent hyperphagia and type 2 diabetes in mice with a mutated serotonin 5-HT2C receptor gene. Nat. Med. 1998;4:1152–1156. doi: 10.1038/2647. [DOI] [PubMed] [Google Scholar]
- 8.Kawahara Y, Grimberg A, Teegarden S, Mombereau C, Liu S, Bale TL, Blendy JA, Nishikura K. Dysregulated editing of serotonin 2C receptor mRNAs results in energy dissipation and loss of fat mass. J. Neurosci. 2008;28:12834–12844. doi: 10.1523/JNEUROSCI.3896-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morabito MV, Abbas AI, Hood JL, Kesterson RA, Jacobs MM, Kump DS, Hachey DL, Roth BL, Emeson RB. Mice with altered serotonin 2C receptor RNA editing display characteristics of Prader-Willi Syndrome. Neurobiol. Dis. 2010;39:169–180. doi: 10.1016/j.nbd.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahoo T, del Gaudio D, German JR, Shinawi M, Peters SU, Person RE, Garnica A, Cheung SW, Beaudet AL. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat. Genet. 2008;40:719–721. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duker AL, Ballif BC, Bawle EV, Person RE, Mahadevan S, Alliman S, Thompson R, Traylor R, Bejjani BA, Shaffer LG, et al. Paternally inherited microdeletion at 15q11.2 confirms a significant role for the SNORD116 C/D box snoRNA cluster in Prader-Willi syndrome. Eur. J. Hum. Genet. 2010;18:1196–1201. doi: 10.1038/ejhg.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soeno Y, Taya Y, Stasyk T, Huber LA, Aoba T, Huttenhofer A. Identification of novel ribonucleo-protein complexes from the brain-specific snoRNA MBII-52. RNA. 2010;16:1293–3000. doi: 10.1261/rna.2109710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen M, Eyras E, Wu J, Khanna A, Josiah S, Rederstorff M, Zhang MQ, Stamm S. Direct cloning of double-stranded RNAs from RNase protection analysis reveals processing patterns of C/D box snoRNAs and provides evidence for widespread antisense transcript expression. Nucleic Acids Res. 2011;39:9720–9730. doi: 10.1093/nar/gkr684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kishore S, Stamm S. Regulation of alternative splicing by snoRNAs. Cold Spring Harb. Symp. Quant. Biol. 2006;71:329–334. doi: 10.1101/sqb.2006.71.024. [DOI] [PubMed] [Google Scholar]
- 15.Xavier KA, Eder PS, Giordano T. RNA as a drug target: methods for biophysical characterization and screening. Trends Biotechnol. 2000;18:349–356. doi: 10.1016/s0167-7799(00)01464-5. [DOI] [PubMed] [Google Scholar]
- 16.Thanaraj TA, Cochet O, Stamm S. Misregulation of alternative splicing as a novel target for drug intervention. Pharm. Visions. 2003;2:4–6. [Google Scholar]
- 17.Schroeder R, Waldsich C, Wank H. Modulation of RNA function by aminoglycoside antibiotics. EMBO J. 2000;19:1–9. doi: 10.1093/emboj/19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu M, Wells KS, Emeson RB. Substrate-dependent contribution of double-stranded RNA-binding motifs to ADAR2 function. Mol. Biol. Cell. 2006;17:3211–3220. doi: 10.1091/mbc.E06-02-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol. Cell. Biol. 2004;24:10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiller M, Zhang Z, Backofen R, Stamm S. pre-mRNA secondary structure and splice site selection. PLoS Genet. 2007;3:2147–2155. doi: 10.1371/journal.pgen.0030204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Novoyatleva T, Heinrich B, Tang Y, Benderska N, Butchbach ME, Lorson CL, Lorson MA, Ben-Dov C, Fehlbaum P, Bracco L, et al. Protein phosphatase 1 binds to the RNA recognition motif of several splicing factors and regulates alternative pre-mRNA processing. Hum. Mol. Genet. 2008:52–70. doi: 10.1093/hmg/ddm284. [DOI] [PubMed] [Google Scholar]
- 22.Anderson ES, Lin CH, Xiao X, Stoilov P, Burge CB, Black DL. The cardiotonic steroid digitoxin regulates alternative splicing through depletion of the splicing factors SRSF3 and TRA2B. RNA. 2012;18:1041–1049. doi: 10.1261/rna.032912.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sumanasekera C, Watt DS, Stamm S. Substances that can change alternative splice-site selection. Biochem. Soc. Trans. 2008;36:483–490. doi: 10.1042/BST0360483. [DOI] [PubMed] [Google Scholar]
- 24.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- 25.Stoilov P, Lin CH, Damoiseaux R, Nikolic J, Black DL. A high-throughput screening strategy identifies cardiotonic steroids as alternative splicing modulators. Proc. Natl Acad. Sci. USA. 2008;105:11218–11223. doi: 10.1073/pnas.0801661105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stoilov P. Screening for Alternative Splicing Modulators. In: Stamm S, Smith C, Luhrmann R, editors. Alternative pre-mRNA splicing: Theory and Protocols. Weinheim: Wiley-Blackwell; 2012. pp. 497–508. [Google Scholar]
- 27.Kapur K, Jiang H, Xing Y, Wong WH. Cross-hybridization modeling on Affymetrix exon arrays. Bioinformatics. 2008;24:2887–2893. doi: 10.1093/bioinformatics/btn571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen S, Warzecha CC, Carstens RP, Xing Y. MADS+: discovery of differential splicing events from Affymetrix exon junction array data. Bioinformatics. 2010;26:268–269. doi: 10.1093/bioinformatics/btp643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 30.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc. Natl Acad. Sci. USA. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azubel M, Habib N, Sperling R, Sperling J. Native spliceosomes assemble with pre-mRNA to form supraspliceosomes. J. Mol. Biol. 2006;356:955–966. doi: 10.1016/j.jmb.2005.11.078. [DOI] [PubMed] [Google Scholar]
- 32.Heinrich B, Zhang Z, Raitskin O, Hiller M, Benderska N, Hartmann AM, Bracco L, Elliott D, Ben-Ari S, Soreq H, et al. Heterogeneous nuclear ribonucleoprotein G regulates splice site selection by binding to CC(A/C)-rich regions in pre-mRNA. J. Biol. Chem. 2009;284:14303–14315. doi: 10.1074/jbc.M901026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cleveland WS, Devlin SJ. Locally weighted regression: an approach to regression analysis by local fitting. J. Am. Stat. Assoc. 1988;83:596–610. [Google Scholar]
- 34.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 35.Dickie P, Morgan AR, Scraba DG, von Borstel RC. The binding of the anthelmintic pyrvinium cation to deoxyribonucleic acid in vitro. Mol. Pharmacol. 1986;29:427–435. [PubMed] [Google Scholar]
- 36.Stockert JC, Trigoso CI, Llorente AR, Del Castillo P. DNA fluorescence induced by polymethine cation pyrvinium binding. Histochem. J. 1991;23:548–552. doi: 10.1007/BF01041182. [DOI] [PubMed] [Google Scholar]
- 37.Saraswati S, Alfaro MP, Thorne CA, Atkinson J, Lee E, Young PP. Pyrvinium, a potent small molecule Wnt inhibitor, promotes wound repair and post-MI cardiac remodeling. PloS One. 2010;5:e15521. doi: 10.1371/journal.pone.0015521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frederick CA, Williams LD, Ughetto G, van der Marel GA, van Boom JH, Rich A, Wang AH. Structural comparison of anticancer drug-DNA complexes: adriamycin and daunomycin. Biochemistry. 1990;29:2538–2549. [PubMed] [Google Scholar]
- 39.Niswender CM, Copeland SC, Herrick-Davis K, Emeson RB, Sanders-Bush E. RNA editing of the human serotonin 5-hydroxytryptamine 2C receptor silences constitutive activity. J. Biol. Chem. 1999;274:9472–9478. doi: 10.1074/jbc.274.14.9472. [DOI] [PubMed] [Google Scholar]
- 40.Sperling J, Azubel M, Sperling R. Structure and function of the Pre-mRNA splicing machine. Structure. 2008;16:1605–1615. doi: 10.1016/j.str.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 41.Sebbag-Sznajder N, Raitskin O, Angenitzki M, Sato TA, Sperling J, Sperling R. Regulation of alternative splicing within the supraspliceosome. J. Struct. Biol. 2012;177:152–159. doi: 10.1016/j.jsb.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raitskin O, Cho DS, Sperling J, Nishikura K, Sperling R. RNA editing activity is associated with splicing factors in lnRNP particles: the nuclear pre-mRNA processing machinery. Proc. Natl Acad. Sci. USA. 2001;98:6571–6576. doi: 10.1073/pnas.111153798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heinrich B, Zhang Z, Raitskin O, Hiller M, Benderska N, Hartmann AM, Bracco L, Elliott D, Ben-Ari S, Soreq H, et al. Heterogeneous nuclear ribonucleoprotein G regulates splice site selection by binding to CC(A/C)-rich regions in pre-mRNA. J. Biol. Chem. 2009;284:14303–14315. doi: 10.1074/jbc.M901026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turner DH, Mathews DH. NNDB: the nearest neighbor parameter database for predicting stability of nucleic acid secondary structure. Nucleic Acids Res. 2010;38:D280–D282. doi: 10.1093/nar/gkp892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Varani L, Spillantini MG, Goedert M, Varani G. Structural basis for recognition of the RNA major groove in the tau exon 10 splicing regulatory element by aminoglycoside antibiotics. Nucleic Acids Research. 2000;28:710–719. doi: 10.1093/nar/28.3.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schirle NT, Goodman RA, Krishnamurthy M, Beal PA. Selective inhibition of ADAR2-catalyzed editing of the serotonin 2c receptor pre-mRNA by a helix-threading peptide. Org. Biomol. Chem. 2010;8:4898–4904. doi: 10.1039/c0ob00309c. [DOI] [PubMed] [Google Scholar]
- 49.Connolly HM, Crary JL, McGoon MD, Hensrud DD, Edwards BS, Edwards WD, Schaff HV. Valvular heart disease associated with fenfluramine-phentermine. N. Engl. J. Med. 1997;337:581–588. doi: 10.1056/NEJM199708283370901. [DOI] [PubMed] [Google Scholar]
- 50.Bocobza S, Adato A, Mandel T, Shapira M, Nudler E, Aharoni A. Riboswitch-dependent gene regulation and its evolution in the plant kingdom. Genes Dev. 2007;21:2874–2879. doi: 10.1101/gad.443907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.