Abstract
Legumes control the nitrogen-fixing root nodule symbiosis in response to external and internal stimuli, such as nitrate, and via systemic autoregulation of nodulation (AON). Overexpression of the CLV3/ESR-related (CLE) pre-propeptide-encoding genes GmNIC1 (nitrate-induced and acting locally) and GmRIC1 (Bradyrhizobium-induced and acting systemically) suppresses soybean nodulation dependent on the activity of the nodulation autoregulation receptor kinase (GmNARK). This nodule inhibition response was used to assess the relative importance of key structural components within and around the CLE domain sequences of these genes. Using a site-directed mutagenesis approach, mutants were produced at each amino acid within the CLE domain (RLAPEGPDPHHN) of GmRIC1. This approach identified the Arg1, Ala3, Pro4, Gly6, Pro7, Asp8, His11, and Asn12 residues as critical to GmRIC1 nodulation suppression activity (NSA). In contrast, none of the mutations in conserved residues outside of the CLE domain showed compromised NSA. Chimeric genes derived from combinations of GmRIC1 and GmNIC1 domains were used to determine the role of each pre-propeptide domain in NSA differences that exist between the two peptides. It was found that the transit peptide and CLE peptide regions of GmRIC1 significantly enhanced activity of GmNIC1. In contrast, the comparable GmNIC1 domains reduced the NSA of GmRIC1. Identification of these critical residues and domains provides a better understanding of how these hormone-like peptides function in plant development and regulation.
Key words: Autoregulation of nodulation, CLE peptides, legumes, nodulation, soybean, symbiosis.
Introduction
In agricultural systems, reduced nitrogen is often limiting and thus requires application of nitrogen fertilizer, which has both cost and environmental concerns (Jensen et al., 2012). Most legume species develop a symbiotic relationship with soil rhizobia that reduces the need for this input due to biological nitrogen fixation. Rhizobia undergo differentiation to bacteroids and are housed in a complex organ, known as a nodule, which maintains the conditions required for nitrogen fixation to occur. The nodule develops on the roots through a re-initiation of cell divisions and concurrent infection events (reviewed by Ferguson et al., 2010; Desbrosses and Stougaard, 2011).
The development of nodules is regulated by the plant in response to internal and external cues, including available reduced nitrogen, and through a systemic regulatory mechanism known as the autoregulation of nodulation (AON; first proposed by Gresshoff and Delves, 1986). AON is established in response to early nodulation signalling events through long-distance signals between the root and shoot (Delves et al., 1986; Li et al., 2009; Reid et al., 2011b ) and is maintained by the nodulation autoregulation receptor kinase (GmNARK) in soybean (Searle et al., 2003). GmNARK is structurally similar to the CLAVATA1 (CLV1) receptor kinase of Arabidopsis (Clark et al., 1997), and is functionally conserved with its homologues from other legume species (Krusell et al., 2002; Nishimura et al., 2002; Schnabel et al., 2005).
In Arabidopsis, the leucine-rich repeat-receptor kinase (LRR-RK) CLAVATA1 regulates the stem cell population of the shoot apical meristem (SAM) through direct perception of the small peptide CLV3 (Fletcher et al., 1999; Ogawa et al., 2008) in a negative feedback loop with the transcription factor WUSCHEL (WUS; Brand et al., 2000; Schoof et al., 2000). Both the CLV3 peptide and WUS protein may act in cells at a distance from their origin of production via intercellular movement (Lenhard and Laux, 2003; Yadav et al., 2011). Many similarities exist between the functional components of the CLV system and AON (reviewed in Reid et al., 2011b ), including the recent identification of a WUSCHEL-RELATED HOMEOBOX gene (WOX5; Osipova et al., 2012), a KINASE-ASSOCIATED PROTEIN PHOSPHATASE gene (KAPP1; Miyahara et al., 2008), CLAVATA2-receptor protein-controlled supernodulation (Krusell et al., 2011), and CLE (CLV3/ESR-related) peptides.
CLE peptides functioning in the Arabidopsis CLV system regulate the stem cell population of the SAM via CLV1, whereas those of AON regulate nodulation in a GmNARK-dependent manner (Gresshoff et al., 2009; Okamoto et al., 2009; Mortier et al., 2010; Reid et al., 2011a ). In soybean, three such CLE peptides have been characterized (Lim et al., 2011; Reid et al., 2011a ). They are induced in response to either nitrate (GmNIC1) or early infection and nodule establishment events in the root zone susceptible to nodulation (GmRIC1 and GmRIC2; Hayashi et al., 2012). GmRIC1 and GmRIC2 inhibit nodulation in a systemic manner when constitutively overexpressed in chimeric plants, with GmRIC1 achieving complete suppression of nodulation in wild-type plants. In contrast, GmNIC1 appears to act locally (as expected for nitrate inhibition of nodulation; see Carroll and Gresshoff, 1983) and only partially, but significantly, suppresses nodulation when overexpressed (Reid et al., 2011a ). The nodulation suppression activity (NSA) of GmRIC1/GmRIC2 is epistatically suppressed in a GmNARK-deficient plant (Reid et al., 2011a ), suggesting that the peptide signals through the LRR-RK (reminiscent of the CLV3–CLV1 situation).
The systemic activity of these CLE peptides, and the shoot involvement through GmNARK, suggests that they may act as long-distance signal ligands for GmNARK in the shoot [NB: similar NSA was detected for CLE overexpression in Lotus japonicus (Okamoto et al., 2009) and Medicago truncatula (Mortier et al., 2010)]. Indeed, it is predicted that perception of GmRIC1/GmRIC2 by GmNARK in the shoot leads to the production of a novel shoot-derived inhibitor (SDI) signal that is transported to the root where it inhibits further nodulation events (Lin et al., 2010, 2011), thus completing the systemic loop of AON (reviewed in Reid et al., 2011b ). Knock-down of the two related CLE genes in M. truncatula resulted in an increase in nodule numbers, further indicating their likely role in inducing AON (Mortier et al., 2012). Despite this evidence, efforts to detect CLE peptides in xylem sap samples or root tissues of GmRIC1-overexpressing plants have not identified CLE peptides to date (Reid et al., 2012).
Genes encoding CLE peptides are widely distributed in plant species, including >20 CLE peptide encoding genes in Arabidopsis and over at least 40 in soybean (Cock and McCormick, 2001; Oelkers et al., 2008; Mortier et al., 2011). CLE peptides possess three basic domains, namely an N-terminal transit (signal) peptide, a variable region which does not share a high degree of homology within the family (beyond predicted hydrophobicity), and a 12–13 amino acid CLE domain (Cock and McCormick 2001). Post-translational modifications and processing are required to produce the 12–13 amino acid mature CLE peptide (Kondo et al., 2006; Ohyama et al., 2009). These modifications include proline hydroxylation and arabinosylation which have been detected for several CLE peptides [CLV3 and CLE2 (Ohyama et al., 2009), CLE9 (Shinohara et al., 2012)]. Amino acid residues neighbouring the CLE domain have been proposed to be required for efficient processing of the mature peptide through proteolytic cleavage (Ni and Clark, 2006; Djordjevic et al., 2011; Ni et al., 2011). The transit peptide has been shown to be critical to the nodule inhibition and secretion of green fluorescent protein (GFP)-tagged GmRIC1 and GmRIC2 peptides (Lim et al., 2011). In addition to the conserved CLE peptide domain, several of the rhizobia-induced CLE peptides in L. japonicus, M. truncatula, and Glycine max, as well as AtCLV3, possess a conserved extension in the C-terminal region of the CLE pre-propeptide comprising dual proline residues. The functional significance of these residues is unclear, but they may assist in processing, protection from degradation, or binding of the CLE peptide.
A number of studies using AtCLV3 have employed site-directed mutagenesis (SDM) constructs in vivo (Ni et al., 2011; Song et al., 2012), and in vitro peptide application (Kondo et al., 2008), or domain swap techniques (Meng et al. 2010), to decipher the role of each of the domains of the CLE pre-propeptides. Alanine replacement mutants at each of the residues within the AtCLV3 domain and surrounding residues have shown that mutants at the R1, P4, G6, D8, P9, L10, H11, and H12 residues of the AtCLV3 peptide were significantly compromised in their ability to complement Atclv3 mutants (Song et al., 2012). Synthetic CLE peptides consisting solely of the CLE domain residues, and engineered genes encoding the transit peptide directly adjacent to the CLE domain, have also been shown to be biologically active and effective at rescuing some Atclv3 phenotypes (Fiers et al., 2005, 2006). In addition to the importance of the CLE domain, the R70 residue of AtCLV3, which immediately precedes the CLE domain, has been identified as a peptilytic processing site; however, mutants at this residue continue to maintain activity (Ni and Clarke, 2006). Up to four residues may be required for cleavage recognition and processing at this residue in the N-terminus of the CLE peptide. A serine protease is likely to be the precise cleaving agent at this location, whereas a progressive carboxypeptidase acts at the C-terminus (Ni et al., 2011). Mutations at the EE residues preceding the AtCLV3 peptide domain blocked processing but continued to rescue mutant phenotypes, which may indicate that the processing is not required to be highly efficient due to high specificity and very low concentrations of the peptide required for activity. It has been shown that the CLE domain and transit peptide determines the function and specificity of the peptide (Fiers et al., 2006; Meng et al., 2010), probably through determining to which tissues and receptors the ligand is presented. This tissue specificity is also a result of the diverse expression patterns that exist for the CLE-encoding genes (Sharma et al., 2003; Jun et al., 2010).
The GmRIC1 CLE domain shares high sequence similarity with the Arabidopsis CLE1–CLE7 peptides, with the consensus sequence only varying at the final residue (H in the Arabidopsis CLE1–CLE7 peptides compared with N in the nodulation CLE peptides). The function of these Arabidopsis CLE peptides remains to be determined; they do not possess the common root apical meristem (RAM)-arresting phenotypes of other CLE peptides and do not induce significant changes in SAM size when applied directly to the plant (Kinoshita et al., 2007). It should be noted that direct plant application may lessen activity because of degradation and possible uptake barriers. Overexpression of genes encoding AtCLE1–AtCLE7 peptides does, however, indicate that they possess biological activity as it results in phenotypes similar to AtCLV3 overexpression, albeit with less severe phenotypes (Strabala et al., 2006). Key residues within GmRIC1 are conserved throughout CLE peptides, including an almost universal conservation of the residues indicated in bold on the GmRIC1 sequence: RLAPEGPD PHHN (italics indicate one of two possible amino acids; Oelkers et al., 2008).
The complete suppression phenotype obtained in GmRIC1-overexpressing roots of soybean (Reid et al., 2011a) provides an excellent system to investigate the contribution of key residues within the CLE peptide to its overall function. To investigate the relative importance of each residue in the CLE domain to the function of GmRIC1, an SDM study was initiated. In addition, chimeric constructs comprised of swapped functional domains of GmRIC1 and GmNIC1 were generated to identify which domains contribute to nodule suppression activity and which are critical for localization of the peptide. These experiments allowed the identification of the key residues required for nodule suppression in soybean and will assist in the identification of biologically active CLE peptides capable of nodule suppression in other legume species. These experiments also provide a basis to compare the function of individual residues within nodulation-suppressing CLE peptides with those identified as critical in different species and developmental processes such as obtained from the study of AtCLV3.
Materials and methods
General plant and bacterial growth conditions and the nodulation assay
Soybean (Glycine max L. Merrill) wild-type Bragg plants were grown in controlled glasshouse conditions (28 °C/26 °C day/night) in 4 litre pots containing grade 3 vermiculite. Plants were watered as required with a modified nutrient solution lacking nitrogen (Herridge, 1982). Plants were inoculated with ~200ml of Bradyrhizobium japonicum CB1809 grown in yeast-mannitol broth (YMB) at 28 °C for 4 d and diluted to ~OD600 0.01. Nodulation was scored 3 weeks after inoculation. Student’s t-tests were used to determine statistical differences.
Generating constructs and transgenic soybean roots
Constructs bearing the coding sequence of GmRIC1 and GmNIC1 cloned in the sense direction only of the pKANNIBAL vector for expression of RNA interference (RNAi) constructs were used as described previously (Wesley et al., 2001; Reid et al., 2011a). Pfu polymerase (Stratagene, La Jolla, CA, USA) was used to amplify PCR products incorporating restriction nuclease sites from these constructs. XhoI/EcoRI (for) and KpnI (rev) restriction sites were included in the primer sequences depending on internal restriction sites of each gene (full primer sequences are included in Supplementary Table S1 available at JXB online). Likely clones were confirmed by direct DNA sequencing and capillary separation. Constructs were subcloned into p15SRK2 integration vectors (A. Kereszt, unpublished) as a NotI fragment before tri-parental mating to introduce them into Agrobacterium rhizogenes K599. Agrobacterium rhizogenes was subsequently used for the induction of transgenic soybean hairy roots according to Kereszt et al. (2007) and Lin et al. (2011).
Site-directed mutagenesis and domain swaps
Point mutants of GmRIC1 were introduced into the cloning vector pKANNIBAL:GmRIC1 using PCR-based SDM. The forward and reverse oligonucleotide primers (Supplementary Table S1 at JXB online) were complementary to the opposite strands of the vector, and contained the desired mutation in the middle of the primers. Extension with these primers generated mutated plasmids at the relevant sites. Each PCR had a total volume of 25 μl containing 5 μl of 5× KAPA HiFi Buffer, 0.75 μl of 10mM dNTPs, 1 μl of each primer (10 μM), 0.25 μl of 25mM MgCl2, 0.5 μl of KAPA HiFi Polymerase (KAPA biosystems, cat. kk2101), and 6ng of pKANNIBAL:GmRIC1 DNA. The reaction program was as follows: one cycle of initial denaturation (95 °C for 3min 30 s), three cycles of amplification with a lower annealing temperature (95 °C for 20 s, 60 °C for 15 s, 72 °C for 3min 30 s), 19 cycles of amplification with a high annealing temperature (65 °C), and one cycle of extension (72 °C for 10min), before the reaction was cooled to 4 °C. The PCR products were treated with the methylation-sensitive endonuclease DpnI (NEB, Ipswich, MA, USA) for 1h at 37 °C to digest the parental DNA template and select for mutation-containing plasmids. The resulting undigested plasmids were used to transform competent TOP10 cells. All constructs were confirmed by direct DNA sequencing.
Chimeric gene domain swaps between regions of GmRIC1 and GmNIC1 were derived from pKANNIBAL:GmRIC1 and pKANNIBAL:GmNIC1. This required using either a long reverse primer method for modifications directly at the C-terminus (Meng et al., 2010) or the generation of overlapping PCR products before amplification with the outermost forward and reverse primers (Heckman and Pease, 2007). Primer sequences are shown in Supplementary Table S2 at JXB online. For the purpose of constructing chimeric genes, the transit peptide was identified through SignalP prediction of the most likely cleavage sites (Bendtsen et al., 2004) and comprises residues 1–29 for GmRIC1 and 1–30 for GmNIC1. The variable region comprises the central residues from 30 to 76 inclusive in GmRIC1 and 31 to 75 in GmNIC1, while the CLE domain comprised the 12 amino acids from 77 to 88 in GmRIC1 and 76 to 87 in GmNIC1. The residues 89–95 in the C-terminal region of GmRIC1 were defined as the C-terminal extension.
Results
Identification of critical GmRIC1 residues for nodule inhibition
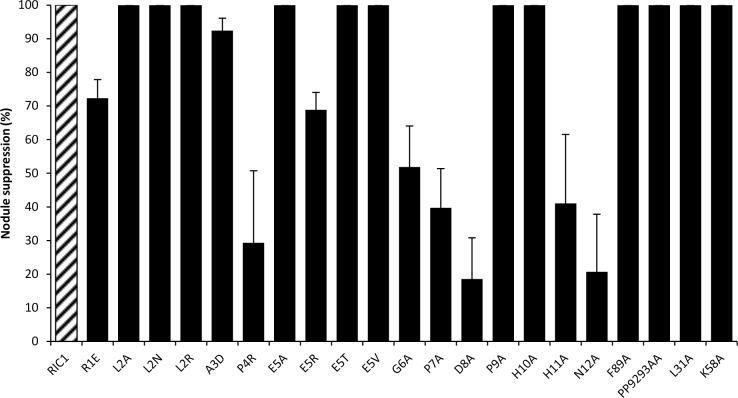
To investigate the functional importance of the amino acids within the CLE domain of GmRIC1, point mutations were produced for each residue (Fig. 1A). The GmRIC1 CLE domain is comprised of 12 amino acids positioned from R77 to N88 in the GmRIC1 sequence, which corresponds to R1–N12 in the CLE domain. Alanine substitutions were created at the L2, E5, G6, P7, D8, P9, H10, H11, and N12 residues, and additional substitutions were also made at R1E, A3D, and P4R. All mutants were derived from a GmRIC1 overexpression construct previously shown to inhibit soybean nodulation completely in a systemic and GmNARK-dependent manner (Reid et al., 2011a).
Fig. 1.
Constructs used in the nodule suppression assay. (A) Site-directed mutants derived from GmRIC1 were created for each residue in the CLE domain as well as several well-conserved residues outside the CLE domain. (B) Chimeric gene constructs were derived from combinations of the GmRIC1 and GmNIC1 signal peptide, variable region, CLE domain, and C-terminal extension. Each construct was named according to the source of each domain (e.g. RRRR for GmRIC1) and was expressed under the 35S promoter in Agrobacterium rhizogenes-induced hairy roots. (This figure is available in colour at JXB online.)
The nodule suppression activity of each construct was determined by comparing nodule numbers on A. rhizogenes-induced hairy roots of the empty vector (114 nodules ±10, n=12 in one representative experiment of four), with GmRIC1 overexpression constructs showing 100% suppression of nodulation (0 nodules ±0, n=12 in each of four replicates). Constructs bearing A substitutions at L2, E5, P9, and H10 retained complete nodule suppression activity, while the A3D mutant showed only a slight disruption in suppression (93% suppression of nodulation, P < 0.05 using Students t-test; Fig. 2). R1E (72%), E5R (69%), G6A (52%), H11A (41%), and P7A (40%) all showed an intermediate loss of suppression, while the most severe loss of suppression was observed in N12A (21%), P4R (29%), and D8A (18%) when compared with RIC1 (P < 0.05).
Fig. 2.
Nodulation suppression activity (NSA) in GmRIC1 site-directed mutants. Suppression of nodulation obtained by overexpression of each of the site-directed mutants was determined relative to the nodule number on empty vector-transformed controls. GmRIC1 and mutants that do not disrupt GmRIC1 function exhibit complete suppression.
Generation of multiple variants at conserved residues to investigate the effect of amino acid charge and size on GmRIC1 function
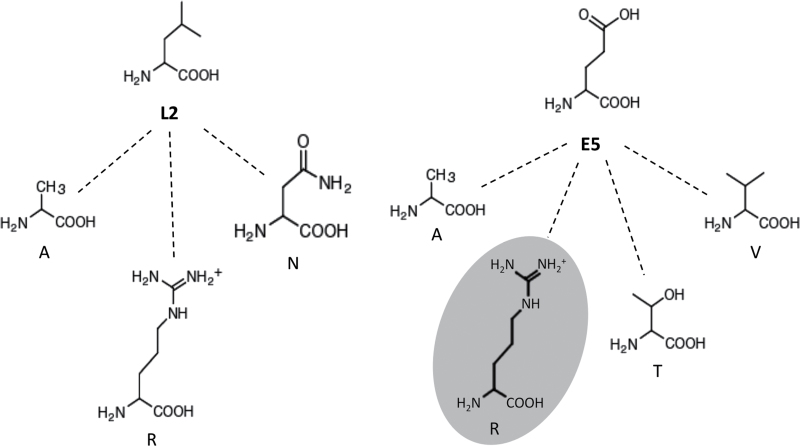
To determine the effect of amino acid charge and size on the function of the GmRIC1 peptide, multiple substitutions were produced at two conserved residues in the CLE domain, L2 and E5 (Fig. 3). A, N, and R mutants were created at L2, while A, T, V, and R mutants were created at E5 (Figs 1A, 3). Each of the three mutations at L2, and A, T, and V mutations at E5 maintained 100% suppression of nodulation. In contrast, the E5R substitution resulted in 69% suppression of nodulation (P < 0.05, Fig. 2).
Fig. 3.
Site-directed mutants of GmRIC1 to determine effects of charge and size of amino acid substitutions. Multiple mutations were produced at two well-conserved residues within the CLE domain, L2 and E5. A, R, and N residues were substituted for L2, while A, R, T, and V were substituted for E5. The shaded R residue represents the only mutation that disrupted the complete suppression of nodulation.
Mutation of residues outside the GmRIC1 CLE domain to determine their role in nodule inhibition
Certain residues located outside of the CLE domain are thought to be critical for the production of the mature peptide, including the signal peptide, the C-terminal region, and potential cleavage sites within the variable domain (Fiers et al., 2006; Ni and Clark 2006; Djordjevic et al., 2011; Ni et al., 2011). Mutations were produced at several of these well-conserved residues to determine their effect on GmRIC1 activity. Alanine substitutions were made at both the L31 and K58 residues of GmRIC1. L31 is located adjacent to the C-terminus of the signal peptide and shares 100% identity across the known nodulation CLE peptides of L. japonicus, M. truncatula, and G. max. K58 is the best conserved residue in the variable region of the nodulation CLE peptides (70% identity between known nodulation CLE peptides). Neither substitution resulted in a reduction in suppression activity of GmRIC1 (Fig. 2).
Alanine substitutions were also produced at two locations in the C-terminal region of GmRIC1, including the residue immediately adjacent to the predicted mature peptide (F89A), and two conserved P residues (92/93, a single construct bearing an AA substitution), which are conserved between 70% of nodulation-related CLE peptides. These substitutions also maintained complete nodule suppression activity.
Amino acid conservation in CLE peptides in relation to functional importance
To determine whether amino acid conservation is a good indicator of functional importance, the results were compared with multiple sequence alignments of all CLE peptides reported to possess nodule suppression activity (Reid et al., 2011a) and with recent AtCLV3 experiments involving site-directed mutagenesis (Song et al., 2012). Within the nodulation-related CLE peptides (Fig. 4), R1, P4, G6, P7, and H11 residues show complete conservation and were all critical to activity in the present study. Additionally, the D8 and N12 residues are conserved in six out of seven nodulation-suppressing CLE peptides and showed reduced suppression activity when mutated. The E5 residue of GmRIC1 differs from the consensus (G in 5/7) in other nodulation CLE peptides and did not alter activity when mutated to A, T, or V, but was compromised by a mutation to R (E81R). Similarly, the GmRIC1 A3 residue differed from the consensus sequence at that position (S in 5/7) and had only a small effect on suppressive activity when mutated. The GmRIC1 L2 residue is completely conserved between the nodulation-suppressing CLE peptides; however, three mutations to A, N, or R indicated that it was not critical to suppression. Neither P9, which is conserved in 70% of nodulation CLE peptides, nor H10, which differs from the consensus (Q in 4/7), was found to alter GmRIC1 activity. None of the alanine mutations in conserved residues outside of the CLE domain, which included L31 (100% conserved), K58 (5/7), and PP92/93 (5/7), was shown to alter activity.
Fig. 4.

Amino acid conservation and importance to GmRIC1 activity. Consensus sequence and level of conservation of each residue in the CLE peptides known to inhibit nodulation are indicated by LOGO alignment (Crooks et al., 2004). Relative conservation of the mutated GmRIC1 residues with reduced (blue) or wild-type (red) nodule suppression activity is shown. The blue E residue represents the activity of the E5R mutation, which most significantly altered activity.
Comparison of GmRIC1 with AtCLV3 showed that seven out of 12 residues in the CLE domain are common between them (see Table 1). Two of these seven conserved residues (P7 and P9) did not share functional importance between the present results and those of Song et al. (2012). The P7 residue was critical to GmRIC1 but not to AtCLV3 activity, whereas the opposite was true at P9, which was critical to AtCLV3 but not to GmRIC1. One of the four non-conserved residues (CLV3-H12/RIC1-N12) was critical to the function of both Atclv3 complementation and legume suppression of nodulation.
Table 1.
Comparison of critical residues in GmRIC1 and AtCLV3 as determined by site-directed mutagenesis by Song et al. (2012).
| GmRIC1 peptide | R | L | A | P | E | G | P | D | P | H | H | N | ||||||||||||
| AtCLV3 peptide | R | T | V | P | S | G | P | D | P | L | H | H | ||||||||||||
| Residue required for GmRIC1 nodule suppression | Y | N | * | Y | ** | Y | Y | Y | N | N | Y | Y | ||||||||||||
| Residue required for complementation of clv3 fifth whorl ovary phenotype | Y | N | N | Y | N | Y | * | Y | Y | * | Y | Y |
*Minor change; **only for E>R change.
Contribution of functional domains to GmRIC1 and GmNIC1 activity
In contrast to GmRIC1 overexpression, which results in 100% suppression of soybean nodulation, overexpression of GmNIC1 only results in an ~50% reduction in nodule numbers (Reid et al., 2011a). To determine which domains contribute to this difference in suppression, several constructs were produced encoding chimeric versions of these two peptides (Fig. 1B). The domains investigated included the transit peptide, variable region, CLE domain, and C-terminal extension. The constructs were named according to the source of their domain. For example, an unmodified GmRIC1 construct was named RRRR, whereas a construct bearing the GmRIC1 signal peptide, GmNIC1 variable region, and CLE domain, and lacking the C-terminal extension, was named RNN, noting that the native GmNIC1 peptide lacks the C-terminal extension domain (Fig. 1B).
Analysis of the role of the signal peptide domain in GmRIC1 and GmNIC1 activity
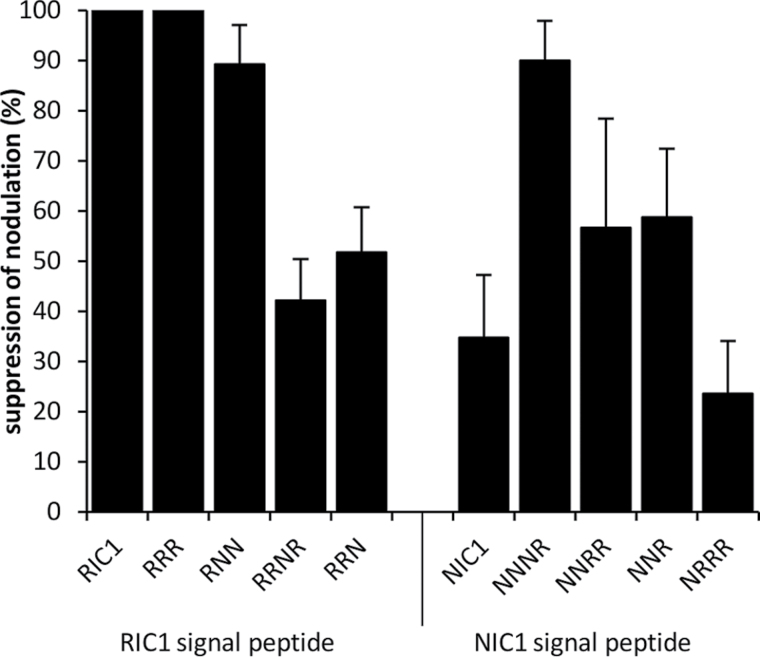
As the signal peptide has been shown to control GmRIC1 secretion (Lim et al., 2011), experiments were conducted to determine whether the GmNIC1 signal peptide was functionally interchangeable with GmRIC1. A GmRIC1 construct was produced where the GmNIC1 signal peptide replaced the native signal peptide (NRRR). The equivalent GmNIC1 construct bearing the native GmRIC1 signal peptide (RNN) was also produced. The RNN construct resulted in significantly increased suppression of nodulation relative to GmNIC1 (P < 0.001; Fig. 5). In contrast, the NRRR construct showed a significantly lower level of suppression relative to the complete suppression by the GmRIC1 peptide (24% suppression, P < 0.001).
Fig. 5.
Suppression of nodulation by chimeric GmNIC1 and GmRIC1 constructs. Chimeric gene constructs were compared with the suppression of nodulation obtained by GmRIC1 and GmNIC1 overexpression. Genes were named according to the source of the transit peptide, variable region, CLE domain, and C-terminal extension, respectively (GmRIC1 would be named RRRR, while GmNIC1 is NNN as it lacks the C-terminal extension).
Analysis of the role of the C-terminal extension domain in GmRIC1 and GmNIC1 activity
GmRIC1 encodes a C-terminal extension after the CLE peptide domain which is conserved in 70% of nodulation-related CLE peptides as well as in AtCLV3, but is absent in GmNIC1. To determine if this domain plays a critical role in nodule suppression activity, both a GmRIC1 construct where this domain was deleted (RRR) and a GmNIC1 construct where the C-terminal domain of GmRIC1 was added (NNNR) were produced. The function of GmRIC1 was not altered by the deletion of the C-terminal extension (RRR) as complete nodule suppression was maintained (Fig. 5). In contrast, the addition of the GmRIC1 C-terminal extension to GmNIC1 (NNNR) resulted in a significant increase in the suppression of nodulation (90% suppression, P < 0.001).
Analysis of the role of the CLE peptide domain in GmRIC1 and GmNIC1 activity
Constructs were created that swapped the CLE domains of GmRIC1 and GmNIC1 (RRNR and NNR) to determine their contribution to the suppression of nodulation. Constructs that swapped the CLE domain regions in the context of the C-terminal region of the donor peptide [the C-terminal extension of GmRIC1 (NNRR) or the stop codon for GmNIC1 (RRN)] were also produced. The GmNIC1 peptide bearing the GmRIC1 CLE domain (NNR) showed no significant change in activity relative to the native GmNIC1 peptide (P=0.11; Fig. 5). Similarly, the construct comprising the GmNIC1 transit peptide and variable region together with GmRIC1 CLE domain in native context adjacent to the GmRIC1 C-terminal extension (NNRR) did not significantly alter suppression activity from that of GmNIC1. Reciprocal swaps, where GmRIC1 contained the GmNIC1 CLE domain (RRNR), showed significantly reduced nodule suppression activity relative to GmRIC1 (P < 0.001; Fig. 5). Likewise, the GmRIC-based construct carrying the CLE domain of GmNIC1 but lacking a C-terminal extension (RRN) showed a significant reduction in suppression activity relative to both native GmRIC1 and the GmRIC1 construct lacking the C-terminal extension (RRR) (P < 0.001; Fig. 5).
Analysis of the role of the variable region domain in GmRIC1 and GmNIC1 activity
The central variable region of CLE peptides is not highly conserved amongst the nodulation-related CLE peptides and has not been unequivocally shown to exert functional specificity in previous CLE domain studies (Ni and Clark, 2006; Meng et al., 2010). Accordingly, specific constructs with direct swaps of this region were not produced. However, to determine any contribution of the variable region to differences in suppression activity, combinations of the constructs that varied only in this region (NNRR versus NRRR and RRN versus RNN) were compared. No significant difference in suppression activity was observed between the two alternative variable domains in the constructs with GmNIC1 transit peptide and the GmRIC1 CLE domain (NNRR versus NRRR; P=0.16; Fig. 5). In contrast, the GmNIC1 variable region enhanced suppression activity when a GmRIC1 transit peptide and GmNIC1 CLE domain were present adjacent to the variable region (RNN versus RRN; P < 0.005; Fig. 5).
Discussion
Although GmNARK and AtCLV1 are closely related homologues with high sequence similarity, little is known about their ligand specificity. The functional importance of the domains and many individual amino acids within AtCLV3 has been well studied and provides an excellent basis to identify functional divergence from GmRIC1 (Kondo et al., 2008; Ni et al., 2011; Song et al., 2012). The experimental system used here, which was employed to identify critical residues within the GmRIC1 peptide through assessment of nodule inhibition, provided a specific and easy to quantify assay having a large and dynamic range of detectable nodule numbers (0 in GmRIC1 compared with an average of 114 nodules in controls). In addition, the chimeric GmRIC1-overexpressing plants do not show any secondary root or shoot developmental phenotypes outside of nodulation. This is not the case for all nodulation-related CLE peptides, as the application or expression of MtCLE12 inhibits root growth in addition to nodulation (Saur et al., 2011). These differences make the use of GmRIC1 in the present assay ideal for analysing the function of CLE peptides in suppressing nodulation.
It was found that the R1, P4, G6, P7, D8, H11, and N12 residues within the GmRIC1 CLE domain were most important to nodule suppression activity. In contrast, mutation of the A3 residue provided only a small disruption of suppression, while mutations in the L2, P9, and H10 residues did not alter activity, suggesting that they play a relatively minor role. The E5 residue was only disrupted by mutation to R, while A, T, and V mutants at E5 were unaffected in nodule suppression activity. Outside of the CLE domain, none of the individual residues tested was found to be critical; however, the GmRIC1 signal peptide, CLE domain, and C-terminal extension region were all found to enhance the activity of GmNIC1.
AtCLV3 and GmRIC1 CLE domain sequences are well conserved, including perfect conservation at seven out of 12 residues. Complementation of Atclv3 plants with alanine substitution mutants showed that the R1, P4, G6, D8, P9, L10, H11, and H12 residues within the CLE domain were all critical to obtaining full complementation, whereas T2, V3, S5, and P7 mutants showed no alteration in activity (Song et al., 2012). Interestingly, the P7 residue which has been identified as a site of post-translational modification (Ohyama et al., 2009) was not critical to function. Of the four residues which were identified as unimportant in AtCLV3, three showed either no or only small loss of suppression activity in the present experiments and none of these was conserved (L2, A3, and E5). The fourth residue found to be unimportant in AtCLV3, P7, was shown to be critical to activity in the present study. The structural effects of these changes and why they show increased tolerance of mutation remains unknown at this stage, but may be further clarified through determination of receptor–ligand structures and binding studies.
Interestingly, the GmRIC1 P9 residue was not identified as important in the present study, despite being critical to AtCLV3 activity (Kondo et al., 2011; Song et al., 2012). Indeed, using peptoid replacement of proline residues, Kondo et al. (2011) showed that substitution could alter activity without altering binding. This difference may indicate an alteration of the post-translational modification site between GmRIC1 and AtCLV3, or a selective preference in the LRR receptor. The GmRIC1 N12/AtCLV3 H12 residue was shown to be critical to both peptides despite the lack of conservation (although both fall into the same charge class). It should be noted that both H and N residues are common at the C-terminus of CLE peptides (Oelkers et al., 2008). The identification of D8 as a critical residue also confirms the finding by Song et al. (2012) of the importance of this residue despite it being identified through in vitro application studies as lacking functional importance (Kondo et al., 2008). In contrast to this, the present finding that the H10 residue of GmRIC1 did not alter activity was different from the minor role identified through in vivo complementation studies using Atclv3 mutants (Song et al., 2012), but was consistent with a lack of activity observed at this position from in vitro peptide application studies (Kondo et al., 2008).
Whether differences in amino acid residue charge and size could affect GmRIC1 activity at two residues that were initially identified as showing no effect when substituted with alanine (L2 and E5) was investigated. Loss of function by the E5R mutation suggests that the chemical properties of this amino acid do play a role, as only this acidic to basic change altered the suppression activity. This indicates that selection at these residues remains important, albeit with a greater tolerance to changes. It is possible that many of the residues determined to be unimportant are indeed critical to activity simply due to their role in establishing the peptide backbone.
Despite high levels of conservation at several amino acids located outside of the CLE domain, none showed a loss of activity in our study using GmRIC1. This is consistent with previous studies (Ni et al., 2011; Song et al., 2012) that failed to identify residues critical to activity outside of the CLE domain, despite several being identified as likely protease cleavage sites. This may indicate that a consensus sequence or motif is sufficient to target protease cleavage, or that sequence within the CLE peptide domain itself directs cleavage of surrounding residues.
Irrespective of the lack of critical residues identified outside of the CLE domain, the domain swap results provided significant evidence for the importance of these domains to the activity of nodulation-related CLE peptides. The importance of the GmRIC1 transit peptide was confirmed to confer a significant functional advantage over the equivalent sequence from GmNIC1. This is consistent with previous results identifying the transit peptide as important for the secretion of GmRIC1 (Lim et al., 2011). Several conserved residues within the transit peptide of nodulation-related peptides of soybean, L. japonicus, and M. truncatula were identified as altered in GmNIC1 (Reid et al., 2011a) and it seems possible that some of these residues could play a role in the localization of the peptide.
Like the transit peptide domain, the CLE domain of GmRIC1 was functionally superior relative to that of GmNIC1. GmNIC1 is the only nodulation-related CLE peptide with a H residue at position 12, where all others possess N12. Our studies demonstrated that N12 is critical to GmRIC1 suppression of nodulation. Interestingly, GmNIC1 is also one of only two identified nodulation-related CLE peptides lacking P9, despite alanine substitutions at this residue maintaining complete activity in the present study. Despite the conservation of two proline residues within the C-terminal tail region of GmRIC1 and other nodulation-related CLE peptides, no functional importance could be determined for the region in GmRIC1 as constructs with the domain deleted retained full activity. Interestingly, however, the domain did confer some advantage to GmNIC1 when it was added. This suggests that the domain possibly enhances processing or protects the mature CLE from breakdown but is not essential to binding. The unchanged activity of the GmRIC1 constructs may suggest that overexpression of GmRIC1 generates a large enough pool of the mature peptide such that protection from cleavage is less important, or can be overcome by saturation of proteases with the peptide. In GmNIC1, reduced breakdown of the mature peptide owing to the C-terminal extension in the NNNR construct may increases the lifetime and therefore the activity of the resulting peptide.
The least conserved region within CLE peptides is the region between the signal peptide and the CLE domain, termed the variable domain. The present results showed that neither the GmRIC1 nor GmNIC1 variable regions conferred a consistent advantage over the other. Of the two comparisons which were made, the GmNIC1 variable region did increase suppression in one case, although it did not restore the activity to control levels and a comparable change was not observed in the reciprocal construct. This probably indicates that some advantage is conferred by different variable regions, especially if a specific protease is involved in cleavage, but that processing is likely to be by a less specific protease or is directed by neighbouring domains.
Taken together, the domain-swap results indicate a significant advantage of the GmRIC1 transit peptide, CLE domain, and, to a lesser extent, the C-terminal tail region, over the corresponding domains in GmNIC1. Although GmNIC1 does inhibit nodulation in a GmNARK-dependent manner, the low efficiency of this suppression may indicate that it is not the only CLE domain peptide in soybean that responds to nitrate, or that other mechanisms that are not dependent on CLE peptide–NARK signalling are more significant players. The data obtained in these studies may provide valuable assistance in identifying possible additional nitrate-responsive CLE peptides with increased nodule suppression efficiency. The low efficiency of GmNIC1 may also indicate that GmNIC1 has a secondary function in another nitrate-dependent signalling pathway via an alternative receptor. The lack of secondary phenotypes resulting in GmNIC1-overexpressing plants, however, indicates that the developmental changes are subtle or are in some way altered in Agrobacterium-induced hairy root systems.
As one of only a few studies to investigate CLE peptide function outside of Arabidopsis and rice, the present results provide useful insight into the functional importance of the protein domains and many individual amino acids of this group of peptides. For example, the critical importance of the GmRIC1 transit peptide identified here may provide a basis for further analysis of this domain to determine key residues for its activity. By identifying the tolerance to mutation at each of the residues of the RIC1 CLE domain and identifying the residues which are critical only in nodulation and not SAM regulation, the findings also provide useful insight into the evolution of CLE peptide signalling in plant development. The results may assist in the identification of CLE peptides in other legumes, particularly those that are likely to possess nodule inhibitory activity. Indeed, the highly sensitive and robust nodule suppression assay used here provides an excellent tool for future nodule suppression studies.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Primer sequences for site-directed mutagenesis.
Table S2. Primers for domain swap constructs.
Acknowledgements
We thank Dr Luguang Wu for assistance in designing the site-directed mutagenesis protocols. Research was funded through an ARC Centre of Excellence grant (CEO348212), UQ Strategic Funds, UQ School of Agriculture and Food Sciences, and Plant Nutrition Trust travel scholarships to DR and BF.
References
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. Journal of Molecular Biology 340, 783–795 [DOI] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. 2000. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619 [DOI] [PubMed] [Google Scholar]
- Carroll BJ, Gresshoff PM. 1983. Nitrate inhibition of nodulation and nitrogen fixation in white clover. Zeitschrift für Pflanzenphysiologie 110, 77–88 [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. 1997. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis . Cell 89, 575–585 [DOI] [PubMed] [Google Scholar]
- Cock J, McCormick S. 2001. A large family of genes that share homology with CLAVATA3 . Plant Physiology 126, 939–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks G, Hon G, Chandonia J, Brenner S. 2004. WebLogo: a sequence logo generator. Genome Research 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delves AC, Mathews A, Day DA, Carter AS, Carroll BJ, Gresshoff PM. 1986. Regulation of the soybean–Rhizobium nodule symbiosis by shoot and root factors. Plant Physiology 82, 588–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbrosses G, Stougaard J. 2011. Root nodulation: a paradigm for how plant–microbe symbiosis influences host developmental pathways. Cell Host and Microbe 10, 348–358 [DOI] [PubMed] [Google Scholar]
- Djordjevic MA, Oakes M, Wong CE, Singh M, Bhalla P, Kusumawati L, Imin N. 2011. Border sequences of Medicago truncatula CLE36 are specifically cleaved by endoproteases common to the extracellular fluids of Medicago and soybean. Journal of Experimental Botany 62, 4649–4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson BJ, Indrasumunar A, Hayashi S, Lin M-H, Lin Y-H, Reid DE, Gresshoff PM. 2010. Molecular analysis of legume nodule development and autoregulation. Journal of Integrative Plant Biology 52, 61–76 [DOI] [PubMed] [Google Scholar]
- Fiers M, Golemiec E, van der Schors R, van der Geest L, Li KW, Stiekema WJ, Liu C-M. 2006. The CLAVATA3/ESR motif of CLAVATA3 is functionally independent from the nonconserved flanking sequences. Plant Physiology 141, 1284–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers M, Golemiec E, Xu J, van der Geest L, Heidstra R, Stiekema W, Liu C. 2005. The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. The Plant Cell 17, 2542–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. 1999. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914 [DOI] [PubMed] [Google Scholar]
- Gresshoff PM, Delves AC. 1986. Plant genetic approaches to symbiotic nodulation and nitrogen fixation in legumes. Plant Gene Research 3, 159–206 [Google Scholar]
- Gresshoff PM, Lohar D, Chan PK, Biswas B, Jiang Q, Reid D, Ferguson B, Stacey G. 2009. Genetic analysis of ethylene regulation of legume nodulation. Plant Signaling and Behaviour 4, 818–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Reid DE, Lorenc MT, Stiller J, Edwards D, Gresshoff PM, Ferguson BJ. 2012. Transient Nod factor-dependent gene expression in the nodulation-competent zone of soybean (Glycine max [L.] Merr.) roots. Plant Biotechnology Journal 10, 995–1010 [DOI] [PubMed] [Google Scholar]
- Heckman KL, Pease LR. 2007. Gene splicing and mutagenesis by PCR-driven overlap extension. Nature Protocols 2, 924–932 [DOI] [PubMed] [Google Scholar]
- Herridge DF. 1982. Relative abundance of ureides and nitrate in plant tissues of soybean as a quantitative assay of nitrogen fixation. Plant Physiology 70, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen ES, Peoples MB, Boddey RM, Gresshoff PM, Hauggaard-Nielsen H, Alves BJR, Morrison MJ. 2012. Legumes for mitigation of climate change and the provision of feedstock for biofuels and biorefineries. A review. Agronomy for Sustainable Development 32, 329–364 [Google Scholar]
- Jun J, Fiume E, Roeder AH, et al. 2010. Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant Physiology 154, 1721–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kereszt A, Li D, Indrasumunar A, Nguyen CDT, Nontachaiyapoom S, Kinkema M, Gresshoff PM. 2007. Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nature Protocols 2, 948–952 [DOI] [PubMed] [Google Scholar]
- Kinoshita A, Nakamura Y, Sasaki E, Kyozuka J, Fukuda H, Sawa S. 2007. Gain-of-function phenotypes of chemically synthetic CLAVATA3/ESR-related (CLE) peptides in Arabidopsis thaliana and Oryza sativa . Plant and Cell Physiology 48, 1821–1825 [DOI] [PubMed] [Google Scholar]
- Kondo T, Nakamura T, Yokomine K, Sakagami Y. 2008. Dual assay for MCLV3 activity reveals structure–activity relationship of CLE peptides. Biochemical and Biophysical Research Communications 377, 312–316 [DOI] [PubMed] [Google Scholar]
- Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y. 2006. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313, 845–848 [DOI] [PubMed] [Google Scholar]
- Kondo T, Yokomine K, Nakagawa A, Sakagami Y. 2011. Analogs of the CLV3 peptide: synthesis and structure–activity relationships focused on proline residues. Plant and Cell Physiology 52, 30–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusell L, Madsen LH, Sato S, et al. 2002. Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 420, 422–426 [DOI] [PubMed] [Google Scholar]
- Krusell L, Sato N, Fukuhara I, et al. 2011. The CLAVATA2 genes of pea and Lotus japonicus affect autoregulation of nodulation. The Plant Journal 65, 861–871 [DOI] [PubMed] [Google Scholar]
- Lenhard M, Laux T. 2003. Stem cell homeostasis in the Arabidopsis shoot meristem is regulated by intercellular movement of CLAVATA3 and its sequestration by CLAVATA1. Development 130, 3163–3173 [DOI] [PubMed] [Google Scholar]
- Li D, Kinkema M, Gresshoff PM. 2009. Autoregulation of nodulation (AON) in Pisum sativum (pea) involves signalling events associated with both nodule primordia development and nitrogen fixation. Journal of Plant Physiology 166, 955–967 [DOI] [PubMed] [Google Scholar]
- Lim CW, Lee YW, Hwang CH. 2011. Soybean nodule-enhanced CLE peptides in roots act as signals in GmNARK-mediated nodulation suppression. Plant and Cell Physiology 52, 1613–1627 [DOI] [PubMed] [Google Scholar]
- Lin M-H, Gresshoff PM, Ferguson BJ. 2011. pHairyRed: a novel binary vector containing the DsRed2 reporter for visual selection of transgenic hairy roots. Molecular Plant 4, 537–545 [DOI] [PubMed] [Google Scholar]
- Lin Y-H, Ferguson BJ, Kereszt A, Gresshoff PM. 2010. Suppression of supernodulation in soybean by a leaf-extracted, NARK- and inoculation-dependent small molecular fraction. New Phytologist 185, 1074–1086 [DOI] [PubMed] [Google Scholar]
- Lin Y-H, Lin M-H, Gresshoff PM, Ferguson BJ. 2011. Petiole feeding as a technique to introduce exogenous solutions into the plant. Nature Protocols 6, 36–45 [DOI] [PubMed] [Google Scholar]
- Meng L, Ruth KC, Fletcher JC, Feldman L. 2010. The roles of different CLE domains in Arabidopsis CLE polypeptide activity and functional specificity. Molecular Plant 3, 760–772 [DOI] [PubMed] [Google Scholar]
- Miyahara A, Hirani TA, Oakes M, Kereszt A, Kobe B, Djordjevic MA, Gresshoff PM. 2008. Soybean nodule autoregulation receptor kinase phosphorylates two kinase-associated protein phosphatases in vitro . Journal of Biological Chemistry 283, 25381–25391 [DOI] [PubMed] [Google Scholar]
- Mortier V, De Wever E, Vuylsteke M, Holsters M, Goormachtig S. 2012. Nodule numbers are governed by interaction between CLE peptides and cytokinin signaling. The Plant Journal 70, 367–376 [DOI] [PubMed] [Google Scholar]
- Mortier V, Den Herder G, Whitford R, Van de Velde W, Rombauts S, D’haeseleer K, Holsters M, Goormachtig S. 2010. CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiology 153, 222–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortier V, Fenta B, Martens C, Rombauts S, Holsters M, Kunert K, Goormachtig S. 2011. Search for nodulation-related CLE genes in the genome of Glycine max . Journal of Experimental Botany 62, 2571–2583 [DOI] [PubMed] [Google Scholar]
- Ni J, Clark SE. 2006. Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiology 140, 726–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Guo Y, Jin H, Hartsell J, Clark S. 2011. Characterization of a CLE processing activity. Plant Molecular Biology 75, 67–75 [DOI] [PubMed] [Google Scholar]
- Nishimura R, Hayashi M, Wu GJ, Kouchi H, Imaizumi-Anraku H, Murakami Y, Kawasaki S, Akao S, Ohmori M, Nagasawa M. 2002. HAR1 mediates systemic regulation of symbiotic organ development. Nature 420, 426–429 [DOI] [PubMed] [Google Scholar]
- Oelkers K, Goffard N, Weiller GF, Gresshoff PM, Mathesius U, Frickey T. 2008. Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biology 8, 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y. 2008. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319, 294 [DOI] [PubMed] [Google Scholar]
- Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. 2009. A glycopeptide regulating stem cell fate in Arabidopsis thaliana . Nature Chemical Biology 5, 578–580 [DOI] [PubMed] [Google Scholar]
- Okamoto S, Ohnishi E, Sato S, Takahashi H, Nakazono M, Tabata S, Kawaguchi M. 2009. Nod factor, nitrate-induced CLE genes that drive systemic regulation of nodulation. Plant and Cell Physiology 50, 67–77 [DOI] [PubMed] [Google Scholar]
- Osipova MA, Mortier V, Demchenko KN, Tsyganov VE, Tikhonovich IA, Lutova LA, Dolgikh EA, Goormachtig S. 2012. WUSCHEL-RELATED HOMEOBOX5 gene expression and interaction of CLE peptides with components of the systemic control add two pieces to the puzzle of autoregulation of nodulation. Plant Physiology 158, 1329–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DE, Ferguson BJ, Gresshoff PM. 2011. a Inoculation- and nitrate-induced CLE peptides of soybean control NARK-dependent nodule formation. Molecular Plant-Microbe Interactions 24, 606–618 [DOI] [PubMed] [Google Scholar]
- Reid DE, Ferguson BJ, Hayashi S, Lin YH, Gresshoff PM. 2011. b Molecular mechanisms controlling legume autoregulation of nodulation. Annals of Botany 108, 789–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DE, Hayashi S, Lorenc M, Stiller J, Edwards D, Gresshoff PM, Ferguson BJ. 2012. Identification of systemic responses in soybean nodulation by xylem sap feeding and complete transcriptome sequencing reveal a novel component of the autoregulation pathway. Plant Biotechnology Journal 10, 680–689 [DOI] [PubMed] [Google Scholar]
- Saur IML, Oakes M, Djordjevic MA, Imin N. 2011. Crosstalk between the nodulation signaling pathway and the autoregulation of nodulation in Medicago truncatula . New Phytologist 190, 865–874 [DOI] [PubMed] [Google Scholar]
- Schnabel E, Journet E-P, Carvalho-Niebel Fd, Duc G, Frugoli J. 2005. The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Molecular Biology 58, 809–822 [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KFX, Jürgens G, Laux T. 2000. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644 [DOI] [PubMed] [Google Scholar]
- Searle IR, Men AE, Laniya TS, Buzas DM, Iturbe-Ormaetxe I, Carroll BJ, Gresshoff PM. 2003. Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299, 109–112 [DOI] [PubMed] [Google Scholar]
- Sharma V, Ramirez J, Fletcher J. 2003. The Arabidopsis CLV3-like (CLE) genes are expressed in diverse tissues and encode secreted proteins. Plant Molecular Biology 51, 415–425 [DOI] [PubMed] [Google Scholar]
- Shinohara H, Moriyama Y, Ohyama K, Matsubayashi Y. 2012. Biochemical mapping of a ligand-binding domain within Arabidopsis BAM1 reveals diversified ligand recognition mechanisms of plant LRR-RKs. The Plant Journal 70, 845–854 [DOI] [PubMed] [Google Scholar]
- Song X-F, Yu D-L, Xu T-T, Ren S-C, Guo P, Liu C-M. 2012. Contributions of individual amino acid residues to the endogenous CLV3 function in shoot apical meristem maintenance in Arabidopsis . Molecular Plant 5, 515–523 [DOI] [PubMed] [Google Scholar]
- Strabala TJ, O’Donnell PJ, Smit AM, Ampomah-Dwamena C, Martin EJ, Netzler N, Nieuwenhuizen NJ, Quinn BD, Foote HC, Hudson KR. 2006. Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiology 140, 1331–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley S, Helliwell C, Smith N, et al. 2001. Construct design for efficient, effective and high-throughput gene silencing in plants. The Plant Journal 27, 581–590 [DOI] [PubMed] [Google Scholar]
- Yadav RK, Perales M, Gruel J, Girke T, Jönsson H, Reddy GV. 2011. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes and Development 25, 2025–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.