Abstract
Lignin is a ubiquitous polymer present in cell walls of all vascular plants, where it rigidifies and strengthens the cell wall structure through covalent cross-linkages to cell wall polysaccharides. The presence of lignin makes the cell wall recalcitrant to conversion into fermentable sugars for bioenergy uses. Therefore, reducing lignin content and modifying its linkages have become major targets for bioenergy feedstock development through either biotechnology or traditional plant breeding. In addition, lignin synthesis has long been implicated as an important plant defense mechanism against pathogens, because lignin synthesis is often induced at the site of pathogen attack. This article explores the impact of lignin modifications on the susceptibility of a range of plant species to their associated pathogens, and the implications for development of feedstocks for the second-generation biofuels industry. Surprisingly, there are some instances where plants modified in lignin synthesis may display increased resistance to associated pathogens, which is explored in this article.
Keywords: plant pathogens, lignin, brown midrib, monolignol pathway, CAD, COMT
INTRODUCTION
In the U.S. and around the world, there are increasing efforts to develop and utilize alternatives to fossil fuels to meet our energy needs, thereby reducing carbon dioxide emissions that potentially impact global warming. Currently, corn grain and sugarcane juice are being converted into ethanol for blending in gasoline. Research efforts have been directed toward developing means to convert plant biomass from a range of sources into liquid fuels for the transportation sector. Cellulosic biofuels rely on chemically and biochemically breaking down cell wall polysaccharides (cellulose and hemicellulose) into their sugar monomers, and converting the sugar into fuels. A third component of cell walls is the phenolic polymer lignin, which structurally fortifies the cell walls making them rigid and resistant to microbial degradation. Lignin content has been shown to negatively impact cellulosic bioenergy conversion via saccharification and fermentation to ethanol (Chen and Dixon, 2007; Dien et al., 2009), which has made reducing lignin and altering lignin composition a major target to improve plants for cellulosic bioenergy. Conversely, increasing the lignin content of herbaceous feedstocks may benefit conversion of biomass to syngas and bio-oil biofuel via pyrolysis. In either case, efforts to manipulate lignin content and composition have primarily focused on the 10 steps of the monolignol pathway (Figure 1), in which lignin monomers are synthesized from the amino acid phenylalanine, then oxidatively polymerized into hydroxyphenol- (H-), guiacyl- (G-), or sinapyl- (S-) lignin. Lignin serves the critical function of reinforcing vascular elements for water transport under negative pressure; in severely lignin deficient plants, vascular collapse has been observed (Piquemal et al., 1998; Jones et al., 2001; Ruel et al., 2009). Thus, there is a lower limit for lignin manipulation. In addition to its roles in fortifying cell walls, lignin deposition has long been implicated as an important defense mechanism against pests and pathogens (Vance et al., 1980). A critical question for bioenergy feedstock development is whether manipulating lignin content and composition will be detrimental to plant defenses against pathogens. Herein, we examine this question and the cause for concern in manipulating lignin, based on current published literature.
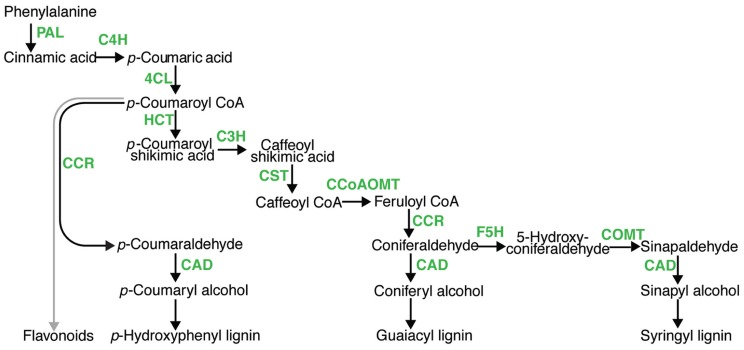
FIGURE 1.
A model for monolignol pathway. In phenylpropanoid metabolism, there are 10 enzymatic steps (green) leading to hydroxycinnamyl alcohols which are polymerized into lignin, namely; phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxylase (C4H), 4-coumarate-CoA ligase (4CL), hydroxycinnamoyl CoA:shikimate transferase (HCT), p-coumarate 3-hydroxylase (C’3H), caffeoyl CoA O-methyltransferase (CCoAOMT), cinnamyl CoA reductase (CRR), ferulate 5-hydroxylase (F5H), caffeic acid O-methyltransferase (COMT), and cinnamyl alcohol dehydrogenase (CAD).
ROLE OF LIGNIN IN PLANT DEFENSE
There is a strong case for the involvement of lignin in plant defense. Lignin provides a physical barrier against initial ingress (Buendgen et al., 1990; Bonello et al., 2003), and in a wide range of plant species lignin or lignin-like phenolic polymers are induced and rapidly deposited in cell walls in response to both biotic and abiotic stresses, which may limit further growth and/or confine invading pathogens (Siegrist et al., 1994; Lange et al., 1995; Baayen et al., 1996; Smit and Dubery, 1997; Bonello et al., 2003; Hudgins et al., 2004; Wuyts et al., 2006; Menden et al., 2007). “Defense” lignin may prevent further ingress or diffusion of pathogen-produced toxins (Carver et al., 1994; Duschnicky et al., 1998). However, “defense” lignin deposition is often only monitored microscopically as cell wall autofluorescence or via histochemical staining techniques (Haegi et al., 2008; Eynck et al., 2009). “Defense” lignin was often shown to have elevated levels of H-subunits as compared to structural lignin in the cases analyzed (Ride, 1975; Hammerschmidt et al., 1985; Doster and Bostock, 1988; Robertsen and Svalheim, 1990; Lange et al., 1995). The phenylpropanoid pathway leads to the synthesis of numerous other phenolic compounds besides monolignols, including phenolic phytoalexins, stilbenes, coumarins, and flavonoids (Lo and Nicholson, 1998; Yu et al., 2000, 2005; Dixon et al., 2002). A number of these compounds have also been implicated in plant defense (Weiergang et al., 1996; Dicko et al., 2005; Lozovaya et al., 2007). For example, the defense signaling hormone salicylic acid (SA) might also be derived from the phenylpropanoid pathway in some plants (Ruuhola and Julkunen-Tiitto, 2003; Pan et al., 2006). Moreover, abiotic or biotic stresses including pathogens have been shown to induce the expression of genes encoding monolignol biosynthetic enzymes in many plant species (Kliebenstein et al., 2002; Truman et al., 2006; Olsen et al., 2008; Zhao et al., 2009). Likewise, the protein levels and enzymatic activities corresponding to these genes were also shown to be elevated under these stresses in a number of plant species (Mitchell et al., 1999). Together these observations indicate that lignin deposition is part of a generalized resistance response to biotic stresses (Nicholson and Hammerschimdt, 1992). Thus, it remains to be determined whether bioenergy crops that are impaired or altered in their ability to synthesize lignin will also be impaired in their ability to induce these defense responses upon pathogen attack. Recent research has suggested that impairing lignin biosynthesis does not lessen resistance to some pathogens (Delgado et al., 2002; Funnell and Pedersen, 2006; Peltier et al., 2009; Quentin et al., 2009; Funnell-Harris et al., 2010). Because very little has been published on the effects of lignin modification on plant–pathogen interactions in bioenergy feedstocks with the exception of maize (Zea mays) and sorghum (Sorghum bicolor), we have also included a review of the literature on effects of impairing steps in the monolignol pathway to pathogen responses in other plant species.
However, several pathogens have been isolated and identified that pose potential threats to some of the perennial grass species being considered as herbaceous bioenergy feedstocks including switchgrass (Panicum virgatum), napiergrass (Pennisetum purpureum), sugarcane/energycane (complex hybrid Saccharum spp.), and miscanthus (Miscanthus × giganteus). Pathogens may pose a greater threat to perennial grasses as compared to annual row crops such as maize and sorghum, because production relies on establishment and harvest across multiple years before replanting, and the continual presence of the plants in the field provides refuge for the pathogens. Another factor that could impact plant–pathogen interactions is the level of genetic diversity within the field setting. Switchgrass varieties are maintained as an outcrossing population (Martïnez-Reyna and Vogel, 2002; Nageswara-Rao et al., 2012), hence maintain level genetic diversity. In contrast, the clonally propagated miscanthus is genetically identical (Lewandowski et al., 2000). Fungal leaf rusts caused by Puccinia spp. have been identified in sugarcane (Dixon et al., 2010) and switchgrass (Gustafson et al., 2003; Wadl et al., 2011). Fungal leaf blights caused by Leptosphaerulina chartarum (Ahonsi et al., 2013) and Leptosphaeria sp. (O’Neill and Farr, 1996) have been identified in Miscanthus × giganteus or related Miscanthus species. Rhizome rot is a significant threat to miscanthus establishment, which is caused by three pathogenic fungal species (Fusarium avenaceum, F. oxysporum, and Mucor hiemalis; Beccari et al., 2010; Covarelli et al., 2012). A fungal smut has been identified on napiergrass caused by Ustilago kamerunensis (Farrell et al., 2001). Anthracnose, a foliar blight caused by Colletotrichum navitas has also been identified in switchgrass (Crouch et al., 2009). Overall, these studies indicated that similar to row crops, fungal pathogens pose a serious threat to these bioenergy crops. In addition, viruses also pose a threat to bioenergy feedstocks, which have been documented in switchgrass and miscanthus (Chatani et al., 1991; Turina et al., 1998; Lamptey et al., 2003).
PHENYLALANINE AMMONIA LYASE
Phenylalanine ammonia lyase (PAL) is the first committed step in monolignol biosynthesis and the phenylpropanoid pathway. Altering the expression of this central gene has been shown to impact plant–pathogen interactions in model systems. In tobacco (Nicotiana tabacum), antisense suppression of this gene led to increased susceptibility to the fungal pathogen Cercospora nicotianae, the causal agent of frogeye disease (Maher et al., 1994; Shadle et al., 2003). Tobacco plants over-expressing this gene had reduced susceptibility to Cercospora nicotianae, but resistance to tobacco mosaic virus (TMV) was unchanged (Shadle et al., 2003). Over-expression of PAL in tobacco resulted in significantly increased levels of the defense signaling compound SA and the defense related compound chlorogenic acid upon induction (Howles et al., 1996; Felton et al., 1999). Furthermore, over-expressing the bacterial salicylate hydroxylase gene (NahG), which degrades SA, in concert with PAL over-expression, increased susceptibility to TMV although resistance to Cercospora nicotianae was unaffected (Shadle et al., 2003). These results indicated that TMV resistance required SA but not chlorogenic acid, while increased resistance to Cercospora nicotianae only required elevated levels of chlorogenic acid and not SA (Shadle et al., 2003). Conversely, PAL over-expression in tobacco resulted in increased susceptibility to the insect Heliothis virescens, and a PAL-suppressed line had increased resistance, which was attributed to the antagonistic relationship between SA signaling and jasmonic acid (JA) signaling (Felton et al., 1999). The Arabidopsis genome contains four PAL genes. T-DNA insertion mutants were isolated for all four genes, and these mutants were crossed to create double, triple, and quadruple pal mutants (Huang et al., 2010). The pal1/2/3/4 quadruple mutant showed increased susceptibility to the bacterial pathogen Pseudomonas syringae relative to WT, and pal1/2 also had increased susceptibility to this pathogen relative to WT and intermediate to pal1/2/3/4 (Huang et al., 2010). SA, lignin and anthocyanin related pigment levels were significantly reduced in pal1/2/3/4 plants (Huang et al., 2010). However, these changes in susceptibility to pathogens cannot be directly attributed to changes in lignin content or composition, because PAL is involved in the synthesis of the full range of phenolic compounds, some of which have been implicated in defense, like chlorogenic acid and flavonoid compounds.
HYDROXYCINNAMOYL CoA: SHIKIMATE TRANSFERASE
In Arabidopsis (Arabidopsis thaliana) and alfalfa (Medicago sativa), antisense/RNAi suppression of hydroxycinnamoyl CoA: shikimate transferase (HCT) one of the initial steps in monolignol biosynthesis, illustrates the potential of genetic/transgenic alterations to this pathway to constitutively activate defenses (Gallego-Giraldo et al., 2011a, b). In both plant species, antisense/RNAi suppression of HCT resulted in significant reductions in lignin content and stunted plants relative to WT (Shadle et al., 2007; Li et al., 2010). In alfalfa, these plants showed increased resistance to the fungal pathogen Colletotrichum trifolli (Gallego-Giraldo et al., 2011b). In the absence of a pathogen, SA levels were highly elevated relative to WT in both species and several defense related genes were also highly induced relative to WT in alfalfa (Gallego-Giraldo et al., 2011a, b). In Arabidopsis, growth was partially restored in NahG HCT-RNAi and SA induction deficient2-2(sid2-2) HCT-RNAi; SID2 encodes an isochorismate synthase required for isochorismate-dependent SA synthesis. These results indicate that the stunted growth phenotype is due to elevated SA, occurring through an isochorismate-dependent pathway, rather than resulting from excess phenylalanine intermediates leading to the synthesis of SA in HCT-RNAi plants. Elevated levels of cold-water extractable pectin were correlated to elevated SA levels in transgenic alfalfa plants, which were RNAi-suppressed for six different genes (steps) in the monolignol pathway (Gallego-Giraldo et al., 2011a). Highest levels of SA and cold-water extractable pectin were observed in HCT-suppressed lines relative to WT or the other five monolignol biosysthetic gene-suppressed lines (Gallego-Giraldo et al., 2011a). Pectic oligosaccharides have been implicated as defense signals in other systems (Darvill and Albersheim, 1984; Roco et al., 1993), and are the potential trigger for the defense responses observed in HCT lines. Thus, the effects observed in the HCT-suppressed lines could potentially result from changes in cell wall structure, the first line of defense for the plant, rather than directly resulting from an alteration in phenylpropanoid metabolism.
CAFFEIC O-METHYLTRANSFERASE
In Arabidopsis and tobacco, antisense/RNAi suppression of caffeic O-methyltransferase (COMT), the penultimate step in monolignol biosynthesis, was reported to increase resistance to pathogens or to have no effect on interaction with pathogens. In Arabidopsis, comt1 mutants show enhanced resistance to the oomycete pathogen Hyaloperonospora arabidopsidis, which is the causal agent of downy mildew (Quentin et al., 2009). There were significantly fewer asexual spores on comt1 plants relative to WT, because sexual sporulation was increased in comt1 plants, resulting in attenuated mycelium growth (Quentin et al., 2009). Exposing the pathogen to the phenolic compound 2-O-5-hydroxyferuloyl-L-malate, which is present in comt1 and absent in WT plants, promoted sexual reproduction (Quentin et al., 2009). However, comt1 plants showed increased susceptibility relative to WT to the bacterial pathogens Xanthomonas campestris pv. campestris and Pseudomonas syringae and a less virulent strain (T4) of the fungal pathogen Botrytis cinerea (Quentin et al., 2009). In tobacco, COMT antisense lines were resistant to Agrobacterium tumefaciens infection, and had reduced tumor area and mass relative to WT (Maury et al., 2010). Bacterial virulence (vir) gene induction was reduced in the COMT-suppressed line likely due to the highly reduced level of the phenolic elicitor of Agrobacterium acetosyringone (Maury et al., 2010). Acetosyringone is probably derived from Coenzyme A dependent β-oxidation of hydroxycinnamoyl-CoA intermediates of monolignol biosynthesis (Blount et al., 2002; Negrel and Javelle, 2010). In Arabidopsis and tobacco, the alteration to phenylpropanoid metabolism by reducing COMT activity appears to directly result in increased resistance to two of the pathogens tested, downy mildew and Agrobacterium, respectively. However, the same Arabidopsis plants showed increased susceptibility to two bacterial pathogens and a less virulent strain of Botrytis cinerea.
CINNAMYL ALCOHOL DEHYDROGENASE
In flax (Linum usitatissimum L.), RNAi suppression of the cinnamyl alcohol dehydrogenase (CAD) gene, the last step in monolignol biosynthesis, increased susceptibility to the pathogenic fungus F. oxysporum. A seedling assay showed the percent of infected seedlings was twofold higher in two CAD RNAi lines relative to WT (Wróbel-Kwiatkowska et al., 2007). In Arabidopsis, the cad-c and cad-d double mutants, which were shown to be required for monolignol biosynthesis (Kim et al., 2004; Sibout et al., 2005), showed increased susceptibility to both a virulent and an avirulent strain of the bacterial pathogen Pseudomonas syringae pv. tomato (Pst;(DC3000, virulent; DC3000/avrPphB, avirulent)) relative to WT based on bacterial growth following inoculation (Tronchet et al., 2010). Together, these results suggest that CAD-deficiency may increase the susceptibility of plants to a range of pathogens. This result might have implications for bioenergy feedstocks, because CAD suppression is often targeted to reduce lignin content.
OTHER STEPS IN MONOLIGNOL SYNTHESIS
In Arabidopsis, the ferulic acid 5-hydroxylase 1 (fah1) mutant, which encodes the ferulic acid 5-hydroxylase (F5H), last hydroxylase in monolignol synthesis, showed increased susceptibility to the fungal pathogen Sclerotinia sclerotiorum relative to WT in leaf assays (Huang et al., 2009). In diploid wheat (Triticum monococcum L.), five genes in monolignol biosynthesis were transiently silenced using particle bombardment of an RNAi vector containing PAL, caffeoyl-CoA O-methyltransferase (CCoAMT), F5H, COMT, or CAD genes (Bhuiyan et al., 2009). The bombarded leaves were inoculated with the powdery mildew fungal pathogens Blumeria graminis f. sp. tritici (host-specific) and Blumeria graminis f. sp. hordei (non-host). The silencing of all five genes individually and in pairs increased the susceptibility to both pathogens relative to the control bombarded with the empty RNAi vector, as determined by penetration efficiency of the fungus (Bhuiyan et al., 2009). However, it is unclear whether this transient approach to gene silencing is relevant to the stable approaches used to impair genes within this pathway for bioenergy feedstock improvement.
BIOENERGY FEEDSTOCKS
There has been very little published on plant pathogen interactions in bioenergy feedstocks with modified lignin content and composition. In hybrid poplar (Populus tremula × Populus alba), it has been reported that no increased disease incidence were observed in antisense COMT or CAD lines relative to WT (Halpin et al., 2007). The one exception where the effects of lignin modification on plant pathogen interactions has been examined are the brown midrib (bmr/bm) mutants of sorghum and maize (Zea mays), which have long been known to have reduced lignin content (Jorgenson, 1931; Porter et al., 1978). There are at least five Bm loci identified in maize (Chen et al., 2012) and at least seven Bmr loci in sorghum (Pedersen et al. unpublished). Three Bmr loci have been cloned and characterized in sorghum. Bmr2, Bmr6, and Bmr12 all encode enzymes in monolignol biosynthesis: a 4-coumarate coenzyme A ligase (4CL), a CAD, and a COMT, respectively (Bout and Vermerris, 2003; Saballos et al., 2009, 2012; Sattler et al., 2009). In maize, the Bm3 locus encodes a COMT protein (Vignols et al., 1995) that is orthologous to Bmr12, and the Bm1 locus encodes a CAD protein that is orthologous to Bmr6 (Halpin et al., 1998; Chen et al., 2012). Lignin deposition and the induction of phenylpropanoid-related genes during pathogen attack (described above) led to the assumption that brown midrib plants are inherently more disease susceptible when challenged. However, studies examining both grain and stalk fungal pathogens, which are the most prevalent and economically significant sorghum pathogens (Chandrashekar and Satyanarayana, 2006), have in general indicated the contrary.
Fungal infection of bm/bmr grain may not appear to be relevant to bioenergy, however, fungal infection of grain can impair seed germination (Raju et al., 1999; Prom et al., 2003), which is critical for all cropping systems. Under field conditions without inoculation, maize bm3 grain showed significantly increased colonization by members of the Gibberella fujikuroi fungal species complex as compared to WT grain (Nicholson et al., 1976). In contrast, studies using uninoculated field-grown sorghum showed that bmr6 and bmr12 grain had the same level of colonization or significantly reduced fungal colonization relative to WT, which included the sorghum pathogen F. hapsinum, a G. fujikuroi species complex member (Funnell and Pedersen, 2006; Funnell-Harris et al., 2010). Other Fusarium spp. colonized both bmr6 and bmr12 grain at similar levels or significantly reduced colonization relative to WT (Funnell and Pedersen, 2006; Funnell-Harris et al., 2010). In particular, two species that commonly infected WT grain were significantly reduced or absent in bmr12 grain, F. proliferatum and a member of the F. incarnatum-F. equiseti species complex (O’Donnell et al., 2007), respectively (Funnell-Harris et al., 2010). Taken together, these results indicated that impairing CAD or COMT activity in sorghum did not increase susceptibility to these Fusarium spp., and bmr12 grain restricted or excluded colonization of two species. These results contradict the single early report from maize where bm3 grain, which is also COMT-deficient, showed increased colonization by the G. fujikuroi species complex (Nicholson et al., 1976).
Studies examining the susceptibility of maize and sorghum bm/bmr mutants to stalk rot pathogens, which impact biomass quality and can contribute to lodging, also showed no change in resistance or increased resistance relative to WT, similar to the grain studies. F. thapsinum was inoculated in the peduncles of bmr6, bmr12 and WT from six near-isogenic backgrounds and disease severity was determined by the length of the purple disease lesion resulting from the fungal infection. Lesion lengths were significantly shorter than corresponding WT background for many bmr6 and bmr12 lines, and lesion lengths were significantly shorter than WT for one or both bmr lines across four different genetic backgrounds (Funnell and Pedersen, 2006; Funnell-Harris et al., 2010). There were no cases where the lesion length was significantly greater in a bmr line relative to the corresponding WT line (Funnell and Pedersen, 2006; Funnell-Harris et al., 2010). Peduncle inoculations of bmr6, bmr12, and wild-type lines with four Fusarium species and Alternaria alternata consistently resulted in decreased lesion lengths on one or both bmr mutants relative to WT for the following pathogens; F. thapsinum, F. verticillioides and Alternaria alternata (Funnell-Harris et al., 2010). Overall, these results consistently indicated that bmr6 and bmr12 were not more susceptible to these pathogens than WT, and in some cases the two bmr mutants appeared to be more resistant to specific pathogens relative to WT. However, fungal viability was assessed within the lesions and outside the lesions, and fungal growth was detected within and outside borders of lesions from bmr12 inoculated peduncles (Funnell-Harris et al., 2010). This result suggests that fungal growth is greater in healthy-appearing tissues outside the necrotic, discolored tissue defined as the “lesion” in bmr12 plants, although these lesions were similar in size or significantly shorter than WT in bmr12 peduncles. Nevertheless, CAD or COMT deficiency in sorghum does not appear to significantly increase susceptibility of plants to these stalk rot pathogens.
A study using inoculations of another fungal stock pathogen Macrophomina phaseolina, which causes charcoal rot, also demonstrated brown midrib mutants were not more susceptible to this pathogen. bmr mutants from sorghum (bmr2, bmr6, bmr7, bmr12, bmr26, and bmr28; three loci, bmr28 is allelic to bmr6, and bmr7 and bmr26 are allelic to bmr12; Saballos et al., 2008) and maize (bm1, bm2, bm3, and bm4; four loci) were inoculated with Macrophomina phaseolina and lesion lengths were compared to corresponding WT lines (Tesso and Ejeta, 2011). Lesion lengths were not significantly different between bm/bmr mutants and the corresponding WT backgrounds for both maize and sorghum (Tesso and Ejeta, 2011). Stalk strength as determined using rind penetration resistance was significantly reduced in maize bm mutants relative to WT (Tesso and Ejeta, 2011). Interestingly, reduced mechanical stalk strength did not appear to increase susceptibility (Tesso and Ejeta, 2011). However, all studies relied on artificial inoculation to ensure a consistent disease response. If decreased rind penetration resistance (stalk strength) increases the ability of the fungi to initially enter and penetrate the stalk, then results from these studies may be misleading. All the bm/bmr mutants examined resulted in similar susceptibility to the charcoal rot pathogen, even though at least three different steps in monolignol biosynthesis were impaired by the corresponding bmr mutation; 4CL (bmr2), COMT (bm3/bmr12), and CAD (bm1/bmr6). The general trend from these studies indicate that maize and sorghum brown midrib mutants are not more susceptible to stalk rot pathogens, and in some cases show increased generalized resistance to specific pathogens.
There are several explanations for the instances of increased generalized resistance observed in the brown midrib mutants. Although the ability of these bmr plants to synthesize structural lignins is decreased and/or altered, there is no evidence bmr plants are impaired in their ability to synthesis “defense” lignin in response to pathogen attack, and the response might even be enhanced. Another explanation is that blocking a step in the lignin biosynthetic pathway would cause accumulation of lignin precursors and other phenolic compounds, because additional substrates would be available for their synthesis. Indeed it has been shown that some of these precursors inhibit the growth of pathogenic fungi or inhibit production of virulence factors (Dowd et al., 1997; Hua et al., 1999; McKeehen et al., 1999; Beekrum et al., 2003). For example, accumulation of ferulic acid, p-coumaric acid, and sinapic acid has been correlated with resistance to Fusarium spp. (McKeehen et al., 1999; Siranidou et al., 2002). We have observed increased soluble phenolic compounds in bmr6 and bmr12 plants relative to WT (Palmer et al., 2008). Alternatively, perturbing the synthesis of lignin, a component of the cell wall which is the first line of defense against pathogens, could trigger generalized cell wall based defense responses similar to HCT-RNAi lines in Arabidopsis and alfalfa (Gallego-Giraldo et al., 2011a). A review focused on the broader role of the cell wall in plant defense was previously published (Underwood, 2012), which documents the significance of the plant cell wall in responses to a wide range of pathogens.
PROSPECTIVE
These studies from a variety of plants indicate that reducing lignin content and altering its composition will not inevitably increase the susceptibility of bioenergy feedstocks to pathogens. There were not any clear trends that indicate that impairing a specific step in monolignol biosynthesis would affect plant susceptibility. In fact, studies from sorghum and maize indicate that impairing CAD or COMT activity in these lignin-modified plants showed more resistance to specific fungal pathogens, albeit these plants are not as resistant to the pathogen as resistant plant germplasm used in breeding efforts. In bioenergy feedstock species, modifications to monolignol biosynthesis will need to be evaluated on a case by case basis to determine the impact of pathogen susceptibility.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Heather Van Buskirk for critically reviewing the manuscript. Funding was provided by National Institute of Food and Agriculture, Grant 2011-67009-30026 (Scott E. Sattler, Deanna L. Funnell-Harris) and additional funding from USDA-ARS, CRIS project 5440-21220-024-00D (Scott E. Sattler, Deanna L. Funnell-Harris).
REFERENCES
- Ahonsi M. O., Ames K. A., Gray M. E., Bradley C. A. (2013). Biomass reducing potential and prospective fungicide control of a new leaf blight of Miscanthus × giganteus caused by Leptosphaerulina chartarum. Bioenergy Res. 10.1007/s12155-012-9293-0 [DOI] [Google Scholar]
- Baayen R. P., Ouellette G. B., Rioux D. (1996). Compartmentalization of decay in carnations resistant to Fusarium oxysporum f. sp. dianthi. Phytopathology 86 1018–1031 [Google Scholar]
- Beccari G., Covarelli L., Balmas V., Tosi L. (2010). First report of Miscanthusgiganteus rhizome rot caused by Fusarium avenaceum, Fusarium oxysporum and Mucor hiemalis. Australas. Plant Dis. Notes 5 28–29 [Google Scholar]
- Beekrum S., Govinden R., Padayachee T., Odhav B. (2003). Naturally occurring phenols: a detoxification strategy for fumonisin B1. Food Addit. Contam. 20 490–493 [DOI] [PubMed] [Google Scholar]
- Bhuiyan N. H., Selvaraj G., Wei Y. D., King J. (2009). Gene expression profiling and silencing reveal that monolignol biosynthesis plays a critical role in penetration defence in wheat against powdery mildew invasion. J. Exp. Bot. 60 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount J. W., Masoud S., Sumner L. W., Huhman D., Dixon R. A. (2002). Over-expression of cinnamate 4-hydroxylase leads to increased accumulation of acetosyringone in elicited tobacco cell-suspension cultures. Planta 214 902–910 [DOI] [PubMed] [Google Scholar]
- Bonello P., Storer A. J., Gordon T. R., Wood D. L., Heller W. (2003). Systemic effects of Heterobasidion annosum on ferulic acid glucoside and lignin of presymptomatic ponderosa pine phloem, and potential effects on bark-beetle-associated fungi. J. Chem. Ecol. 29 1167–1182 [DOI] [PubMed] [Google Scholar]
- Bout S., Vermerris W. (2003). A candidate-gene approach to clone the sorghum Brown midrib gene encoding caffeic acid O-methyltransferase. Mol. Genet. Genomics 269 205–214 [DOI] [PubMed] [Google Scholar]
- Buendgen M. R., Coors J. G., Grombacher A. W., Russell W. A. (1990). European corn borer resistance and cell wall composition of three maize populations. Crop Sci. 30 505–510 [Google Scholar]
- Carver T. L. W., Zeyen R. J., Robbins M. P., Vance C. P., Boyles D. A. (1994). Suppression of host cinnamyl alcohol dehydrogenase and phenylalanine ammonia lyase increases oat epidermal cell susceptibility to powdery mildew penetration. Physiol. Mol. Plant Pathol. 44 243–259 [Google Scholar]
- Chandrashekar A., Satyanarayana K. V. (2006). Disease and pest resistance in grains of sorghum and millets. J. Cereal Sci. 44 287–304 [Google Scholar]
- Chatani M., Matsumoto Y., Mizuta H., Ikegami M., Boulton M. I., Davies J. W. (1991). The nucleotide sequence and genome structure of the geminivirus miscanthus streak virus. J. Gen. Virol. 72 2325–2331 [DOI] [PubMed] [Google Scholar]
- Chen F., Dixon R. A. (2007). Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 25 759–761 [DOI] [PubMed] [Google Scholar]
- Chen Y., Liu H., Ali F., Scott M. P., Ji Q., Frei U. K., et al. (2012). Genetic and physical fine mapping of the novel brown midrib gene bm6 in maize (Zea mays L.) to a 180 kb region on chromosome 2. Theor. Appl. Genet. 1–13 [DOI] [PubMed] [Google Scholar]
- Covarelli L., Beccari G., Tosi L. (2012). Miscanthus rhizome rot: a potential threat for the establishment and the development of biomass cultivations. Biomass Bioenergy 46 263–269 [Google Scholar]
- Crouch J. A., Beirn L. A., Cortese L. M., Bonos S. A., Clarke B. B. (2009). Anthracnose disease of switchgrass caused by the novel fungal species Colletotrichum navitas. Mycol. Res. 113 1411–1421 [DOI] [PubMed] [Google Scholar]
- Darvill A. G., Albersheim P. (1984). Phytoalexins and their elicitors – a defense against microbial infection in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 35 243–275 [Google Scholar]
- Delgado N. J., Casler M. D., Grau C. R., Jung H. G. (2002). Reactions to smooth bromegrass clones with divergent lignin or etherified ferulic acid concentration to three fungal pathogens. Crop Sci. 42 1824–1831 [Google Scholar]
- Dicko M. H., Gruppen H., Barro C., Traore A. S., Van Berkel W. J. H, Voragen A. G. J. (2005). Impact of phenolic compounds and related enzymes in sorghum varieties for resistance and susceptibility to biotic and abiotic stresses. J. Chem. Ecol. 31 2671–2688 [DOI] [PubMed] [Google Scholar]
- Dien B. S., Sarath G., Pedersen J. F., Sattler S. E., Chen H., Funnell-Harris D. L., et al. (2009). Improved sugar conversion and ethanol yield for forage sorghum (Sorghum bicolor L. Moench) lines with reduced lignin contents. Bioenergy Res. 2 153–164 [Google Scholar]
- Dixon L. J., Castlebury L. A., Aime M. C., Glynn N. C., Comstock J. C. (2010). Phylogenetic relationships of sugarcane rust fungi. Mycol. Prog. 9 459–468 [Google Scholar]
- Dixon R. A., Achnine L., Kota P., Liu C. J., Reddy M. S. S., Wang L. (2002). The phenylpropanoid pathway and plant defence – A genomics perspective. Mol. Plant Pathol. 3 371–390 [DOI] [PubMed] [Google Scholar]
- Doster M. A., Bostock R. M. (1988). Quantification of lignin formation in almond bark in response to wounding and infection by Phytophthora species. Phytopathology 78 473–477 [Google Scholar]
- Dowd P. F., Duvick J. P., Rood T. (1997). Comparative toxicity of allelochemicals and their enzymatic oxidation products to maize fungal pathogens, emphasizing Fusarium graminearum. Nat. Toxins 5 180–185 [DOI] [PubMed] [Google Scholar]
- Duschnicky L. G., Ballance G. M., Sumner M. J., Macgregor A. W. (1998). The role of lignification as a resistance mechanism in wheat to a toxin-producing isolate of Pyrenophora tritici-repentis. Can. J. Plant Pathol. 20 35–47 [Google Scholar]
- Eynck D., Koopman B., Karlovsky P, Von Tiedemann A. (2009). Internal resistance in winter oilseed rape inhibits systemic spread of the vascular pathogen Verticillium longisporum. Phytopathology 99 802–811 [DOI] [PubMed] [Google Scholar]
- Farrell G., Simons S. A., Hillocks R. J. (2001). Aspects of the biology of Ustilago kamerunensis, a smut pathogen of Napier grass (Pennisetum purpureum). J. Phytopathol. 149 739–744 [Google Scholar]
- Felton G. W., Korth K. L., Bi J. L., Wesley S. V., Huhman D. V., Mathews M. C., et al. (1999). Inverse relationship between systemic resistance of plants to microorganisms and to insect herbivory. Curr. Biol. 9 317–320 [DOI] [PubMed] [Google Scholar]
- Funnell D. L., Pedersen J. F. (2006). Reaction of sorghum lines genetically modified for reduced lignin content to infection by Fusarium and Alternaria spp. Plant Dis. 90 331–338 [DOI] [PubMed] [Google Scholar]
- Funnell-Harris D. L., Pedersen J. F., Sattler S. E. (2010). Alteration in lignin biosynthesis restricts growth of Fusarium spp. in brown midrib sorghum. Phytopathology 100 671–681 [DOI] [PubMed] [Google Scholar]
- Gallego-Giraldo L., Escamilla-Trevino L., Jackson L. A., Dixon R. A. (2011a). Salicylic acid mediates the reduced growth of lignin down-regulated plants. Proc. Natl. Acad. Sci. U.S.A. 108 20814–20819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Giraldo L., Jikumaru Y., Kamiya Y., Tang Y. H., Dixon R. A. (2011b). Selective lignin downregulation leads to constitutive defense response expression in alfalfa (Medicago sativa L.). New Phytol. 190 627–639 [DOI] [PubMed] [Google Scholar]
- Gustafson D. M., Boe A., Jin Y. (2003). Genetic variation for Puccinia emaculata infection in switchgrass. Crop Sci. 43 755–759 [Google Scholar]
- Haegi A., Bonardi V., Dall’aglio E., Glissant D., Tumoni G., Collins N. D., et al. (2008). Histological and molecular analysis of RDG2a barley resistance to leaf stripe. Mol. Plant Pathol. 9 463–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpin C., Holt K., Chojecki J., Oliver D., Chabbert B., Monties B., et al. (1998). Brown-midrib maize (bm1) –a mutation affecting the cinnamyl alcohol dehydrogenase gene. Plant J. 14 545–553 [DOI] [PubMed] [Google Scholar]
- Halpin C., Thain S. C., Tilston E. L., Guiney E., Lapierre C., Hopkins D. W. (2007). Ecological impacts of trees with modified lignin. Tree Genet. Genomes 3 101–110 [Google Scholar]
- Hammerschmidt R., Bonnen A. M., Bergstrom G. C., Baker K. K. (1985). Association of Epidermal lignification with nonhost resistance of cucurbits to fungi. Can. J. Bot. 63 2393–2398 [Google Scholar]
- Howles P. A., Sewalt V. J. H., Paiva N. L., Elkind Y., Bate N. J., Lamb C., et al. (1996). Overexpression of L-phenylalanine ammonia-lyase in transgenic tobacco plants reveals control points for flux into phenylpropanoid biosynthesis. Plant Physiol. 112 1617–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua S. S. T., Grosjean O. K., Baker J. L. (1999). Inhibition of aflatoxin biosynthesis by phenolic compounds. Lett. Appl. Microbiol. 29 289–291 [DOI] [PubMed] [Google Scholar]
- Huang J., Bhinu V. S., Li X., Bashi Z. D., Zhou R., Hannoufa A. (2009). Pleiotropic changes in Arabidopsis f5h and sct mutants revealed by large-scale gene expression and metabolite analysis. Planta 230 1057–1069 [DOI] [PubMed] [Google Scholar]
- Huang J. L., Gu M., Lai Z. B., Fan B. F., Shi K., Zhou Y. H., et al. (2010). Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 153 1526–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudgins J. W., Christiansen E., Franceschi V. R. (2004). Induction of anatomically based defense responses in stems of diverse conifers by methyl jasmonate: a phylogenetic perspective. Tree Physiol. 24 251–264 [DOI] [PubMed] [Google Scholar]
- Jones L., Ennos A. R., Turner S. R. (2001). Cloning and characterization of irregular xylem4 (irx4): A severely lignin-deficient mutant of Arabidopsis. Plant J. 26 205–216 [DOI] [PubMed] [Google Scholar]
- Jorgenson L. R. (1931). Brown midrib in maize and its linkage relations. J. Am. Soc. Agron. 23 549–557 [Google Scholar]
- Kim S. J., Kim M. R., Bedgar D. L., Moinuddin S. G. A., Cardenas C. L., Davin L. B., et al. (2004). Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101 1455–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D. J., Lim J. E., Landry L. G., Last R. L. (2002). Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol. 130 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamptey J. N. L., Plumb R. T., Shaw M. W. (2003). Interactions between the grasses Phalaris arundinacea, Miscanthus sinensis and Echinochloa crus-galli, and Barley and Cereal yellow dwarf viruses. J. Phytopathol. 151 463–468 [Google Scholar]
- Lange B. M., Lapierre C, Sandermann Jnr H. (1995). Elicitor-induced spruce stress lignin. Structural similarity to early developmental lignins. Plant Physiol. 108 1277–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowski I., Clifton-Brown J. C., Scurlock J. M. O., Huisman W. (2000). Miscanthus: European experience with a novel energy crop. Biomass Bioenergy 19 209–227 [Google Scholar]
- Li X., Bonawitz N. D., Weng J. K., Chapple C. (2010). The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids. Plant Cell 22 1620–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo S. C. C., Nicholson R. L. (1998). Reduction of light-induced anthocyanin accumulation in inoculated sorghum mesocotyls – implications for a compensatory role in the defense response. Plant Physiol. 116 979–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozovaya V. V., Lygin A. V., Zernova O. V., Ulanov A. V., Li S. X., Hartman G. L., et al. (2007). Modification of phenolic metabolism in soybean hairy roots through down regulation of chalcone synthase or isoflavone synthase. Planta 225 665–679 [DOI] [PubMed] [Google Scholar]
- Maher E. A., Bate N. J., Ni W. T., Elkind Y., Dixon R. A., Lamb C. J. (1994). Increased disease susceptibility of transgenic tobacco plants with suppressed levels of preformed phenylpropanoid products. Proc. Natl. Acad. Sci. U.S.A. 91 7802–7806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martïnez-Reyna J. M., Vogel K. P. (2002). Incompatibility systems in switchgrass. Crop Sci. 42 1800–1805 [Google Scholar]
- Maury S., Delaunay A., Mesnard F., Cronier D., Chabbert B., Geoffroy P., et al. (2010). O-methyltransferase(s)-suppressed plants produce lower amounts of phenolic vir inducers and are less susceptible to Agrobacterium tumefaciens infection. Planta 232 975–986 [DOI] [PubMed] [Google Scholar]
- McKeehen J. D., Busch R. H., Fulcher R. G. (1999). Evaluation of wheat (Triticum aestivum L.)phenolic acids during grain development and their contribution to Fusarium resistance. J. Agric. Food Chem. 47 1476–1482 [DOI] [PubMed] [Google Scholar]
- Menden B., Kohlhoff M., Moerschbacher B. M. (2007). Wheat cells accumulate a syringyl-rich lignin during the hypersensitive resistance response. Phytochemistry 68 513–520 [DOI] [PubMed] [Google Scholar]
- Mitchell H. J., Hall S. A., Stratford R., Hall J. L., Barber M. S. (1999). Differential induction of cinnamyl alcohol dehydrogenase during defensive lignification in wheat (Triticum aestivum L.): characterisation of the major inducible form. Planta 208 31–37 [Google Scholar]
- Nageswara-Rao M., Stewart C. N., Jr., Kwit C. (2012). Genetic diversity and structure of natural and agronomic switchgrass (Panicum virgatum L.) populations. Genet. Res. Crop Evol. 1–12 [Google Scholar]
- Negrel J., Javelle F. (2010). The biosynthesis of acetovanillone in tobacco cell-suspension cultures. Phytochemistry 71 751–759 [DOI] [PubMed] [Google Scholar]
- Nicholson R. L., Bauman L. F., Warren H. L. (1976). Association of Fusarium moniliforme with brown midrib maize. Plant Dis. Rep. 60 908–910 [Google Scholar]
- Nicholson R. L., Hammerschimdt R. (1992). Phenolic compounds and their role in disease resistance. Annu. Rev. Phytopathol. 30 369–389 [Google Scholar]
- O’Donnell K., Sarver B. A., Brandt M., Chang D. C., Noble-Wang J., Park B. J., et al. (2007). Phylogenetic diversity and microsphere array-based gentyping of human pathogenic Fusaria, including isolates from the multistate contact lens-associated U.S. keratitis outbreaks of 2005 and 2006. J. Clin. Microbiol. 45 2235–2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen K. M., Lea U. S., Slimestad R., Verheul M., Lillo C. (2008). Differential expression of four Arabidopsis PAL genes; PAL1 and PAL2 have functional specialization in abiotic environmental-triggered flavonoid synthesis. J. Plant Physiol. 165 1491–1499 [DOI] [PubMed] [Google Scholar]
- O’Neill N. R., Farr D. F. (1996). Miscanthus blight, a new foliar disease of ornamental grasses and sugarcane incited by Leptosphaeria sp. and its anamorphic state Stagonospora sp. Plant Dis. 80 980–987 [Google Scholar]
- Palmer N. A., Sattler S. E., Saathoff A. J., Funnell D., Pedersen J. F., Sarath G. (2008). Genetic background impacts soluble and cell wall-bound aromatics in brown midrib mutants of sorghum. Planta 229 115–127 [DOI] [PubMed] [Google Scholar]
- Pan Q., Zhan J., Liu H., Zhang J., Chen J., Wen P., et al. (2006). Salicylic acid synthesized by benzoic acid 2-hydroxylase participates in the development of thermotolerance in pea plants. Plant Sci. 171 226–233 [Google Scholar]
- Peltier A. J., Hatfield R. D., Grau C. R. (2009). Soybean stem lignin concentration relates to resistance to Sclerotinia sclerotiorum. Plant Dis. 93 149–154 [DOI] [PubMed] [Google Scholar]
- Piquemal J., Lapierre C., Myton K., O’connell A., Schuch W., Grima-Pettenati J., et al. (1998). Down-regulation of cinnamoyl-CoA reductase induces significant changes of lignin profiles in transgenic tobacco plants. Plant J. 13 71–83 [Google Scholar]
- Porter K. S., Axtell J. D., Lechtenberg V. L., Colenbrander V. F. (1978). Phenotype, fiber composition, and in vitro dry matter disappearance of chemically induced brown midrib (bmr) mutants of sorghum. Crop Sci. 18 205–208 [Google Scholar]
- Prom L. K., Waniska R. D., Kollo A. I., Rooney W. L. (2003). Response of eight sorghum cultivars inoculated with Fusarium thapsinum, Curvularia lunata, and a mixture of the two fungi. Crop Prot. 22 623–628 [Google Scholar]
- Quentin M., Allasia V., Pegard A., Allais F., Ducrot P.-H., Favery B., et al. (2009). Imbalanced lignin biosynthesis promotes the sexual reproduction of homothallic oomycete pathogens. PLoS Pathog. 5:e1000264 10.1371/journal.ppat.1000264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju N. S., Niranjana S. R., Janardhana G. R., Prakash H. S., Shekar Shetty H., Mathur S. B. (1999). Improvement of seed quality and field emergence of Fusarium moniliforme infected sorghum seeds using biological agents. J. Sci. Food Agric. 79 206–212 [Google Scholar]
- Ride J. P. (1975). Lignification in wounded wheat leaves in response to fungi and its possible role in resistance. Physiol. Plant Pathol. 5 125–134 [Google Scholar]
- Robertsen B., Svalheim O. (1990). The nature of lignin-like compounds in cucumber hypocotyls induced by alpha-1,4-linked oligogalacturonides. Physiol. Plant. 79 512–518 [Google Scholar]
- Roco A., Castaneda P., Perez L. M. (1993). Oligosaccharides released by pectinase treatment of citrus-limon seedlings are elicitors of the plant-response. Phytochemistry 33 1301–1306 [Google Scholar]
- Ruel K., Berrio-Sierra J., Derikvand M. M., Pollet B., Thévenin J., Lapierre C., et al. (2009). Impact of CCR1 silencing on the assembly of lignified secondary walls in Arabidopsis thaliana. New Phytol. 184 99–113 [DOI] [PubMed] [Google Scholar]
- Ruuhola T., Julkunen-Tiitto R. (2003). Trade-off between synthesis of salicylates and growth of micropropagated Salix pentandra. J. Chem. Ecol. 29 1565–1588 [DOI] [PubMed] [Google Scholar]
- Saballos A., Ejeta G., Sanchez E., Kang C., Vermerris W. (2009). A Genomewide analysis of the cinnamyl alcohol dehydrogenase family in sorghum [Sorghum bicolor (L.) Moench] Identifies SbCAD2 as the Brown midrib6 Gene. Genetics 181 783–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saballos A., Sattler S. E., Sanchez E., Foster T. P., Xin Z., Kang C., et al. (2012). Brown midrib2 (Bmr2) encodes the major 4-coumarate: coenzyme A ligase involved in lignin biosynthesis in sorghum (Sorghum bicolor (L.) Moench). Plant J. 70 818–830 [DOI] [PubMed] [Google Scholar]
- Saballos A., Vermerris W., Rivera L., Ejeta G. (2008). Allelic association, chemical characterization and saccharification properties of brown midrib mutants of sorghum (Sorghum bicolor (L.) Moench). Bioenergy Res. 1 193–204 [Google Scholar]
- Sattler S. E., Saathoff A. J., Haas E. J., Palmer N. A., Funnell-Harris D. L., Sarath G., et al. (2009). A Nonsense mutation in a cinnamyl alcohol dehydrogenase gene is responsible for the sorghum brown midrib6 phenotype. Plant Physiol. 150 584–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadle G., Chen F., Reddy M. S. S., Jackson L., Nakashima J., Dixon R. A. (2007). Down-regulation of hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase in transgenic alfalfa affects lignification, development and forage quality. Phytochemistry 68 1521–1529 [DOI] [PubMed] [Google Scholar]
- Shadle G. L., Wesley S. V., Korth K. L., Chen F., Lamb C., Dixon R. A. (2003). Phenylpropanoid compounds and disease resistance in transgenic tobacco with altered expression of L-phenylalanine ammonia-lyase. Phytochemistry 64 153–161 [DOI] [PubMed] [Google Scholar]
- Sibout R., Eudes A., Mouille G., Pollet B., Lapierre C., Jouanin L., et al. (2005). cinnamyl alcohol dehydrogenase-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17 2059–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist J., Jeblick W., Kauss H. (1994). Defense responses in infected and elicited cucumber (Cucumis sativus L.) hypocotyl segments exhibiting acquired resistance. Plant Physiol. 105 1365–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siranidou E., Kang Z., Buchenauer H. (2002). Studies on symptom development, phenolic compounds and morphological defence responses in wheat cultivars differing in resistance to Fusarium head blight. J. Phytopathol. 150 200–208 [Google Scholar]
- Smit F., Dubery I. A. (1997). Cell wall reinforcement in cotton hypocotyls in response to a Verticillium dahliae elicitor. Phytochemistry 44 811–815 [Google Scholar]
- Tesso T., Ejeta G. (2011). Stalk strength and reaction to infection by Macrophomina phaseolina of brown midrib maize (Zea mays) and sorghum (Sorghum bicolor). Field Crops Res. 120 271–275 [Google Scholar]
- Tronchet M., Balague C., Kroj T., Jouanin L., Roby D. (2010). Cinnamyl alcohol dehydrogenases-C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis. Mol. Plant Pathol. 11 83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman W., Torres De Zabala M., Grant M. (2006). Type III effectors orchestrate a complex interplay between transcriptional networds to modify basal defence responses during pathogenesis and resistance Plant J. 46 14–33 [DOI] [PubMed] [Google Scholar]
- Turina M., Maruoka M., Monis J., Jackson A. O, Scholthof K.-B. G. (1998). Nucleotide sequence and infectivity of a full-length cDNA clone of panicum mosaic virus. Virology 241 141–155 [DOI] [PubMed] [Google Scholar]
- Underwood W. (2012). The plant cell wall: a dynamic barrier against pathogen invasion. Front. Plant Sci. 3:85 10.3389/fpls.2012.00085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C. P., Kirk T. K., Sherwood R. T. (1980). Lignification as a mechanism of disease resistance. Annu. Rev. Phytopathol. 18 259–288 [Google Scholar]
- Vignols F., Rigau J., Torres M. A., Capellades M., Puigdomenech P. (1995). The brown midrib3 (Bm3) mutation in maize occurs in the gene encoding caffeic acid O-methyltransferase. Plant Cell 7 407–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadl P. A., Dean D., Li Y., Vito L. M., Scheffler B. E., Hadziabdic D., et al. (2011). Development and characterization of microsatellites for switchgrass rust fungus (Puccinia emaculata). Conserv. Genet. Res. 3 185–188 [Google Scholar]
- Weiergang I., Hipskind J. D., Nicholson R. L. (1996). Synthesis of 3-deoxyanthocyanidin phytoalexins in sorghum occurs independent of light. Physiol. Mol. Plant Pathol. 49 377–388 [Google Scholar]
- Wróbel-Kwiatkowska M., Starzycki M., Zebrowski J., Oszmiański J., Szopa J. (2007). Lignin deficiency in transgenic flax resulted in plants with improved mechanical properties. J. Biotechnol. 128 919–934 [DOI] [PubMed] [Google Scholar]
- Wuyts N., Lognay G., Swennen R, De Waele D. (2006). Nematode infection and reproduction in transgenic and mutant Arabidopsis and tobacco with an altered phenylpropanoid metabolism. J. Exp. Bot. 57 2825–2835 [DOI] [PubMed] [Google Scholar]
- Yu C. K. Y., Springob K., Schmidt J. R., Nicholson R. L., Chu I. K., Yip W. K., et al. (2005). A stilbene synthase gene (SbSTS1) is involved in host and nonhost defense responses in Sorghum. Plant Physiol. 138 393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu O., Jung W., Shi J., Croes R. A., Fader G. M., Mcgonigle B., et al. (2000). Production of the isoflavones genistein and daidzein in non-legume dicot and monocot tissues. Plant Physiol. 124 781–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Buchwaldt L., Rimmer S. R., Sharpe A., Mcgregor L., Bekkoui D., et al. (2009). Patterns of differential gene expression in Brassica napus cultivars infected with Sclerotinia sclerotiorum. Mol. Plant Pathol. 10 635–649 [DOI] [PMC free article] [PubMed] [Google Scholar]