Abstract
MicroRNAs (miRNAs) are endogenous non-coding ~21 nucleotide RNAs that regulate gene expression at the transcriptional and post-transcriptional levels in plants and animals. They play an important role in development, abiotic stress, and pathogen responses. miRNAs with their targets have been widely studied in model plants, but limited knowledge is available on the small RNA population of cotton (Gossypium hirsutum)—an important economic crop, and global identification of related targets through degradome sequencing has not been developed previously. In this study, small RNAs and their targets were identified during cotton somatic embryogenesis (SE) through high-throughput small RNA and degradome sequencing, comparing seedling hypocotyl and embryogenic callus (EC) of G. hirsutum YZ1. A total of 36 known miRNA families were found to be differentially expressed, of which 19 miRNA families were represented by 29 precursors. Twenty-five novel miRNAs were identified. A total of 234 transcripts in EC and 322 transcripts in control (CK) were found to be the targets of 23 and 30 known miRNA families, respectively, and 16 transcripts were targeted by eight novel miRNAs. Interestingly, four trans-acting small interfering RNAs (tas3-siRNAs) were also found in degradome libraries, three of which perfectly matched their precursors. Several targets were further validated via RNA ligase-mediated rapid amplification of 5’ cDNA ends (RLM 5’-RACE). The profiling of the miRNAs and their target genes provides new information on the miRNAs network during cotton SE.
Key words: Degradome, Gossypium hirsutum, high throughput small RNA sequencing, miRNAs, miRNA target genes, somatic embryogenesis, tas3-siRNA.
Introduction
MicroRNAs (miRNAs) are endogenous non-coding RNAs of ~21 nucleotides (nt) in length, derived from single-stranded RNA hairpin precursors cleaved by a double-stranded-specific RNase (Dicer) in animals and Dicer-like1 (DCL1) in plants (Hutvagner et al., 2001; Tang et al., 2003). They were initially identified in Caenorhabditis elegans as developmental timing regulators and subsequently emerged in almost all eukaryotes (Fire et al., 1998). miRNAs are highly conserved among species based on comparisons between different plant species (Axtell et al., 2006). There also exist non-conserved or species-specific miRNAs in plants. As non-conserved miRNAs are often expressed at very low levels, many are not found in small-scale sequencing projects. Recently developed high-throughput sequencing technology has allowed the identification of low-abundance miRNAs in several species (Szittya et al., 2008; Pantaleo et al., 2010; Song et al., 2011). More recently, several other classes of small regulatory RNAs have also been identified. One specialized class of small RNA is the trans-acting small interfering RNAs (ta-siRNAs), which are similar to miRNAs. They act in trans to regulate mRNAs at the post-transcriptional level (Vazquez et al., 2004). Ta-siRNAs were generated from TAS gene transcripts (trans-acting siRNA genes), which are cleaved by a miRNA, resulting in the production of 21nt small RNAs that are in phase with the miRNA cleavage site (Allen et al., 2005).
miRNAs regulate gene expression through partially or fully complementary sequence homology with their targets, acting at transcriptional and post-transcriptional levels in animals and higher plants. They have been implicated in the control of diverse cellular, physiological, and developmental processes including developmental regulation, hormone response, and stress adaptation. For example, miR156 regulates the phase transition of plants from the juvenile to the adult during shoot development by targeting SPL genes (Wu and Poethig, 2006). miR172 acts downstream of miR156 and mediates regulation of APETALA2 (AP2) and AP2-like genes which are needed for proper specification of organs during flower development (Aukerman and Sakai, 2003). miR393 targets mRNAs encoding TIR1 and three closely related F-box proteins (Sunkar and Zhu, 2004) which are auxin receptors (Dharmasiri et al., 2005), and targets short-lived repressors of ARF transcriptional activators for ubiquitin-mediated degradation in response to auxin. miR160 targets ARF10, ARF16, and ARF17 (Mallory et al., 2005; Wang et al., 2005; P.P. Liu et al., 2007), miR167 targets ARF6 and ARF8 (Wu et al., 2006), and miR390 directs cleavage of TAS3, leading to the production of ta-siRNAs that target ARF3 and ARF4 mRNAs (Allen et al., 2005).
To date, large numbers of small RNAs have been identified by direct cloning and/or deep sequencing, with some targets being originally predicted via bioinformatics, based on either the perfect or nearly perfect sequence complementarity between a miRNA and the target mRNA or sequence conservation among different species in higher plants (Archak and Nagaraju, 2007; Alves-Junior et al., 2009). However, due to the existence of some mismatches in miRNA–target pairing, target prediction was subjected to challenge (Jones-Rhoades and Bartel, 2004). To date, only a small proportion of targets have been validated experimentally (Yang, 2006; P.P. Liu et al., 2007), and validated targets of the conserved small RNAs largely remain elusive. Recently, a new high-throughput method called parallel analysis of RNA ends (PARE), also known as degradome sequencing, has been successfully adapted to screen miRNA targets, resulting in the identification of the previously validated/predicted new targets of miRNAs and ta-siRNAs. Combined with small RNA sequencing, many new miRNAs with low abundance and their targets have been identified (Addo-Quaye et al., 2008; Pantaleo et al., 2010; Song et al., 2011).
Cotton is one of the most important economic crops, providing excellent natural fibre, and is also a source of oil. Somatic embryogenesis (SE), which is well established for certain genotypes in cotton, is a well-known illustration of cell totipotency, involving dedifferentiation and redifferentiation through restructuring to generate embryogenic cells, somatic embryos, and, ultimately, the regeneration of new plants. It is a complicated biological process involving finely coordinated molecular signalling pathways (Yang and Zhang, 2010).
The SE system has been used for cotton genetic improvement by transgenic research and genetic engineering. However, a reproducible and highly efficient plant regeneration system is required for diverse cotton genotypes, many of which remain recalcitrant. The study of regulatory molecules and associated gene networks during SE is of great importance for the longer term understanding of embryogenic competence and regenerability necessary for crop improvement via biotechnology. An elite genotype for in vitro cellular manipulation has been identified, and has been well characterized at both morphological and molecular levels (Zhu et al., 2008; Yang et al., 2012). Attention has been paid to the possible regulation of SE by small RNAs using undifferentiated and differentiated rice callus, resulting in the identification of a unique set of miRNAs only expressed or differentially expressed in embryogenic cells (Luo et al., 2006; Chen et al., 2011). In a different study, miRNA expression during SE was characterized in citrus, by analysing conserved miRNAs derived from miRBase (Griffiths-Jones et al., 2008) using quantitative real-time PCR (qRT-PCR) (Wu et al., 2011). Since the initial report on cotton miRNAs (Zhang et al., 2007), many conserved and cotton-specific miRNAs have been identified from different tissues involved in several biological processes in cotton (Kwak et al., 2009; Pang et al., 2009; Ruan et al., 2009; M. Wang et al., 2012; Z.M. Wang et al., 2012; Yin et al., 2012). However, the roles of miRNAs during cotton SE remain unknown.
To investigate small RNA targets comprehensively and provide basic information for further understanding the miRNA-mediated post-transcriptional regulation network during cotton SE, two small RNA libraries and two degradome libraries were constructed using RNA from hypocotyl tissue (a non-embryonic control) and embryonic callus (EC) from G. hirsutum genotype YZ1, and sequenced using a Illumina Genome Analyzer. A total of 36 differentially expressed known miRNA families and 25 novel miRNAs, with 476 genes as targets, were identified. The expression patterns of miRNAs and their targets were confirmed experimentally. Some conserved and cotton-specific miRNAs showed a tissue-specific expression pattern. The profiling of the miRNAs and their target genes provides novel information about the regulatory network of miRNAs during SE in cotton, and the data presented here will serve as foundation for future studies addressing fundamental molecular and developmental mechanisms that govern plant embryogenesis.
Materials and methods
Plant material and RNA isolation
Seeds of G. hirsutum YZ1 were decoated, surface-sterilized, and germinated on half-strength Murashige and Skoog (MS) medium (1/2 macro salts plus 15g of glucose, pH 6.0) at 28 °C in the dark for 5–7 d. The hypocotyls of the seedlings were cut into 5–7mm sections, some of which were immediately sampled as control (CK, 0h). Explants from different time points/stages during cotton SE [6, 12, 24, and 48h, and 5d after induction, non-embryogenic callus (NEC), embryogenic callus (EC), globular-stage somatic embryo (GE), torpedo-stage somatic embryo (TE), and cotyledon-stage somatic embryo (CE)] were sampled as previously described (Yang et al., 2012). Two samples (CK and EC) were used for small RNA and degradome sequencing, and all the samples were used for qRT-PCR to verify the expression patterns. Total RNA was isolated from each sample by using a modified guanidine thiocyanate method (Zhu et al., 2005).
Construction of small RNA and degradome libraries
Small RNA library construction and deep sequencing were carried out as described previously (Hafner et al., 2008). A 20 µg aliquot of total RNA was sent to the Beijing Genomics Institute (Shenzhen) where the libraries were constructed and sequenced using an Illumina Genome Analyzer. Briefly, the small RNAs (~18–30 nt) were purified from 10 µg of total RNA by polyacrylamide gel electrophoresis, and ligated first to a 5’ RNA adaptor and then to a 3’ RNA adaptor. A reverse transcription reaction was followed by several cycles of PCR to obtain sufficient product for Sequencing by Synthesis (SBS) sequencing via Solexa technology.
Two cotton degradome libraries were constructed as previously described (Addo-Quaye et al., 2008). In brief, poly(A+) RNA molecules were isolated from 200 µg of total RNA using the Oligotex mRNA kit (Qiagen), and then a 5’ RNA oligonucleotide adaptor containing an MmeI recognition site was ligated to the 5’-phosphate of the poly(A+) RNA by T4 RNA ligase. This was followed by purification of the ligated products using the Oligotex kit. Subsequently, five PCR cycles were performed on the products of a reverse transcription reaction which were then digested with MmeI and ligated to a 3’ double DNA adaptor. Finally, the ligation products were amplified with 20 PCR cycles, gel-purified, and subjected to SBS sequencing by the Illumina Genome analyzer.
Bioinformatics analysis of sequencing data
Small RNA reads and degradome reads were both generated from Illumina HiSeq™ analysis. The raw data were pre-processed by the Fastx-toolkit pipeline (http://hannonlab.cshl.edu/fastx_toolkit/) to remove low quality reads, reads smaller than 18 nt, trim adaptor sequences, and contamination formed by adaptor–adaptor ligation. As for the small RNA sequencing data, clean reads ranging from 18 to 44 nt were mapped to the 91 713 cotton unique transcript sequences (defined as Zhuref-2), which cleans and assembles all the cotton expressed sequence tags (ESTs) from NCBI using TGICL (Li et al., 2009). Sequences matching non-coding rRNA, tRNA, small nuclear RNA (snRNA), and small nucleolar RNA (snoRNA) in the GenBank and Rfam database (Altschul et al., 1990), reads matching repeat sequences and reads matching the known miRNAs of all plants loaded in the miRBase17.0 database (Griffiths-Jones et al., 2008) were each annotated, and then reannotated based on the rule GenBank > Rfam, and rRNA etc. > known miRNA > repeat to give every small RNA a unique category. The miRNA categories (tag copy number ≥2) were used for identifying the known/conserved miRNAs, with the criterion that the tags were similar to their homologues within two mismatches and without gaps. Reads that were potential siRNAs were excluded based on their characteristic 2 nt overhang on each 3’ end of the double-stranded sequence, by pairing between two random sequences. Finally, the unannotated reads were used for prediction of novel miRNAs.
For the degradome sequencing data, 20–21 nt sequences of high quality were collected for subsequent analysis. The unique reads that perfectly matched cotton contigs were retained. Approximately 15 nt upstream and downstream of 5’ cotton EST sequences, mapped by degradome reads, were extracted to generate 31 nt target signatures as ‘t-signatures’ (German et al., 2008), which were collected to align with the newly identified miRNAs and all mature known miRNAs from eight plant species including G. hirsutum, present in miRBase17.0 using the CleaveLand pipeline (Addo-Quaye et al., 2009). Alignments with scores up to four where G:U pairs scored 0.5 and no mismatches were found at the site between the 10th and 11th nucleotides of the corresponding miRNAs were considered potential targets.
Identification of conserved miRNAs and prediction of novel miRNAs in cotton
Sequences in the small RNA libraries with no more than two mismatches and >16 matches to currently known miRNAs from all plant species were regarded as potential reference miRNAs of a known miRNA family. Precursors of known miRNAs and novel miRNA candidates were identified by extracting 200 nt of the sequence flanking the contig sequences matching the known miRNAs and unannotated small RNAs by analysing their secondary structural features using the MIREAP pipeline (https://source-forge.net/projects/mireap/). RNAs with the characteristic hairpin structure, with minimal matched nucleotide pairs of miRNA and miRNA* exceeding 16 nt and with maximal size differences of miRNA and miRNA* up to 4 nt, were retained as precursors of known miRNAs or novel miRNAs. The filtered pre-miRNA sequences were folded again using MFOLD and selected manually (Zuker, 2003).
qRT-PCR
QRT-PCR was carried out to determine the validity of RNA-Seq technology for expression profile analysis. All primers (Supplementary Table S1 available at JXB online) were designed according to the miRNA mature sequence and cDNAs sequences using Primer Premier 5 (http://www.premierbiosoft.com/crm/jsp/com/pbi/crm/clientside/ProductList.jsp) and synthesized commercially (Genscript Bioscience, Nanjing). qRT-PCR for miRNAs and targets were performed using 3 µg of total RNA from 11 cotton samples (0, 6, 12, 24, and 48h, and 5 d explants after induction, NEC, EC, GE, TE, and CE) using the One Step Primer Script miRNA cDNA Synthesis Kit (Takara) and SuperScript® III Reverse Transcriptase (Invitrogen), respectively.. PCR was performed using 2 µl of 10× diluted cDNA products, 2× SsoFast EvaGreen Supermix With Low ROX (BIO-RAD), and 0.25 µM forward and reverse primers in a 20 µl system. The reactions were incubated in a 7500 Thermocycler for 30 s at 95 °C, followed by 40 cycles of 5 s at 95 °C and 35 s at 60 °C. All reactions were run in three replicates, and ubiquitin7 (GhUBQ7) was used as the endogenous reference gene (Tu et al., 2007). All qRT-PCRs for miRNAs were performed using the most common sequence in each miRNA family as the reference sense primers (longer or shorter depending on the T m of the mature miRNA sequence); the reversed primer was supplied in the kit.
RNA ligase-mediated rapid amplification of 5’ cDNA ends (RLM-5’ RACE)
A 5 µg aliquot of total RNA from CK was used to purify mRNA using the GeneRacer kit (Invitrogen). 5’ RNA adaptor was ligated to the degraded mRNA with a 5’ free phosphoric acid by T4 RNA ligase, followed by a reverse transcription reaction. Subsequently, 1 µl of 10× diluted reverse transcription product was used to amplify the 5’ end of the corresponding targets using a 5’ RNA adaptor primer and a 3’ gene-specific primer. Thirty-five more cycles of PCR were further performed using a 5’ nested adaptor primer and a 3’ gene-specific nested primer. The final PCR product was detected by gel electrophoresis and cloned into a pGEM-T Easy vector for sequencing. The 3’ primers are listed in Supplementary Table S1 at JXB online.
Results
Overview of small RNA library sequencing
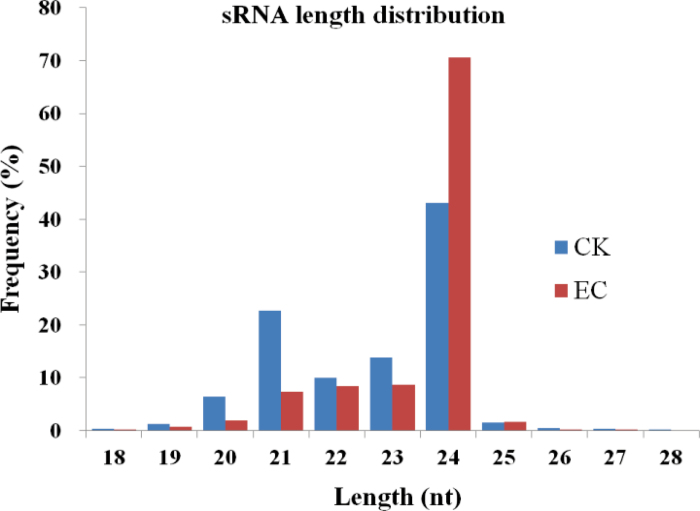
Two small RNA libraries were constructed using total RNA obtained from CK and EC of G. hirsutum YZ1, and sequenced by Solexa SBS technology. After removing adaptor contaminations and low quality tags, a total of 24 948 581 clean reads from CK (representing 9 773 591 unique sequences) and 25 984 975 from EC (representing 12 930 499 unique sequences) were obtained (Table 1), ranging from 18 nt to 44 nt in length. The sequences of 18–28 nt in length, which accounted for ~99.9% of all the clean reads, were extracted to analyse small RNA length distribution (Fig. 1). The size of the small RNAs was not evenly distributed in each library, and a high abundance of 21–24 nt sequences was found, which accounted for 89% and 95% of the total reads in CK and EC, respectively, representing the typical length of plant mature miRNAs. The number of 24 nt sequences was significantly greater than that of other sequences. The result was consistent with that found for rice (Morin et al., 2008), peanut (Zhao et al., 2010), soybean (Song et al., 2011), Medicago truncatula (Szittya et al., 2008), and a previous study in cotton (Yin et al., 2012). In addition, the fractions of these tags were substantially different between the two libraries. For the CK data set, the small RNA distribution showed a major peak at 24 nt (43%); the next largest fractions were 21 (23%), 23 (14%), and 22 nt (10%). However, the small RNA distribution was 24 (71%), 23 (9%), 22 (8%), and 21 nt (7%) in the EC data set. These observations highlighted differences in the complexity of the two small RNA pools, indicative of differential regulation of gene expression during cotton SE. Small RNAs were analysed by BLASTN against the known non-coding RNAs deposited in the Rfam and NCBI GenBank databases in order to remove rRNA, tRNA, snRNA, snoRNA, and small cytoplasmic RNA (scRNA)—these accounted for 1 817 529 (91 671 unique sequences) in CK and 1 423 175 reads (59 178 unique sequences) in EC (Table 1), respectively. A total of 279 391 (2.86%) and 261 738 (2.02%) unique reads, respectively, were mapped to Zhuref-2 (cotton unique transcript sequences database). Subsequently, siRNAs were identified and removed using their characteristic 2 nt ovhang on each 3’ end of the double-stranded sequence, by pairing between two random sequences. The raw and processed data are available in GEO (accession no. GSE41132).
Table 1.
Data set summary of sequencing of two (EC and CK) small RNA and degradome libraries.
| Category | Unique reads | Total reads | |||
|---|---|---|---|---|---|
| EC | CK | EC | CK | ||
| Small RNA data | Clean reads | 12 930 499 | 9 773 591 | 25 984 975 | 24 948 581 |
| Match Rfam/GenBank | 59 178 (0.46%) | 91 671 (0.95%) | 1 423 175 (5.48%) | 1 817 529 (7.28%) | |
| Match siRNA | 324 928 (2.51%) | 281 310 (2.88%) | 1 293 326 (4.98%) | 1 832 324 (7.34%) | |
| Match miRNA | 27 874 (0.22%) | 40 282 (0.41%) | 1 158 162 (4.46%) | 4 727 372 (18.95%) | |
| Match Zhuref-2 | 261 738 (2.02%) | 279 391 (2.86%) | 2 404 867 (9.25%) | 3 920 275 (15.71%) | |
| Unann | 12 518 519 (96.81%) | 9 360 328 (95.77%) | 22 110 312 (85.09%) | 16 571 356 (66.42%) | |
| Degradome data | Clean reads | 501 061 | 446 590 | 15 519 691 | 15 098 505 |
| Match Rfam/GenBank | 1306 (0.27%) | 712 (0.17%) | 99 457 (0.64%) | 32 773 (0.22%) | |
| Match repeats | 1184 (0.24%) | 687 (0.15%) | 18 806 (0.12%) | 9824 (0.07%) | |
| Match Zhuref-2 | 219 317 (43.77%) | 231 452 (51.83%) | 9 990 673 (64.37%) | 11 254 030 (74.53%) | |
| Unann | 279 254 (55.73%) | 213 739 (47.86%) | 5 410 755 (34.86%) | 3 801 878 (25.18%) | |
Fig. 1.
The length distribution of small RNAs in CK and EC libraries. (This figure is available in colour at JXB online.)
Degradome sequencing and data summary
After RNA extraction and poly(A+) RNA purification from CK and EC of G. hirsutum YZ1, two degradome libraries were constructed, which were then subjected to PARE sequencing using the Solexa Analyzer (Addo-Quaye et al., 2008; German et al., 2008). In total, ~15.1 million (CK) and 15.5 million (EC) clean reads were obtained (Table 1), with ~70% sequences of 20–21 nt in length. To remove the structural RNAs (rRNAs, tRNAs, snRNAs, and snoRNAs) representing 0.17% and 0.27% of clean unique reads in CK and EC, respectively, a BLASTN was applied against the Rfam and GenBank database. Likewise, the signatures in each library corresponding to repeats/transposons, representing 0.15% and 0.24% of clean unique reads, respectively, were also removed from the unique data set by performing searches against the repeat database (http://www.girinst.org/server/RepBase/). The remaining sequences were mapped to a cotton transcript reference database. Finally, 231 452 (51.83%) and 219 317 (43.77%) unique reads were mapped to the reference database. All these mapped sequences were analysed to detect candidate targets of miRNAs.
Identification and expression analysis of known miRNAs during cotton SE
Known cotton miRNAs were identified by mapping the tags to miRBase (release 17.0), which contains 34 known miRNAs in G. hirsutum. Finally, 30 known cotton miRNAs, containing three cotton-specific miRNA families [miR2948, miR2949 (miR2949a, miR2949b, and miR2949c), and miR3476] were identified, belonging to 18 families in each of the two libraries. Their corresponding miRNA*—the complementary strands of functional mature miRNAs—were also discovered (except miR399d*; Supplementary Table S2 at JXB online). After normalizing the reads of the tags of each miRNA family as ‘reads per million’ (RPM), miRNAs in 15 families were identified as being differentially expressed (i.e. a ≥2-fold change) between EC and CK, with miR390, miR394, and miR827 up-regulated in EC compared with CK, while nine conserved miRNA families (miR156, miR162, miR166, miR167, miR172, miR2950, miR396, miR479, and miR482) and three cotton-specific miRNA families (miR2948, miR2949, and miR3476) were down-regulated in EC.
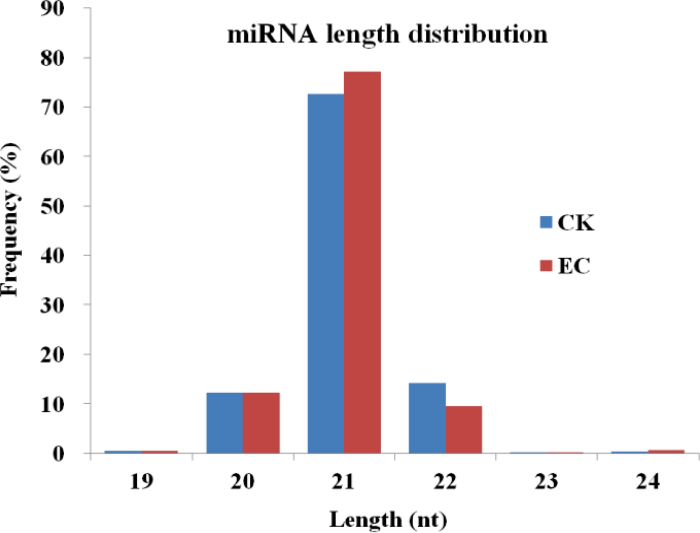
To identify plant-conserved miRNAs in cotton, all the small RNA tags were further aligned to known plant miRNA database using the mature miRNAs of all plant species loaded in miRBase (release 17.0). Also 1468 and 655 unique tags from CK and EC, representing 44 and 35 known miRNA families, were obtained (Supplementary Table S2 at JXB online), covering almost all the plant conserved miRNA families. Among these tags, 21 nt sequences were the most abundant fractions, accounting for ~73% and 77% in CK and EC, respectively, followed by ~10% of 20 nt and 22 nt RNAs in both samples (Fig. 2). Among these tags, 131 from CK and 97 from EC (Supplementary Table S3) were perfectly matched to known miRNAs or miRNA*s in other plant species, indicating the high sequence conservation of plant miRNAs. Twenty-nine precursors (Supplementary Table S4, Supplementary File S1) were identified using the mfold web server (Zuker, 2003)—23 precursors were known, while miR156e, miR156f, miR159, miR171, miR172a, and miR2937 (defined sequentially following the known miRNAs loaded in miRBase17.0) were newly identified in this study (as marked in red in Supplementary File S1). Twenty-four miRNA*s were also detected for the known miRNAs with precursors in this study (Supplementary Table S4). These sequences are rarely found via conventional sequencing because of their rapid degradation in cells. The detection of miRNA*s represented further evidence for the existence of mature miRNAs.
Fig. 2.
The length distribution of known miRNAs during cotton SE. (This figure is available in colour at JXB online.)
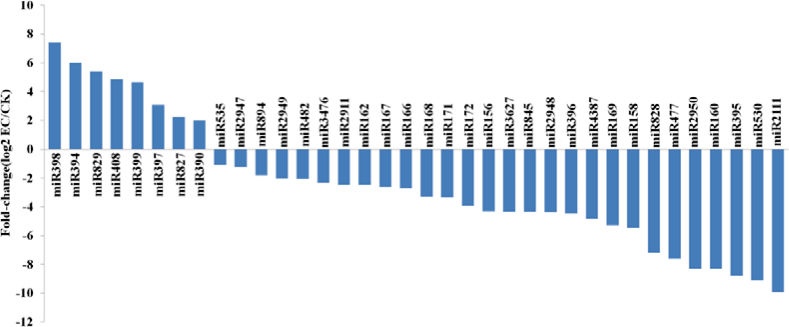
Among these families, 36 differentially expressed miRNA families (≥2 fold-change) were identified (Fig. 3). Of these, 28 miRNA families were down-regulated in EC compared with CK, whereas eight miRNA families (miR390, miR394, miR397, miR398, miR399, miR408, miR827, and miR829) showed up-regulated patterns. The miRNA families miR156, miR166, miR168, miR3476, and miR535 showed high expression levels in both libraries (Supplementary Table S2 at JXB online). These results suggest a possible miRNA-mediated mechanism for gene expression control in cotton SE.
Fig. 3.
Differentially expressed known miRNA families during cotton SE. (This figure is available in colour at JXB online.)
Identification and expression analysis of novel miRNAs during cotton SE
Novel miRNAs were identified by folding the sequences of potential miRNA precursors using the mfold web server (Zuker, 2003)—the formation of stable hairpin structures is an essential feature of miRNAs. Because a completely sequenced genome of tetraploid cotton is not yet available, unique small RNA sequences were pooled and mapped to Zhuref-2 to identify potentially novel miRNAs. In total, 25 novel miRNAs representing 23 unique miRNA sequences were predicted (Table 2; Supplementary Table S5, Supplementary File S1 at JXB online). These novel miRNA candidates were given names in the form of ‘ghr-miR_N plus number’, e.g., ghr-miR_N1 and using a, b, or c to differentiate miRNAs from the same family. Besides hairpin structure prediction, detection of miRNA*s is another way to confirm novel miRNAs (Meyers et al., 2008). Complementary sequences of two novel miRNAs (ghr-miR_N9 and ghr-miR_N17) were identified (Supplementary Table S5), further confirming the identity of these miRNAs.
Table 2.
Novel miRNAs identified in either library.
| miRNA | Mature sequence | Length of mature miRNA (nt) | RPM | Log2 (EC/CK) | MFE | |
|---|---|---|---|---|---|---|
| ECa | CKb | |||||
| ghr-miR_N1a | TGGACCCAGAAGTATGCCATG | 21 | 0.27 | 0.01 | 4.75 | –101.99 |
| ghr-miR_N1b | TGGACCCAGAAGTATGCCATG | 21 | 0.27 | 0.01 | 4.75 | –106.79 |
| ghr-miR_N2a | TCTTCAAAGTAGTAGATCATT | 21 | 0.01 | 0.36 | –5.17 | –28.4 |
| ghr-miR_N2b | TCTTCAAAGTAGTAGATCATT | 21 | 0.01 | 0.36 | –5.17 | –28.4 |
| ghr-miR_N3 | TCTTGTACTGCATCATAACTT | 21 | 0.50 | 0.72 | –0.53 | –55.9 |
| ghr-miR_N4 | TCCCTTTGGATGTCTTCTTGC | 21 | 0.31 | 0.01 | 4.94 | –27.6 |
| ghr-miR_N5 | TTCGATTCGGCTTGGATGAAAC | 22 | 1.15 | 0.48 | 1.26 | –22.6 |
| ghr-miR_N6 | TGGACTGAGATTTGAGGGTAT | 21 | 0.23 | 0.01 | 4.53 | –21.86 |
| ghr-miR_N7 | GTTGATCCCGGCATCTCACCC | 21 | 0.46 | 0.40 | 0.20 | –162.1 |
| ghr-miR_N8 | GTAGTTGAACGACGTTTATCTA | 22 | 0.50 | 0.01 | 5.64 | –35.4 |
| ghr-miR_N9 | TTACTTTAGATGTCTCCTTCA | 21 | 34.21 | 160.69 | –2.23 | –48.92 |
| ghr-miR_N10 | CTCGGTTGTACAGCGGAGCGG | 21 | 0.42 | 0.01 | 5.40 | –53.02 |
| ghr-miR_N11 | AGGATGTGGAAAGGTTATCAATG | 23 | 0.01 | 0.24 | –4.59 | –32.4 |
| ghr-miR_N12 | TAACTGAAGAGTTTGATCATGG | 22 | 0.01 | 12.02 | –10.23 | –90.5 |
| ghr-miR_N13 | AAGAGAGAAAGAGAGGCCTGGA | 22 | 0.01 | 3.01 | –8.23 | –30.62 |
| ghr-miR_N14 | CCGGAATAGGGTATGAGGCATT | 22 | 0.01 | 1.72 | –7.43 | –21.3 |
| ghr-miR_N15 | TGCGTGCTAGACTGAATTCGGGT | 23 | 0.01 | 0.36 | –5.17 | –33.2 |
| ghr-miR_N16 | TAGAGATTGCATTTCCTCTTC | 21 | 0.89 | 4.85 | –2.45 | –29.4 |
| ghr-miR_N17 | AAGGCAAAGGAAGAAAAAGAGTGA | 24 | 0.01 | 1.12 | –6.81 | –42 |
| ghr-miR_N18 | CTTTGGAGGGGAGATTAGAGC | 21 | 0.01 | 0.32 | –5.00 | –61.05 |
| ghr-miR_N19 | CAGGGCTTCTCTGCATTGGCA | 21 | 0.01 | 0.48 | –5.59 | –32.1 |
| ghr-miR_N20 | AACCAATGACTATTCATGATTCC | 23 | 0.01 | 4.17 | –8.70 | –25.3 |
| ghr-miR_N21 | AGTGAATTAAGAACAAACTTT | 21 | 0.01 | 0.28 | –4.81 | –41.5 |
| ghr-miR_N22 | GAAGTGGAAGTGGAAGTGGAAGT | 23 | 0.01 | 0.20 | –4.32 | –63.06 |
| ghr-miR_N23 | TTCAGAGACCATCCCTTCCTT | 21 | 0.01 | 0.28 | –4.81 | –47.6 |
RPM, Reads per million; MFE, minimum free energy.
a, bCount of tags from the dominant arm of the precursor.
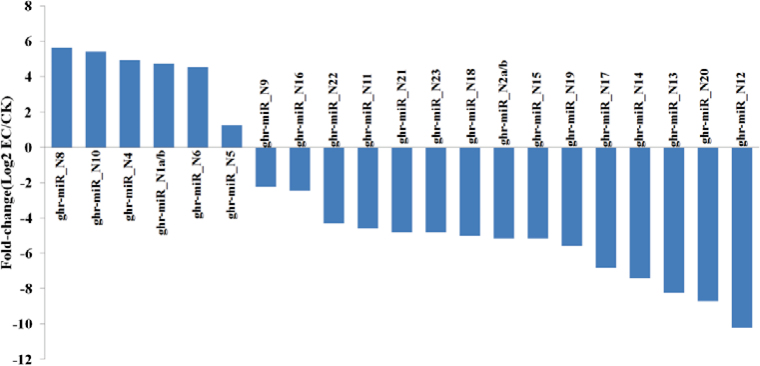
To analyse the expression profiles of these novel miRNAs, we normalized the reads of these miRNAs as RPM. Among these 23 novel unique miRNAs, 21 miRNAs were differentially expressed (≥2 fold-change) between EC and CK (Table 2, Fig. 4), with six miRNAs (ghr-miR_N1, ghr-miR_N4, ghr-miR_N5, ghr-miR_N6, ghr-miR_N8, and ghr-miR_N10) being up-regulated, 15 miRNAs being down-regulated, and another two miRNAs (ghr-miR_N3 and ghr-miR_N7) being expressed equally in EC and CK tissues.
Fig. 4.
Differentially expressed novel miRNAs during cotton SE. (This figure is available in colour at JXB online.)
Identification and annotation of targets for miRNAs during cotton SE
The target genes of the miRNAs during cotton SE were further studied by degradome library sequencing. In contrast to other mRNA degradation mechanisms, miRNA-mediated mRNA cleavage possesses special features. Cleavage occurs between the 10th and 11th nucleotides from the 5’ end of the miRNA which has near perfect complementarity to its target. These features were used to identify targets of miRNAs. As a result, 322 genes in CK and 234 genes in EC (Supplementary Table S6 at JXB online) were identified as targets of 30 (CK) and 23 (EC) annotated known miRNA families. Twenty conserved targets were identified for nine known miRNA families (Table 3). These included two SBP transcription factor genes targeted by miR156; one ARF16 gene targeted by miR160; six ARF6 and one ARF8 gene targeted by miR167; one RNase H-like superfamily protein gene targeted by miR168; one NUCLEAR FACTOR Y gene (NF-YA3) targeted by miR169; two Myb/SANT-like family protein genes targeted by miR319; three F-box/RNI-like superfamily protein (TIR1) genes targeted by miR393; two growth-regulating factor (GRF) genes targeted by miR396; and one copper zinc superoxide dismutase gene (CZSOD) targeted by miR398. Moreover, large amounts of non-conserved targets were also identified. Oxidoreductase genes were targeted by miR158, tubulin alpha-5 genes were found to be targeted by miR167, zinc finger (C2H2 type) family protein genes were targeted by miR168, one ethylene-responsive element-binding protein gene and one DNA methyltransferase 1-associated protein gene were found to be targeted by miR169, and ACTIN7 genes were targeted by miR399 (Supplementary Table S6). Interestingly, two plant-specific GATA-type zinc finger transcription factor family protein genes emerged as the targets of miR482 and miR2949a*, indicating that miR2949a* might be a functional miRNA, and there might be some relationship between these two miRNAs. Moreover, three genes were identified as the targets of miR3476* (Supplementary Table S6).
Table 3.
Conserved targets of known miRNAs identified during cotton SE.
| miRNA | Target ID | Target annotation | Cleavage site | Abundance | Score | |
|---|---|---|---|---|---|---|
| EC | CK | |||||
| miR156 | zhu2_gi|164260183|gb|ES799263.1|ES799263# | SPL 9 | 493 | 1035 | 83 | 1 |
| zhu2_CL15525Contig1 | SPL | 567 | 0 | 320 | 1 | |
| miR160 | zhu2_gi|164303952|gb|ES831580.1|ES831580# | ARF16 | 31 | 0 | 374 | 2.5 |
| miR167 | zhu2_CL3970Contig2# | ARF6 | 659 | 482 | 1518 | 4 |
| zhu2_CL11135Contig1 | ARF6 | 471 | 482 | 1518 | 4 | |
| zhu2_CL3970Contig1 | ARF 6 | 79 | 482 | 1518 | 4 | |
| zhu2_CL15585Contig1# | ARF 6 | 524 | 482 | 1518 | 4 | |
| zhu2_CL4794Contig1# | ARF 8 | 905 | 380 | 230 | 4 | |
| zhu2_gi|164348622|gb|ES831296.1|ES831296 | ARF 6 | 577 | 0 | 1306 | 4 | |
| zhu2_CL3001Contig1 | ARF 6 | 1004 | 0 | 1306 | 4 | |
| miR168 | zhu2_CL2858Contig3 | Ribonuclease H-like superfamily protein | 980 | 56 | 0 | 4 |
| miR169 | zhu2_CL17360Contig1# | NF-YA3 | 1282 | 125 | 0 | 2 |
| miR319 | zhu2_gi|164329837|gb|ES808978.1|ES808978 | Myb/SANT-like family protein | 662 | 0 | 23 | 4 |
| zhu2_gi|164369589|gb|ES805414.1|ES805414 | Myb/SANT-like family protein | 663 | 0 | 23 | 4 | |
| miR393 | zhu2_CL2451Contig2 | TIR1 | 192 | 83 | 88 | 2.5 |
| zhu2_CL2451Contig1 | TIR1 | 1784 | 83 | 88 | 2.5 | |
| zhu2_CL14972Contig1# | TIR1 | 46 | 83 | 0 | 2.5 | |
| miR396 | zhu2_CL3508Contig2# | GRF8 | 1001 | 18 | 0 | 1 |
| zhu2_CL9598Contig1 | GRF5 | 512 | 0 | 374 | 1 | |
| miR398 | zhu2_gi|164324123|gb|ES849495.1|ES849495# | CZSOD | 39 | 30 | 0 | 4 |
‘#’ miRNA targets validated by RLM-5’ RACE.
The cleavage site is the nucleotide number from the 5’ end of cDNA.
Besides the targets annotated for conserved miRNAs and species-specific miRNAs, targets for novel miRNAs were also identified (Table 4). Sixteen target genes for eight novel miRNAs were identified. In contrast to conserved miRNAs, low splicing frequency targets were found for the novel cotton miRNAs. Some target genes were involved in signal transduction pathways, such as leucine-rich repeat protein kinase-like protein and protein–protein interaction regulator family protein, indicating that the corresponding new miRNAs might participate in specific developmental processes during cotton SE. A cysteine-rich receptor-like protein kinase (RLK8) was found as the target of ghr-miR_N16 with a relatively low score. The detection of miRNA targets provides further evidence for the existence of novel miRNAs in cotton.
Table 4.
Targets for novel miRNAs identified during cotton SE.
| miRNA | Target ID | Target annotation | Cleavage site(nt) | Abundance | Score | |
|---|---|---|---|---|---|---|
| EC | CK | |||||
| ghr-miR_N2 | zhu2_CL2290Contig2 | API5 | 694 | 0 | 55 | 4 |
| ghr-miR_N3 | zhu2_gi|164336128|gb|ES836641.1| ES836641 | Ribosomal protein L7Ae/L30e /S12e/Gadd45 family protein | 485 | 0 | 224 | 4 |
| zhu2_CL531Contig1 | Alpha-helical ferredoxin | 1505 | 0 | 224 | 4 | |
| zhu2_gi|164336793|gb|ES811631.1| ES811631 | Protein–protein interaction regulator family protein | 327 | 24 | 0 | 4 | |
| ghr-miR_N7 | zhu2_CL9546Contig1 | Sucrase/ferredoxin-like family protein | 440 | 53 | 0 | 4 |
| ghr-miR_N9 | zhu2_CL2051Contig2 | Emp24/gp25L/p24 family/GOLD family protein | 353 | 0 | 29 | 4 |
| zhu2_gi|164314833|gb|ES846918.1| ES846918 | class II aminoacyl-tRNA and biotin synthetases superfamily protein | 507 | 0 | 43 | 3 | |
| zhu2_gi|164245414|gb|EV494500.1| EV494500 | Emp24/gp25L/p24 family/GOLD family protein | 302 | 0 | 29 | 4 | |
| ghr-miR_N13 | zhu2_CL1659Contig1 | Zinc knuckle (CCHC-type) family protein | 33 | 0 | 29 | 4 |
| ghr-miR_N16 | zhu2_gi|164307981|gb|ES832690.1| ES832690 | CRK8|cysteine-rich RLK8 | 378 | 0 | 32 | 0.5 |
| ghr-miR_N19 | zhu2_CL16787Contig1 | Unknown protein | 702 | 0 | 24 | 3.5 |
| ghr-miR_N23 | zhu2_gi|164264211|gb|ES806413.1| ES806413 | Leucine-rich repeat protein kinase family protein | 281 | 0 | 64 | 4 |
| zhu2_gi|164350949|gb|ES815762.1| ES815762 | Leucine-rich repeat protein kinase family protein | 279 | 0 | 64 | 4 | |
| zhu2_CL416Contig2 | Leucine-rich repeat protein kinase family protein | 578 | 0 | 64 | 4 | |
| zhu2_gi|164342258|gb|ES832378.1| ES832378 | Leucine-rich repeat protein kinase family protein | 250 | 0 | 64 | 4 | |
| zhu2_gi|164258795|gb|ES844390.1| ES844390 | ADP/ATP carrier 2 | 617 | 0 | 36 | 4 | |
Identification of tas3-siRNAs and their transcripts
Ta-siRNAs form a class of plant-specific endogenous small RNAs, whose biogenesis requires an initial miRNA-mediated cleavage of their precursors. Like miRNAs, ta-siRNAs repress gene expression through mRNA degradation. Of the four ta-siRNAs precursors (TAS1–TAS4) identified in Arabidopsis, cleavage of TAS3 is unique as it requires the specific action of the miR390/ARGONAUTE7 (AGO7) complex for ta-siRNA production (Montgomery et al., 2008). Features of the TAS3 family and the characteristic targets of miR390 are highly conserved in plants (Allen et al., 2005).
In this study, a blast search was carried out against the unannotated groups of the two small RNA sequencing libraries, using the first 12 bases which were identical in TAS3-5’ D7 and D8 in both Arabidopsis and rice (B. Liu et al., 2007). Four tas3-siRNAs were obtained in both libraries, with an abundance of 2889.94, 9.39, 335.81, and 34.02 in EC, and 121.21, 8.49, 18.28, and 2 in CK, respectively (Table 5), namely ghr-TAS3a D6 (+), ghr-TAS3b D7 (+), ghr-TAS3c D8 (+), and ghr-TAS3d D7 (+). They are identical to or have some mismatches with their corresponding homologues (the mismatched nucleotides are indicated in bold in Table 5). The reference database zhuref-2 was searched using these sequences, and four transcripts were found (zhu2_CL11974Contig1, zhu2_gi|164318918|gb|ES828745.1|ES828745, zhu2_gi|164333334|gb|ES843623.1|ES843623, and zhu2_CL11405ConTig1). Correspondingly, they have two target sites for miR390 (Fig. 5). In order to determine its authenticity, the four TAS3-derived ta-siRNAs, ghr-TAS3a D6 (+), ghr-TAS3b D7 (+), ghr-TAS3c D8 (+), and ghr-TAS3d D7 (+) were submitted to the CleaveLand pipeline. Two ARF gene homologues (zhu2_CL19867Contig1, ARF4 and zhu2_CL14460Contig1, ARF3) were shown to be cleaved by all those four tas3-siRNAs (Table 5).
Table 5.
TAS3 genes found in the degradome during cotton SE.
| Name | Length (nt) | RPM | Sequence | Homologue | Target ID | Cleavage site | Abundance (EC/CK) | |
|---|---|---|---|---|---|---|---|---|
| EC | CK | |||||||
| ghr-TAS3a D6(+) | 21 | 2889.94 | 121.21 | TTCTTGACCTTGTAAGACCCA | Os TAS3 5’D6 | zhu2_CL19867 Contig1(ARF4) | 198 | 37/184 |
| ghr-TAS3b D7(+) | 21 | 9.39 | 8.94 | TTCTTGACCTTGTAAGACCCC | At TAS3 5’D7 | |||
| ghr-TAS3c D8(+) | 21 | 335.81 | 18.28 | TTCTTGACCTTGTAAGACCTT | At TAS3 5’D8 | zhu2_CL14460 Contig1(ARF3)# | 402 | 0/40 |
| ghr-TAS3d D7(+) | 21 | 34.02 | 2 | TTCTTGACCTTGTAAGACCCT | At TAS3 5’D7 | |||
The bases in bold are the mismatched nucleotides of cotton TAS3 genes with their homlogs in Arabidopsis.
The cleavage site is the nucleotide number from the 5’ end of cDNA.
‘#’ Validated by RLM-5’ RACE.
Fig. 5.
TAS3 transcripts identified during cotton SE. (A) Target sites of miR390 and tas3-siRNA genes on zhu2_CL11974Contig1. (B) Target sites of miR390 and tas3-siRNA genes on zhu2_gi|164318918|gb|ES828745.1|ES828745. (C) Target sites of miR390 and tas3-siRNA genes on zhu2_gi|164333334|gb|ES843623.1|ES843623. (D) Target sites of miR390 and tas3-siRNA genes on zhu2_CL11405Contig1. Alignment of miR390 with a portion of its target sequence. Two target sites of ghr-miR390 are indicated which were upstream and downstream of the of the tas3-siRNA-producing site respectively. The solid line indicates matched RNA base pairs, ‘.’ Indicates G:U pairs, ‘:’ represents mismatches. ghr-TAS3a D6 (+), ghr-TAS3b D7 (+), and ghr-TAS3c D8 (+) are indicated by underlining (This figure is available in colour at JXB online.)
Expression pattern analysis of miRNAs and their targets during cotton SE
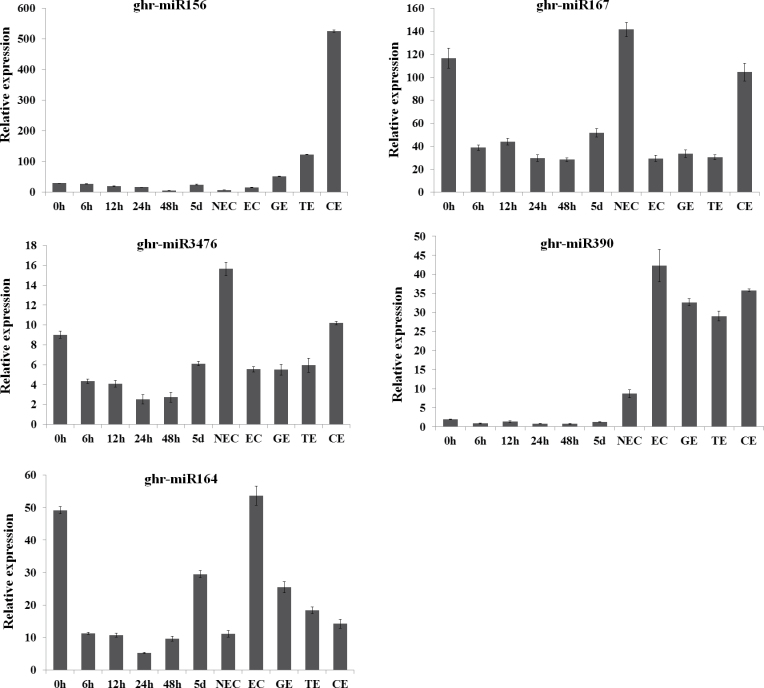
The expression patterns of five miRNAs during SE were validated by qRT-PCR. The abundances of these miRNAs, namely miR156, miR164, miR167, miR390, and miR3476, were relatively high in both libraries. All the five miRNAs tested showed perfect agreement with the expression profiles analysed by small RNA sequencing between the two samples (EC versus CK), with ghr-miR156, ghr-miR167, and ghr-miR3476 down-regulated, ghr-miR390 up-regulated, and ghr-miR164 almost equally expressed. The expression patterns of the five miRNAs during cotton SE, in which 11 typical time points/stages during SE were sampled (0, 6, 12, 24, and 48h, and 5 d explants after induction, NEC, EC, GE, TE, and CE), were further analysed. All the five miRNAs showed an accumulated peak at the CE stage (Fig. 6). ghr-miR156 showed an extremely low expression level during the dedifferentiation stage and EC stage, but gradually increased during somatic embryo development, and reached a relatively high expression level at the CE stage, while ghr-miR167 and ghr-miR3476 also exhibited a relatively very high expression level at 0h, then was down-regulated during the initial dedifferentiation stage (6–48h post-induction), and up-regulated during the late dedifferentiation stage (48h; NEC) to reach the highest level at NEC. However, ghr-miR390 was expressed at a relatively low level during the dedifferentiation stage, and reached the highest expression level at the EC stage, and then remained at moderate levels during embryo development. Although there was no significant difference between the expression levels of ghr-miR164 in the embryonic and hypocotyl samples (EC versus CK), there was differential expression during other time points/stages during cotton SE (Fig. 6).
Fig. 6.
Relative expression levels of five known miRNAs during cotton SE. (A) miR156, (B) miR167, (C) miR3476, (D) miR390, (E) miR164. The equation ratio=2-ΔCt was applied to calculate the relative expression level using GhUBQ7 as the reference gene; three biological replicates and three technical replicates were performed. 0h, hypocotyls; 6h, 12h, 24h, 48h, 5d: hypocotyls induced for 6, 12, 24, and 48h, and 5 d, respectively; NEC, non-embryonic callus; EC, embryonic callus; GE, globular-stage somatic embryo; TE, torpedo-stage somatic embryo; CE. cotyledon-stage somatic embryo.
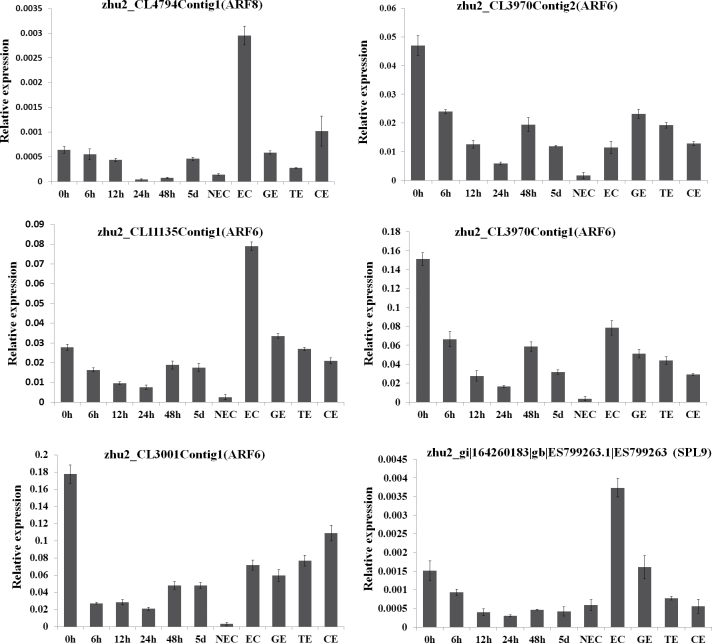
The expression pattern of five miR167 targets (zhu2_CL4794Contig1, ARF8; zhu2_CL3970Contig2, ARF6; zhu2_CL11135Contig1, ARF6; zhu2_CL3970Contig1, ARF6; and zhu2_CL3001Contig1, ARF6) and one miR156 target (zhu2_gi|164260183|gb|ES799263.1|ES799263, SPL9) (Fig. 7) was also analysed during cotton SE by qRT-PCR. All the six targets tested showed perfect agreement with the expression profiles analysed by degradome sequencing between the two samples (EC versus CK). Similar expression patterns were observed for four ARF6 genes (zhu2_CL3970Contig2, zhu2_CL11135Contig1, zhu2_CL3970Contig1, and zhu2_CL3001Contig1) during the initial and late dedifferentiation stages.
Fig. 7.
Relative expression levels of six target genes (five ARF genes and one SPL gene) during cotton SE. (A) zhu2_CL4794Contig1 (ARF8) targeted by miR167. (B) zhu2_CL3970Contig2 (ARF6) targeted by miR167. (C) zhu2_CL11135Contig1 (ARF6) targeted by miR167. (D) zhu2_CL3970Contig1 (ARF6) targeted by miR167. (E) zhu2_CL3001Contig1 (ARF6) targeted by miR167. (F) zhu2_gi|164260183|gb|ES799263.1|ES799263 (SPL9) targeted by miR156. The equation ratio=2-ΔCt was applied to calculate the relative expression level using GhUBQ7 as the reference gene; three biological replicates and three technical replicates were performed. 0h, hypocotyls; 6h, 12h, 24h, 48h, 5d, hypocotyls induced for 6, 12, 24, and 48h, and 5 d, respectively; NEC, non-embryonic callus; EC, embryonic callus; GE, globular-stage somatic embryo; TE, torpedo-stage somatic embryo; CE, cotyledon-stage somatic embryo.
As expected, a perfect inverse expression pattern was found for the SPL transcript (zhu2_gi|164260183|gb|ES799263.1|ES799263) and miR156 during embryo development, ARF6 (zhu2_CL3970Contig2, zhu2_CL11135Contig1 and zhu2_CL3970Contig1) and miR167 during the late dedifferentiation stage (48h; NEC) and embryo development. The comparative analysis of target expression and cleavage frequency between CK and EC showed that high cleavage frequency was found when the expression levels were also higher for SPL9, ARF6 (zhu2_CL3970Contig2 and zhu2_CL3970Contig2), and ARF8 transcripts, consistent with the possibility of a regulatory role for those miRNAs during cotton SE.
Validation of target genes by RLM-5’ RACE in cotton
For several multicopy genes in G. hirsutum, targets were matched by the same read. To examine whether the targets mapped by the same read were sliced by the same miRNA, RLM 5’-RACE experiments were carried out (experimentally identified targets are indicated with ‘#’ in Table 3). Among two SPL genes targeted by miR156, only one (zhu2_gi|164260183|gb|ES799263.1|ES799263) could be further validated (Fig. 8A), which was confirmed by degradome sequencing (Supplementary File S1 at JXB online); for seven targets annotated as ARF genes which were matched by the identical read of ghr-miR167, three (zhu2_CL4794Contig1, zhu2_CL3970Contig2, and zhu2_CL15585Contig1) were found to be cleaved by ghr-miRNA167 after sequencing several clones (Fig. 8B–D), as indicated in degradome sequencing (Supplementary File S1); one TIR1 gene (zhu2_CL14972Contig1) was validated as a target of miR393 (Fig. 8E; Supplementary File S1) among three TIR1 genes; and, in addition, an ARF16 (zhu2_gi|164303952|gb|ES831580.1|ES831580) targeted by miR160, a NF-YA3 (zhu2_CL17360Contig1) targeted by miR169, and a CZSOD (zhu2_gi|164324123|gb|ES849495.1|ES849495) targeted by miR398 were also validated (Fig. 8F–8H; Supplementary File S1). Furthermore, one TAS3 transcript (zhu2_CL11974Contig1) was verified as the target of miR390 (Fig. 8I). Surprisingly, an mRNA encoding CZSOD2 (zhu2_CL2800Contig1, Fig. 8J, Supplementary File S1) and a SurE-like phosphatase gene (zhu2_CL3163Contig2; Fig. 8K, Supplementary File S1) was detected as the target of miR159 and miR3476* respectively. Unexpectedly, the validated cleavage site was lagging by 6 nt and 9 nt, respectively, compared with that from the degradome libraries (Supplementary File S1). By degradome sequencing, two cleavage sites were detected in the SPL9 gene (only the dominant one is shown in Table 4); however, only one cleavage site could be further validated by RLM 5’-RACE. For the validated GRF8 (zhu2_CL3508Contig2) and ARF3 (zhu2_CL14460Contig1), the cleavage sites were one base ahead and one base behind, respectively, compared with the dominant one from the degradome libraries (Fig. 8L, M; Supplementary File S1), which were exactly between the 10th and 11th nucleotides from the 5’ end of the miRNA.
Fig. 8.
Validation of miRNA target genes by RLM 5’-RACE. Cleavage site for SPL9 (zhu2_gi|164260183|gb|ES799263.1|ES799263) targeted by miR156 (A); ARF8 (zhu2_CL4794Contig1) and ARF6 (zhu2_CL3970Contig2, zhu2_CL15585Contig1) targeted by miR167 (B–D); TIR1 (zhu2_CL14972Contig1) targeted by miR393 (E); ARF16 (zhu2_gi|164303952|gb|ES831580.1|ES831580) targeted by miR160 (F); NF-YA3 (zhu2_CL17360Contig1) targeted by miR169 (G); CZSOD (zhu2_gi|164324123|gb|ES849495.1|ES849495) targeted by miR398 (H); TAS3 (zhu2_CL11974Contig1) targeted by miR390 (I); CZSOD2 (zhu2_CL2800Contig1) targeted by miR159 (J); SurE-like phosphatase transcript (zhu2_CL3163Contig2) targeted by miR3476* (K); GRF8 (zhu2_CL3508Contig2) transcript targeted by miR396 (L); and ARF3 (zhu2_CL14460Contig1) transcript targeted by TAS3a D6 (+) (M). The solid lines indicate matched RNA base pairs, ‘.’ indicates G:U pairs, ‘:’ represent other types of mismatch. The black arrow indicates a cleavage site verified by RLM 5’-RACE, with the frequency of cloned RACE products shown above the alignment.
Discussion
The miRNAs and relative targets during cotton SE
To date, 299 and 591 miRNAs have been uploaded to the miRBase database from Arabidopsis and rice, respectively (Griffiths-Jones et al., 2008). However, prior to this study, only 34 miRNAs from cotton had been annotated in the miRBase database (release 17.0). In this study, 36 differentially expressed known miRNAs families, most of which were conserved with other species and predicted previously, were identified by sequence similarity of the mature/miR* with miRNAs of all plants. In addition, 25 novel miRNAs, some of which may be cotton specific, were identified, using universal rules for miRNA annotation (Meyers et al., 2008), with lower abundance than that of conserved miRNAs. It is reasonable to propose that the conserved miRNAs are probably responsible for controlling the basic cellular and developmental processes in many eukaryotes, while the non-conserved or species-specific miRNAs are involved in the regulation of the species-specific regulatory pathways and functions (Glazov et al., 2008). Limited by the degree of coverage of reference genes, tags that were similar to their homologues within two mismatches without gaps were annotated as the sequences of an miRNA family. It is recognized that this analysis might miss other less conserved miRNA families which had more mismatches with their homologues. About 62% of the known miRNAs and 54% of novel miRNAs started with a 5’ uridine, a signature of miRNA (Yao et al., 2007). The minimum free energy (MFE) for hairpin structures of known and novel miRNA precursors was from –22.1 to –78 kcal/mol and from –21.3 to –162.1 kcal/mol, respectively (Supplementary Tables S5, S6 at JXB online). This feature was in accordance with previous studies (Bonnet et al., 2004). The length of known and novel miRNA precursors varied from 80 nt to 273 nt and from 77 nt to 361 nt in CK and EC, respectively (Supplementary Tables S4, S5). These results are consistent with those reported in other plants (Wang et al., 2011).
Some conserved miRNA targets have been previously predicted in cotton (Zhang et al., 2007; Kwak et al., 2009; Pang et al., 2009), but only a few miRNA targets have been identified experimentally (Pang et al., 2009). The recently developed high-throughput experimental approach (Addo-Quaye et al., 2008; German et al., 2008) has allowed the identification of target genes for known and novel miRNAs. As expected, a number of predicted targets for highly conserved and non-conserved miRNAs were identified during cotton SE (Supplementary Table S6 at JXB online). However, some conserved miRNAs (miR162, miR164, miR2911, miR2947, miR2948, miR3954, miR408, miR4387, miR828, and miR894; Supplementary Table S6) and more novel miRNAs (68%, Table 4) did not have detectable sliced targets in the degradomes examined. It is possible that the levels of some conserved/known non-conserved miRNAs (e.g. miR4387 and miR828) or their sliced targets are too low to detect in the samples analysed here, or they may be present in other tissues that have not yet been analysed. Alternatively, some miRNAs might inhibit target gene expression through translational repression (Huang et al., 2007; Fabian et al., 2010). Furthermore, targets were identified using miRNAs from eight plant species which had large amounts of known miRNAs, but this was only on the basis of sequence conservation, which might mean that the threshold score 4 between miRNA–target pairs is too rigorous. Additionally, for the known miRNAs, many target genes may be missed since the known miRNAs were annotated by similarity and mismatches in a very strict range (no more than two). To obtain more integrated information on miRNA targets, degradome libraries from different tissues, organs, and developmental stages should be constructed and integrated with a complete cotton genome sequence.
In most cases, targets of the same miRNAs belong to the same gene family (Table 3); however, some unexpected targets were also identified in both libraries. Two genes coding for RNase H-like superfamily proteins were found to be targeted by miR156 with low frequency. Several α-tubulin genes were found to be the targets of miR167, with a much lower cutting frequency than that of ARF genes. The expression patterns of two of those α-tubulin genes (zhu2_gi|164304473|gb|EV495182.1|EV495182 and zhu2_gi|84164246|gb|DR452894.1|DR452894) were analysed during cotton SE (data not shown), and the differential expression levels among different stages of cotton SE imply that these genes might also be the targets of miRNAs.
Most miRNA families identified were differentially expressed (Supplementary Table S2 at JXB online). Of the identified differentially expressed miRNAs, the expression profiles of miR156, miR164, miR167, miR390, and miR3476 were validated by qRT-PCR. The expression profiles of miR156, miR167, and miR3476 were up-regulated from GE to CE, while miR164 was down-regulated from EC to CE, implying that these five miRNAs may participate in the process of redifferentiation during cotton SE. This was consistent with a previous study in rice, which showed that the expression of miR156 was notably higher in differentiating callus than in the dedifferentiated tissues (Luo et al., 2006). The expression pattern of miR167 was similar to that found during SE in Valencia sweet orange (Wu et al., 2011).
To detect the negative regulation of target genes by miRNAs, the expression pattern of specific miRNAs (miR156 and miR167) and their target genes was validated by qRT-PCR during cotton SE. Most target genes (four ARF genes and one SPL gene) showed perfect agreement between the two tissue samples and inverse expression patterns compared with miR167 and miR156 during somatic embryo development. This suggests that miR156 and miR167 may play a role in GE formation and the succeeding conversion to CE. Further studies are needed to clarify the regulatory networks associated with different miRNAs during cotton SE.
Regulation networks of miRNAs during cotton SE
miRNAs mediate gene silencing generally by two mechanisms: mRNA cleavage and translation repression. In higher plants, miRNAs regulate gene expression mainly by slicing mRNAs (Addo-Quaye et al., 2008). miRNA-directed cleavage leaves a 5’-uncapped 3’ fraction of the sliced genes. The present results suggest that conserved miRNAs silence more targets than cotton-specific miRNAs, implying that conserved miRNAs play crucial roles in universal mechanisms of regulation across plant species. Conserved miRNAs mainly regulate genes encoding transcription factors, whereas cotton-specific miRNAs regulate a wider range of genes, suggesting a new feature of miRNA regulation in cotton.
As the small RNA and degradome libraries were prepared from hypocotyl and embryogenic callus, it has been possible to identify miRNAs with detected target genes associated with cotton SE. In this study, many transcription factors targeted by miRNAs were identified, such as SPL genes targeted by miR156, ARF genes targeted by miR160 and miR167, NF-YA3 targeted by miR169, IAA genes targeted by miR159 and miR166, zinc finger family genes targeted by miR168, miR482, and miR2949, and homeobox-related WOX genes targeted by miR 482. These transcription factors are involved in plant growth and/or responses to environmental changes. SPLs function in a broad range of developmental processes in Arabidopsis. SPL is regulated by miR156, and acts as a pleiotropic regulator of plant development (Wang et al., 2009) and can promote vegetative phase transition by activating another miRNA (miR172). In addition, SPL genes play critical roles in regulating zygotic embryo development (Nodine and Bartel, 2010), with similarities to SE. A no apical meristem (NAC) domain transcriptional regulator gene (zhu2_CL3735Contig1) was found to be the target of miR394 and miR482, implying that NAC domain transcriptional regulators might also be regulated by miRNAs other than miR164 (Supplementary Table S6 at JXB online). Interestingly, ghr-miR2949* and miR482 were found to target plant-specific GATA-type zinc finger transcription factor family protein genes (Supplementary Table S6), the products of which may participate in hypocotyl and petiole elongation in Arabidopsis (Shikata et al., 2004); and those targets were found in the hypocotyl tissues in this study. Therefore, miR2949* and miR482 might regulate hypocotyl elongation via their targets. miR2949* was also identified in EC, but at a lower expression level. The regulation of plant-specific GATA-type zinc finger transcription factors by miR2949* and miR482 was not found in previous studies. Further study of the relationship between these targets and the two miRNAs should reveal the function of these partners in the regulation of miRNA biogenesis and cotton SE.
Auxin plays an important role during cotton SE (Yang and Zhang, 2010). miR160, miR167, and miR390 are involved in the auxin signalling transduction pathway through regulating the expression of ARF10, ARF16, ARF17, ARF6, ARF8, ARF2, ARF3, and ARF4 in Arabidopsis and rice (Yang, 2006; P.P. Liu et al., 2007; Marin et al., 2010). miR167 is known to regulate plant fertility through participating in the auxin signalling pathway, by targeting ARF6 and ARF8 genes (Wu et al., 2006). In Arabidopsis, miR167 mediates transcript cleavage of ARF8, but not ARF6 (Ru et al., 2006). However, in cotton, both ARF6 and ARF8 were shown to be cleaved by miR167, which was consistent with previous data from citrus (Wu et al., 2011). In the present study, ghr-miR167 was found to be expressed at higher levels in CK than in EC (Fig. 6), and two of the predicted targets (zhu2_CL4794Contig1, ARF8; zhu2_CL11135Contig1, ARF6) were expressed at lower levels in CK than in EC (Fig. 7), indicating that the regulation of target genes by miRNAs was not absolutely quantitative but time and space specific. The organized expression patterns of miR156, miR167 and their targets during somatic embryo development suggest their specific functions during cotton SE. Three TIR1 genes, which were also involved in the auxin signalling pathway (Si-Ammour et al., 2011), were identified as targets of miR393. In addition, miR390, which can direct formation of ta-siRNAs to target ARF genes, resulting in modulating developmental timing and patterning in Arabidopsis (Fahlgren et al., 2006), was found to be expressed during SE in the present study. Those differentially expressed miRNAs and their auxin signalling-related targets might indicate their function during cotton SE, and further analysis of their relationship and regulation will illustrate the networks of auxin regulation during plant embryogenesis.
In conclusion, it was illustrated that the combination of degradome sequencing and small RNA sequencing can be used for genome-wide profiling of miRNA regulation networks during cotton SE. The global identification of miRNAs and their related targets during cotton SE in this study provides new information about the regulatory network of miRNAs in cotton, which will contribute to the understanding of miRNA function during plant embryogenesis.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Primers used in this study.
Table S2. Known miRNAs identified during cotton SE.
Table S3. Reads which perfectly matched the known mature miRNAs in CK and EC.
Table S4. Precusors of known miRNAs.
Table S5. Novel miRNA identified in two libraries.
Table S6 miRNA targets identified from the degradome of CK and EC.
Supplementary File S1. The precursor structures for 29 known miRNAs and 25 novel miRNAs.
Acknowledgements
Funding by the National Natural Science Foundation of China (nos 30810103911, 30871560, 31101185) and the National Basic Research Program of China (973 Program) (no. 2010CB126001) is greatly appreciated.
Glossary
Abbreviations:
- AGO7
argonaute-like protein 7
- CE
cotyledon-stage somatic embryo
- Dicer
double-stranded-specific RNase
- EC
embryogenic callus
- GE
globular-stage somatic embryo
- GRF
growth-regulating factor
- miRNA
microRNA
- miRNA*
the complementary strands of functional mature miRNA
- MFE
minimum free energy
- NF-YA3
NUCLEAR FACTOR Y gene
- PARE
parallel analysis of RNA ends
- qRT-PCR
quantitative real-time PCR
- RLK8
cysteine-rich receptor-like protein kinase 8
- RLM 5’-RACE
RNA ligase-mediated rapid amplification of 5’ cDNA ends
- SE
somatic embryogenesis
- ta-siRNA
trans-acting small interfering RNA
- TE
torpedo-stage somatic embryo.
References
- Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ. 2008. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Current Biology 18, 758–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addo-Quaye C, Miller W, Axtell MJ. 2009. CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25, 130–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. 2005. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. Journal of Molecular Biology 215, 403–410 [DOI] [PubMed] [Google Scholar]
- Alves-Junior L, Niemeier S, Hauenschild A, Rehmsmeier M, Merkle T. 2009. Comprehensive prediction of novel microRNA targets in Arabidopsis thaliana. Nucleic Acids Research 37, 4010–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archak S, Nagaraju J. 2007. Computational prediction of rice (Oryza sativa) miRNA targets. Genomics, Proteomics and Bioinformatics 5, 196–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H. 2003. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. The Plant Cell 15, 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Jan C, Rajagopalan R, Bartel DP. 2006. A two-hit trigger for siRNA biogenesis in plants. Cell 127, 565–577 [DOI] [PubMed] [Google Scholar]
- Bonnet E, Wuyts J, Rouze P, Van de Peer Y. 2004. Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics 20, 2911–2917 [DOI] [PubMed] [Google Scholar]
- Chen CJ, Liu Q, Zhang YC, Qu LH, Chen YQ, Gautheret D. 2011. Genome-wide discovery and analysis of microRNAs and other small RNAs from rice embryogenic callus. RNA Biology 8, 538–547 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. 2005. The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445 [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. 2010. Regulation of mRNA translation and stability by microRNAs. Annual Review of Biochemistry 79, 351–379 [DOI] [PubMed] [Google Scholar]
- Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC. 2006. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Current Biology 16, 939–944 [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 [DOI] [PubMed] [Google Scholar]
- German MA, Pillay M, Jeong DH, et al. 2008. Global identification of microRNA–target RNA pairs by parallel analysis of RNA ends. Nature Biotechnology 26, 941–946 [DOI] [PubMed] [Google Scholar]
- Glazov EA, Cottee PA, Barris WC, Moore RJ, Dalrymple BP, Tizard ML. 2008. A microRNA catalog of the developing chicken embryo identified by a deep sequencing approach. Genome Reseach 18, 957–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. 2008. miRBase: tools for microRNA genomics. Nucleic Acids Research 36, D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, Lin C, Holoch D, Lim C, Tuschl T. 2008. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods 44, 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Liang Z, Yang B, Tian H, Ma J, Zhang H. 2007. Derepression of MicroRNA-mediated protein translation inhibition by apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G (APOBEC3G) and its family members. Journal of Biological Chemistry 282, 33632–33640 [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293, 834–838 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. 2004. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Molecular Cell 14, 787–799 [DOI] [PubMed] [Google Scholar]
- Kwak P, Wang Q, Chen X, Qiu C, Yang Z. 2009. Enrichment of a set of microRNAs during the cotton fiber development. BMC Genomics 10, 457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. 2009. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967 [DOI] [PubMed] [Google Scholar]
- Liu B, Chen Z, Song X, et al. 2007. Oryza sativa dicer-like4 reveals a key role for small interfering RNA silencing in plant development. The Plant Cell 19, 2705–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PP, Montgomery TA, Fahlgren N, Kasschau KD, Nonogaki H, Carrington JC. 2007. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. The Plant Journal 52, 133–146 [DOI] [PubMed] [Google Scholar]
- Luo YC, Zhou H, Li Y, Chen JY, Yang JH, Chen YQ, Qu LH. 2006. Rice embryogenic calli express a unique set of microRNAs, suggesting regulatory roles of microRNAs in plant post-embryogenic development. FEBS Letters 580, 5111–5116 [DOI] [PubMed] [Google Scholar]
- Mallory AC, Bartel DP, Bartel B. 2005. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. The Plant Cell 17, 1360–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin E, Jouannet V, Herz A, Lokerse AS, Weijers D, Vaucheret H, Nussaume L, Crespi MD, Maizel A. 2010. miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. The Plant Cell 22, 1104–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Axtell MJ, Bartel B, et al. 2008. Criteria for annotation of plant microRNAs. The Plant Cell 20, 3186–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC. 2008. Specificity of ARGONAUTE7–miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133, 128–141 [DOI] [PubMed] [Google Scholar]
- Morin RD, Aksay G, Dolgosheina E, Ebhardt HA, Magrini V, Mardis ER, Sahinalp SC, Unrau PJ. 2008. Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. Genome Research 18, 571–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodine MD, Bartel DP. 2010. MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes and Development 24, 2678–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang M, Woodward AW, Agarwal V, Guan X, Ha M, Ramachandran V, Chen X, Triplett BA, Stelly DM, Chen ZJ. 2009. Genome-wide analysis reveals rapid and dynamic changes in miRNA and siRNA sequence and expression during ovule and fiber development in allotetraploid cotton (Gossypium hirsutum L.). Genome Biology 10, R122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaleo V, Szittya G, Moxon S, Miozzi L, Moulton V, Dalmay T, Burgyan J. 2010. Identification of grapevine microRNAs and their targets using high-throughput sequencing and degradome analysis. The Plant Journal 62, 960–976 [DOI] [PubMed] [Google Scholar]
- Ru P, Xu L, Ma H, Huang H. 2006. Plant fertility defects induced by the enhanced expression of microRNA167. Cell Research 16, 457–465 [DOI] [PubMed] [Google Scholar]
- Ruan MB, Zhao YT, Meng ZH, Wang XJ, Yang WC. 2009. Conserved miRNA analysis in Gossypium hirsutum through small RNA sequencing. Genomics 94, 263–268 [DOI] [PubMed] [Google Scholar]
- Shikata M, Matsuda Y, Ando K, Nishii A, Takemura M, Yokota A, Kohchi T. 2004. Characterization of Arabidopsis ZIM, a member of a novel plant-specific GATA factor gene family. Journal of Experimental Botany 55, 631–639 [DOI] [PubMed] [Google Scholar]
- Si-Ammour A, Windels D, Arn-Bouldoires E, Kutter C, Ailhas J, Meins F, Vazquez F. 2011. miR393 and secondary siRNAs regulate expression of the TIR1/AFB2 auxin receptor clade and auxin-related development of Arabidopsis leaves. Plant Physiology 157, 683–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song QX, Liu YF, Hu XY, Zhang WK, Ma B, Chen SY, Zhang JS. 2011. Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biology 11, 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Zhu JK. 2004. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. The Plant Cell 16, 2001–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szittya G, Moxon S, Santos DM, Jing R, Fevereiro MPS, Moulton V, Dalmay T. 2008. High-throughput sequencing of Medicago truncatula short RNAs identifies eight new miRNA families. BMC Genomics 9, 593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Reinhart BJ, Bartel DP, Zamore PD. 2003. A biochemical framework for RNA silencing in plants. Genes and Development 17, 49–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu L, Zhang X, Liu D, Jin S, Cao J, Zhu L, Deng F, Tan J, Zhang C. 2007. Suitable internal control genes for qRT-PCR normalization in cotton fiber development and somatic embryogenesis. Chinese Science Bulletin 52, 3110–3117 [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crété P. 2004. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Molecular Cell 16, 69–79 [DOI] [PubMed] [Google Scholar]
- Wang JW, Czech B, Weigel D. 2009. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138, 738–749 [DOI] [PubMed] [Google Scholar]
- Wang JW, Wang LJ, Mao YB, Cai WJ, Xue HW, Chen XY. 2005. Control of root cap formation by MicroRNA-targeted auxin response factors in Arabidopsis. The Plant Cell 17, 2204–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Liu H, Li D, Chen H. 2011. Identification and characterization of maize microRNAs involved in the very early stage of seed germination. BMC Genomics 12, 154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wang QL, Wang BM. 2012. Identification and characterization of microRNAs in Asiatic cotton (Gossypium arboreum L.). PLoS One 7, e33696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZM, Xue W, Dong CJ, Jin LG, Bian SM, Wang C, Wu XY, Liu JY. 2012. A comparative miRNAome analysis reveals seven fiber initiation-related and 36 novel miRNAs in developing cotton ovules. Molecular Plant 5, 889–900 [DOI] [PubMed] [Google Scholar]
- Wu G, Poethig RS. 2006. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133, 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MF, Tian Q, Reed JW. 2006. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133, 4211–4218 [DOI] [PubMed] [Google Scholar]
- Wu XM, Liu MY, Ge XX, Xu Q, Guo WW. 2011. Stage and tissue-specific modulation of ten conserved miRNAs and their targets during SE of Valencia sweet orange. Planta 233, 495–505 [DOI] [PubMed] [Google Scholar]
- Yang JH. 2006. Evidence of an auxin signal pathway, microRNA167–ARF8–GH3, and its response to exogenous auxin in cultured rice cells. Nucleic Acids Research 34, 1892–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XY, Zhang XL, Yuan DJ, Jin FY, Zhang YC, Xu J. 2012. Transcript profiling reveals complex auxin signalling pathway and transcription regulation involved in dedifferentiation and redifferentiation during SE in cotton. BMC Plant Biology 12, 110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XY, Zhang XL. 2010. Regulation of SE in higher plants. Critical Reviews in Plant Science 29, 36–57 [Google Scholar]
- Yao Y, Guo G, Ni Z, Sunkar R, Du J, Zhu JK, Sun Q. 2007. Cloning and characterization of microRNAs from wheat (Triticum aestivum L.). Genome Biology 8, R96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Li Y, Han X, Shen F. 2012. Genome-wide profiling of miRNAs and other small non-coding RNAs in the Verticillium dahliae inoculated cotton roots. PLoS One 7, e35765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wang Q, Wang K, Pan X, Liu F, Guo T, Cobb GP, Anderson TA. 2007. Identification of cotton microRNAs and their targets. Gene 397, 26–37 [DOI] [PubMed] [Google Scholar]
- Zhao CZ, Xia H, Frazier T, Yao YY, Bi YP, Li AQ, Li MJ, Li CS, Zhang BH, Wang XJ. 2010. Deep sequencing identifies novel and conserved microRNAs in peanuts (Arachis hypogaea L.). BMC Plant Biology 10, 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu HG, Tu LL, Jin SX, Xu L, Tan JF, Deng FL, Zhang XL. 2008. Analysis of genes differentially expressed during initial cellular dedifferentiation in cotton. Chinese Science Bulletin 53, 3666–3676 [Google Scholar]
- Zhu L, Tu L, Zeng F, Liu D, Zhang X. 2005. An improved simple protocol for isolation of high quality RNA from Gossypium spp. suitable for cDNA library construction. Acta Agronomica Sinica 31, 1657–1659 [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research 31, 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.