Abstract
Human mesenchymal stem/stromal cells (hMSC) are increasingly used in advanced cellular therapies. The clinical use of hMSCs demands sequential cell expansions. As it is well established that membrane glycerophospholipids (GPL) provide precursors for signaling lipids that modulate cellular functions, we studied the effect of the donor's age and cell doublings on the GPL profile of human bone marrow MSC (hBMSC). The hBMSCs, which were harvested from five young and five old adults, showed clear compositional changes during expansion seen at the level of lipid classes, lipid species, and acyl chains. The ratio of phosphatidylinositol to phosphatidylserine increased toward the late-passage samples. Furthermore, 20:4n-6-containing species of phosphatidylcholine and phosphatidylethanolamine accumulated while the species containing monounsaturated fatty acids (FA) decreased during passaging. Additionally, in the total FA pool of the cells, 20:4n-6 increased, which happened at the expense of n-3 polyunsaturated FAs, especially 22:6n-3. The GPL and FA correlated with the decreased immunosuppressive capacity of hBMSCs during expansion. Our observations were further supported by alterations in the gene expression levels of several enzymes involved in lipid metabolism and immunomodulation. The results show that extensive expansion of hBMSCs harmfully modulates membrane GPLs, affecting lipid signaling and eventually impairing functionality.
Keywords: glycerophospholipid profile, mesenchymal stem cell, arachidonic acid, docosahexaenoic acid, lipid signaling, mass spectrometry
Human mesenchymal stem/stromal cells (hMSC) are currently being studied in a number of clinical applications, for example, to improve the engraftment of hematopoietic stem cell transplant, to promote myocardial repair, and to control immunological responses in graft versus host diseases, autoimmune diseases, and solid organ transplantations (1–5). In addition to being immunologically privileged, these cells can modulate both innate and adaptive immune responses in vitro and in vivo. hMSCs have been shown to be able to inhibit T-cell proliferation, inhibit dendritic cell maturation (6), recruit regulatory T-cells (7, 8), and modulate B-cell functions (9). The mechanisms by which these cells exert their immune modulatory functions are still unclear, but it is likely that both direct cell-cell contacts and the secretion of soluble factors are needed. Several cytokines, growth factors, enzymes, and lipid mediators, such as transforming growth factor β1, the tryptophan degrading enzyme indoleamine 2,3-dioxygenase, and prostaglandin E2 (PGE2), have been indicated as key players in the immunomodulatory process (7, 10, 11).
A long expansion period has been considered to have an adverse effect on the proliferation and differentiation potential and other functional properties of progenitor cells (12, 13). One of the key questions in MSC therapy is, how many cell doublings the cells can undergo before the risks of cellular malfunction or even malignant transformation surface? The number of safe cell doublings may be dependent on the cell source, donor age, and culture conditions.
To date, only a few studies on stem cell membrane lipids have been performed (13, 14), and the changes in lipid composition during the expansion have not been studied systematically. Although glycosphingolipids and signaling lipids have been studied in relation to the cell identity and functions (15–18), an important group of membrane lipids, the glycerophospholipids (GPL), composing the main part of membrane lipids has received less attention. The most prominent class, phosphatidylcholine (PC), makes up approximately one half of all membrane phospholipids. Phosphatidylethanolamine (PE), phosphatidylserine (PS), and phosphatidylinositol (PI) are the other common mammalian GPL classes. Each class consists of numerous different molecular species with different acyl, alkyl, or alkenyl chain assemblies (19). Consequently, a single cell contains more than a thousand different GPL molecular species (20, 21). However, the ultimate reasons for the structural diversity of cellular lipids, apparently of great functional importance, are still largely unknown. Modern mass spectrometry and bioinformatics tools offer new ways to analyze the compositional changes in complex stem cell lipidomes and link those with the altered potency and quality of the cells (22–24).
Membrane lipids not only form a protective layer around the cell but also mediate biological signals through several pathways employing G-protein activators, second messengers, and nuclear receptor activators (25, 26). The signaling pathways utilizing phosphorylated PIs (PIP), eicosanoids, or sphingolipids are well understood. Cellular mediators, such as inositol phosphates, diacylglycerols (DAG), lysophospholipids, ceramides, cleaved fatty acids (FA) [e.g., arachidonic acid (AA, 20:4n-6) and docosahexaenoic acid (DHA, 22:6n-3)], or their derivatives, are released from membrane lipids and play an important role in many physiological processes including the regulation of inflammation. The essential polyunsaturated fatty acid (PUFA) 20:4n-6 is found primarily at the sn-2 position of most membrane phospholipids. It is a precursor for the synthesis of prostaglandins (PG), tromboxanes, and leukotrienes (27), one of which, a specific PGE2, has been proposed as one of the soluble factors mediating the MSC immune function (7, 28) The n-3 PUFAs, especially 22:6n-3, are precursors for resolvins, protectins, and maresins which, in contrast to 20:4n-6 and most n-6 PUFAs, exert their functions during the resolution phase of inflammation (29)
The aim of our study was to compare the GPL profiles of hBMSCs from young and old donors, study the effects of sequential expansion of the cells on these profiles, determine the expression of genes related to lipid metabolism and immunomodulation, and gain a better understanding of the possible relations of cell lipidome changes and functionality. Knowledge of the connections of lipid alterations during expansion and their effect on the immunosuppressive capacity of the therapeutic cells may have a profound influence on clinical cell expansion protocols.
MATERIALS AND METHODS
Ethics and bone marrow donors
All patient protocols were approved by the Ethical Committee of Northern Ostrobothnia Hospital District or Ethical Committee of Hospital District of Helsinki and Uusimaa. The hBMSCs were obtained from bone marrow aspirates taken from the iliac crest or upper femur methaphysis of adult patients after written informed consent. The hBMSCs from donors of different ages (anonymous coding) were isolated as previously described (30, 31).
Cell culture of hBMSC
The cells were cultured in minimum essential α-medium (αMEM) supplemented with 20 mM HEPES, 10% fetal bovine serum (FBS), 2 mM L-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin (all from Gibco, Invitrogen, Paisley, UK). The same serum lot was used throughout the study. The medium was renewed twice a week, the cells were harvested when 70–80% confluent and plated at a density of 1,000 cells/cm2. Population doublings for every passage were calculated using the formula NH = 2PD × N1, where NH is the number of cells harvested and N1 is the number of cells plated. For lipid analysis, the cells were washed with ice-cold PBS scraped on ice pelleted in silylated vials and stored at −70°C for later use. The expression of surface antigens CD13, CD14, CD19, CD29, CD34, CD44, CD45, CD49e, CD73, CD90, CD105, CD106, CD166, CD271, HLA-ABC, and HLA-DR were analyzed by flow cytometry (FACSAria, Becton Dickinson, San Jose, CA, and FlowJo 7.6.1 software, Treestar, Asland, OR) (supplementary Table I).
Phospholipid mass spectrometry and fatty acid gas chromatography
Total lipids of the hBMSCs were extracted according to Folch et al. (32), spiked with internal standards, and dissolved in chloroform/methanol 1:2 for direct infusion experiments. Several internal standards were used to correct for the effects of the polar head group and acyl chain length on the instrument response according to previously reported procedures (33–35). Just prior to the mass spectrometry analysis, 1% NH4OH was added, and the lipid extracts with the internal standards were infused to the electrospray source of a Quattro Micro triple quadrupole mass spectrometer (Micromass, Manchester, UK) at the flow rate of 8 μl/min. The collision energy of the instrument was set to 25–65 eV, and negative and positive ion modes were used. Argon was used as the collision gas. The PC and lysoPC (precursor of 184), PE (neutral loss of 141), PS (neutral loss of 87), and PI (precursor of 241) species were selectively detected using head group-specific MS/MS scanning modes (36, 37). The acyl chain assembly of the major lipid species was confirmed by MS/MS techniques. The mass spectra were processed by MassLynx software (Micromass, Manchester, UK), and the individual lipid species were quantified by using the internal standards and LIMSA software (38). The lipid species were abbreviated as follows: [total carbon number in the chains]:[total number of double bonds in the chains]. The relative concentrations of the lipid classes were obtained by summing the concentrations of the individual molecular species in a class.
The FAs in the total lipids of the cells were determined as methyl ester derivatives by gas chromatography as detailed in Käkelä et al. (39). The FA composition was calculated as molar percentage, and the FAs were marked by using the following abbreviations: [carbon number]:[number of double bonds] n-[position of the first double bond calculated from the methyl end] (e.g., 22:6n-3).
Co-culture assay
Peripheral blood mononuclear cells (PBMC) were isolated from buffy coats from healthy anonymous blood donors (Finnish Red Cross Blood Service) by density gradient centrifugation (Ficoll-Pague plus, GE Healthcare, Piscataway, NJ) and cryo-preserved for later use. Prior to use, the PBMCs were thawed gently and labeled with 5 µM 5(6)-carboxyfluorescein diacetate N-succinimidyl ester (CFSE) solution (Molecular probes, OR). The CFSE-labeled PBMCs were cultured in triplicates at 1.5 × 106 cells/well in a 48-well plate with hBMSCs in RPMI growth medium (RPMI, 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin). hBMSCs were allowed to attach for 2 h before PBMCs were added. To activate the T-cell proliferation, 100 ng/ml of antihuman CD3 antibody clone Hit3a (BioLegend, San Diego, CA) was added to the coculture. T-cell proliferation was recorded after four days of incubation as a dilution of fluorescent dye by flow cytometry.
Telomere length analysis
Telomere length were analyzed by the southern blot analysis of terminal restriction fragment (TRF) lengths (40) Genomic DNA from snap-frozen cell pellets was purified using the Qiagen DNeasy Blood and Tissue Kit and extracted with ethanol. Quality of purified DNA was evaluated by 1% agarose gel electrophoresis. Telomere length analysis was performed using TeloTAGGG Telomere Length Assay Kit (Roche, Basel, Switzerland). DNA was digested using RsaI and HinfI enzymes and electrophoresed on a 0.8% agarose gel 5 V/cm. Southern blotting was performed using 20× salium sodium citrate (SSC) buffer. The blot was hybridized overnight using a digoxigenin (DIG)-labeled telomere-specific probe (TTAGGG) and incubated with alkaline phosphatase-labeled anti-DIG antibody. The blot was then incubated with CDP-Star chemiluminescent substrate and exposed to autoradiography film (GE Healthcare). The autoradiogram was scanned by densitometry, and TRF length was calculated using ImageJ analysis software (41) according to TRF = ΣOD/Σ (ODi/Li), where ODi is optical density and Li is the length of the TRF at position i. TRF signals between 3 and 20 kb were used for telomere length measurements (40).
Western blotting
Snap-frozen cell pellets were lysed using RIPA buffer (Thermo Scientific, Rockford, IL) containing protease inhibitor cocktail (Sigma, St. Louis, MO). Protein concentrations were determined using BCA protein assay kit (Pierce, Rockford, IL). Total protein (20 µg) was run on a 12% SDS-PAGE gel (Bio-Rad Laboratories, Hercules, CA) and electrotransferred to Hybond ECL Nitrocellulose membrane (GE Healthcare). The membrane was then blocked with 5% milk in PBS containing 0.1% Tween-20 and immunoblotted using anti-p16INK4A (1: 800, clone DCS-50, Sigma) and anti-p21 (1:250, Clone SXM30, BD Pharmingen). β-actin (1: 8000, monoclonal anti-β-actin, clone AC-74, Sigma) was used as a loading control. Polyclonal anti-mouse horseradish peroxidase (HRP)-conjugated antibody was used as a secondary antibody (1:1000, Dako Cytomation, Glostrup, Denmark). Detection was performed using enhanced chemiluminescent detection system (ECL, GE Healthcare). Quantification of band intensities were performed using GS 800 densitometer and Quantity One software (both from Bio-Rad Laboratories).
Microarrays
RNA was extracted using Qiagen AllPrep DNA/RNA Mini Kit (Qiagen, CA) and a Qiagen supplementary protocol, Purification of total RNA containing miRNA from animal cells using the RNAeasy Plus Mini Kit, according to the vendor's instructions. Extracted DNA was stored for later use and was not used in this study.
Labeled RNAs (800 ng/sample) were hybridized onto Agilent SurePrint G3 Human GE 8 × 60 K, and then the slides were washed and scanned according to the manufacturer's recommendations. The raw data files (.txt files) were imported into R version 2.13 software (42) and preprocessed by the BioConductor package limma version 3.4.5 (43). After quality control of the data, the median probe intensities were log2-transformed and normalized according to the method of the quantiles (44). The probes for the same Entrez Genes or lincRNAs (as of 1 January 2012) were averaged.
A linear model including the AGE*PASSAGE + SUBJECT + DYE terms followed by a moderated t-test was utilized for finding the differentially expressed genes (log2 fold change > 0.58) in the comparisons of interest (nominal P-value < 0.01). The differentially expressed genes were illustrated with violin plot by using R package vioplot with height argument 0.4 and analyzed for significant enrichments of GO-BP classes by using R library GOSim version 1.2.5 (45). Enrichments with P-value < 0.01 were considered significant. Default parameters were used in the GO analysis.
Statistical analysis
The data were analyzed in both univariate and multivariate ways. In the univariate approach, each GPL species or individual FA was analyzed separately and presented in figures as mean + SD. Statistical significances for the differences between hBMSCs from the young and old donors or early and late passages were calculated using Student paired t-test (*P < 0.05, **P < 0.01, ***P < 0.001). In addition, the changes in each lipid percentage were analyzed with linear mixed-effects models, fitting a model with fixed terms for age and passage and a random effect for cell line (a repeated measures type analysis). The effect of each term for every lipid was estimated using normal F-test P-values and visualized with interaction plots depicting the mean values in each passage and group. Software for statistical analysis was R version 2.12, using package “nlme” for a mixed-effects analysis.
In the multivariate approach, the lipidome data were subjected to principal components analysis (PCA) to study differences between the samples in terms of the whole GPL profile and to find out which GPL species were mainly responsible for the variation in the data. PCA was computed using log10 normalized data, and the relative positions of the samples and variables were plotted using the first two principal components. In addition, quantitative multivariate measures of the differences among the sample groups were determined by soft independent modeling of class analogy (SIMCA) (46).
RESULTS
Cultivated hBMSCs reached replicative senescence, and the cells from old donors showed larger variation in their growth potential
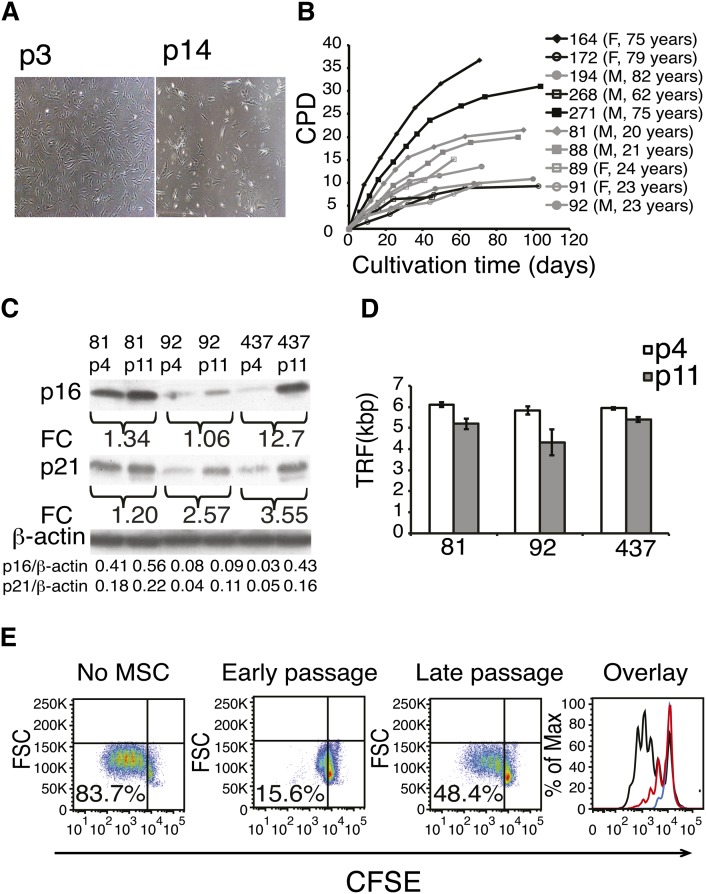
hBMSCs isolated from five elderly donors (62–82 years of age, mean 74.6 years) and five young adult donors (20–24 years of age, mean 22.2 years) were characterized and shown to fulfill the established minimal criteria for MSCs (47) (supplementary Fig. I). At the end of the cell culture (45–108 days, depending on the donor), the hBMSCs adopted typical senescent morphology (“fried egg”) (Fig. 1A, right). The proliferation capacity of the primary hBMSCs was assessed by determining the cumulative population doublings as cells were passaged until senescence. The hBMSCs from old donors showed greater individual variation in their growth kinetics than those from young donors (Fig. 1B). It should be noted that the cell doublings before passage 4 were not included in the calculations. The replicative senescence was further studied from selected samples and passages. The replicative senescence enhanced the expression of cell-cycle components p16INK4A and p21CIP1/WAF1 (Fig. 1C). Additional proof was obtained from telomere length measurements. Telomere length in passage 11 cells was 1.0 ± 0.5 kbp shorter than that in passage 4 (Fig. 1D, supplementary Fig. II).
Fig. 1.
Long-term cell culture of hBMSCs. A: A representative example of cell morphology at passage 3 and in senescence (p14) at a magnification of 40×. B: hBMSCs were isolated from the human bone marrow of ten donors and cultured until senescence was reached. Cumulative population doublings (CPD) was calculated in relation to the starting point of the cell culture at every passage (F, female; M, male). C: Western plot analysis of cell cycle components p16 and p21 of early and late passages of three different hBMSC donors. Fold change (FC) represents the increase of band intensities in late-passage cells compared with early-passage cells. Intensities of the p16 and p21 bands normalized to β-actin are listed on the last rows. D: Telomere lengths (TRF, terminal restriction factor) were determined in early- and late-passage cells from three different cell lines. Results are show as mean + SD of three technical replicates. Telomere lengths were shortened on average 1 kb ± 0.5 (Student t-test P = 0.03, n = 3). E: Immunosuppressive capacity of early and late passages analyzed by coculture assay. 5(6)-Carboxyfluorescein diacetate N-succinimidyl ester (CSFE)-labeled peripheral blood mononuclear cells (PBMC) were cultured with hBMSC in ratio of 1:10 for four days. T-cell proliferation was stimulated with monoclonal CD3 antibody, and proliferation was analyzed as CFSE dilution by flow cytometry. In the overlay panel, black line indicates no hBMSCs, red line indicates late-passage hBMSCs, and blue line indicates early-passage hBMSC. X-axis shows the fluorescence intensity of CFSE on a logarithmic scale, and Y-axis shows the cell count. Experiment repeated three times with similar results using hBMSCs and PBMCs from independent donors.
GPL profiles of hBMSCs from young and old donors
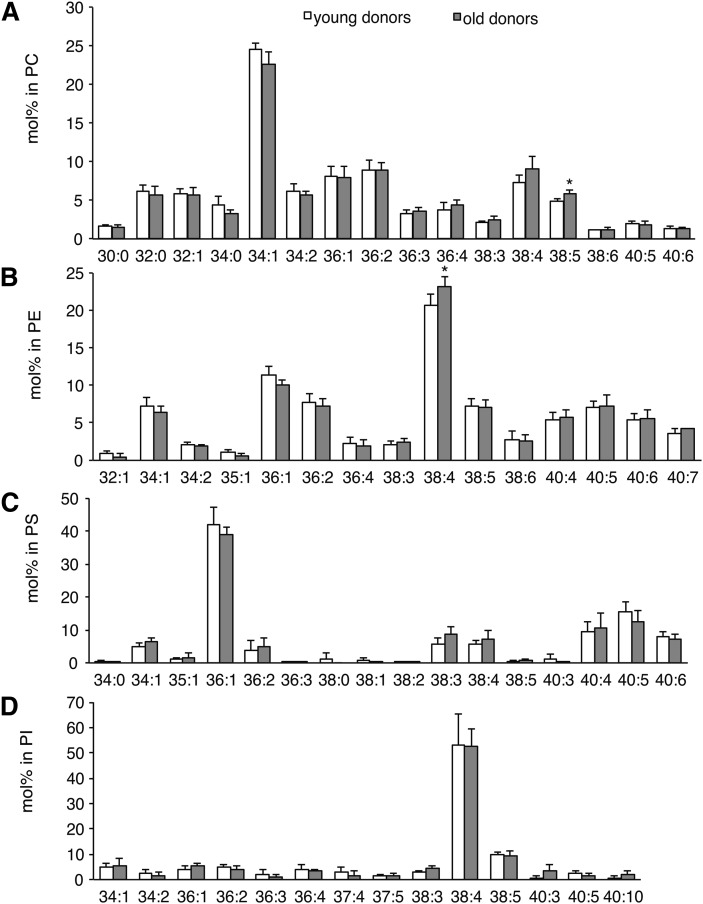
The average values for GPL class totals for hBMSCs were PC, 41–46%; PE, 34–38%; PS, 5–8%; and PI, 4–8% (Table 1). In general, the hBMSCs from the young and old donors had very similar class profiles. The GPL species of the hBMSCs (Fig. 2) showed that the major PC species (in descending order) were 34:1, 36:2, 36:1, and 38:4 (Fig. 2A), whereas the most abundant PE species were 38:4, 36:1, 36:2, 40:5, 38:5, and 34:1 (Fig. 2B). The predominant PS species were 36:1, 40:5, and 40:6 (Fig. 2C), and a single species, 38:4, accounted for almost 50% of the total PI (Fig. 2D). In the early-passage samples, statistically significant differences (P < 0.05) due to the donor age were found for PC38:5 and PE38:4 (Fig. 2A, B). The levels of PE alkenyl and PC alkyl species also changed during the long-term passaging (Table 1).
TABLE 1.
Molecular percentage of lipid classes determined by ESI-MS
| Young Donors | Old Donors | |||
| Lipid Class | Early Passage | Late Passage | Early Passage | Late Passage |
| PC diacyl | 37.3 ± 2.7 | 37.4 ± 3.4 | 35.5 ± 2.4 | 39.0 ± 3.7 |
| PC alkyl | 5.8 ± 0.3 | 5.1 ± 0.4** | 5.3 ± 0.5 | 6.6 ± 0.8* |
| PE diacyl | 20.4 ± 1.1 | 15.1 ± 2.1* | 19.9 ± 3.8 | 17.2 ± 3.4 |
| PE alkenyl | 16.1 ± 2.1 | 22.7 ± 4.7* | 16.8 ± 2.6 | 16.5 ± 1.9 |
| PI | 3.9 ± 1.2 | 7.7 ± 1.7* | 5.4 ± 1.3 | 8.2 ± 3.4 |
| PS | 8.0 ± 4.8 | 4.9 ± 1.1 | 5.3 ± 1.1 | 5.9 ± 2.1 |
| PI/PS | 0.5 ± 0.2 | 1.6 ± 0.4* | 1.0 ± 0.1 | 1.4 ± 0.5 |
| lysoPC | 0.5 ± 0.1 | 1.5 ± 0.9 | 1.2 ± 0.5 | 1.2 ± 0.6 |
The totals for lipid classes were calculated by summing individual lipid species of each class. The statistical difference between the young and the old donors was calculated using Student two-tailed t-test (paired). *P < 0.05, **P < 0.01.
Fig. 2.
Species composition in GPL classes of hBMSCs as determined by ESI-MS. The individual lipid species were quantified using appropriate internal standards. The values for GPL species were expressed as molar percentage per the total of detected lipids in a given class. PC (A), PE (B), PS (C), and PI (D) species from passage 4 are shown. Results are the mean + SD (n = 5) for both the young (white bars) and old (gray bars) donors (*P < 0.05).
Expansion of hBMSCs increases membrane PI content in relation to PS
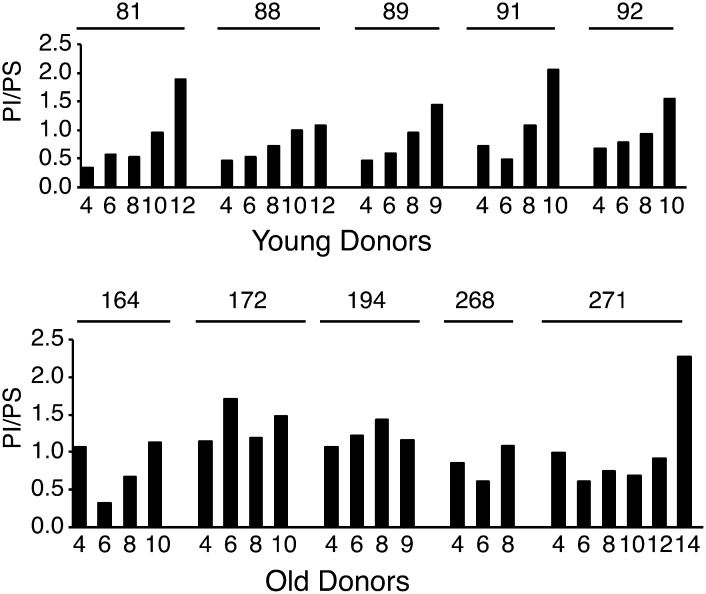
During long-term cultivation, PI totals increased, especially in the cells from young donors (Table 1). This increase of PI became even clearer when the PI totals were studied in relation to the PS totals. In the hBMSCs from the old donors, the PI/PS ratio was already high at the earliest passage (Fig. 3). Interestingly, lysoPC totals also increased consistently during cell expansion of the hBMSCs from young donors but showed varying responses in the hBMSC from old donors (supplementary Fig. III).
Fig. 3.
The ratio of PI to PS from early to late passages of the hBMSC. hBMSCs were cultured from passage 4 to senescence. Cells were harvested for GPL analysis from every other passage. The GPL species were analyzed after extraction by direct infusion ESI-MS. The PI and PS class totals were obtained by summing the amounts of individual species in each class, and subsequently the ratio of PI to PS was calculated. Young donors, n = 5. Old donors, n = 5.
Expansion of hBMSCs increases 20:4n-6-containing GPL species at the expense of MUFA and n-3 PUFA-containing species
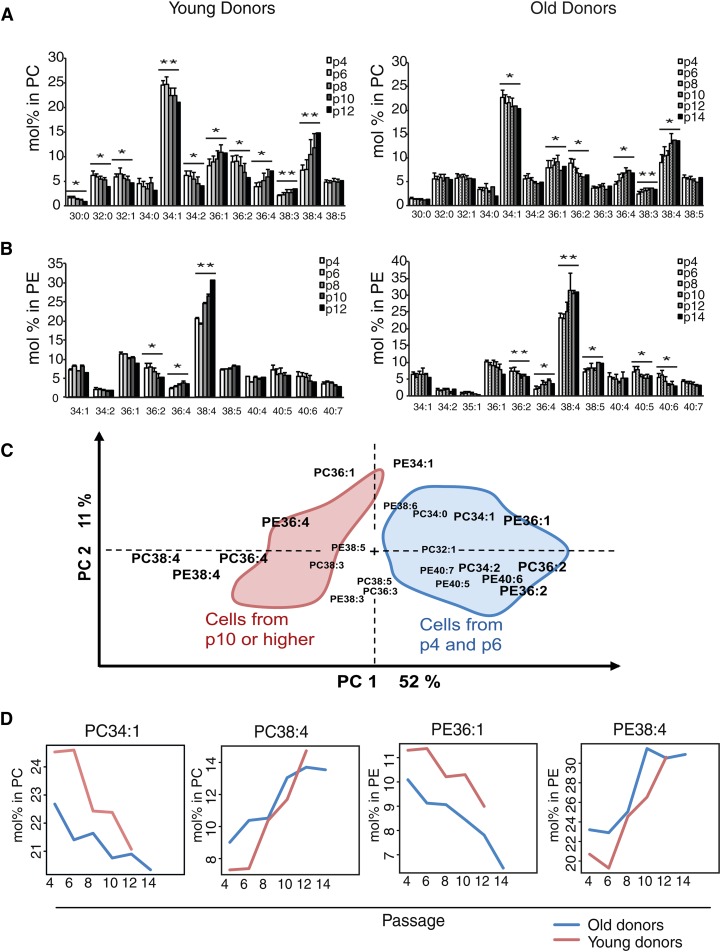
Expansion of the hBMSCs induced alterations in the relative amounts of numerous individual lipid species, as revealed by ESI-MS. For instance, in the PC diacyl species, the molar percentage of 38:4, 36:4, and 36:1 increased toward the latest passages, while the molar percentage of 34:1, 34:2, and 36:2 decreased (Fig. 4A, B). The PE diacyl species profiles exhibited an increase in 38:4 and 36:4 species and a decline in the monounsaturated species (34:1 and 36:1) and in those containing n-3 PUFAs (40:5, 40:6, and 40:7). The most dramatic change was seen in the PC and PE diacyl species 38:4. Over all the passages, the levels of PC34:1 and PE36:1 species in the hBMSCs from the young donors were consistently higher than those from the old donors (Fig. 4C). These changes were further highlighted when multivariate statistical analysis (PCA) indicated the 38:4 and 36:4 species of PC and PE (with 20:4n-6) as the ones explaining the greatest part of the compositional variation among all the studied hBMSCs (Fig. 4D). In the PCA biplot, these 20:4n-6-containing species had an inverse relationship with the mono- and diunsaturated ones. The principal component 1 explained up to 52% of the variation among the samples.
Fig. 4.
Effect of long-term cell culture on the PC and PE species of hBMSC. hBMSCs were cultured from passage 4 (p4) to senescence. Cells were harvested for GPL analysis from every other passage. The GPL species were analyzed after extraction by direct infusion ESI-MS, and their values were expressed as molar percentage per the total of detected lipids in a given class. Results are the mean + SD of hBMSCs from young (left panel) and old (right panel) donors. Paired Student t-test was performed between the earliest and last passages. (*P < 0.05, **P < 0.01). A: PC. B: PE. C: Multivariate PCA using PC and PE data as loadings. For simplicity, the areas covering the positions of the individual sample points are shown. D: Mean values of PC34:1, PC38:4, PE36:1, and PE38:4 in both groups over passages.
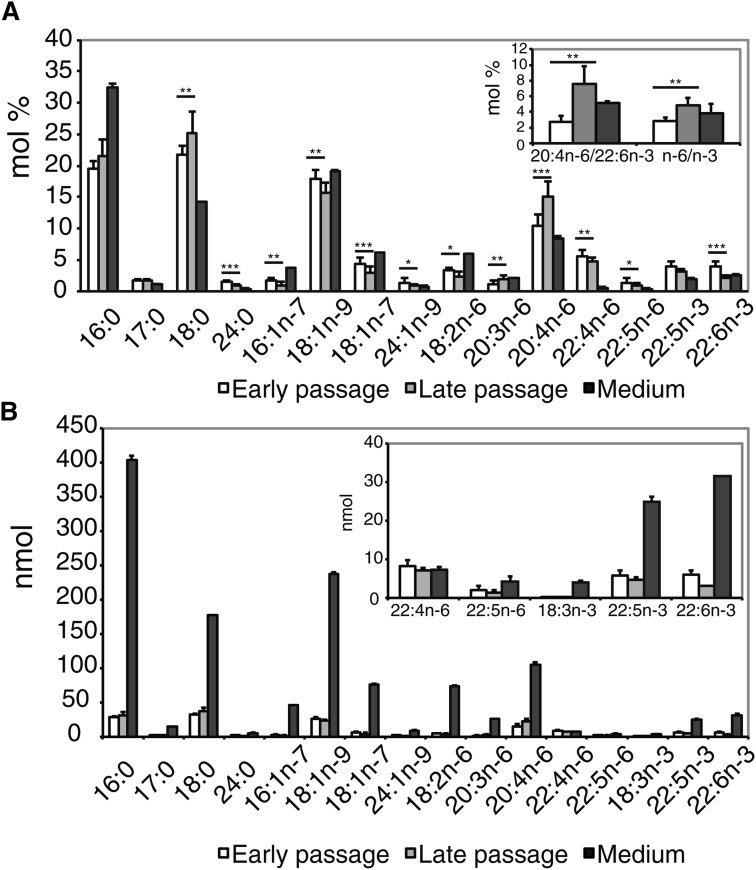
The FA profiles by GC showed that the proportions of individual saturated FAs (SFA), such as 18:0, increased toward the late passages, whereas individual monounsaturated FAs (MUFA) decreased (Fig. 5A). Among n-6 PUFAs, 20:4n-6 increased clearly and constantly throughout the expansion along with 20:3n-6, while other principal n-6 PUFAs decreased (Fig. 5A, supplementary Fig. IV). At the same time, the two prominent n-3 PUFAs, 22:5n-3 and 22:6n-3, decreased, the response being clearer for 22:6n-3 (Fig. 5A, supplementary Fig. IV). Consequently, the ratios of 20:4n-6 to 22:6n-3 and the total n-6 PUFA to the total n-3 PUFA were significantly increased from passage 4 to the last passage studied (Fig. 5A, insert). Thus, the clearest finding in the PC and PE species profiles, the increases of 38:4 and 36:4, was supported by the observed clear accumulation of 20:4n-6 in the cellular total FA pool. Compared with the medium in the early-passage samples, the relative levels (mol%) of 16:0 and MUFAs (16:1n-7, 18:1n-7, 18:1n-9) were lower, but those of 18:0 and PUFAs (except linoleic acid, 18:2n-6) were higher (Fig. 5A). In addition to the relative levels, we further studied the total amount of FAs in the cells on the culture dish and the amount of FAs available for the cells in the surrounding medium. The results demonstrated that, in the medium, FAs (including the essential PUFAs of the n-6 and n-3 families) were available in excessive amounts compared with the cells (Fig. 5B).
Fig. 5.
Effect of long-term culture on the FA composition of hBMSC. Samples for gas chromatographic FA analysis were collected as described in Fig. 4. FAs were analyzed as methyl esters. Results for the cells from the earliest and last passages were expressed as mean + SD, n = 10 (*P < 0.05, **P < 0.01, ***P < 0.001). For the medium, three replicates were analyzed. A: Molar percentages of FAs from the earliest passage (white bar) and the last passage (light-gray bar) compared with those of the medium (dark-gray bar). Insert shows the ratios of 20:4n-6 to 22:6n-3 and n-6 PUFAs to n-3 PUFAs in the earliest and last passages compared with values of the medium. B: Total amounts (nmol) for each FA in the cells and surrounding culture medium. Insert shows the total amounts (nmol) of some PUFA species in enlarged scale.
Immunosuppressive capacity of hBMSCs decreases during cell expansion and correlates with the lipid changes
Using a coculture assay, we next studied the ability of expanded hBMSCs to suppress T-cell proliferation. The early-passage hBMSCs inhibited T-cell proliferation almost completely (Fig. 1E, blue line); the suppression was less complete when the late-passage hBMSCs were used (Fig. 1E, red line). To further study the correlation between immunosuppressive capacity and the identified lipid or fatty acid indicators, we analyzed the GPLs and FAs of hBMSCs from eight donors. These cells (from passage 4) had diverse functionality. Convincing correlation with the functionality was observed for PC species (38:4, 38:5, 34:1 and the ratio of 38:4/34:1) and fatty acids (20:4n-6, 22:6n-3 and the ratio of the two) (Table 2).
TABLE 2.
Regression analysis of correlation between immunosuppression capacity and the best lipid indicators in hBMSCs
| Lipid or Fatty Acid Indicator | Correlation Coefficient | R2 | P | Value Range (mol%) |
| PC38:4(chains 18:0, 20:4n-6) | −0.91 | 0.84 | 0.001 | 4.7–7.7 |
| PC38:5(chains 18:1, 20:4n-6) | −0.82 | 0.68 | 0.012 | 4.2–6.3 |
| PC34:1(chains 16:0, 18:1) | +0.80 | 0.64 | 0.017 | 23.5–27.0 |
| PC38:4/PC34:1 | −0.90 | 0.81 | 0.003 | 0.17–0.33 |
| 20:4n-6 | −0.71 | 0.50 | 0.050 | 6.1–9.6 |
| 22:6n-3 | +0.75 | 0.56 | 0.032 | 2.0–3.3 |
| 20:4n-6/22:6n-3 | −0.76 | 0.58 | 0.029 | 2.2–4.9 |
The hBMSCs (n = 8, all cultured until passage 4) suppressed T- cell proliferation in a coculture assay (degree of inhibition with hBMSC:PBMC ratio 1:20; range, 70–88%) and relative amounts of selected lipid and fatty acid indicators or their ratios in the cells.
Gene expression differences related to donor age and cell expansion
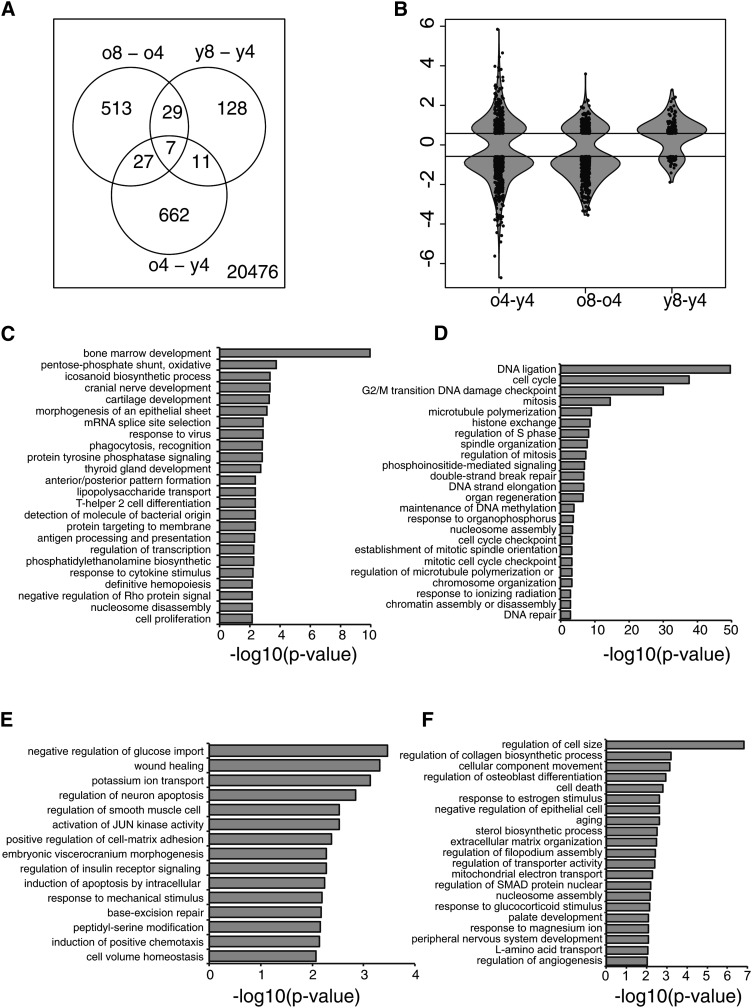
Next, we made a gene expression analysis of the early-passage (4) and late-passage (8) cells. Measuring the compositional distances for the samples from different passages by SIMCA showed that, in terms of the GPL profile, all passages 4 and 6 samples were statistically significantly different from all samples from passages 10 and higher (P < 0.05). As passage 8 represented a time point preceding changes in the metabolite profile, cells from this passage were chosen for the gene expression analysis. Altogether, 707 genes were found to be differently expressed between the old and young donor samples in passage 4 (Fig. 6A). Interestingly, during expansion, the hBMSCs from the old donors showed almost five times more gene expression changes (576 genes) than the cells of the young donors (175 genes). Only 36 were shared between the young and old donors (Fig. 6A). In addition, the variance in the fold changes was the greatest (i.e., 2.3) when comparing the old and young donors. The fold change variances due to cell expansion were 1.4 for the old donors and 0.7 for the young donors. Thus, cell expansion seemed to affect gene expression more in the cells of the old donors than in those of the young donors (Fig. 6B).
Fig. 6.
Differential gene expression of hBMSCs. Differentially expressed genes between old and young donor hBMSCs in the earliest passage 4 (o4-y4) and between passages 8 and 4 of old (o8-o4) and young (y8-y4) donor hBMSCs were compared. Cut-off values 0.58 (upregulated genes) and −0.58 (downregulated genes) were used. A: Venn diagram demonstrating the separate and overlapping genes in each group. B: Violin plot combines boxplot with kernel density plot and is used to illustrate fold change variations and the amount of differentially expressed genes in the above-mentioned comparisons. C–F: GO enrichment analysis of differentially expressed genes. C: Genes with higher expression level in old (o4-y4). D: Genes in which expression decreased during cultivation (o8-o4). E: Genes with higher expression level in young (o4-y4). F: Genes in which expression induced during cultivation (o8-o4). The Y axis shows the GO term while the X axis shows −log10 P-values for the enriched GO terms. For clarity, several GO terms have been excluded based on their similarity. mRNA data are available in GEO database http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE39035 with GEO accession number GSE39035.
We further investigated the enrichment of differentially expressed genes in different biological processes. In the old donors, gene ontologies (GO) relating to lipid metabolism and immunological processes as well as differentiation and developmental functions were downregulated (Fig. 6C), whereas GO related to adhesion, signal transduction, and apoptosis were upregulated (Fig. 6D). The expansion-induced changes (old donors) relating to cell division, aging, and apoptosis were upregulated (Fig. 6E), whereas the downregulated GO terms included sterol biosynthetic processes, development, and aging (Fig. 6F). Results from the young donor group were similar but less distinct (supplementary Table VI).
As revealed by GO analysis, the expression of several genes related to lipid metabolism changed. Both ethanolamine kinase 2 and choline kinase β had lower expression levels in the hBMSCs from the old donors than in those from the young donors. Scavenger receptor B1 had lower expression level and scavenger receptor B3 (also known as CD36) had higher expression level in old donors. Interestingly gene expression of phospholipid scramblase 1 was lower and putative aminophospholipid translocase (ATP10A) was higher in the hBMSCs of the old donors. Further, the gene expression of phospholipid scramblase 4 was elevated during expansion. Gene expression of several other putative aminophospholipid translocases differed between young and old donors and further during expansion (supplementary Tables III–V). Further, inositol triphosphate receptor 1 gene expression was elevated during expansion. The gene expression of enzymes 1-acylglycerol-3-phosphate-O-acyltransferase 2 and serine palmitoyltransferase were elevated during expansion. Five genes for FA metabolizing enzymes were differently expressed (Table 3). Among them, the expression levels of three FA desaturase genes were lower in the cells of the old donors (i.e., FADS1, FADS2,and SCD, coding for fatty acid Δ5-, Δ6-, and Δ9-desaturases, respectively). LysoPC acyl transferase genes LPCAT2 and LPCAT3 also had lower expression levels in old donors. In addition, the gene producing cyclooxygenase 1 enzyme [COX1, also known as prostaglandin endoperoxide synthase (PTGS1)] and gene producing prostacyclin receptor (PTGIR, also known as prostaglandin I2 receptor) were more expressed in passage 8 compared with passage 4. Finally, leukotriene C4 synthase and ELOVL fatty acid elongase 3 were expressed at lower levels in the cells of the old donors compared with those of the young donors.
TABLE 3.
Differentially expressed genes
| Mean Fold Change | ||||
| Description | HGNC | Gene ID | o4-y4 | o8-o4 |
| Lipid metabolism | ||||
| 1-acylglycerol-3-phosphate O-acyltransferase 2 | AGPAT2 | 10555 | 0.6 | |
| ATPase, class V, type 10A (aminophospholipid translocase) | ATP10A | 57194 | 0.7 | |
| Choline kinase β | CHKB | 1120 | −1.2 | |
| CD36 molecule (thrombospondin receptor)] | CD36 | 948 | 4.4 | |
| ELOVL fatty acid elongase 3 | ELOVL3 | 83401 | −1.0 | |
| Ethanolamine kinase 2 | ETNK2 | 55224 | −0.8 | |
| Fatty acid desaturase 1(delta -5–desaturase) | FADS1 | 3992 | −1.4 | |
| Fatty acid desaturase 2(delta-6-desaturase) | FADS2 | 9415 | −1.6 | |
| Fat storage-inducing transmembrane protein 2 | FITM2 | 128486 | 0.7 | |
| Inositol 1,4,5-trisphosphate receptor, type 1 | ITPR1 | 3708 | 1.0 | |
| Leukotriene C4 synthase | LTC4S | 4056 | −1.3 | |
| Lysophosphatidylcholine acyltransferase 2 | LPCAT2 | 54947 | −2.2 | |
| Lysophosphatidylcholine acyltransferase 3 | LPCAT3 | 10162 | −0.7 | |
| Phospholipid scramblase 1 | PLSCR1 | 5359 | −0.9 | |
| Phospholipid scramblase 4 | PLSCR4 | 57088 | 1.1 | |
| Prostaglandin-endoperoxide synthase 1 | PTGS1 | 5742 | −0.7 | |
| Prostaglandin I2 (prostacyclin) receptor (IP) | PTGIR | 5739 | 0.7 | |
| Scavenger receptor class B, member 1 | SCARB1 | 949 | −1.9 | |
| Serine palmitoyltransferase, long chain base subunit 3 | SPTLC3 | 55304 | −0.6 | |
| Stearoyl-CoA desaturase (delta-9-desaturase) | SCD | 6319 | −2.4 | 1.1 |
| Immune response | ||||
| Alpha-2-macroglobulin | 2 | −3.8 | ||
| Ankyrin repeat and SOCS box containing 2 | ASB2 | 51676 | −1.2 | |
| Ankyrin repeat and SOCS box containing 5 | ASB5 | 140458 | 1.0 | |
| Ankyrin repeat and SOCS box containing 10 | ASB10 | 136371 | −0.9 | |
| Bradykinin receptor B1 | BDKRB1 | 623 | −3.9 | |
| CD74 molecule, major histocompatibility complex, class II invariant chain | CD74 | 972 | −0.7 | |
| Complement component 3a receptor 1 | C3AR1 | 719 | 1.0 | |
| Interferon (α, β and omega) | IFNAR2 | 3455 | −0.9 | |
| Interleukin 6 | IL6 | 3569 | 0.7 | |
| Interleukin 26 | IL26 | 55801 | −1.4 | |
| Linker for activation of T cells family member 2 | LAT2 | 7462 | −0.9 | |
| Major histocompatibility complex, class II, DM α | HLA-DMA | 3108 | −2.0 | |
| Major histocompatibility complex, class II, DM β | HLA-DMB | 3109 | −2.0 | |
| Major histocompatibility complex, class II, DR α | HLA-DRA | 3122 | −2.1 | |
| Suppressor of cytokine signaling 1 | SOCS1 | 8651 | −0.6 | |
| Suppressor of cytokine signaling 3 | SOCS3 | 9021 | −1.0 | −0.8 |
| Transforming growth factor, β receptor 1 | TGFBR1 | 7046 | 1.2 | |
Differentially expressed genes related to lipid metabolism and immune response. o4-y4, comparison between old and young donor samples in passage 4; o8-o4, comparison between old donor samples in passages 8 and 4.
As an indicator for the modulation of immune functions, the genes for the suppressors of cytokine signaling (SOCS1 and SOCS2) and several genes from ASB family (Ankyrin repeat and SOCS box-containing proteins) were less expressed in the old donors; interestingly, ASB5 expression was increased during cell expansion (Table 3). The expression of the immunologically important COX-1 gene PTGS1 was downregulated in the late-passage cells. Interestingly, both TGFBR1 and IL6 were upregulated in the late-passage cells. Other important genes related to the immune system, such as bradykinin receptor 1, HLA-DMA, HLA–DMB, HLA-DRA, CD74, and LAT2, were less expressed in the early-passage cells of the old donors.
During passaging in the hBMSCs of both young and old donors, the predominant gene expression changes were related to cell cycle, chromatin assembly, and DNA integrity (i.e., histone cluster genes; see supplementary Table II). Importantly, of those 36 genes changing during passaging in both young and old donors, 12 were histone cluster genes (Fig. 6A).
DISCUSSION
This study focused on the membrane GPL and total FA composition of hBMSCs. The role of the donor's age, individual variation, cell expansion, and FA composition of the surrounding medium were taken into account. Since no previous studies on MSC phospholipid profiles have been reported, we compared our results on the lipid profiles of hBMSCs with the previously published profiles of functionally very similar human primary fibroblasts (21, 48). Our study demonstrates that at the lipid class level (Table 1), the relative amounts of PC, PS, and PI of the hBMSCs were almost identical to that described for human fibroblasts. However, the relative amount of PE was much higher in the hBMSCs than was the amount reported for fibroblasts (21) or mammalian cells on average (49). The predominant PE species of the hBMSCs was 38:4 (18:0/20:4n-6), whereas in human fibroblasts, PE species 36:1, 38:4, and 36:2 were present in equal amounts (21). This implies that biosynthetic or remodeling pathways of the GPLs in these two cell types have differences. These differences can be due to altered activities of CoA-independent transacylase (CoA-IT), which in immunologically activated cells promotes transfer of 20:4n-6 from PC to PE, from which different PLA2 readily release 20:4n-6 to be used for production of lipid mediators (50, 51). In addition, the higher expression level of the PE biosynthetic enzyme ethanolamine kinase 2, found in the cells of the young donors (Table 3), may be linked to an increase in the relative amounts of PE ether lipids (Table 1).
Expansion caused further enrichment of 20:4n-6-containing PC and PE species (38:4 and 36:4), which happened at the expense of n-3 PUFA-containing species (40:5, 40:6, and 40:7) and short-chain saturated and monounsaturated species of PC and PE (Fig. 4A, B). It is possible that this increase in the ratio of GPL species with n-6 PUFA to those with n-3 PUFA had consequences for the functionality of the cells (i.e., their responsiveness to inflammatory signals and capability for immunomodulation) (52). If cleaved by activated phospholipases, the different PUFAs give rise to signaling molecules with different, even opposing functional consequences (53, 54). This hypothesis was supported by the results obtained from the T-cell suppression assay in which we clearly showed that the cells lose part of their immunosuppressive potential when expanded (Fig. 1E).
The analysis of FA composition confirmed the central feature that aging hBMSCs gained 20:4n-6 and lost 22:6n-3 and other n-3 PUFAs. The 20:4n-6 is a precursor for several pro- and also anti-inflammatory molecules, such as PGs [cyclooxygenase (COX) pathway] or lipoxins [lipoxygenase (LOX) pathway] (29). Because the rate of COX catalyzed synthesis of PGs is known to be affected by the local availability of 20:4n-6 and as the expression level of the COX-1 gene (PTGS1) was elevated in the passage 8 samples, it is plausible that the production of 20:4n-6-derived proinflammatory eicosanoids is facilitated in aging cells (55). As described earlier by others, the inducible form of cyclooxygenase, COX-2 gene (PTGS2), is highly expressed in hBMSCs (56). Our microarray data confirmed PTGS2 expression but did not show statistically significant differences. Due to the low gene expression levels of LPCAT2 and 3, the recycling of 20:4n-6 to PC may have been impaired in the cells of the old donors, thus strengthening the shift to eicosanoid production. The connection to eicosanoid production is in line with the previously proposed role of eicosanoids in hMSC immunomodulative properties (7) and further supported by our observation that during expansion and simultaneous increment of 20:4n-6 content, hBMSCs lose part of their capacity to suppress T-cell proliferation (Fig. 1E). Similarly, we found convincing negative correlations between the immunomodulatory capacity of hBMSCs from eight donors and the relative amounts of 20:4n-6-containing species in PC or the relative amount of 20:4n-6 in total FAs of the cells (Table 2). This encourages for further studies of the immunomodulative effector lipids affecting stem cells.
The composition of the total PUFA pool available in the cells (but not in the medium) (Fig. 5A, B) dictated the species profiles of membrane phospholipids and may also have affected the proportions of different GPL classes in the membranes (e.g., the ratio of PI/PS) (Fig. 3). The increase of the PI/PS ratio during expansion may be a direct consequence of the change in the cellular PUFA status due to the substrate preference in the PS biosynthesis for 22:6n-3 and in PI synthesis for 20:4n-6 (57, 58).The change in PI and PS levels is biologically highly important as both of these anionic GPLs are concentrated on the inner leaflets of membranes where they activate proteins and take part in signaling events (59). Anionic lipids and PIPs act as potent activators of cPLA2 releasing 20:4n-6 and other PUFAs (60). In addition, PI, PIPs, and PS normally function in concert to target protein kinases, such as PKC, Akt, sphingosine kinase, and 3-phosphoinositide-dependent kinase-1, to membranes for activation and signaling (61–64). PIP-derived and phospholipase C-liberated DAG and PS activate the PKC, but their roles are different. Even small concentrations of DAG turn PKC on, but full enzymatic activity is achieved by PS (65). Thus, changes in the ratio of PI/PS may have profound effects on hBMSC therapeutic functions via the regulation of the activities of kinases, phospholipases, and/or other proteins. We also emphasize that PIPs regulate the scavenger receptor B1 (SR-B1) of lipoprotein trafficking (66), and in our study, SR-B1 was expressed at a higher level in the hBMSCs of the young donors. Interestingly, we found that gene expression of several ATPases was altered. ATP10A, one of the putative aminophospholipid translocases, and bidirectional phospholipid translocator scramblase 1 were expressed at higher levels in the hBMSCs of the old donors. Furthermore, the gene expression of scramblase 4 was increased during expansion (67, 68) Thus our data indicate physiologically important GPL remodeling with functional consequences in the hBMSCs.
In resting cells, the amount of free PUFAs is strictly regulated by lysophospholipid acyltransferases that recycle most of the cleaved PUFAs back to the membrane GPLs. Only a small fraction of PUFAs is used for the synthesis of the eicosanoids and other signaling molecules (69, 70). However, the stimulation of the cells results in the activation of cPLA2, and due to the high rates of PUFA liberation, the shift to the production of the signaling molecules with biological activity is enforced (71). The increased concentrations of lysoPC, detected especially in the late-passage hBMSC samples of the young donors, are likely to reflect enhanced PLA2 activity (several forms of the enzyme may be activated) and represent the pool of acceptor lysophospholipids (72, 73), thus giving lysophospholipid acyltransferases a role in the regulation of inflammation (74). In this respect, the hBMSCs from the young and the old donors had different metabolism: lysoPC acyltransferases had higher expression levels and lysoPC totals also increased during expansion more noticeably in the hBMSCs of the young donors compared with the old donors (supplementary Fig. III). Additionally, lysoPC contributes to cellular signaling by activating G-protein-coupled receptors, ERK and MAPK signaling, and phospholipase C (75–77). The latter releases DAG and inositol triphosphate with the liberation of intracellular Ca2+ and activation of PKC. The COX enzymes reside in endoplasmic reticulum close to cPLA2 and are likely to convert mobilized 20:4n-6 and other PUFAs into eicosanoids (54). Consequently, less free FAs may have been recycled to PC by the lysoPC acyltransferase, which could then be observed as the lysoPC acceptor accumulation.
If the relative increase of 20:4n-6-containing GPLs of the hBMSCs have functional consequences, we can assume that the simultaneous decrease of n-3 PUFA content is of immunological importance as well. Recently, the so-called traditional eicosanoids have been found to be accompanied by resolvins, protectins, and maresins, which are derivatives of 22:6n-3 and 20:5n-3 and play a role in the resolution phase of the inflammation (29).The enzymes COX and LOX synthesize from the n-3 PUFA counterparts to the n-6 PUFA-derived eicosanoids, which, however, have mainly anti-inflammatory properties (78). In T-cells, the n-3 PUFAs have been found to suppress immune functions by modulating interleukin-2 expression and exerting their effects via PKC isoforms and nuclear factor pathways (79). Thus, the shift from a more balanced n-6/n-3 PUFA ratio toward the dominance of n-6 PUFAs found in the total FA pool of the aging hBMSCs may have altered the lipid signaling pathways from an anti- to proinflammatory direction (Fig. 5). Due to the constant excess amount of FAs in the surrounding medium compared with the cells (Fig. 5B), we can assume that the observed changes in hBMSC lipidome were due to an active process of the replicative senescence verified by several different markers (Fig. 1A–D). However, it is possible that supplementing the culture medium with even more n-3 PUFAs than present in standard growth medium might support hBMSCs growth and maintain their physiological functionality for longer without manifestations of senescence. Indeed, it is fascinating and important to highlight that different GPL/FA compositions may cause the hBMSCs to respond very differently to the same external stimuli and that, depending on the GPL/FA substrate, the same enzymatic pathways produce different end-product eicosanoids with completely different, even opposing, biological functions (55). The PUFA alterations reported here (Figs. 4 and 5) presumably affect the composition of end products involved in activation or resolution of inflammation, linked to the changes in T-cell suppression capacity (Fig. 1E). Thus, our results will serve as a basis for further studies aiming to understand the lipidomic aspects of the MSC immunosuppression.
This study is the first to show major changes in phospholipid profiles and total FAs during expansion and senescence of therapeutically important hBMSCs. We demonstrate that the 20:4n-6 contents of cellular lipids increased while the n-3 PUFA contents decreased during long-term cultivation. The gene expression differences were most notable between the early-passage cells from the old and the young donors and supported our lipid findings. Our results suggest that the free PUFAs derived from membrane lipids are of high importance in hBMSC immunological functionality. Because PUFA-derived signaling lipids are known to be involved in MSC functionality but are extremely challenging to use as biomarkers, biosynthetic precursors in membranes could be used instead as indicators of cell functionality. In combination with additional markers, such as mRNA of lipid enzymes and/or other functional factors, membrane lipids can serve as powerful new biomarkers for the functionality of therapeutic cells.
Supplementary Material
Acknowledgments
The authors thank Birgitta Rantala and Lotta Sankkila for excellent technical assistance and Tanja Kaartinen for advice in T-cell proliferation assay.
Footnotes
Abbreviations:
- AA
- arachidonic acid
- CFSE
- 5(6)-Carboxyfluorescein diacetate N-succinimidyl ester
- CoA-IT
- CoA-independent transacylase
- COX
- cyclooxygenase
- cPLA2
- cytosolic phospholipases A2
- DAG
- diacylglycerol
- DHA
- docosahexaenoic acid
- GPL
- glyserophospholipid
- GO
- gene ontology
- hBMSC
- human bone marrow mesenchymal stromal cell
- hMSC
- human mesenchymal stromal cell
- LOX
- lipoxygenase
- miRNA
- micro ribonucleic acid
- MSC
- mesenchymal stromal cell
- PBMC
- peripheral blood mononuclear cell
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PG
- prostaglandin
- PGE2
- prostaglandin E2
- PI
- phosphatidylinositol
- PIP
- phosphorylated PI
- PS
- phosphatidylserine
- SFA
- saturated fatty acid, TRF, terminal restriction fragment
This work was partly supported by the SalWe Research Program of Intelligent Monitoring of Health and Well-being (IMO) (Tekes Grant 648/10) and the EVO Medical Research Fund of the Finnish Red Cross Blood Service.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of six figures and four tables.
REFERENCES
- 1.Ball L. M., Bernardo M. E., Roelofs H., Lankester A., Cometa A., Egeler R. M., Locatelli F., Fibbe W. E. 2007. Cotransplantation of ex vivo–expanded mesenchymal stem cells accelerates lymphocyte recovery and may reduce the risk of graft failure in haploidentical hematopoietic stem-cell transplantation. Blood. 110: 2764–2767 [DOI] [PubMed] [Google Scholar]
- 2.Le Blanc K., Samuelsson H., Gustafsson B., Remberger M., Sundberg B., Arvidson J., Ljungman P., Lonnies H., Nava S., Ringden O. 2007. Transplantation of mesenchymal stem cells to enhance engraftment of hematopoietic stem cells. Leukemia. 21: 1733–1738 [DOI] [PubMed] [Google Scholar]
- 3.Le Blanc K., Frassoni F., Ball L., Locatelli F., Roelofs H., Lewis I., Lanino E., Sundberg B., Bernardo M. E., Remberger M., et al. , and Developmental Committee of the European Group for Blood and Marrow Transplantation 2008. Mesenchymal stem cells for treatment of steroid-resistant, severe, acute graft-versus-host disease: a phase II study. Lancet. 371: 1579–1586 [DOI] [PubMed] [Google Scholar]
- 4.Perico N., Casiraghi F., Introna M., Gotti E., Todeschini M., Cavinato R. A., Capelli C., Rambaldi A., Cassis P., Rizzo P., et al. 2011. Autologous mesenchymal stromal cells and kidney transplantation: a pilot study of safety and clinical feasibility. Clin. J. Am. Soc. Nephrol. 6: 412–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hare J. M., Traverse J. H., Henry T. D., Dib N., Strumpf R. K., Schulman S. P., Gerstenblith G., DeMaria A. N., Denktas A. E., Gammon R. S., et al. 2009. A randomized, double-blind, placebo-controlled, dose-escalation study of intravenous adult human mesenchymal stem cells (prochymal) after acute myocardial infarction. J. Am. Coll. Cardiol. 54: 2277–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiesa S., Morbelli S., Morando S., Massollo M., Marini C., Bertoni A., Frassoni F., Bartolome S. T., Sambuceti G., Traggiai E., et al. 2011. Mesenchymal stem cells impair in vivo T-cell priming by dendritic cells. Proc. Natl. Acad. Sci. USA. 108: 17384–17389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aggarwal S., Pittenger M. F. 2005. Human mesenchymal stem cells modulate allogeneic immune cell responses. Blood. 105: 1815–1822 [DOI] [PubMed] [Google Scholar]
- 8.English K., Ryan J. M., Tobin L., Murphy M. J., Barry F. P., Mahon B. P. 2009. Cell contact, prostaglandin E2 and transforming growth factor beta 1 play non-redundant roles in human mesenchymal stem cell induction of CD4+CD25Highforkhead box P3+ regulatory T cells. Clin. Exp. Immunol. 156: 149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corcione A., Benvenuto F., Ferretti E., Giunti D., Cappiello V., Cazzanti F., Risso M., Gualandi F., Mancardi G. L., Pistoia V., et al. 2006. Human mesenchymal stem cells modulate B-cell functions. Blood. 107: 367–372 [DOI] [PubMed] [Google Scholar]
- 10.Di Nicola M., Carlo-Stella C., Magni M., Milanesi M., Longoni P. D., Matteucci P., Grisanti S., Gianni A. M. 2002. Human bone marrow stromal cells suppress T-lymphocyte proliferation induced by cellular or nonspecific mitogenic stimuli. Blood. 99: 3838–3843 [DOI] [PubMed] [Google Scholar]
- 11.Meisel R., Zibert A., Laryea M., Göbel U., Däubener W., Dilloo D. 2004. Human bone marrow stromal cells inhibit allogeneic T-cell responses by indoleamine 2,3-dioxygenase–mediated tryptophan degradation. Blood. 103: 4619–4621 [DOI] [PubMed] [Google Scholar]
- 12.Tekkatte C., Gunasingh G. P., Cherian K. M., Sankaranarayanan K. 2011. “Humanized” stem cell culture techniques: the animal serum controversy. Stem Cells Int. 2011: 504723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuchs B., Schiller J., Cross M. A. 2007. Apoptosis-associated changes in the glycerophospholipid composition of hematopoietic progenitor cells monitored by 31P NMR spectroscopy and MALDI-TOF mass spectrometry. Chem. Phys. Lipids. 150: 229–238 [DOI] [PubMed] [Google Scholar]
- 14.Park H., Haynes C. A., Nairn A. V., Kulik M., Dalton S., Moremen K., Merrill A. H., Jr 2010. Transcript profiling and lipidomic analysis of ceramide subspecies in mouse embryonic stem cells and embryoid bodies. J. Lipid Res. 51: 480–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu R. K., Suzuki Y., Yanagisawa M. 2010. Membrane glycolipids in stem cells. FEBS Lett. 584: 1694–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freund D., Fonseca A. V., Janich P., Bornhauser M., Corbeil D. 2010. Differential expression of biofunctional GM1 and GM3 gangliosides within the plastic-adherent multipotent mesenchymal stromal cell population. Cytotherapy. 12: 131–142 [DOI] [PubMed] [Google Scholar]
- 17.Suila H., Pitkänen V., Hirvonen T., Heiskanen A., Anderson H., Laitinen A., Natunen S., Miller-Podraza H., Satomaa T., Natunen J., et al. 2011. Are globoseries glycosphingolipids SSEA-3 and -4 markers for stem cells derived from human umbilical cord blood? J. Mol. Cell. Biol. 3: 99–107 [DOI] [PubMed] [Google Scholar]
- 18.Ratajczak M. Z., Kim C. H., Abdel-Latif A., Schneider G., Kucia M., Morris A. J., Laughlin M. J., Ratajczak J. 2012. A novel perspective on stem cell homing and mobilization: review on bioactive lipids as potent chemoattractants and cationic peptides as underappreciated modulators of responsiveness to SDF-1 gradients. Leukemia. 26: 63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Meer G. 2005. Cellular lipidomics. EMBO J. 24: 3159–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hicks A. M., DeLong C. J., Thomas M. J., Samuel M., Cui Z. 2006. Unique molecular signatures of glycerophospholipid species in different rat tissues analyzed by tandem mass spectrometry. Biochim. Biophys. Acta. 1761: 1022–1029 [DOI] [PubMed] [Google Scholar]
- 21.Blom T. S., Koivusalo M., Kuismanen E., Kostiainen R., Somerharju P., Ikonen E. 2001. Mass spectrometric analysis reveals an increase in plasma membrane polyunsaturated phospholipid species upon cellular cholesterol loading. Biochemistry. 40: 14635–14644 [DOI] [PubMed] [Google Scholar]
- 22.Wenk M. R. 2010. Lipidomics: new tools and applications. Cell. 143: 888–895 [DOI] [PubMed] [Google Scholar]
- 23.Shevchenko A., Simons K. 2010. Lipidomics: coming to grips with lipid diversity. Nat. Rev. Mol. Cell Biol. 11: 593–598 [DOI] [PubMed] [Google Scholar]
- 24.Galle J., Bader A., Hepp P., Grill W., Fuchs B., Käs J. A., Krinner A., Marquaß B., Müller K., Schiller J., et al. 2010. Mesenchymal stem cells in cartilage repair: state of the art and methods to monitor cell growth, differentiation and cartilage regeneration. Curr. Med. Chem. 17: 2274–2291 [DOI] [PubMed] [Google Scholar]
- 25.Bensinger S. J., Tontonoz P. 2008. Integration of metabolism and inflammation by lipid-activated nuclear receptors. Nature. 454: 470–477 [DOI] [PubMed] [Google Scholar]
- 26.Wymann M. P., Schneiter R. 2008. Lipid signalling in disease. Nat. Rev. Mol. Cell Biol. 9: 162–176 [DOI] [PubMed] [Google Scholar]
- 27.Funk C. D. 2001. Prostaglandins and leukotrienes: advances in eicosanoid biology. Science. 294: 1871–1875 [DOI] [PubMed] [Google Scholar]
- 28.Cutler A. J., Limbani V., Girdlestone J., Navarrete C. V. 2010. Umbilical cord-derived mesenchymal stromal cells modulate monocyte function to suppress T cell proliferation. J. Immunol. 185: 6617–6623 [DOI] [PubMed] [Google Scholar]
- 29.Stables M. J., Gilroy D. W. 2011. Old and new generation lipid mediators in acute inflammation and resolution. Prog. Lipid Res. 50: 35–51 [DOI] [PubMed] [Google Scholar]
- 30.Leskelä H. V., Risteli J., Niskanen S., Koivunen J., Ivaska K. K., Lehenkari P. 2003. Osteoblast recruitment from stem cells does not decrease by age at late adulthood. Biochem. Biophys. Res. Commun. 311: 1008–1013 [DOI] [PubMed] [Google Scholar]
- 31.Peura M., Bizik J., Salmenperä P., Noro A., Korhonen M., Pätilä T., Vento A., Vaheri A., Alitalo R., Vuola J., et al. 2009. Bone marrow mesenchymal stem cells undergo nemosis and induce keratinocyte wound healing utilizing the HGF/c-Met/PI3K pathway. Wound Repair Regen. 17: 569–577 [DOI] [PubMed] [Google Scholar]
- 32.Folch J., Lees M., Sloane Stanley G. H. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226: 497–509 [PubMed] [Google Scholar]
- 33.Koivusalo M., Haimi P., Heikinheimo L., Kostiainen R., Somerharju P. 2001. Quantitative determination of phospholipid compositions by ESI-MS: effects of acyl chain length, unsaturation, and lipid concentration on instrument response. J. Lipid Res. 42: 663–672 [PubMed] [Google Scholar]
- 34.Käkelä R., Somerharju P., Tyynelä J. 2003. Analysis of phospholipid molecular species in brains from patients with infantile and juvenile neuronal-ceroid lipofuscinosis using liquid chromatography-electrospray ionization mass spectrometry. J. Neurochem. 84: 1051–1065 [DOI] [PubMed] [Google Scholar]
- 35.Hermansson M., Uphoff A., Käkelä R., Somerharju P. 2005. Automated quantitative analysis of complex lipidomes by liquid chromatography/mass spectrometry. Anal. Chem. 77: 2166–2175 [DOI] [PubMed] [Google Scholar]
- 36.Brügger B., Erben G., Sandhoff R., Wieland F. T., Lehmann W. D. 1997. Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc. Natl. Acad. Sci. USA. 94: 2339–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sullards M. C., Merrill A. H., Jr 2001. Analysis of sphingosine 1-phosphate, ceramides, and other bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Sci. STKE. 2001: pl1. [DOI] [PubMed] [Google Scholar]
- 38.Haimi P., Uphoff A., Hermansson M., Somerharju P. 2006. Software tools for analysis of mass spectrometric lipidome data. Anal. Chem. 78: 8324–8331 [DOI] [PubMed] [Google Scholar]
- 39.Käkelä R., Käkelä A., Kahle S., Becker B. H., Kelly A., Furness R. W. 2005. Fatty acid signatures in plasma of captive herring gulls as indicators of demersal or pelagic fish diet. Mar. Ecol. Prog. Ser. 293: 191–200 [Google Scholar]
- 40.Kimura M., Stone R. C., Hunt S. C., Skurnick J., Lu X., Cao X., Harley C. B., Aviv A. 2010. Measurement of telomere length by the Southern blot analysis of terminal restriction fragment lengths. Nat. Protocols. 5: 1596–1607 [DOI] [PubMed] [Google Scholar]
- 41.Schneider C. A., Rasband W. S., Eliceiri K. W. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.R Development Core Team 2011. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria.
- 43.Smyth G. K. 2005. Limma: linear models for microarray data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor. R. Gentleman, V. Carey, S. Duboit, R. Irizarry, and W. Huber, editors. Springer, NY. 397–420. [Google Scholar]
- 44.Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 19: 185–193 [DOI] [PubMed] [Google Scholar]
- 45.Fröhlich H., Speer N., Poustka A., Beissbarth T. 2007. GOSim--an R-package for computation of information theoretic GO similarities between terms and gene products. BMC Bioinformatics. 8: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Svante W., Sjöström M. 1977. SIMCA: a method for analyzing chemical data in terms of similarity and analogy. In Chemometrics: Theory and Application. B. Kowalski, editor. American Chemical Society, Washington, DC. 243–282. [Google Scholar]
- 47.Dominici M., Le Blanc K., Mueller I., Slaper-Cortenbach I., Marini F., Krause D., Deans R., Keating A., Prockop D., Horwitz E. 2006. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 8: 315–317 [DOI] [PubMed] [Google Scholar]
- 48.Haniffa M. A., Wang X., Holtick U., Rae M., Isaacs J. D., Dickinson A. M., Hilkens C. M. U., Collin M. P. 2007. Adult human fibroblasts are potent immunoregulatory cells and functionally equivalent to mesenchymal stem cells. J. Immunol. 179: 1595–1604 [DOI] [PubMed] [Google Scholar]
- 49.Vance J. E., Steenbergen R. 2005. Metabolism and functions of phosphatidylserine. Prog. Lipid Res. 44: 207–234 [DOI] [PubMed] [Google Scholar]
- 50.Chilton F. H., Fonteh A. N., Surette M. E., Triggiani M., Winkler J. D. 1996. Control of arachidonate levels within inflammatory cells. Biochim. Biophys. Acta. 1299: 1–15 [DOI] [PubMed] [Google Scholar]
- 51.Diez E., Chilton F. H., Stroup G., Mayer R. J., Winkler J. D., Fonteh A. N. 1994. Fatty acid and phospholipid selectivity of different phospholipase A2 enzymes studied by using a mammalian membrane as substrate. Biochem. J. 301: 721–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schoeniger A., Adolph S., Fuhrmann H., Schumann J. 2011. The impact of membrane lipid composition on macrophage activation in the immune defense against Rhodococcus equi and Pseudomonas aeruginosa. Int. J. Mol. Sci. 12: 7510–7528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Russo G. L. 2009. Dietary n-6 and n-3 polyunsaturated fatty acids: from biochemistry to clinical implications in cardiovascular prevention. Biochem. Pharmacol. 77: 937–946 [DOI] [PubMed] [Google Scholar]
- 54.Norris P. C., Dennis E. A. 2012. Omega-3 fatty acids cause dramatic changes in TLR4 and purinergic eicosanoid signaling. Proc. Natl. Acad. Sci. USA. 109: 8517–8522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown J. M., Nemeth K., Kushnir-Sukhov N. M., Metcalfe D. D., Mezey E. 2011. Bone marrow stromal cells inhibit mast cell function via a COX2-dependent mechanism. Clin. Exp. Allergy. 41: 526–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kalinski P. 2012. Regulation of immune responses by prostaglandin E2. J. Immunol. 188: 21–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim H. Y., Bigelow J., Kevala J. H. 2004. Substrate preference in phosphatidylserine biosynthesis for docosahexaenoic acid containing species. Biochemistry. 43: 1030–1036 [DOI] [PubMed] [Google Scholar]
- 58.Chilton F. H., Murphy R. C. 1986. Remodeling of arachidonate-containing phosphoglycerides within the human neutrophil. J. Biol. Chem. 261: 7771–7777 [PubMed] [Google Scholar]
- 59.Fadeel B., Xue D. 2009. The ins and outs of phospholipid asymmetry in the plasma membrane: roles in health and disease. Crit. Rev. Biochem. Mol. Biol. 44: 264–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leslie C. C., Channon J. Y. 1990. Anionic phospholipids stimulate an arachidonoyl-hydrolyzing phospholipase A2 from macrophages and reduce the calcium requirement for activity. Biochim. Biophys. Acta. 1045: 261–270 [DOI] [PubMed] [Google Scholar]
- 61.Kochs G., Hummel R., Fiebich B., Sarre T. F., Marme D., Hug H. 1993. Activation of purified human protein kinase C alpha and beta I isoenzymes in vitro by Ca2+, phosphatidylinositol and phosphatidylinositol 4,5-bisphosphate. Biochem. J. 291: 627–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stahelin R. V., Hwang J. H., Kim J., Park Z., Johnson K. R., Obeid L. M., Cho W. 2005. The mechanism of membrane targeting of human sphingosine kinase 1. J. Biol. Chem. 280: 43030–43038 [DOI] [PubMed] [Google Scholar]
- 63.Huang B. X., Akbar M., Kevala K., Kim H. 2011. Phosphatidylserine is a critical modulator for Akt activation. J. Cell Biol. 192: 979–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lucas N., Cho W. 2011. Phosphatidylserine binding is essential for plasma membrane recruitment and signaling function of 3-phosphoinositide-dependent kinase-1. J. Biol. Chem. 286: 41265–41272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Newton A. C., Koshland D. E., Jr 1990. Phosphatidylserine affects specificity of protein kinase C substrate phosphorylation and autophosphorylation. Biochemistry. 29: 6656–6661 [DOI] [PubMed] [Google Scholar]
- 66.Shetty S., Eckhardt E. R., Post S. R., van der Westhuyzen D. R. 2006. Phosphatidylinositol-3-kinase regulates scavenger receptor class B type I subcellular localization and selective lipid uptake in hepatocytes. Arterioscler. Thromb. Vasc. Biol. 26: 2125–2131 [DOI] [PubMed] [Google Scholar]
- 67.Bevers E. M., Comfurius P., Dekkers D. W. C., Zwaal R. F. A. 1999. Lipid translocation across the plasma membrane of mammalian cells. Biochim. Biophys. Acta. 1439: 317–330 [DOI] [PubMed] [Google Scholar]
- 68.Sahu S. K., Aradhyam G. K., Gummadi S. N. 2009. Calcium binding studies of peptides of human phospholipid scramblases 1 to 4 suggest that scramblases are new class of calcium binding proteins in the cell. Biochim. Biophys. Acta. 1790: 1274–1281 [DOI] [PubMed] [Google Scholar]
- 69.Perez-Chacon G., Astudillo A. M., Balgoma D., Balboa M. A., Balsinde J. 2009. Control of free arachidonic acid levels by phospholipases A2 and lysophospholipid acyltransferases. Biochim. Biophys. Acta. 1791: 1103–1113 [DOI] [PubMed] [Google Scholar]
- 70.Pérez-Chacón G., Astudillo A. M., Ruiperez V., Balboa M. A., Balsinde J. 2010. Signaling role for lysophosphatidylcholine acyltransferase 3 in receptor-regulated arachidonic acid reacylation reactions in human monocytes. J. Immunol. 184: 1071–1078 [DOI] [PubMed] [Google Scholar]
- 71.Cao Y., Pearman A. T., Zimmerman G. A., McIntyre T. M., Prescott S. M. 2000. Intracellular unesterified arachidonic acid signals apoptosis. Proc. Natl. Acad. Sci. USA. 97: 11280–11285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fuentes L., Hernandez M., Fernandez-Aviles F. J., Crespo M. S., Nieto M. L. 2002. Cooperation between secretory phospholipase A2 and TNF-receptor superfamily signaling: implications for the inflammatory response in atherogenesis. Circ. Res. 91: 681–688 [DOI] [PubMed] [Google Scholar]
- 73.Shi Y., Zhang P., Zhang L., Osman H., Mohler E. R., 3rd, Macphee C., Zalewski A., Postle A., Wilensky R. L. 2007. Role of lipoprotein-associated phospholipase A2 in leukocyte activation and inflammatory responses. Atherosclerosis. 191: 54–62 [DOI] [PubMed] [Google Scholar]
- 74.Jackson S. K., Abate W., Tonks A. J. 2008. Lysophospholipid acyltransferases: novel potential regulators of the inflammatory response and target for new drug discovery. Pharmacol. Ther. 119: 104–114 [DOI] [PubMed] [Google Scholar]
- 75.Meyer zu Heringdorf D., Jakobs K. H. 2007. Lysophospholipid receptors: signalling, pharmacology and regulation by lysophospholipid metabolism. Biochim. Biophys. Acta. 1768: 923–940 [DOI] [PubMed] [Google Scholar]
- 76.Sasaki Y., Asaoka Y., Nishizuka Y. 1993. Potentiation of diacylglycerol-induced activation of protein kinase C by lysophospholipids. Subspecies difference. FEBS Lett. 320: 47–51 [DOI] [PubMed] [Google Scholar]
- 77.Tan M., Hao F., Xu X., Chisolm G. M., Cui M. 2009. Lysophosphatidylcholine activates a novel PKD2-mediated signaling pathway that controls monocyte migration. Arterioscler. Thromb. Vasc. Biol. 29: 1376–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wall R., Ross R. P., Fitzgerald G. F., Stanton C. 2010. Fatty acids from fish: the anti-inflammatory potential of long-chain omega-3 fatty acids. Nutr. Rev. 68: 280–289 [DOI] [PubMed] [Google Scholar]
- 79.Denys A., Hichami A., Khan N. A. 2005. n-3 PUFAs modulate T-cell activation via protein kinase C-α and -ϵ and the NF-κB signaling pathway. J. Lipid Res. 46: 752–758 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.