Abstract
Many viruses package their genomes concomitant with assembly. Here, we show that this reaction can be described by three coefficients: association of capsid protein (CP) to nucleic acid (NA), KNA; CP-CP interaction, ω; and α, proportional to the work required to package NA. The value of α can vary as NA is packaged. A phase diagram of average lnα versus lnω identifies conditions where assembly is likely to fail or succeed. NA morphology can favor (lnα > 0) or impede (lnα < 0) assembly. As lnω becomes larger, capsids become more stable and assembly becomes more cooperative. Where (lnα + lnω) < 0, the CP is unable to contain the NA, so that assembly results in aberrant particles. This phase diagram is consistent with quantitative studies of cowpea chlorotic mottle virus, hepatitis B virus, and simian virus 40 assembling on ssRNA and dsDNA substrates. Thus, the formalism we develop is suitable for describing and predicting behavior of experimental studies of CP assembly on NA.
Introduction
Viruses are self-replicating molecular machines and obligate parasites that package a genome and deliver it to a host. In many cases, the shell (or capsid) of a spherical virus self-assembles around the viral genome. This is the case for spherical viruses such as hepatitis B virus (HBV, a member of the Hepadnaviridae family), cowpea chlorotic mottle virus (CCMV, family Bromoviridae), and simian virus 40 (SV40, polyomaviridae), three viruses that are unrelated but have well characterized assembly systems. Like HBV, CCMV, and SV40, most spherical virions that package their nucleic acids concomitant with assembly have capsid proteins (CPs) with conspicuously basic nucleic acid (NA) binding domains. Of course, for some spherical viruses, the genome is pumped into an empty procapsid through a special portal (e.g., bacteriophage phi 29 and herpes simplex virus). For a few virus families (e.g., Picornaviridae and Comoviridae), which have neither a discernible NA binding domain nor a portal, the mechanism(s) by which NA is packaged is unknown.
NA plays a structural role in the assembly of many viruses, but there is surprisingly little information about this role, nor is there a quantitative description of its assembly-related functions (1). Substantial amounts of NA are seen in only a few (e.g., Gouet et al. (2), Tang et al. (3), Larson et al. (4), and Chen et al. (5)) of the many virus crystal structures. Cryo-electron microscopy (cryo-EM) image reconstructions, which include all low-resolution terms, frequently show NA density, which often has considerable fine structure (e.g., Tihova et al. (6)). In an x-ray structure of Pariacoto virus, the viral ssRNA is clearly visualized as a dodecahedral cage with dsRNA segments running through protein-defined grooves to generate a dodecahedral cage (3), even though there is no evidence for a periodic organization of complementary segments intrinsic to the viral genome. Even more striking is that when packaging essentially random RNA in a closely related nodavirus, flock house virus, capsid structure still induced the packaged RNA to conform to a dodecahedral conformation (6). An unrelated virus, satellite tobacco necrosis virus, has a similar RNA organization (4); chemical analysis of its viral RNA suggests that it may have a fold that favors dodecahedral organization (7). A crystal structure of beanpod mottle virus showed a trefoil of ∼30 ssRNA nucleotides visible on the 20 icosahedral threefolds (5), where the viral RNA had evidence of patches of pyrimidine-rich sequences (8). In bluetongue virus (family Reoviridae), with a multipartite dsRNA genome, the genome appears to be a liquid-crystal phase; however, the outermost layer is sufficiently ordered by contact with the capsid that the paths (but not the sequence) of the RNA can be traced (2). In HBV, packaged random ssRNA and genomic dsDNA (9,10) can be seen in cryo-EM image reconstructions; NA density is concentrated near basic C termini (localized to fivefold and quasi-sixfold vertices). There are two fundamental points to be made from this very short catalog. 1), There is interplay between the viral capsid and packaged NA that appears to be driven by the capsid structure. 2), The majority of packaged NA shows much less order than the protein shell. Theoretical simulations of encapsidated dsDNA also underscore the importance of interaction with CPs in NA organization (11–14).
We (and many others) have hypothesized that NA organization before and during capsid assembly can affect the capsid-assembly reaction itself (12,15–21). Using coarse-grained simulations, Hagan and Elrad posited two extremes for self-assembly: stepwise addition of CPs where protein-protein interactions are strong and initial formation of a disordered nucleoprotein complex that subsequently anneals to a virus-like particle where protein-protein interactions are weak (Fig. 1 B) (18). Formation of a disordered micellelike structure followed by reorganization of subunits was also proposed by McPherson (22). Neutralization of charge by CP subunits has been proposed as a mechanism for collapse of such micelles (23). Elaborating on the theme of stepwise addition of subunits, assembly may be a highly cooperative reaction where the addition of the first subunit creates an environment that preferentially completes a capsid, or it may be a low-cooperativity system where subunits distribute evenly over all NAs. Solution studies of coordinated assembly and packaging have mainly relied upon electrophoretic mobility-shift assays (Fig. 2) to demonstrate assembly of CPs that have a very strong electrostatic association with NA. The three posited assembly paths (Fig. 1 B) have predictable and distinct electrophoretic results (Fig. 2 C). The disordered-complex path will show no electrophoretic shift if subunits dissociate from the NA during electrophoresis, whereas it will result in a gradual shift of the NA band if the CP remains NA-associated (15). The low-cooperativity stepwise path will result in a gradual shift of NA migration (15), whereas the high-cooperativity path, where one NA is coated with CP before the next NA begins to get encapsidated, resulting in a bimodal distribution (21). Implicit in these simplified models is that the nature of the NA will make a defining contribution to the path of capsid assembly.
Figure 1.
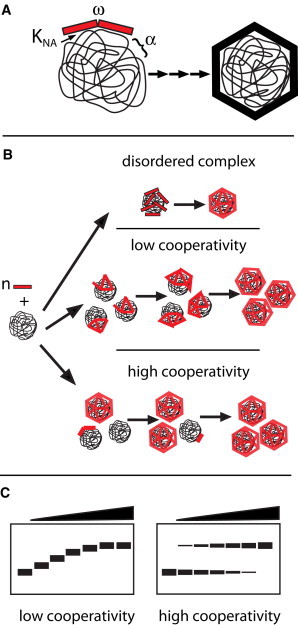
Schematic of the assembly of an NA-filled virus-like particle. (A) Capsid protein (CP) interaction with nucleic acid (NA) is defined by the CP-NA association constant, KNA, the CP-CP interaction, ω, and the work of fitting the NA into the constraints of the growing capsid, α. (B) Three general paths of assembly. In collapse of disordered complexes, subunits adsorb to the NA to form a micellelike structure and then rearrange to form a capsid; this path has been associated with weak CP-CP interaction (19). A low-cooperativity assembly path has a more ordered addition of subunits, where the subunits are uniformly distributed over the many NA polymers. A high-cooperativity path, also based on ordered addition of subunits, results from preferential addition of subunits to NA where growth is already nucleated. (C) Hypothetical observations of different assembly reactions in which products are electrophoretically separated on an agarose gel, after which bound and free NA are visualized by ethidium bromide staining. Disordered complex formation and low-cooperativity assembly lead to a gradual shift of NA migration through a gel, so long as the CP remains NA-associated. High-cooperativity assembly leads to a bimodal distribution of NA into free and encapsidated fractions.
Figure 2.
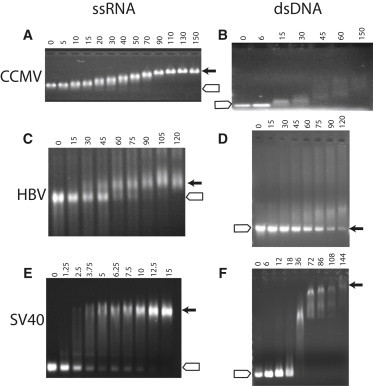
Titrations of ssRNA or dsDNA by different CPs showing the ratio of CP oligomer per NA polymer. Agarose gels run at neutral show electrophoretic migration of NA in the presence of CP at the stated ratio per NA polymer. (A and B) CCMV dimeric CP titrating 3100-nucleotide viral RNA1 (A) (15) or a dsDNA 600-mer (B) (49). (C and D) HBV CP (Cp183-EEE) titrating a 3200-nucleotide viral RNA (C) (21) or an 1800-base pair dsDNA (D) (12). Note that HBV capsids run almost at the same position as the dsDNA but are not evident. (E and F) SV40 VP1 pentamers titrating an 850-nucleotide ssRNA (E) (33) or a 600-mer dsDNA that is ∼1/10 the length of the SV40 genome (F). In all cases (A–F), the expected position for free NA is denoted by the open arrow and the capsid (where known) by the closed arrow. Of these different CPs, only SV40 consistently forms dsDNA-filled capsids (16). Gel were 0.8% or 1% agarose run using a Tris-acetate buffer system.
A formalism for describing protein assembly on NAs remains elusive. von Hippel and co-workers described a model of nonspecific binding of proteins to a linear NA lattice (24,25), which has been qualitatively applied to virus assembly (15,21) but does not explicitly account for polyvalent proteins and the more complex patterns of protein-NA interaction possible in a virus. Physical models with simplified representations of NA have provided insights into the process of assembly, particularly kinetics. Indeed, the relationship between charge and volume of packaged NA has been considered a determinant of particle size (26). RNA has also been considered an antenna that both attracts protein to an assembly site and facilitates one-dimensional diffusion along the RNA (27), analogous to suppressor diffusion on DNA (28). In previous works, we described a formalism for assembly of empty spherical capsids (29,30). Here, we extend it to assembly of NA-filled particles. We observe that the synergism between NA packaging and protein-protein interaction leads to testable predictions consistent with our observations.
Results and Discussion
Development of an assembly model
A basic scheme has been used to describe the equilibrium of a series of concurrent reactions in which one subunit at a time is assembled to form an empty capsid, and, by analogy, an NA-filled capsid (Fig. 1 A). In both cases, each subunit is multivalent, often tetravalent dimers (e.g., HBV and CCMV) or penta- and hexavalent pentamers (e.g., SV40). For the empty capsid (30,31), each intersubunit contact has a microscopic association constant of Kcontact. The first two subunits interact with Kcontact; the third subunit may then interact with both subunits of the growing complex with Kcontact2. The microscopic association constants must be modified by a statistical term, s, particular to the geometry of each intermediate, to account for reaction degeneracy. This is sufficient information to describe the equilibrium concentration of an N-subunit capsid and each n-subunit intermediate. It is important to note that this model of empty-capsid formation predicts a steeply descending energy surface (29) such that free subunits (n = 1) and complete N-subunit capsids are the dominant species with infinitesimal concentrations of intermediates (29,30).
The mass-action laws describing assembly of an empty capsid and n-subunit intermediates (n-mers) are
| (1) |
| (2) |
The equilibrium constants can be broken down to components
| (3) |
| (4) |
In Eqs. 3 and 4, for a capsid of N subunits or an intermediate of n subunits, ∏sj is the product of the j degeneracy terms peculiar to each intermediate, Kcontact is the microscopic association constant common to every protein-protein contact, and the exponent (the superscript) is the total number of contacts made, where each multivalent subunit makes up to c contacts. There are critical simplifications explicit in these equations, namely, that off-path complexes make no contribution to the reaction and that all assembly reactions eventually reach equilibrium. However, assembly can go off path, stop short of equilibrium, or otherwise run into kinetic barriers (29,31,32). Kinetic simulations consistent with the above formalism do a remarkable job of matching kinetic and equilibrium data (see, e.g., Kler et al. (33) and Zlotnick and Mukhopadhyay (34)).
Adding NA to the assembly reaction requires recasting of the capsid-assembly formalism (Fig. 1). Association of CPs to form an NA-filled virus needs to include terms for protein-protein interaction, protein-NA interaction, and the energy of packaging the NA. The approach taken by McGhee and von Hippel (24,25) to describe assembly of protein on a linear NA substrate is instructive: KNA (in units of M−1) was used to describe the association of one protein to substrate and the interaction between adjacent proteins was defined by the unitless association coefficient ω. McGhee and von Hippel also made the assumption that proteins would bind several consecutive nucleotides, and they rigorously considered the effects of bound protein occluding several sites. In low-cooperativity reactions (low-ω reactions), the frequent gaps between proteins led to apparent negative cooperativity of association, a barrier that arose because the gaps could not fit an integral number of subunits. However, such exclusion effects are not appropriate for virus CPs, where a protein may bind a discontinuous segment of NA and the nucleoprotein contacts may be much more fluid.
We consider that assembly of a virus must include terms for 1), binding NA, 2), protein-protein interaction, and 3), constraining NA in a shell. Following McGhee and von Hippel, we define the association constant for NA, KNA, in units of M−1. The protein-protein interaction is closely related to Kcontact but is now unit-less, because it describes the intramolecular interaction between two or more NA-bound proteins; it is thus appropriate to use the McGhee-von Hippel term ω. The ω term is not identical to Kcontact as it does not include the entropic costs of diffusion and loss of rotational freedom inherent in Kcontact. These costs are now built into KNA. For NA-based assembly, the first protein binds to NA with an association of KNA. When a second protein is added to the NA, if it is adjacent to the first, the binding constant is ωKNA. If the third protein binds to a doubly contiguous site, its association constant will be based on ω2KNA. Of course, these products of microscopic association constants must also have a degeneracy component, s.
Unlike the one-dimensional McGhee-von Hippel DNA lattice, viral CPs must also constrain NA that would normally occupy a volume much larger than the capsid interior (14,17,18). In the case of dsDNA, the substrate favors a volume substantially greater than typical for a virus capsid (13,35). To account for the effect of NA conformation on assembly, we introduce the coefficient α as a component of the assembly association constant. As the growing capsid will progressively change the conformational space available to the NA, there must be a unique α for each added subunit. For simplicity of expression, we will only singly subscript α with the number of subunits in the intermediate, though ideally it may be appropriate to acknowledge the many possible arrangements of subunits and conformations of NA. When the NA impedes assembly, α will have a value between 0 and 1, attenuating association; when the NA promotes assembly, α will be ≥1.
Considering an assembly reaction at equilibrium, the mass-action expressions for a virus-like particle consisting of a single NA molecule packaged by a capsid of N subunits (VLP) and for n-subunit intermediates are
| (5) |
| (6) |
The corresponding evaluations of the association constant include α and ω terms. Since both degeneracy and α terms are unique to each intermediate, they are included within the product. The contribution of ω, the same for all contacts, is readily described with an exponent of the number contacts.
| (7) |
| (8) |
These simple equations describe complex behavior. Most of the tradeoff is between the conflicting forces described by α and ω (and ωcontacts). When α is ∼1, the resulting binding isotherm shows cooperativity based entirely on ω; because a large exponent is applied to ω, binding curves under conditions of α ∼ 1 will always show a strong positive cooperativity. To generate low-cooperativity, as seen with CCMV CP binding to RNA (15), the overall value of α must be <1. Low cooperativity will be accentuated when α decreases as n approaches N, so that the product of α and ωcontacts is nearly constant throughout the binding isotherm. This is reminiscent of the closure difficulties observed in coarse-grained simulations where the encapsidated NA was too large for the particle (19). The hypothesized counterpoint between α and ω in CCMV disguises the effect of multivalent interactions between CP subunits during capsid closure and results in a binding curve that appears hyperbolic.
Now consider reactions where the substrate hinders initial steps but supports subsequent assembly. When 1/α is similar to but smaller than ω, at low CP concentrations there will be weak CP-CP interaction and little evidence of capsid formation (the concentration dependence of binding to NA is defined only by KNA). However, as the CP concentration increases, the exponential effect defined in Eqs. 5 and 6 will become dominant, resulting in a steeply sigmoidal binding curve, as seen with SV40 binding to dsDNA (16).
Because virus-assembly reactions are prone to kinetic traps, it is necessary to expand Eqs. 7 and 8 to allow consideration of kinetic effects. Assembly kinetics of a capsid population can be simulated with a series of ordinary differential equations for each intermediate assuming addition of one subunit at a time. The mathematical approach is consistent with high and low cooperativity models (Fig. 1); the result reasonably mimics formation of empty capsids (36,37) and capsids assembled on RNA (33). For empty capsids, such ordinary differential equation simulations closely match coarse-grained dynamic simulations (38,39). Equation 9 describes assembly of an n-mer with only one possible predecessor and scion (a simplified form of Eq. 9, lacking an explicit α, was shown to fit observed assembly kinetics for SV40 CP binding to a short RNA (33)):
| (9) |
In the first two, forward, terms of this equation, f is the microscopic forward rate that is modified by the forward degeneracy term sf and the time-dependent NA conformational coefficient α‡; although α‡ is different from the equilibrium α, the values for the two terms are likely to be similar. The backward rate terms, b, are based on the forward rate modified by the energetic cost of dissociation, that is, the number of CP-CP contacts broken (ω to an appropriate exponent) and breaking the CP-NA interaction (KNA). It is clear that a small number of CP-CP contacts or apparent-novel contacts with weak ω values will lead to dissociation of fragile complexes. The multiplicity of possible paths has a modest effect for assembly of empty capsids (40). However, for assembly on NA, each incoming subunit has unique α‡ that may vary substantially depending on the particular assembly path and thus has a unique ability to redirect assembly. For simplicity of exposition, we use a single indexing scheme in Eq. 9. The important point is that assembly on NA may be particularly sensitive to local energy minima. As with empty particles, kinetic traps fall into categories of 1), overnucleation leading to accumulation of partial capsids with insufficient CP to allow completion; 2), high association energy, which leads to stalling reactions short of equilibration due to paucity of intermediates; or 3), failure of subunits incorporated with incorrect geometry to dissociate. Strong CP-CP association makes assembly more prone to the first two forms of kinetic trap. Due to antagonism between α and ω, the last sort of failure is of particular importance for NA-directed assembly.
A phase diagram of assembly
A phase diagram is a natural way to view correlated parameters. Siber and co-workers developed phase diagrams to investigate the effects of RNA and protein charge on capsid size but did not explicitly consider the CP-CP interaction or the work required to constrain the NA in a small volume (see 62). Equations 5–8 indicate tradeoffs and synergism between the suitability of an NA for packaging, α, and the strength of protein-protein interaction, ω; these are globally illustrated by a phase diagram (Fig. 3). The major effect of KNAN is to shift assembly to lower subunit concentration. As α and ω are unitless association constants, they are easily scaled and understood in terms of their natural logs, proportional to their free energies. Because ω is constant throughout assembly, whereas α changes discontinuously, we will construct the phase diagram comparing lnω to the average term ln<α>, acknowledging that particularly in the earliest steps of assembly α and <α> may differ substantially. The free energy for assembly of an N-subunit VLP would thus be
| (10) |
Considering ω independently, strong protein-protein interactions are expected to result in highly cooperative assembly (where high cooperativity is a term relative to other examples of cooperative capsid assembly). The weakest reasonable value for ω is 1 (lnω = 0), indicating negligible interaction. The value of ω is analogous to Kcontact. Because subunits are multivalent, making several equivalent independent contacts, the pseudocritical concentration of assembly of an empty capsid is approximately Kcontact−(contacts/2), in the micromolar range for HBV and the nanomolar range for SV40. Assembly of empty capsids is inhibited when Kcontact is too strong due to kinetic traps that lead to errors in assembly and overnucleation. At modest values, assembly simulations indicate that reversibility is critical for releasing improperly bound subunits (29,39). ω is expected to behave in much the same way as Kcontact, correlating with increased cooperativity until, at high ω values, irreversibly incorporated defects and overnucleation lead to defective assembly.
Figure 3.
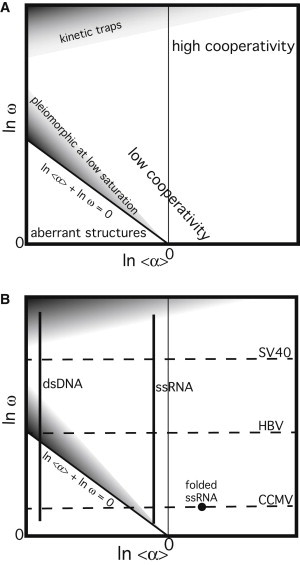
Phase diagrams of virus assembly. (A) Diagram highlighting stable and metastable phases. Low cooperativity may arise from weak CP-CP interaction, NA that is an unsuitable substrate for assembly, or (as described by McGhee and von Hippel (25)) the inability of CP to redistribute on NA to optimize interaction. High cooperativity is driven by high lnω and high ln<α>. (B) Overlay of assembly properties of three viruses and two NAs on the virus-assembly phase diagram. The deduction that ssRNA has a negative ln<α> is based on the volume normally occupied by a viral RNA (48,61) and observations that CCMV assembly on RNA normally shows low cooperativity, and that substoichiometric concentrations of CCMV CP cause RNA to compact to a form that is encapsidated with high cooperativity (15).
The effects of <α> on assembly, and the structural basis for effect, are more complex. When the NA (or non-NA) substrate favors assembly, ln<α> will be greater than zero. A spherical nanoparticle may be an ideal embodiment of a high ln<α> substrate (41,42). When the substrate disfavors assembly, as stiff linear dsDNA would, ln<α> will be less than zero. The evaluation of <α> is possible empirically. We suggest that the underlying physics for α is related to the work done on NA by capsid protein. The work done on the NA may include folding, restraining movement, and compaction. In general, viral ssRNA is flexible, highly branched, and relatively compact, but when unconstrained, it is still larger than the interior of its cognate capsid (17,43). dsDNA, on the other hand, is a stiff linear polymer (with a persistence length greater than or similar to the capsids diameter of most viruses (13,35)) that must be bent to conform to the capsid of a small virus, which may involve substantial work distributed over a small number of subunits. Work done by the NA includes restraining and orienting the capsid proteins and also possibly folding their RNA-binding domains. In addition, α may also include the energetic expense of distorting protein-protein interactions (i.e., ω) from ideality. Thus, at each step, α contributes to the free energy of assembly:
| (11) |
The phase diagram of virus assembly, lnω versus ln<α>, has several distinct zones (Fig. 3 and Table 1). Where ln<α> is positive, increased lnω corresponds to increasingly cooperative assembly. As ln<α> increases, the KVLP product increases, also enhancing cooperativity. There is a low-cooperativity region of the phase diagram; low cooperativity can result from a combination of low lnω (i.e., from weak CP-CP interaction) and negative ln<α> (NA that is a poor substrate for assembly). Not on this phase diagram, the inability of CP to redistribute on NA to optimize interaction will result in unfillable gaps giving rise to the appearance of negative cooperativity (25).
Table 1.
Interplay of α and ω
| Condition | Response |
|---|---|
| α ≥ 1 and ω ≫ 1 | Assembly is highly cooperative |
| α > 1 | NA structure promotes assembly (increased cooperativity) |
| α < 1 | NA structure inhibits assembly |
| α < 1 and αω > 1 | CP-CP interaction trumps NA structure |
| α < 1 and αω < 1 | NA structure dominates assembly |
Where ln<α> is less than zero, the phase diagram is more complex. The line described by (ln<α> + lnω) = 0 is a demarcation between VLP assembly and aberrant particles. Below this line it is energetically more favorable either to have subunits bind NA independent of one another or to find an alternative, noncapsid protein-protein interaction. Even above the (ln<α> + lnω) = 0 line, assembly may not yield VLPs.
Although it will not be investigated in detail in this article, the generalized description of assembly (Fig. 1) implies a third dimension of ln(KNA). One can expect ln(KNA) to be responsive to ionic strength or to CP charge changed by mutation. As ln(KNA) decreases, the tendency to assemble empty capsids can be expected to increase where the sum (lnω + lnα) is close to or less than 0. In accordance with the predictions of CP adsorbing to the surface of a sphere (44), one can predict that weaker ln(KNA) will tend to lead to stepwise cooperative assembly of capsids; strong ln(KNA), particularly in the presence of weak lnω, will favor formation of a disordered complex of CP associated with NA.
Kinetic effects on assembly make their appearance on the phase diagram in two regions due to predicted defects in assembly: where (ln<α> + lnω) is slightly greater than zero and where lnω is very high. Near the (ln<α> + lnω) = 0 border, early intermediates in assembly may be dominated by a relatively negative lnα, so it is likely that they will not resemble on-path complexes. Furthermore, if subsequent subunits are not able to participate in remodeling the complex, the result will be a region where, although (ln<α> + lnω) > 0, aberrant structures form a metastable state. The shape of this region will be dependent on assembly path but is likely to be broader at progressively more negative values of lnα and larger values of lnω.
In a similar way, at high values of lnω, necessarily, the nature of the kinetic trap depends on the path where subunits are added to the growing polymer. At high values of ω, assembly reactions of empty capsids can be trapped, because the reaction runs out of subunits to complete capsids, because improperly bound subunits do not dissociate, and because reactions become starved for intermediates (29,32,34,39). Defects in assembly due to high lnω will be path-dependent and will be further stabilized by the strength of KNA. Like reactions at the (ln<α> + lnω) = 0 border, we suggest that high lnω defects are more likely to form and be trapped at negative values of ln<α>. Although they are not thermodynamic phenomena, these traps have a place in demarcating the phase space of successful assembly.
The behavior of NA or other packaged cargo will vary almost continuously during assembly. The value of α can be thought of as a complex order parameter so that α and the phase diagram will be unique for each possible intermediate. Nonetheless, the phase diagrams outlined above allow a straightforward analysis of real assembly data below.
Three examples of assembly: CCMV, HBV, and SV40
Experimental observations of virus assembly can be understood in terms of an αωKNA product. We will consider three examples of virus assembly: CCMV, HBV, and SV40 (summarized in Table 2). In each case, we know something about assembly of empty particles, assembly on ssRNA, and assembly on dsDNA. Where apparent cooperativity is low, electrophoresis shows that all of the NA in a given sample has a similar amount of bound CP. Where cooperativity is high, we expect, and find, a bimodal distribution of free and bound NA (Fig. 1, B and C).
Table 2.
Summary of CCMV, HBV, and SV40 assembly properties
| CCMV | HBV | SV40 | |
|---|---|---|---|
| KDapparent | >75 μM (46) | ∼10 μM (51) | ≪0.1 μM (53) |
| Kcontact∗ | >9 mM | ∼3mM | ≪1 mM |
| Substrate† | |||
| ssRNA | Low cooperativity (15) | High cooperativity (21) | High cooperativity |
| dsDNA (low ν) | Condensate (49) | Condensate (12) | Condensate (16) |
| dsDNA (high ν) | Tubes (49) | T = 4, aberrant (12) | T = 7 (16) |
Kcontact is proportional to the c/2 root of KDapparent, where c is the number of contacts/subunit (29).
For dsDNA, results are reported for low and high occupancy, ν.
CCMV was the first spherical virus assembled in vitro, forming both empty capsids and infectious, RNA-filled particles (45). Virions are comprised of 28-nm-diameter T = 3 capsids assembled from 90 dimers. Empty particles readily assemble from purified CP in a pH-dependent reaction. At pH 5, assembly is observed at >2 μM CP dimer (36); assembly is not observed at neutral pH at concentrations up to 75 μM (45). Assembly of empty particles depends on formation of pentamers of dimers (46,47). CCMV assembly on RNA shows low cooperativity (Fig. 2 A). Viral RNA mixed with CP and electrophoresed through a native agarose gel 20 min after mixing shows a heterogeneous mixture of partial capsids at partial saturation, indicating low cooperativity (15). After 24 h incubation, the same reactions show a bimodal distribution of free RNA and VLP, indicative of a reorganization of CP to a lower energy state due to a larger number of CP-CP interactions, analogous to Ostwald ripening. Indeed, small amounts of CCMV CP cause RNA to fold into a more compact structure that migrates faster on a gel and supports highly cooperative assembly of capsids; that is, substoichiometric concentrations of CP are able to compact the RNA to prime further assembly (15); a similar, but sequence-specific, NA compaction was recently reported for two other RNA viruses (bacteriophage MS2 and Sstellite tobacco necrosis virus) (48). Thus, compaction may be an evolved mechanism for increasing α. Indeed, NA encoding high-affinity sites for CP, especially if those sites are strategically spaced, is a specific realization of NA with a (relatively) positive lnα. Conversely, when CCMV CP was mixed with low-α dsDNA at neutral pH, it assembled into 17-nm-diameter tubes, not capsids at all (Fig. 4) (49). When tubes are acidified, strengthening protein-protein interaction, the subunits reorganized, dissociating from the dsDNA to form empty T = 1 capsids (47).
Figure 4.
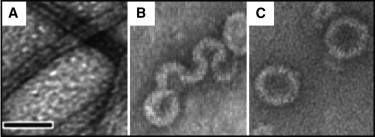
Electron micrographs showing the diversity of assembly products when different viral coat proteins associate with a 600-base pair dsDNA (lnα ≪ 0). (A) CCMV CP (low lnω) assembly is completely redirected to form 17-nm-diameter tubes with a core of parallel overlapping dsDNA molecules (49). (B) HBV CP has a moderate lnω, similar to the lnα of dsDNA, resulting in aberrant particles showing conformational influences from both macromolecules as the CP unsuccessfully tries to form 35-nm-diameter spheres (12). (C) The CP of SV40 (high lnω) overcomes the stiffness of dsDNA to yield virus-like 45-nm-diameter capsids (from Mukherjee et al. (16)). All micrographs were stained with uranyl acetate. Scale bar, 50 nm.
CCMV can be put in the context of the αω phase diagram (Fig. 3 B). The very weak association energy at neutral pH, demonstrated by failure to form empty capsids, indicates that ω is small, <80 based on extrapolating its low-pH pseudocritical concentration to neutral pH (36,46). However, when substoichiometric CP assembly refolds the ssRNA into a more compact form, assembly becomes cooperative. As even a weak ω leads to high cooperativity of assembly, and CP is the same with both unfolded and compacted RNA, this leads to the conclusion that unfolded ssRNA must have a lnα value that is <0, whereas when RNA is compact, lnα becomes >0, allowing ω to drive cooperative assembly. With either RNA state, the αωKNA product is sufficient to drive capsid assembly. On the other hand, for dsDNA the lnα term must be substantial and negative, resulting in a (lnα + lnω) sum that is <0, favoring a noncapsid alternative protein-protein interaction, so that dsDNA redirects assembly.
HBV is a major cause of human disease, contributing to ∼600,000 deaths each year. HBV has an unusual lifecycle in that it is a dsDNA virus with an ssRNA intermediate (50): its 120-dimer T = 4 capsid assembles on viral RNA, which is then reverse-transcribed within the capsid. HBV is also one of the best-characterized virus assembly systems (34), where ω, approximated by Kcontact, under physiological conditions is ∼150–300 (51). The association of the HBV CP for ssRNA and ssDNA is driven by an RNA-binding domain with a net charge of +30/dimer and is very strong (12,21). When assembled on RNA, CP binding is highly cooperative and yields capsids with virus-like morphology, ∼35 nm in diameter (21). However, when dimeric HBV CP is added to dsDNA, the resulting complexes are heterogeneous (12). In electron micrographs it can be seen that the majority of HBV CP-DNA particles appear to be spirals and aggregates of capsid fragments, with possibly a few normal capsids (Fig. 4). Nonetheless, in vivo, all infectious HBV virions contain a circular, almost entirely double-stranded DNA genome.
HBV CP crosses the αω phase diagram along a different line than CCMV. The lnω for HBV is substantially greater than lnω for CCMV, corresponding to highly cooperative assembly on ssRNA (lnα ≤ 0 based on analysis of CCMV assembly). Because of its higher lnω, because infectious HBV does contain dsDNA, and because at least some HBV assembled on dsDNA appears to form VLPs, HBV CP with a dsDNA substrate therefore has an overall lnαω > 0. However, the heterogeneity of dsDNA assembly products (Fig. 4) indicates that lnαω < 0 for some early intermediates in the assembly process. The structure of any given assembly product therefore depends on its stochastic starting point and assembly path.
Unlike HBV or CCMV, SV40 is a dsDNA virus that assembles on a dsDNA substrate in vivo, albeit NA is compacted by bound histones. SV40 has unusual architecture, basing a 45-nm-diameter T = 7 capsid of 72 CP pentamers. In vitro, SV40 is capable of packaging bare dsDNA, up to twice the length of its 5200-bp genome; these complexes have been used as vectors for transfection (20,52). Assembly of empty SV40 capsids can be induced by calcium, high ionic strength, or pH, but this rapid and quantitative reaction yields pleiomorphic results consistent with its very high association energy (53). Assembly can also be catalyzed by chaperones in vitro, indicating the presence of a kinetic, not thermodynamic, barrier to formation of virus-like capsids (54). Assembly of SV40 on ssRNA is highly cooperative and yields mainly 22-nm-diameter T = 1 particles; with longer RNA (≥1200 nucleotides), a few 35-nm particles are also observed (33). On dsDNA, SV40 has a sigmoidal assembly curve, suggesting that there is a barrier to initiating assembly but that it proceeds cooperatively thereafter (16). Assembly products with DNA are uniformly 45 nm in diameter (Fig. 4).
The very strong association constant of SV40 CP indicates a high value for ω; the ω value of the structurally similar Human Papillomavirus 16 is on the order of 1000 (55). With an RNA substrate, assembly is aggressive and pleiomorphic, suggesting the presence of kinetic traps, errors in assembly that could not be corrected, presumably because of slow dissociation and rearrangement of subunits. In terms of the phase diagram, this places SV40 near the border high cooperativity and kinetic traps. However, even with lnα ≪ 0, the lnα + lnω sum is still substantially greater than 0, and normal capsids arise with a sufficiently high CP concentration.
An important confirmatory observation is that with a dsDNA substrate (with lnα ≪ 0), low concentrations of CPs from CCMV, HBV, and SV40 all induced formation of large cylindrical DNA condensates (12,15,16). Condensates are a stable or metastable phase of DNA, typically induced by multivalent cations, where DNA strands are approximately parallel, with a minimal spacing of typically 2.7 nm (13,35); these condensates usually are toroidal but may also be rodlike. Such complexes are incompatible with capsid-like CP-CP interactions. In these complexes, the CPs may be noncovalently cross-linked and bundle many dsDNA polymers. The assembly conditions of these condensates, low lnα and low [CP], are consistent with conditions where negative lnα and low CP concentration effectively eliminate the role of ω in assembly. Thus, the condensate assembly reaction is based on KNA.
Conclusions
A substantial number of spherical viruses package their genomes during the process of capsid assembly. Besides the examples discussed in this article (Bromoviridae, Hepadnaviridae, and Polyomaviridae), families of viruses that assemble in conjunction with packaging their genomes include Flaviviridae, Togaviridae, Papillomaviridae, and possibly Retroviridae. Although the biological details are unique to each virus, the underlying physics will remain the same. The challenge we address in this work is to place the sometimes-conflicting factors governing assembly and NA packaging into a unified framework. Here, we consider assembly as a reaction where each step is the product of terms of binding NA (KNA), protein-protein interaction (ω), and the work of encapsidating the NA (α). From this analysis of assembly, it can be argued that NA plays a particularly dominant role in directing at the earliest stages in the process. RNA compaction by CP (observed in CCMV and bacteriophage MS2 (15,48)) and DNA compaction by histones (in SV40) are viral mechanisms for increasing α. Later in a virion’s assembly, the assembly energy related to ω becomes dominant. Where assembly is able to equilibrate, conflicts between NA and CP can resolve. The phase diagram indicates conditions where assembly can be trapped due to the stability of an early assembly product. In those regions, in vitro assembly will be frustrated and in vivo assembly will require chaperones or a multiphase assembly process to avoid energetic bottlenecks (e.g., HBV assembly on an RNA form of its genome). This logic suggests that evolutionary pressures select for coordination between ω and α. Indeed, this coordination may be useful for understanding the speciation of viruses and for learning how to manipulate them.
The description of assembly of an NA-filled capsid is directly applicable to experimental data. The value of ω for the interaction between two subunits is approximately Kcontact. The two terms differ because Kcontact includes entropic loss of freedom due to protein association (56). The entropic cost of association, implicit in Kcontact and KNA, but not in ω, has been experimentally estimated to be −5 ± 4 cal M−1 deg−1, about +1.5 kcal/mol under physiological conditions (56), which is substantial compared to the −3.5 kcal/mol energy/contact determined for HBV (51).
Consider the role of ω in assembly. A series of HBV CP mutations (observed in chronic infections) that modestly enhance ω may provide an advantage to the virus when production of the CP is suppressed by the host (29,57). Small alterations at the HBV CP-CP interface can have catastrophic results. Small molecules that enhance HBV assembly, increasing ω, disrupt the timing and organization of virion formation (34). Thus, we suggest that virus viability is very sensitive to ω. The phase diagrams of Fig. 3 provide a predictive guideline to effects of ω.
Determination of α will be difficult. The average value of α, <α>, can be estimated from the difference between association of an empty capsid, association of capsid protein to NA in the absence of assembly, and assembly of an NA-filled capsid. Perhaps a more powerful method of evaluating α is to compare NA substrates to a standard. The variant of length has been examined in silico (18) and to some degree in vitro (20). However, the effects on α of NA length, sequence, local, and higher-order structure have not been examined systematically in solution, though presumably these factors will be critical for in vivo NA packaging and for taking advantage of the NA capacity of viruses as NA carriers in culture and gene therapy. Though <α> can be determined experimentally from bulk assembly experiments, understanding the basis for α and how it changes during assembly will almost certainly require single-molecule experiments and coarse-grained simulations.
Consider the role of α. Viral RNAs tend to be more compact than random RNA (17,43). The term α provides a quantifiable index of the selective pressure for different NA tertiary structures (7,48). Besides the basic biology of infection, viruses can find use in bio- and nanotechnology. This application makes the work required to package nonviral NAs a question of great importance (20,52). This question can also be applied to non-NA polymers (14) and solid substrates (42,58,59).
The role of KNA is critical to assembly. Without a strong CP-NA interaction there will certainly be a strong entropic barrier to NA-packaging, as predictable from the explicit assumptions developed in Eqs. 5–10. The strength of KNA will contribute to the ability of a CP to package any NA and will be absolutely necessary for NA with α < 1. Values for KNA are not generally known. An SV40 VP1 pentamer has a nanomolar dissociation constant for DNA (60). The KNA of HBV and CCMV for different NAs has not been quantified, but it appears to be strong given their affinity for dsDNA despite the fact that dsDNA induces unusual CP-CP interactions (12,49).
The ability of a virus to package its own genome is central to biology. This same process can be used to package nonviral cargo and harness viruses to designed tasks. The ability of a virus to assemble on a given substrate requires either adapting the substrate to the virus, a function of α, or adapting the virus CP to the substrate, a function of the product αω. Learning how to regulate α and ω will open new opportunities to develop viruses as tools.
Acknowledgments
A 2011 meeting at the Kavli Institute for Theoretical Physics provided the impetus for first description of these concepts. We acknowledge discussions with Fred Mackintosh, Robijn Bruinsma, and Boris Shklovskii on the physics of assembly, David Reguera on the effect of KNA on the phase diagram, and Ariella Oppenheim on the biochemistry of SV40.
This work was supported by National Institutes of Health grant R01-AI077688 to A.Z.
Footnotes
J. Zachary Porterfield’s present address is Department of Internal Medicine, 330 Cedar St., Boardman 110, P.O. Box 208056, Yale University, New Haven, CT 06520-8056.
References
- 1.Schneemann A. The structural and functional role of RNA in icosahedral virus assembly. Annu. Rev. Microbiol. 2006;60:51–67. doi: 10.1146/annurev.micro.60.080805.142304. [DOI] [PubMed] [Google Scholar]
- 2.Gouet P., Diprose J.M., Mertens P.P. The highly ordered double-stranded RNA genome of bluetongue virus revealed by crystallography. Cell. 1999;97:481–490. doi: 10.1016/s0092-8674(00)80758-8. [DOI] [PubMed] [Google Scholar]
- 3.Tang L., Johnson K.N., Johnson J.E. The structure of pariacoto virus reveals a dodecahedral cage of duplex RNA. Nat. Struct. Biol. 2001;8:77–83. doi: 10.1038/83089. [DOI] [PubMed] [Google Scholar]
- 4.Larson S.B., Koszelak S., McPherson A. Double-helical RNA in satellite tobacco mosaic virus. Nature. 1993;361:179–182. doi: 10.1038/361179a0. [DOI] [PubMed] [Google Scholar]
- 5.Chen Z.G., Stauffacher C., Johnson J.E. Protein-RNA interactions in an icosahedral virus at 3.0 Å resolution. Science. 1989;245:154–159. doi: 10.1126/science.2749253. [DOI] [PubMed] [Google Scholar]
- 6.Tihova M., Dryden K.A., Schneemann A. Nodavirus coat protein imposes dodecahedral RNA structure independent of nucleotide sequence and length. J. Virol. 2004;78:2897–2905. doi: 10.1128/JVI.78.6.2897-2905.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schroeder S.J., Stone J.W., Mathews D.M. Ensemble of secondary structures for encapsidated satellite tobacco mosaic virus RNA consistent with chemical probing and crystallography constraints. Biophys. J. 2011;101:167–175. doi: 10.1016/j.bpj.2011.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacFarlane S.A., Shanks M., Lomonossoff G.P. Analysis of the nucleotide sequence of bean pod mottle virus middle component RNA. Virology. 1991;183:405–409. doi: 10.1016/0042-6822(91)90155-5. [DOI] [PubMed] [Google Scholar]
- 9.Dryden K.A., Wieland S.F., Yeager M. Native hepatitis B virions and capsids visualized by electron cryomicroscopy. Mol. Cell. 2006;22:843–850. doi: 10.1016/j.molcel.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 10.Wang J.C., Dhason M.S., Zlotnick A. Structural organization of pregenomic RNA and the carboxy-terminal domain of the capsid protein of hepatitis B virus. PLoS Pathog. 2012;8:e1002919. doi: 10.1371/journal.ppat.1002919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petrov A.S., Harvey S.C. Packaging double-helical DNA into viral capsids: structures, forces, and energetics. Biophys. J. 2008;95:497–502. doi: 10.1529/biophysj.108.131797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhason M.S., Wang J.C.-Y., Zlotnick A. Differential assembly of Hepatitis B Virus core protein on single- and double-stranded DNA suggests the dsDNA core is spring-loaded. Virology. 2011;430:20–29. doi: 10.1016/j.virol.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tzlil S., Kindt J.T., Ben-Shaul A. Forces and pressures in DNA packaging and release from viral capsids. Biophys. J. 2003;84:1616–1627. doi: 10.1016/S0006-3495(03)74971-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Y., Zandi R., Gelbart W.M. Packaging of a polymer by a viral capsid: the interplay between polymer length and capsid size. Biophys. J. 2008;94:1428–1436. doi: 10.1529/biophysj.107.117473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson J.M., Willits D.A., Zlotnick A. Interaction with capsid protein alters RNA structure and the pathway for in vitro assembly of cowpea chlorotic mottle virus. J. Mol. Biol. 2004;335:455–464. doi: 10.1016/j.jmb.2003.10.059. [DOI] [PubMed] [Google Scholar]
- 16.Mukherjee S., Kler S., Zlotnick A. Uncatalyzed assembly of spherical particles from SV40 VP1 pentamers and linear dsDNA incorporates both low and high cooperativity elements. Virology. 2010;397:199–204. doi: 10.1016/j.virol.2009.10.050. [DOI] [PubMed] [Google Scholar]
- 17.Yoffe A.M., Prinsen P., Ben-Shaul A. Predicting the sizes of large RNA molecules. Proc. Natl. Acad. Sci. USA. 2008;105:16153–16158. doi: 10.1073/pnas.0808089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elrad O.M., Hagan M.F. Encapsulation of a polymer by an icosahedral virus. Phys. Biol. 2010;7:045003. doi: 10.1088/1478-3975/7/4/045003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kivenson A., Hagan M.F. Mechanisms of capsid assembly around a polymer. Biophys. J. 2010;99:619–628. doi: 10.1016/j.bpj.2010.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimchi-Sarfaty C., Arora M., Gottesman M.M. High cloning capacity of in vitro packaged SV40 vectors with no SV40 virus sequences. Hum. Gene Ther. 2003;14:167–177. doi: 10.1089/104303403321070865. [DOI] [PubMed] [Google Scholar]
- 21.Porterfield J.Z., Dhason M.S., Zlotnick A. Full-length HBV core protein packages viral and heterologous RNA with similarly high cooperativity. J. Virol. 2010;84:7174–7184. doi: 10.1128/JVI.00586-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McPherson A. Micelle formation and crystallization as paradigms for virus assembly. Bioessays. 2005;27:447–458. doi: 10.1002/bies.20196. [DOI] [PubMed] [Google Scholar]
- 23.Devkota B., Petrov A.S., Harvey S.C. Structural and electrostatic characterization of pariacoto virus: implications for viral assembly. Biopolymers. 2009;91:530–538. doi: 10.1002/bip.21168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kowalczykowski S.C., Paul L.S., von Hippel P.H. Cooperative and noncooperative binding of protein ligands to nucleic acid lattices: experimental approaches to the determination of thermodynamic parameters. Biochemistry. 1986;25:1226–1240. doi: 10.1021/bi00354a006. [DOI] [PubMed] [Google Scholar]
- 25.McGhee J.D., von Hippel P.H. Theoretical aspects of DNA-protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 1974;86:469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- 26.Zandi R., van der Schoot P. Size regulation of ss-RNA viruses. Biophys. J. 2009;96:9–20. doi: 10.1529/biophysj.108.137489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu T., Shklovskii B.I. Kinetics of viral self-assembly: role of the single-stranded RNA antenna. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2007;75:051901. doi: 10.1103/PhysRevE.75.051901. [DOI] [PubMed] [Google Scholar]
- 28.von Hippel P.H., Berg O.G. Facilitated target location in biological systems. J. Biol. Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- 29.Katen S.P., Zlotnick A. The thermodynamics of virus capsid assembly. Methods Enzymol. 2009;455:395–417. doi: 10.1016/S0076-6879(08)04214-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zlotnick A. To build a virus capsid. An equilibrium model of the self assembly of polyhedral protein complexes. J. Mol. Biol. 1994;241:59–67. doi: 10.1006/jmbi.1994.1473. [DOI] [PubMed] [Google Scholar]
- 31.Katen S.P., Chirapu S.R., Zlotnick A. Trapping of hepatitis B virus capsid assembly intermediates by phenylpropenamide assembly accelerators. ACS Chem. Biol. 2010;5:1125–1136. doi: 10.1021/cb100275b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morozov A.Y., Bruinsma R.F., Rudnick J. Assembly of viruses and the pseudo-law of mass action. J. Chem. Phys. 2009;131:155101. doi: 10.1063/1.3212694. [DOI] [PubMed] [Google Scholar]
- 33.Kler S., Asor R., Raviv U. RNA encapsidation by SV40-derived nanoparticles follows a rapid two-state mechanism. J. Am. Chem. Soc. 2012;134:8823–8830. doi: 10.1021/ja2110703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zlotnick A., Mukhopadhyay S. Virus assembly, allostery and antivirals. Trends Microbiol. 2011;19:14–23. doi: 10.1016/j.tim.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purohit P.K., Kondev J., Phillips R. Mechanics of DNA packaging in viruses. Proc. Natl. Acad. Sci. USA. 2003;100:3173–3178. doi: 10.1073/pnas.0737893100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson J.M., Tang J., Zlotnick A. Regulating self-assembly of spherical oligomers. Nano Lett. 2005;5:765–770. doi: 10.1021/nl050274q. [DOI] [PubMed] [Google Scholar]
- 37.Porterfield J.Z., Zlotnick A. An overview of capsid assembly kinetics. In: Stockley P.G., Twarock R., editors. Emerging Topics in Physical Virology. Imperial College Press; London: 2010. pp. 131–158. [Google Scholar]
- 38.Hagan M.F., Chandler D. Dynamic pathways for viral capsid assembly. Biophys. J. 2006;91:42–54. doi: 10.1529/biophysj.105.076851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rapaport D.C. Studies of reversible capsid shell growth. J. Phys. Condens. Matter. 2010;22:104115. doi: 10.1088/0953-8984/22/10/104115. [DOI] [PubMed] [Google Scholar]
- 40.Endres D., Miyahara M., Zlotnick A. A reaction landscape identifies the intermediates critical for self-assembly of virus capsids and other polyhedral structures. Protein Sci. 2005;14:1518–1525. doi: 10.1110/ps.041314405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hagan M.F. Controlling viral capsid assembly with templating. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2008;77:051904. doi: 10.1103/PhysRevE.77.051904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen C., Kwak E.-S., Dragnea B. Packaging of gold particles in viral capsids. J. Nanosci. Nanotechnol. 2005;5:2029–2033. doi: 10.1166/jnn.2005.506. [DOI] [PubMed] [Google Scholar]
- 43.Kuznetsov Y.G., Daijogo S., McPherson A. Atomic force microscopy analysis of icosahedral virus RNA. J. Mol. Biol. 2005;347:41–52. doi: 10.1016/j.jmb.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 44.Hagan M.F. A theory for viral capsid assembly around electrostatic cores. J. Chem. Phys. 2009;130:114902. doi: 10.1063/1.3086041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bancroft J.B. The self-assembly of spherical plant viruses. Adv. Virus Res. 1970;16:99–134. doi: 10.1016/s0065-3527(08)60022-6. [DOI] [PubMed] [Google Scholar]
- 46.Zlotnick A., Aldrich R., Young M.J. Mechanism of capsid assembly for an icosahedral plant virus. Virology. 2000;277:450–456. doi: 10.1006/viro.2000.0619. [DOI] [PubMed] [Google Scholar]
- 47.Burns K., Mukherjee S., Zlotnick A. Altering the energy landscape of virus self-assembly to generate kinetically trapped nanoparticles. Biomacromolecules. 2010;11:439–442. doi: 10.1021/bm901160b. [DOI] [PubMed] [Google Scholar]
- 48.Borodavka A., Tuma R., Stockley P.G. Evidence that viral RNAs have evolved for efficient, two-stage packaging. Proc. Natl. Acad. Sci. USA. 2012;109:15769–15774. doi: 10.1073/pnas.1204357109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mukherjee S., Pfeifer C.M., Zlotnick A. Redirecting the coat protein of a spherical virus to assemble into tubular nanostructures. J. Am. Chem. Soc. 2006;128:2538–2539. doi: 10.1021/ja056656f. [DOI] [PubMed] [Google Scholar]
- 50.Nassal M. Hepatitis B viruses: reverse transcription a different way. Virus Res. 2008;134:235–249. doi: 10.1016/j.virusres.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 51.Ceres P., Zlotnick A. Weak protein-protein interactions are sufficient to drive assembly of hepatitis B virus capsids. Biochemistry. 2002;41:11525–11531. doi: 10.1021/bi0261645. [DOI] [PubMed] [Google Scholar]
- 52.Mukherjee S., Abd-El-Latif M., Oppenheim A. High cooperativity of the SV40 major capsid protein VP1 in virus assembly. PLoS ONE. 2007;2:e765. doi: 10.1371/journal.pone.0000765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kanesashi S.N., Ishizu K., Handa H. Simian virus 40 VP1 capsid protein forms polymorphic assemblies in vitro. J. Gen. Virol. 2003;84:1899–1905. doi: 10.1099/vir.0.19067-0. [DOI] [PubMed] [Google Scholar]
- 54.Chromy L.R., Pipas J.M., Garcea R.L. Chaperone-mediated in vitro assembly of Polyomavirus capsids. Proc. Natl. Acad. Sci. USA. 2003;100:10477–10482. doi: 10.1073/pnas.1832245100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mukherjee S., Thorsteinsson M.V., Zlotnick A. A quantitative description of in vitro assembly of human papillomavirus 16 virus-like particles. J. Mol. Biol. 2008;381:229–237. doi: 10.1016/j.jmb.2008.05.079. [DOI] [PubMed] [Google Scholar]
- 56.Tamura A., Privalov P.L. The entropy cost of protein association. J. Mol. Biol. 1997;273:1048–1060. doi: 10.1006/jmbi.1997.1368. [DOI] [PubMed] [Google Scholar]
- 57.Yuan T.T., Sahu G.K., Shih C. The mechanism of an immature secretion phenotype of a highly frequent naturally occurring missense mutation at codon 97 of human hepatitis B virus core antigen. J. Virol. 1999;73:5731–5740. doi: 10.1128/jvi.73.7.5731-5740.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dixit S.K., Goicochea N.L., Dragnea B. Quantum dot encapsulation in viral capsids. Nano Lett. 2006;6:1993–1999. doi: 10.1021/nl061165u. [DOI] [PubMed] [Google Scholar]
- 59.Sun J., DuFort C., Dragnea B. Core-controlled polymorphism in virus-like particles. Proc. Natl. Acad. Sci. USA. 2007;104:1354–1359. doi: 10.1073/pnas.0610542104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li P.P., Nakanishi A., Kasamatsu H. Simian virus 40 Vp1 DNA-binding domain is functionally separable from the overlapping nuclear localization signal and is required for effective virion formation and full viability. J. Virol. 2001;75:7321–7329. doi: 10.1128/JVI.75.16.7321-7329.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gopal A., Zhou Z.H., Gelbart W.M. Visualizing large RNA molecules in solution. RNA. 2012;18:284–299. doi: 10.1261/rna.027557.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Siber A., Zandi R., Podgornik R. Thermodynamics of nanospheres encapsulated in virus capsids. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2010;81:051919. doi: 10.1103/PhysRevE.81.051919. [DOI] [PubMed] [Google Scholar]


