Abstract
Polymicrogyria is a disorder of neuronal development resulting in structurally abnormal cerebral hemispheres characterized by over-folding and abnormal lamination of the cerebral cortex. Polymicrogyria is frequently associated with severe neurologic deficits including intellectual disability, motor problems, and epilepsy. There are acquired and genetic causes of polymicrogyria, but most patients with a presumed genetic etiology lack a specific diagnosis. Here we report using whole-exome sequencing to identify compound heterozygous mutations in the WD repeat domain 62 (WDR62) gene as the cause of recurrent polymicrogyria in a sibling pair. Sanger sequencing confirmed that the siblings both inherited 1-bp (maternal allele) and 2-bp (paternal allele) frameshift deletions, which predict premature truncation of WDR62, a protein that has a role in early cortical development. The probands are from a non-consanguineous family of Northern European descent, suggesting that autosomal recessive PMG due to compound heterozygous mutation of WDR62 might be a relatively common cause of PMG in the population. Further studies to identify mutation frequency in the population are needed.
Keywords: malformations of cortical development, high-throughput nucleotide sequencing, genetic testing, epilepsy, intellectual disability
INTRODUCTION
Polymicrogyria (PMG) is a CNS malformation characterized pathologically by numerous abnormal small gyri and shallow sulci and abnormal lamination of the cerebral cortex possibly due to impaired neuronal migration or proliferation during cortical development [Leventer et al., 2010]. Clinically, PMG is diagnosed when cranial magnetic resonance imaging (MRI) identifies an irregular appearing cortical surface, a thickened or overfolded cortex, and irregularity at the grey-white matter boundary [Leventer et al., 2010]. Polymicrogyria is a heterogeneous malformation that can be focal or diffuse, unilateral or bilateral, isolated or occurring with other cortical abnormalities [Jansen and Andermann, 2005]. Clinical manifestations are also variable but frequently include cognitive impairment, motor deficits, and epilepsy. Known causes of PMG include intrauterine insults such as infection or ischemia as well as genetic causes which include recurrent gene deletion syndromes or genetically linked loci in which the specific contributing gene has not been identified, including regions on chromosomes 1p36.3, 2p16.1–p23.1, 4q21.21–q22.1, 6q16–q22, 6q26–q27, 21q2, 22q11.2, and Xq28 [Villard et al., 2002; Robin et al., 2006; Dobyns et al., 2008; Ben Cheikh et al., 2009]. Specific gene mutations in RAB3GAP1/2, TBR2, KIAA1279, PAX6, TUBB2B, SRPX2, and GPR56 have also been associated with PMG [Roll et al., 2006; Dobyns et al., 2008; Jaglin et al., 2009; Bahi-Buisson et al., 2010; Borck et al., 2011]. Recent reports have also implicated homozygous mutations in the WDR62 gene in diverse brain malformations which often include polymicrogyria [Bilguvar et al., 2010; Nicholas et al., 2010; Yu et al., 2010]. Unlike the majority of cases of PMG that appear to be spontaneous, these genetic alterations were identified in consanguineous kindreds.
We recently demonstrated the use of high-throughput sequencing to identify a novel causative gene in a family with an autosomal recessive form of Charcot–Marie–Tooth disease [Lupski et al., 2010]. We also showed the utility of DNA capture-sequencing of the human exome in rare variant discovery [Bainbridge et al., 2010]. Here we describe the use of high-throughput whole-exome sequencing to identify compound heterozygous mutations in WDR62 in two brothers from a non-consanguineous family with PMG.
METHODS
Clinical Ascertainment
Informed consent for participation, sample collection, and medical records review was obtained using protocols approved by the Institutional Review Board for Baylor College of Medicine (BCM) and affiliated hospitals. Clinical histories and physical examinations were obtained by one of the authors (board certified pediatric neurologist). DNA was extracted from peripheral blood using the Puregene DNA isolation kit (Gentra System, Minneapolis, MN).
Clinical Case Reports
Patient II-1 was born to a 37-year-old G1P0–1 mother at term via induced vaginal delivery. The pregnancy was complicated by gestational diabetes. Microcephaly was diagnosed in utero at 30 weeks gestation, and he was apparently microcephalic at birth (birth measurements were not available). Cranial MRI on day 13 of life demonstrated bilateral parietal polymicrogyria and an otherwise immature cortical gyral pattern (Fig. 1A–C). Developmentally, he was globally delayed. He developed infantile spasms at age four months, and he continues to suffer with medically intractable symptomatic generalized epilepsy. At age 8.8 years, head circumference was 46.5 cm (<5th centile), weight was 19.6 kg (<5th centile), and height was 109.2 cm (<5th centile). At his current age of 9 years, he also has intellectual disabilities and spastic quadriparesis.
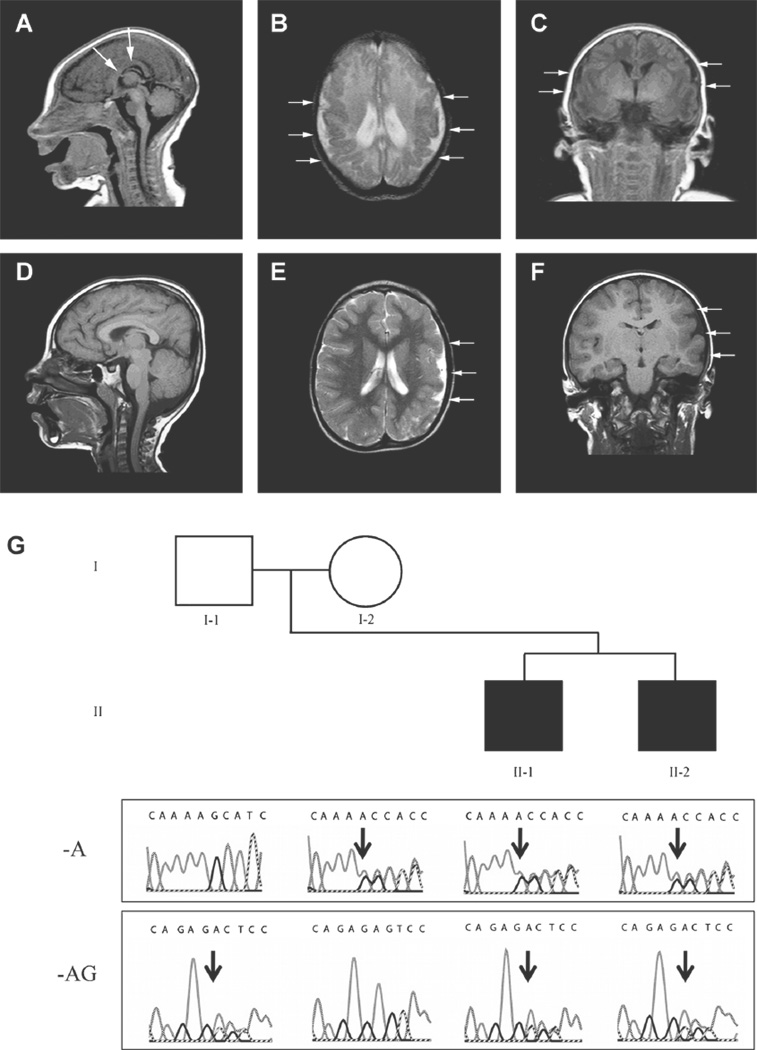
FIG. 1.
Magnetic resonance imaging of the head in patients II-1 (A–C) and II-2 (D–F). A: Sagittal T1 image demonstrates microcephaly and an abnormal corpus callosum (depicted by white arrows). B: Axial T2 and [C] coronal T1 images demonstrate bilateral parietal polymicrogyria and an abnormal sulcal and gyral pattern. Patient II-2: D: Sagittal T1 image demonstrates overall preservation of midline structures. E: Axial T1 and (F) coronal T2 images demonstrate extensive polymicrogyria in the left cerebral hemisphere. White arrows in panels B, C, E, and F depict regions of polymicrogyria. G: Pedigree with Sanger traces below demonstrating WDR62 mutation family segregation. The affected patients (black squares) inherited both the maternal c.2083delA and paternal c.2472_2473delAG alleles. In the traces, black arrows show the beginning of each deletion. Following each heterozygous deletion there are overlapping traces representing the wild-type and shifted sequences. [Color figure can be seen in the online version of this article, available at http://onlinelibrary.wiley.com/journal/10.1002/(ISSN)1552–4833]
Patient II-2 was born to the then 39-year-old G2P1–2 mother at term via planned caesarean. There were no complications during the pregnancy. Birth measurements were not available. Cranial MRI at age 4 years demonstrated an asymmetrically small left cerebral hemisphere with extensive polymicrogyria and a focus of gray matter heterotopia in the right parietal region (Fig. 1D–F); growth parameters at this age included head circumference 45 cm (<5th centile) and weight 13.7 kg (~5th centile). At his current age of 7 years, he has delayed social development and a mild right hemiparesis; but he is ambulatory, uses age-appropriate language, is cognitively on target with his peers, and he has not developed epilepsy. Refer to Figure 1G for the pedigree.
Array comparative genomic hybridization using the BCM Medical Genetics Laboratories 180k oligonucleotide array V8.1 demonstrated normal genomic copy-number in both brothers.
Exome Capture and Sequencing, Read Mapping, and Variant Annotation
A whole exome-enriched library was prepared from genomic DNA isolated from the peripheral blood of the two patients using a Roche NimbleGen (Madison, WI) solution-based capture reagent that included the exons of high-confidence protein-coding genes from CCDS, RefSeq, and Vega gene models as well as microRNAs [Ashurst et al., 2005; Pruitt et al., 2007, 2009; Griffiths-Jones et al., 2008]. The library was sequenced with 50-bp reads on the Applied Biosystems SOLiD platform (Carlsbad, CA), and reads were aligned to the March 2006 human reference assembly (NCBI36/hg18). After sequencing, alignment of raw colorspace reads to the human reference assembly was done using BFAST (version 0.6.4d) [Homer et al., 2009], and base quality was then recalibrated using the Genome Analysis Toolkit (GATK) [McKenna et al., 2010]. A mean of 5.3 Gb of sequencing data aligned to the targeted regions of each sample representing 86.8% of bases having ≥10× coverage. Variant calling of Single Nucleotide Polymorphisms (SNPs) and insertions/deletions was done with SAMtools [Li et al., 2009]. Subsequent variants were then filtered using the following criteria: Phred-like variant quality score of at least 40 (or 30 if observed on both strands) and a minor allele frequency of 15%. Common mutations were excluded by filtering high-quality variants against dbSNP130 [Sherry et al., 2001], 1000 Genomes Pilot 1 [Durbin et al., 2010], and a BCM internal database composed of approximately 600 control exomes. We annotated the list of novel mutations for frameshift insertions/deletions, splice-site, missense, and nonsense variants using the ANNOVAR annotation package [Wang et al., 2010]. Functional consequences of nonsynonymous mutations were predicted using the SIFT [Kumar et al., 2009] and PolyPhen-2 [Adzhubei et al., 2010] algorithms.
Causative Mutation Identification
We hypothesized that an X-linked or an autosomal recessive inheritance pattern was responsible for recurrent polymicrogyria in this family given that both patients were male and the first affected members of their pedigree, however, a dominant inheritance mechanism due to germline mosaicism was also a possibility. For the X-linked model we required both brothers to share at least one novel variant in the same X chromosome gene. The parents were of Northern European descent and did not endorse any known consanguinity. As such, we looked for deleterious compound heterozygous mutations shared by both brothers as the most likely explanation for autosomal recessive inheritance. Due to the differences in imaging and in clinical severity in the brothers, we also generated a list of discordant variants not shared by both brothers that may represent genetic modifiers of the PMG phenotype(s).
RESULTS
Exome Sequencing Identifies Compound Heterozygous Mutations in WDR62
The number of shared variants present under different filtering criteria and inheritance models are demonstrated in Table I. Using the X-linked model, we found a single novel shared variant at chrX:77,800,349 in exon 2 of ZCCHC5 (NM_152694.2), a small two-exon gene encoding 475 amino acids. This missense mutation predicts p.Glu75Asp (c.225G>C) and is expected to be benign by the PolyPhen-2 algorithm. The function of ZCCHC5 is not well known, and the protein has not been associated with known neuronal development pathways. Furthermore, a recent study involving sequencing of the coding exons of the X chromosome in families with X-linked mental retardation identified recurrent, common mutations in ZCCHC5 [Tarpey et al., 2009]. Since common variants are less likely to be deleterious, the identified mutations in ZCCHC5 were not interpreted to be disease-causing.
TABLE I.
Total and Shared Variants in Siblings with PMG
| Nucleotide variants | Patient II-1 | Patient II-2 | Shared |
|---|---|---|---|
| Coding | 17,449 | 17,264 | 13,387 |
| NS/SS/I | 8,571 | 8,439 | 6,411 |
| Rare, non-benigna | 191 | 199 | 106 |
| X-linked | 1 | 3 | 1 |
| Compound heterozygous | 4 | 7 | 3 |
Rare, non-benign variants were not in dbSNP130, 1000 Genomes Pilot 1, or the BCM internal database and were predicted to be deleterious by the SIFT algorithm.
NS, nonsynonymous; SS, splice-site; I, frameshift insertion/deletion.
Under an autosomal recessive model, we identified three genes with compound heterozygous variants common to both probands: ZNF717, EPB41L4A, and WDR62. There were two frameshift insertions in exon 5 of ZNF717 (NM_001128223.1) at chr3:75,869,245 and chr3:75,870,842 that predict p.Cys740SerfsX4 (c.2219_2220insCA) and p.Thr208AspfsX5 (c.622_623insG), respectively. Both mutations are expected to cause early protein truncation. However, we excluded ZNF717 because it exhibits a high-frequency of nonsynonymous mutations in our internal control database, a finding which suggests that ZNF717 is not a highly evolutionarily conserved gene. We also identified compound heterozygous mutations in EPB41L4A (NM_022140.3). The first mutation at chr5:111,528,715 (c.1933-2_1933-1insATTTT) was located in the 3′ splice site upstream of exon 23 and changes the AG acceptor site to a TG, possibly causing exon skipping. The second event was a missense mutation at chr5:111,629,882 in exon 5 encoding p.Arg127His (c.380G>A) predicted to be deleterious by Polyphen-2. The EPB41L4A gene encodes an erythrocyte membrane protein that has no known role in neurogenesis or cortical development, and therefore it was also excluded.
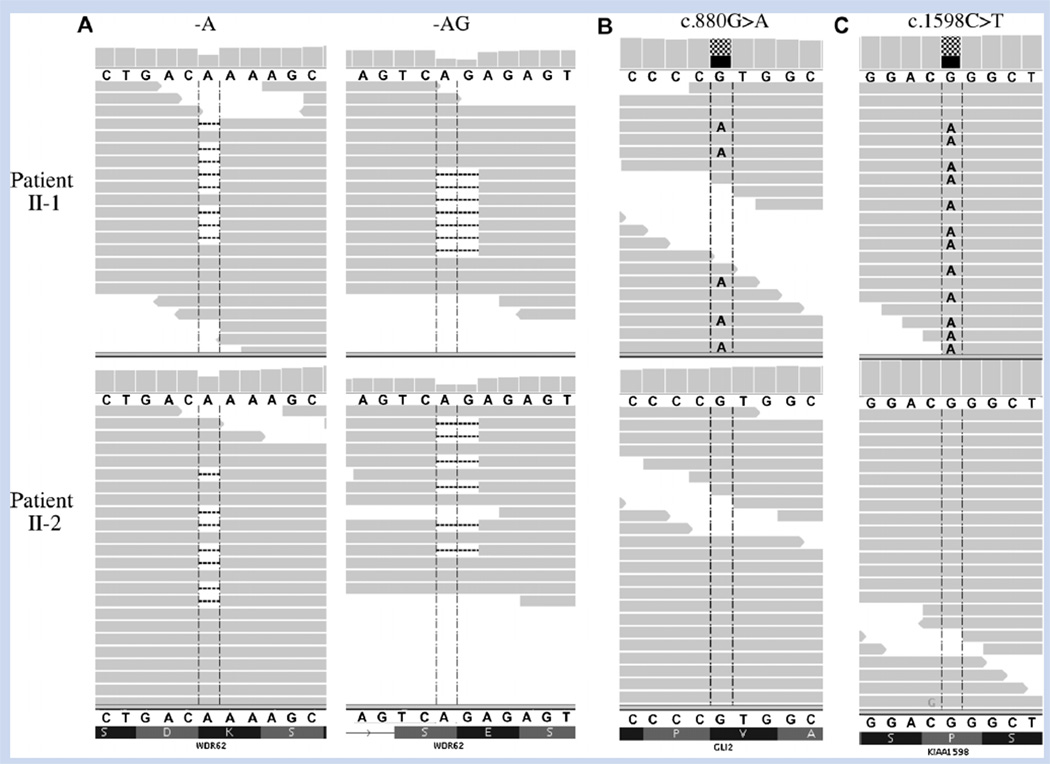
The remaining candidate gene WDR62 (MIM: #600176) was recently implicated in cortical malformations occurring in consanguineous kindreds via an autosomal recessive inheritance mechanism [Bilguvar et al., 2010; Nicholas et al., 2010; Yu et al., 2010]. Affected individuals in those pedigrees were homozygous for deleterious mutations inWDR62 whereas the brothers in our study shared compound heterozygous mutations in the gene (Fig. 2A). The first shared mutation at chr19:41,273,990 in exon 17 of WDR62 (NM_173636.4) predicts p.Ser696AlafsX4 (c.2083delA) while the second shared mutation at chr19:41,283,492 in exon 23 predicts p.Gln918GlyfsX18 (c.2472_2473delAG). Both deletions are expected to cause nonsense mediated decay resulting in reduced or null protein activity. Sanger sequencing validated the probands’ mutations and demonstrated that the parents were unaffected carriers of the 1-bp (mother) and 2-bp (father) deletions (Fig. 1G).
FIG. 2.
A: Sequence alignment visualization showing heterozygous WDR62 mutations in both patient II-1 (upper panel) and patient II-2 (lower panel). Individual sequencing reads are represented by the horizontal grey bars. Both c.2083delA and c.2472_2473delAG deletions are present in approximately half of each patient’s reads, representing a heterozygous state for each mutation. Alignment also shows heterozygous missense variants in (B) GLI2 and (C) KIAA1598 in patient II-1 but not in patient II-2. [Color figure can be seen in the online version of this article, available at http://onlinelibrary.wiley.com/journal/10.1002/(ISSN)1552–4833]
DISCUSSION
This study represents the first report of novel compound heterozygous mutations in the WDR62 gene as a cause of autosomal recessive polymicrogyria. The WDR62 gene on chromosome 19q13.12 contains 32 exons and normally encodes 1,523 amino acids. Its exact function is unknown, but several studies have suggested an important role in neuronal proliferation and migration [Bilguvar et al., 2010; Nicholas et al., 2010; Yu et al., 2010]. One such study noted increased WDR62 expression during early mouse brain development specifically in neural crest cells and ventricular and subventricular zones [Bilguvar et al., 2010]. The latter cortical regions are especially important to neurogenesis [Lim and Alvarez-Buylla, 1999]. Proper cortical development depends on the coordinated processes of neuronal proliferation and differentiation, and defects can result in a number of brain malformations [Squier and Jansen, 2010].
A recent report proposed that WDR62 acts as a spindle pole protein required for proper neuroprogenitor cell division [Nicholas et al., 2010]. Mutations in WDR62 may therefore interrupt this process leading to abnormal cortical development. Similarly, mutations in another spindle pole gene ASPM cause autosomal recessive primary microcephaly (MCPH5; MIM: #608716), which has features overlapping with PMG [Fish et al., 2006; Hutchins et al., 2010]. Mutations in another gene GPR56 cause bifrontoparietal polymicrogyria (BFPP), and expression of GPR56 is required for structural integrity of the pial basement membrane during neuronal migration [Bahi-Buisson et al., 2010]. Thus, it is likely that defects in other structural proteins essential to cortical development also lead to a wide array of brain malformations including PMG.
WDR62 mutations are associated with a wide spectrum of cortical malformations in addition to PMG. Affected individuals often have variable levels of cortical thickening and structural defects that may include the hippocampus and cerebellum and involve one or both brain hemispheres [Bilguvar et al., 2010]. One report suggested that such phenotypic variability may be due to reduced protein function caused by missense mutations versus complete absence of function resulting from nonsense mutations [Nicholas et al., 2010]. However, despite identical frameshift mutations causing premature stop codons in WDR62, patient II-1 in our study had a more severe brain malformation characterized by extensive bilateral PMG, an abnormal corpus callosum, and a much more severe clinical phenotype compared to patient II-2who had asymmetric PMG and was less severely affected clinically. Furthermore, no detectable genomic copy number variation was present in either patient to explain their phenotypic differences. We propose several explanations for the phenotypic discordance.
The pregnancy resulting in patient II-1 was complicated by gestational diabetes. Infants of diabetic mothers are at significant risk for major neurologic anomalies including anencephaly, arrhinencephaly, microcephaly, holoprosencephaly, and neural tube defects [Becerra et al., 1990; Tyrala, 1996]. It is possible that the abnormal metabolic environment during fetal development superimposed on the WDR62 mutations caused the more severe phenotype in patient II-1.
Another possibility is that variation in other genes involved in cortical development could have a synergistic effect on patient II-1’s phenotype. Of the 85 rare, non-benign variants unique to patient II-1, we identified two that may have a role in brain development. The first was a novel heterozygous missense mutation at chr2:121,444,473 in GLI2 (NM_005270.4: c.880G>A) predicting a p.Val294Met change that was not observed in control exomes (Fig. 2B). This gene has been associated with dominantly inherited pituitary anomalies and holoprosencephaly through its role in the developing ventral brain and facial structures [Roessler et al., 2003]. GLI2 is a transcription factor that mediates Sonic Hedgehog signaling in vertebrates and like patient II-1, carriers of GLI2 mutations may have severe structural brain abnormalities, epilepsy, and developmental delay. Unlike patient II-1, however, pituitary anomalies and polydactyly are often observed with GLI2 loss-of-function mutations [Roessler et al., 2003]. Patient II-1 (but not patient II-2) also carried a heterozygous mutation at chr10:118,651,341 in KIAA1598 (NM_001127211.1: c.1598C>T) predicting a p.Pro533Leu change (Fig. 2C). This gene is highly expressed in the brain and encodes a protein with a role in axon guidance via its interaction with actin filament retrograde flow and L1-CAM [Shimada et al., 2008]. To date however, mutations in this gene have not been associated with any clinical phenotypes. Sanger sequencing showed that both the GLI2 and KIAA1598 variants were inherited from the phenotypically normal father which decreases the likelihood that they contributed significantly to patient II-1’s condition. Further work is required to understand the precise function of WDR62 in brain development and how other genetic and/or non-genetic factors affect the spectrum of phenotypes observed in patients with WDR62 alterations.
In conclusion, we report novel compound heterozygous mutations in WDR62 causing autosomal recessive polymicrogyria in a non-consanguineous kindred. These results support sequence analysis of WDR62 in the clinical diagnostic testing of individuals with PMG, as families with an affected child in whom both parents are carriers will have a25%risk of recurrence; and pre-implantation genetic diagnosis is possible.
ACKNOWLEDGMENTS
We are grateful to the family for participation in this study. We thank Robert McNeil for image compilation. This study was funded by NIH/NINDS grant 5K08NS062711-03 (M.B.R.). We thank the BCM Medical Genetics Laboratories for technical assistance. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NINDS or the National Institutes of Health.
Grant sponsor: NIH/NINDS; Grant number: 5K08NS062711-03.
REFERENCES
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashurst JL, Chen CK, Gilbert JG, Jekosch K, Keenan S, Meidl P, Searle SM, Stalker J, Storey R, Trevanion S, Wilming L, Hubbard T. The Vertebrate Genome Annotation (Vega) database. Nucleic Acids Res. 2005;33:D459–D465. doi: 10.1093/nar/gki135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahi-Buisson N, Poirier K, Boddaert N, Fallet-Bianco C, Specchio N, Bertini E, Caglayan O, Lascelles K, Elie C, Rambaud J, Baulac M, An I, Dias P, des Portes V, Moutard ML, Soufflet C, El Maleh M, Beldjord C, Villard L, Chelly J. GPR56-related bilateral frontoparietal polymicrogyria: Further evidence for an overlap with the cobblestone complex. Brain. 2010;133:3194–3209. doi: 10.1093/brain/awq259. [DOI] [PubMed] [Google Scholar]
- Bainbridge MN, Wang M, Burgess DL, Kovar C, Rodesch MJ, D’Ascenzo M, Kitzman J, Wu YQ, Newsham I, Richmond TA, Jeddeloh JA, Muzny D, Albert TJ, Gibbs RA. Whole exome capture in solution with 3 Gbp of data. Genome Biol. 2010;11:R62. doi: 10.1186/gb-2010-11-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra JE, Khoury MJ, Cordero JF, Erickson JD. Diabetes mellitus during pregnancy and the risks for specific birth defects: A population-based case-control study. Pediatrics. 1990;85:1–9. [PubMed] [Google Scholar]
- Ben Cheikh BO, Baulac S, Lahjouji F, Bouhouche A, Couarch P, Khalili N, Regragui W, Lehericy S, Ruberg M, Benomar A, Heath S, Chkili T, Yahyaoui M, Jiddane M, Ouazzani R, LeGuern E. A locus for bilateral occipital polymicrogyria maps to chromosome 6q16-q22. Neurogenetics. 2009;10:35–42. doi: 10.1007/s10048-008-0143-3. [DOI] [PubMed] [Google Scholar]
- Bilguvar K, Ozturk AK, Louvi A, Kwan KY, Choi M, Tatli B, Yalnizoglu D, Tuysuz B, Caglayan AO, Gokben S, Kaymakcalan H, Barak T, Bakircioglu M, Yasuno K, Ho W, Sanders S, Zhu Y, Yilmaz S, Dincer A, Johnson MH, Bronen RA, Kocer N, Per H, Mane S, Pamir MN, Yalcinkaya C, Kumandas S, Topcu M, Ozmen M, Sestan N, Lifton RP, State MW, Gunel M. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010;467:207–210. doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borck G, Wunram H, Steiert A, Volk AE, Korber F, Roters S, Herkenrath P, Wollnik B, Morris-Rosendahl DJ, Kubisch C. A homozygous RAB3GAP2 mutation causes Warburg Micro syndrome. Hum Genet. 2011;129:45–50. doi: 10.1007/s00439-010-0896-2. [DOI] [PubMed] [Google Scholar]
- Dobyns WB, Mirzaa G, Christian SL, Petras K, Roseberry J, Clark GD, Curry CJ, McDonald-McGinn D, Medne L, Zackai E, Parsons J, Zand DJ, Hisama FM, Walsh CA, Leventer RJ, Martin CL, Gajecka M, Shaffer LG. Consistent chromosome abnormalities identify novel polymicrogyria loci in 1p36.3, 2p16.1–p23.1, 4q21.21–q22.1, 6q26-q27, and 21q2. Am J Med Genet Part A. 2008;146A:1637–1654. doi: 10.1002/ajmg.a.32293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish JL, Kosodo Y, Enard W, Paabo S, Huttner WB. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc Natl Acad Sci. 2006;103:10438–10443. doi: 10.1073/pnas.0604066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homer N, Merriman B, Nelson SF. BFAST: An alignment tool for large scale genome resequencing. PLoS ONE. 2009;4:e7767. doi: 10.1371/journal.pone.0007767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins JR, Toyoda Y, Hegemann B, Poser I, Heriche JK, Sykora MM, Augsburg M, Hudecz O, Buschhorn BA, Bulkescher J, Conrad C, Comartin D, Schleiffer A, Sarov M, Pozniakovsky A, Slabicki MM, Schloissnig S, Steinmacher I, Leuschner M, Ssykor A, Lawo S, Pelletier L, Stark H, Nasmyth K, Ellenberg J, Durbin R, Buchholz F, Mechtler K, Hyman AA, Peters JM. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–599. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglin XH, Poirier K, Saillour Y, Buhler E, Tian G, Bahi-Buisson N, Fallet-Bianco C, Phan-Dinh-Tuy F, Kong XP, Bomont P, Castelnau-Ptakhine L, Odent S, Loget P, Kossorotoff M, Snoeck I, Plessis G, Parent P, Beldjord C, Cardoso C, Represa A, Flint J, Keays DA, Cowan NJ, Chelly J. Mutations in the beta-tubulin gene TUBB2B result in asymmetrical polymicrogyria. Nat Genet. 2009;41:746–752. doi: 10.1038/ng.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen A, Andermann E. Genetics of the polymicrogyria syndromes. J Med Genet. 2005;42:369–378. doi: 10.1136/jmg.2004.023952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Leventer RJ, Jansen A, Pilz DT, Stoodley N, Marini C, Dubeau F, Malone J, Mitchell LA, Mandelstam S, Scheffer IE, Berkovic SF, Andermann F, Andermann E, Guerrini R, Dobyns WB. Clinical and imaging heterogeneity of polymicrogyria: A study of 328 patients. Brain. 2010;133:1415–1427. doi: 10.1093/brain/awq078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DA, Alvarez-Buylla A. Interaction between astrocytes and adult subventricular zone precursors stimulates neurogenesis. Proc Natl Acad Sci. 1999;96:7526–7531. doi: 10.1073/pnas.96.13.7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR, Reid JG, Gonzaga-Jauregui C, Rio Deiros D, Chen DC, Nazareth L, Bainbridge M, Dinh H, Jing C, Wheeler DA, McGuire AL, Zhang F, Stankiewicz P, Halperin JJ, Yang C, Gehman C, Guo D, Irikat RK, Tom W, Fantin NJ, Muzny DM, Gibbs RA. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med. 2010;362:1181–1191. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas AK, Khurshid M, Desir J, Carvalho OP, Cox JJ, Thornton G, Kausar R, Ansar M, Ahmad W, Verloes A, Passemard S, Misson JP, Lindsay S, Gergely F, Dobyns WB, Roberts E, Abramowicz M, Woods CG. WDR62is associated with the spindle pole and is mutated inhuman microcephaly. Nat Genet. 2010;42:1010–1014. doi: 10.1038/ng.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, Hart E, Suner MM, Landrum MJ, Aken B, Ayling S, Baertsch R, Fernandez-Banet J, Cherry JL, Curwen V, Dicuccio M, Kellis M, Lee J, Lin MF, Schuster M, Shkeda A, Amid C, Brown G, Dukhanina O, Frankish A, Hart J, Maidak BL, Mudge J, Murphy MR, Murphy T, Rajan J, Rajput B, Riddick LD, Snow C, Steward C, Webb D, Weber JA, Wilming L, Wu W, Birney E, Haussler D, Hubbard T, Ostell J, Durbin R, Lipman D. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin NH, Taylor CJ, McDonald-McGinn DM, Zackai EH, Bingham P, Collins KJ, Earl D, Gill D, Granata T, Guerrini R, Katz N, Kimonis V, Lin JP, Lynch DR, Mohammed SN, Massey RF, McDonald M, Rogers RC, Splitt M, Stevens CA, Tischkowitz MD, Stoodley N, Leventer RJ, Pilz DT, Dobyns WB. Polymicrogyria and deletion 22q11.2 syndrome: Window to the etiology of a common cortical malformation. Am J Med Genet Part A. 2006;140A:2416–2425. doi: 10.1002/ajmg.a.31443. [DOI] [PubMed] [Google Scholar]
- Roessler E, Du YZ, Mullor JL, Casas E, Allen WP, Gillessen-Kaesbach G, Roeder ER, Ming JE, Ruiz I, Altaba A, Muenke M. Loss-of-function mutations in the human GLI2 gene are associated with pituitary anomalies and holoprosencephaly-like features. Proc Natl Acad Sci. 2003;100:13424–13429. doi: 10.1073/pnas.2235734100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll P, Rudolf G, Pereira S, Royer B, Scheffer IE, Massacrier A, Valenti MP, Roeckel-Trevisiol N, Jamali S, Beclin C, Seegmuller C, Metz-Lutz MN, Lemainque A, Delepine M, Caloustian C, de Saint Martin A, Bruneau N, Depetris D, Mattei MG, Flori E, Robaglia-Schlupp A, Levy N, Neubauer BA, Ravid R, Marescaux C, Berkovic SF, Hirsch E, Lathrop M, Cau P, Szepetowski P. SRPX2 mutations in disorders of language cortex and cognition. Hum Mol Genet. 2006;15:1195–1207. doi: 10.1093/hmg/ddl035. [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Toriyama M, Uemura K, Kamiguchi H, Sugiura T, Watanabe N, Inagaki N. Shootin1 interacts with actin retrograde flow and L1-CAM to promote axon outgrowth. J Cell Biol. 2008;181:817–829. doi: 10.1083/jcb.200712138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squier W, Jansen A. Abnormal development of the human cerebral cortex. J Anat. 2010;217:312–323. doi: 10.1111/j.1469-7580.2010.01288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpey PS, Smith R, Pleasance E, Whibley A, Edkins S, Hardy C, O’Meara S, Latimer C, Dicks E, Menzies A, Stephens P, Blow M, Greenman C, Xue Y, Tyler-Smith C, Thompson D, Gray K, Andrews J, Barthorpe S, Buck G, Cole J, Dunmore R, Jones D, Maddison M, Mironenko T, Turner R, Turrell K, Varian J, West S, Widaa S, Wray P, Teague J, Butler A, Jenkinson A, Jia M, Richardson D, Shepherd R, Wooster R, Tejada MI, Martinez F, Carvill G, Goliath R, de Brouwer AP, van Bokhoven H, Van Esch H, Chelly J, Raynaud M, Ropers HH, Abidi FE, Srivastava AK, Cox J, Luo Y, Mallya U, Moon J, Parnau J, Mohammed S, Tolmie JL, Shoubridge C, Corbett M, Gardner A, Haan E, Rujirabanjerd S, Shaw M, Vandeleur L, Fullston T, Easton DF, Boyle J, Partington M, Hackett A, Field M, Skinner C, Stevenson RE, Bobrow M, Turner G, Schwartz CE, Gecz J, Raymond FL, Futreal PA, Stratton MR. A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat Genet. 2009;41:535–543. doi: 10.1038/ng.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrala EE. The infant of the diabetic mother. Obstet Gynecol Clin North Am. 1996;23:221–241. doi: 10.1016/s0889-8545(05)70253-9. [DOI] [PubMed] [Google Scholar]
- Villard L, Nguyen K, Cardoso C, Martin CL, Weiss AM, Sifry-Platt M, Grix AW, Graham JM, Jr, Winter RM, Leventer RJ, Dobyns WB. A locus for bilateral perisylvian polymicrogyria maps to Xq28. Am J Hum Genet. 2002;70:1003–1008. doi: 10.1086/339433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TW, Mochida GH, Tischfield DJ, Sgaier SK, Flores-Sarnat L, Sergi CM, Topcu M, McDonald MT, Barry BJ, Felie JM, Sunu C, Dobyns WB, Folkerth RD, Barkovich AJ, Walsh CA. Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture. Nat Genet. 2010;42:1015–1020. doi: 10.1038/ng.683. [DOI] [PMC free article] [PubMed] [Google Scholar]