Abstract
Group IVA cytosolic phospholipase A2 (cPLA2α), which harbors an N-terminal lipid binding C2 domain and a C-terminal lipase domain, produces arachidonic acid from the sn-2 position of zwitterionic lipids such as phosphatidylcholine. The C2 domain has been shown to bind zwitterionic lipids, but more recently, the anionic phosphomonoester sphingolipid metabolite ceramide-1-phosphate (C1P) has emerged as a potent bioactive lipid with high affinity for a cationic patch in the C2 domain β-groove. To systematically analyze the role that C1P plays in promoting the binding of cPLA2α-C2 to biological membranes, we employed biophysical measurements and cellular translocation studies along with mutagenesis. Biophysical and cellular translocation studies demonstrate that C1P specificity is mediated by Arg59, Arg61, and His62 (an RxRH sequence) in the C2 domain. Computational studies using molecular dynamics simulations confirm the origin of C1P specificity, which results in a spatial shift of the C2 domain upon membrane docking to coordinate the small C1P headgroup. Additionally, the hydroxyl group on the sphingosine backbone plays an important role in the interaction with the C2 domain, further demonstrating the selectivity of the C2 domain for C1P over phosphatidic acid. Taken together, this is the first study demonstrating the molecular origin of C1P recognition.
Keywords: calcium, cytosolic phospholipase A2α, eicosanoids, lipid binding, membrane binding
Group IVA cytosolic phospholipase A2 (cPLA2α) is an 85 kDa enzyme, which liberates arachidonic acid (AA) from the sn-2 position of membrane phospholipids in response to inflammatory agonists (1–3). cPLA2α consists of an N-terminal lipid binding C2 domain and a C-terminal catalytic or lipase domain that is separated by a flexible linker (2, 4). The C2 domain is ∼120 amino acid module, binds to zwitterionic lipids such as phosphatidylcholine (PC), and docks to PC-rich internal membranes in mammalian cells (5–9) in a Ca2+-dependent manner. Following the membrane binding and penetration of the C2 domain (10, 11), the ∼600 residue catalytic domain releases AA from zwitterionic lipids (3, 12). The generation of AA initiates pathways leading to eicosanoid synthesis, which has been implicated in heart disease (13), asthma (14), arthritis (15), cancers (16), and Alzheimer's disease (17).
The spatial and temporal translocation of cPLA2α to the nuclear envelope, endoplasmic reticulum, and Golgi apparatus is controlled by both cell-specific and agonist-dependent events. Recently, two anionic lipids, ceramide-1-phosphate (C1P) (18, 19) and PI(4,5)P2 (20–23) have been found to bind and activate cPLA2α. The molecular mechanisms regulating cPLA2α binding to C1P and PI(4,5)P2 are only beginning to unravel with the C1P (24) and PI(4,5)P2 (21, 25) binding sites being identified in the C2 domain and catalytic domain, respectively. Although the cationic site in the catalytic domain is promiscuous in anionic lipid binding (21, 23, 26), C1P is the only membrane-embedded anionic lipid that has been shown to increase membrane affinity of the cPLA2α C2 domain (19). Thus, it is thought that C1P acts as a coincidence detector (27) to promote the membrane docking of the C2 domain through elongating the membrane residence time (19, 24, 28). It is known that the C2 domain is responsible for the translocation of the catalytic domain to internal membranes; thus, C1P binding probably contributes to this functionality. Although the binding to C1P has not been observed at supraphysiological calcium levels, these types of binding experiments must be done at physiologically relevant cytoplasmic calcium concentrations, because C1P is completely shielded by calcium above 300 μM (29).
To date, cPLA2α (19) and TNF-α-converting enzyme (TACE) (30) are the only documented proteins that have been shown to bind to C1P selectively over other anionic lipids. Moreover, C1P can regulate the translocation of cPLA2α as well as increase its biological activity (31). These data are intriguing , inasmuch as one may expect other phosphomonoesters to bind with some affinity. For instance, how does the C2 domain recognize C1P over phosphatidic acid (PA) (see Fig. 6A)? Mutagenesis of residues in the C2 domain cationic patch (Arg57, Lys58, Arg59) (see Fig. 3) abrogate the C1P-dependent translocation and activity in cells (31), as well as the 10-fold increase in binding affinity C1P provides when incorporated in lipid vesicles. Thus far, these data are a bit muddled, owing to the double and triple mutants that were employed in full-length cPLA2α (24, 31).
Fig. 6.
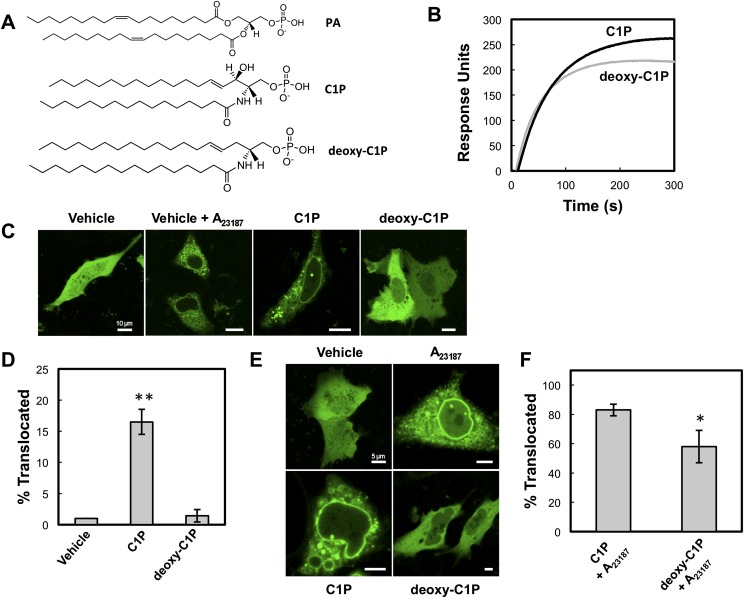
cPLA2α coordinates the sphingosine backbone hydroxyl group to bind specifically to C1P. A: Chemical structures of PA, C1P, and deoxy-C1P. B: SPR responses for POPC-C1P (97:3) (black) and POPC-deoxy-C1P (97:3) (gray) at 50 nM cPLA2α-C2 in buffer containing 20 mM HEPES, 160 mM KCl, pH 7.4. C: A549 cells were seeded, transfected with EGFP-cPLA2α for 24 h, and treated with vehicle (98% dodecane:2% ethanol), 500 nM C1P or 500 nM deoxy-C1P for 2 h, then subsequently imaged with confocal microscopy. D: Statistical analysis of cells imaged in C. E: A549 cells were treated as stated in C, but were additionally treated with 10 μM A23187 in DMSO for 15 min. Vehicle corresponds to dodecane-ethanol (98:2) plus DMSO. F: Statistical analysis of cells imaged in E. Data were collected in triplicate and quantified using a Student's t-test. Error bars represent standard deviation. ** = P < 0.0002, * = P < 0.01.
Fig. 3.
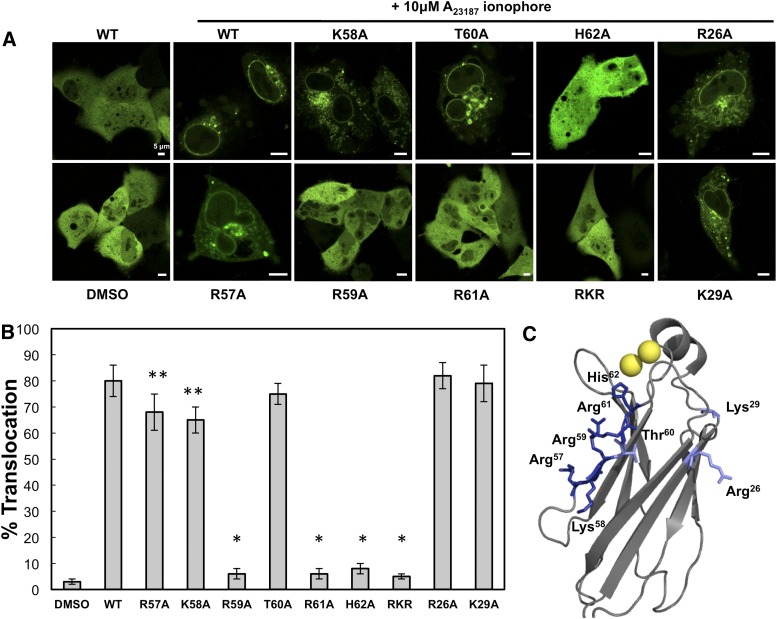
Mutation of C1P binding residues abrogates cPLA2α’s ability to translocate in A549 cells. Cells were seeded, transfected at 24 h, and imaged at 48 h with confocal microscopy. A: Cells expressing EGFP-WT-cPLA2α or respective mutations were treated with 10 μM A23187 in DMSO to induce cPLA2α WT and mutant translocation. B: The percent translocation and standard deviation were calculated from three independent experiments, where 26–66 cells were counted in each experiment. The data are shown as an average ± the standard deviation. Statistical analysis was completed by employing a Student's t-test to determine the P value for each mutation with respect to WT. * = P < 0.0001, ** = P < 0.05. C: A display of the single point mutations mapped to the gray cPLA2α-C2 crystal structure (PDB 1CJY), where the cationic β-groove is indicated by dark blue residues, control mutations in light blue, and the Ca2+ ions in yellow.
To clarify the basis of C1P binding, single mutations of all five of the basic residues of the cationic patch (RKRTRH) were made in the C2 domain and full-length enzyme. To gain insight into the mechanism of the cPLA2α interaction with cell membranes containing C1P, single point mutations in the full-length protein as well as in the C2 domain were constructed to examine the importance of each residue in the basic patch in vitro and in cells. Subsequently, molecular dynamics (MD) simulations were performed, with the C2 domain docking to a bilayer containing PC or PC-C1P to further elucidate the origin of C1P specificity. Taken together, this experimental and computational investigation demonstrates that an RxRH sequence adjacent to the calcium binding loops of the C2 domain mediates C1P specificity.
MATERIALS AND METHODS
Materials
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), and N-palmitoyl-ceramide-1-phosphate (C1P) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL) and used without further purification. N-hexadecanoyl-3-deoxy-sphingosine-1-phosphate (deoxy-C1P), synthesized by Avanti Polar Lipids, Inc. was a kind gift from Walt Shaw and Stephen Burgess. Octyl glucoside, (3-[3-cholamidopropyl) dimethylammonio]1-propane-sulfonate (CHAPS), and bicinchoninic acid (BCA) protein assay kit were from Fisher Scientific (Hampton, NH). L1 sensor chips were from GE Healthcare (Piscataway, NJ). Phospholipid concentrations were determined by a modified Bartlett analysis (32). Restriction endonucleases and enzymes for molecular biology were obtained from New England Biolabs (Beverly, MA). A549 transfection reagents (PLUS™ reagent and Lipofectamine LTX) were from Life Technologies (Grand Island, NY).
DNA mutagenesis and protein purification
The QuikChange site-directed mutagenesis kit (Agilent Technologies; Santa Clara, CA) was used to introduce mutations into the pET28a vector with a His6 tag engineered into the N-terminus of the cPLA2α C2 domain gene. The manufacturer's instructions were used to perform temperature cycling using Pfu DNA polymerase. This replicates both strands with high fidelity without displacing the mutagenic primers. A mutated plasmid containing staggered nicks was generated and treated with DpnI endonuclease. This enzyme specifically digests methylated and hemimethylated parental DNA templates and selects for mutations containing synthesized DNA. The nicked DNAs were then transformed into Escherichia coli XL-10 Gold cells. All mutated constructs were sequenced to ensure presence of the desired mutation. The C2 domain and respective mutations were expressed and purified from E. coli BL21(DE3) cells as previously described (10). Protein concentrations were determined by the BCA method, and aliquots of 3 mg/ml were made using storage buffer (10 mM HEPES, pH 7.4, 0.16 M KCl).
Surface plasmon resonance measurements
All surface plasmon resonance (SPR) measurements were performed at 25°C. A detailed protocol for coating the L1 sensor chip has been described elsewhere (33, 34). Briefly, after washing the sensor chip surface, 90 μl of vesicles containing either POPC or POPC-C1P (97:3) were injected at 5 μl/min to give a response of 6,200 resonance units. An uncoated flow channel was used as a control surface. Under our experimental conditions, no binding was detected to this control surface beyond the refractive index change for the C2 domain or cPLA2α as previously reported (9, 11, 34). Each lipid layer was stabilized by injecting 10 μl of 50 mM NaOH three times at 100 μl/min. SPR measurements were done at the flow rate of 5 μl/min. 50–90 μl of protein in 10 mM HEPES, pH 7.4, containing 0.16 M KCl and 10 μM Ca2+, was injected to give a sufficient association time for each binding signal to reach saturation (Req) (Fig. 1A, C). The lipid surface on the L1 chip, which is composed of intact lipid vesicles (34, 35), was regenerated using 10 μl of 50 mM NaOH. After sensorgrams were obtained for five or more different concentrations of each protein within a 10-fold range of Kd, each of the sensorgrams was corrected for refractive index change by subtracting the control surface response. Req values were then plotted versus protein concentrations (C), and the Kd value was determined by a nonlinear least-squares analysis of the binding isotherm using an equation, Req = Rmax/(1 + Kd/C) (36). Each data set was repeated three times to calculate a standard deviation value.
Fig. 1.
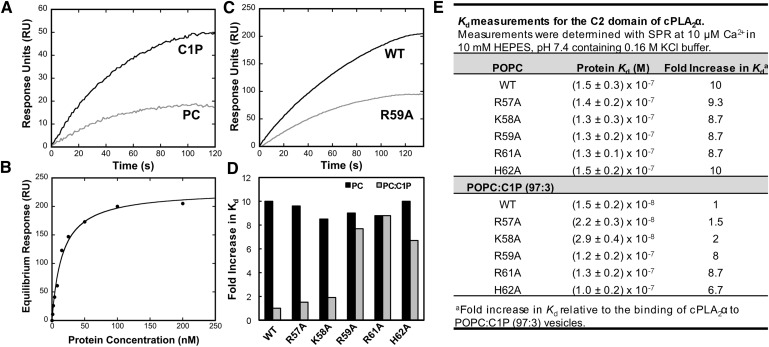
Lipid binding properties of cPLA2α-C2 and mutations. Quantitative binding analysis was performed for the cPLA2α-C2 and respective mutations to POPC or POPC-C1P (97:3) vesicles at 10 μM Ca2+. A: SPR sensorgrams are shown for 10 nM WT cPLA2α-C2 binding to POPC (light gray) or POPC:C1P (97:3) (black) vesicles. B: The equilibrium response (Req) from WT cPLA2α-C2 binding at each respective protein concentration was plotted versus [cPLA2α-C2] to fit with a nonlinear least-squares analysis of the binding isotherm (Req = Rmax/(1 + Kd/C) to determine the Kd (see 1E). C: SPR sensorgrams are shown for 100 nM WT cPLA2α-C2 (black) or 100 nM R59A binding to POPC-C1P (97:3) (gray) vesicles. D: Fold increase in Kd was normalized for each respective protein to the Kd value for WT cPLA2α-C2 for POPC-C1P (97:3) -containing vesicles. The filled bars depict binding to POPC vesicles, and the gray bars display binding to POPC-C1P (97:3) vesicles. E: Kd values for WT and respective mutations binding to POPC or POPC-C1P (97:3) vesicles. The binding experiments were completed from independent experiments in triplicate and are listed with their respective standard deviation.
C1P stoichiometry measurements
The cPLA2α-C2 domain contains only one endogenous tryptophan, Trp71, which lies beneath the cationic patch that has previously (24, 31) and herein been shown to bind C1P. The stoichiometric ratio of C1P to cPLA2α-C2 was determined through modification of the previously established methodology as described for the C2 domain of PKCα and PI(4,5)P2 (37). HEPES, pH 7.4, 10 mM, containing 0.16 M KCl and 500 nM Ca2+, where the Ca2+/EGTA was calculated according to Maxchelator software (www.stanford.eu/∼cpatton/-maxc.html) was used as the assay buffer. Briefly, cPLA2α-C2 was held constant at 1 μM and measured for maximal tryptophan fluorescence on an Aminco Bowman Series 2 luminescence spectrophotometer using excitation and emission wavelengths of 284 nm and 340 nm and bandwidths of 4 and 8 nm, respectively. Subsequently, small unilamellar vesicles containing POPC-POPE-C1P (50:40:10) were added at increasing molar C1P concentrations until the fluorescence was no longer quenched. The data were normalized, where the maximum tryptophan quenched was set to 1, then the rise and saturation phases of the data were fit using a linear least-squares equation (37). The intersection of the two lines defines the stoichiometry of cPLA2α-C2 to C1P. The stoichiometry experiment was performed in duplicate with 10 mol% C1P and confirmed in triplicate using vesicles containing 5 mol% C1P [POPC-POPE-C1P (55:40:5)].
Cellular protein expression and confocal microscopy imaging
A549 lung adenocarcinoma cells were transfected with enhanced green fluorescent protein (EGFP)-cPLA2α wild type (WT) or mutants using PLUS™ reagent and lipofectamine LTX according to the manufacturers’ protocols (Life Technologies; Grand Island, NY). Cells were grown in 50:50 DMEM-RPMI with 10% FBS and 1% penicillin-streptomycin as described previously (31). Cells were treated with 10 μM A23187 Ca2+ ionophore to induce translocation to cellular membranes, then imaged via confocal microscopy (Zeiss LSM 710) on Nunc Lab-Tek II chambered cover glasses, 8-well (Thermo Fisher Scientific; Waltham, MA) using an oil 63× 1.4 numerical aperture objective.
Dodecane-ethanol lipid delivery in A459 cells
Lipids were prepared as previously described (38). Briefly, lipid mixtures were prepared by drying the allotted volume of chloroform-methanol (2:1) solubilized lipids under N2 gas to make a 2.5 mM stock solution. Lipids were resuspended in dodecane-ethanol (98:2) at 37°C and subsequently sonicated and heated to 37°C for 20 min. Lipid solutions were diluted to the indicated concentrations using the dodecane-ethanol mixture. Cells expressing EGFP-cPLA2α were treated 24 h post transfection with 500 nM C1P or deoxy-C1P in dodecane-ethanol (98:2) for 2 h at 37°C and imaged with confocal microscopy.
Molecular dynamics set-up
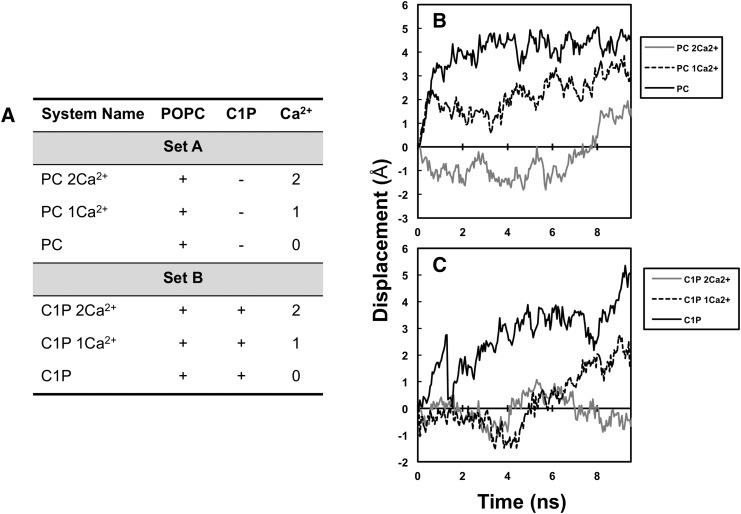
All simulations reported in this study were done on an all-atoms scheme. The first set of simulations was performed without C1P (with only POPC) while reducing the number of Ca2+ ions from 2 to 0 (system names according to the number of Ca2+ ions present: PC 2Ca2+, PC 1Ca2+, PC) (see Fig. 4A). In the second set of simulations, 1 C1P molecule was introduced with a different number of Ca2+ ions bound (system names C1P 2Ca2+, C1P 1Ca2+, C1P).
Fig. 4.
MD simulations of cPLA2α-C2 docking PC- and C1P-containing bilayers. A: The MD simulations were performed on six different systems: POPC with both Ca2+ ions present (PC 2Ca2+), POPC and one Ca2+ ion present (PC 1Ca2+), POPC without Ca2+ present (PC), POPC-C1P with both Ca2+ ions present (C1P 2Ca2+), POPC-C1P with one Ca2+ ion present (C1P 1Ca2+), and POPC-C1P without Ca2+ present (C1P). B: The MD output of cPLA2α-C2 docked to a POPC bilayer; or C: a POPC-C1P bilayer under variable conditions as defined in A.
System model
First, a patch of POPC bilayer was created using the ‘membrane’ plugin in visual molecular dynamics (39) in the xy plane such that the z-axis formed the membrane normal. The dimension of the patch (100 Å × 100 Å) was sufficient to cover the protein's area in the xy plane and to leave an additional margin of at least 15 Å on each side. The layer that interacted with the protein was called the ‘positive layer.’ For the bilayer where C1P was introduced, one POPC molecule was replaced with one C1P molecule. The C1P molecule was placed directly below the β-groove cationic patch previously shown to bind C1P (24). The surrounding POPC molecules were rotated and translated to remove any large steric clashes with C1P. A layer of 15 Å of water on the top and the bottom of the bilayer was used to ensure sufficient hydration.
Statistical ensemble
Periodic boundary conditions were applied in the xy directions to simulate an infinite planar layer and in the z direction to simulate a multilayer system. First, the membrane system was equilibrated without the protein in both sets beginning with 10,000 steps of conjugate-gradient energy minimization to remove remaining steric overlaps. This was followed by 100 ps of dynamical run. Due to a lack of homogeneity of the system (presence of two types of lipids) and unavailability of experimental data about the area per lipid for C1P, simulations of the bilayers were performed with isothermal-isobaric ensemble with the temperature fixed at 300° K. This method is appropriate for inhomogeneous systems such as a lipid bilayer of heterogeneous composition, for two reasons. First, for such systems, no large temperature difference between the components of the system is created, and therefore it is not necessary to couple the different components to separate heat baths. Second, the method has the advantage of not being critically dependent on the choice of the piston parameters and allows the area per lipid to fluctuate and stabilize to an optimum value (40). The Langevin piston method (41) was used to impose a constant pressure P = 1 atm with a damping coefficient of 5 ps–1. The particle mesh Ewald (PME) method was used for computation of the electrostatic forces (42, 43) with the grid spacing below 1.0 Å. All hydrogen bonds were restrained, allowing a time step of 2 fs.
Docking of protein on the membrane surface
In this study, the starting orientation of the protein with respect to the membrane in both the sets was the same as the experimentally validated binding orientation with POPC (44, 45). Ca2+ ions were also kept intact, as found in the crystal structure (46). Ca2+ ion 2 was removed in simulations involving one Ca2+ ion, inasmuch as Ca2+ ion 2 has been shown to regulate enzyme activity and Ca2+ ion 1 has been shown to regulate membrane binding (11). The penetration of protein into the membrane was also carefully emulated by translation of the protein along the z-axis. Some translations of the lipids in the xy plane and rigid-body rotations around the z-axis were also performed to eliminate unfavorable contacts and atomic overlaps. Additional water was added on the top of the protein-lipid system to create a layer of 15 Å above the uppermost atom (with the highest z-coordinate) of the protein. Counter ions were added to ensure the electroneutrality of the system. The remaining poor contacts or overlaps between the protein and lipid atoms were removed by minimizing the energy of the system by 10,000 steps of conjugate-gradient minimization followed by a dynamical simulation run of 10 ns.
Force field parameters
The antechamber tool (47) from Amber 7 (48) was used to parameterize C1P along with POPC. Antechamber is a set of auxiliary programs that can be used to efficiently identify bond and atom types, judge atomic equivalences, assign partial atomic charges, generate residue topology files, and, finally, create force field parameters on the basis of the above information. Generalized amber force field (49) parameters were used, and AM1-bond charge correction was used for generation of high-quality atomic charges. After the preparation of the input coordinate and topology files with Amber, NAMD 2.5 (50) was used for carrying out all molecular dynamics simulations. An explicit atom representation for all atoms, including both heavy atoms and hydrogens, was implemented. This representation yields a force field that does not require an artificial nonisotropic dielectric constant to remain stable and accurately represents all atoms over the course of the simulations.
Computing observables from MD simulation
System coordinates were saved every 2 ps during the course of the simulation to analyze the following properties under different conditions. To monitor the movement of the protein, ‘membrane surface’ was defined as the surface layer in the xy plane with z-coordinate equal to the average of z-coordinates of phosphorus atoms present in the positive layer of that system. The change in the difference of the z-coordinate of the center of mass of the protein and the z-coordinate of the membrane surface was used to monitor the movement of the protein with respect to the membrane. For analysis, the initial value of this difference was translated to a value of zero such that a difference in positive value would indicate the movement of the protein away from the layer as compared with the initial configuration, and a negative value would indicate its movement toward the membrane. To compare the binding orientation of the domain with and without C1P, tilting of the protein with respect to the membrane surface was also investigated. The first principal axis of the domain was calculated with the Cα coordinates of its atoms. Owing to the shape of the domain, its first principal axis passes through the binding and the nonbinding loops, and its movement is around the x axis in the plane of the paper. Therefore, the rotation of the first principal axis would directly demonstrate the rotation of the C2 domain.
Homology and sequence alignments
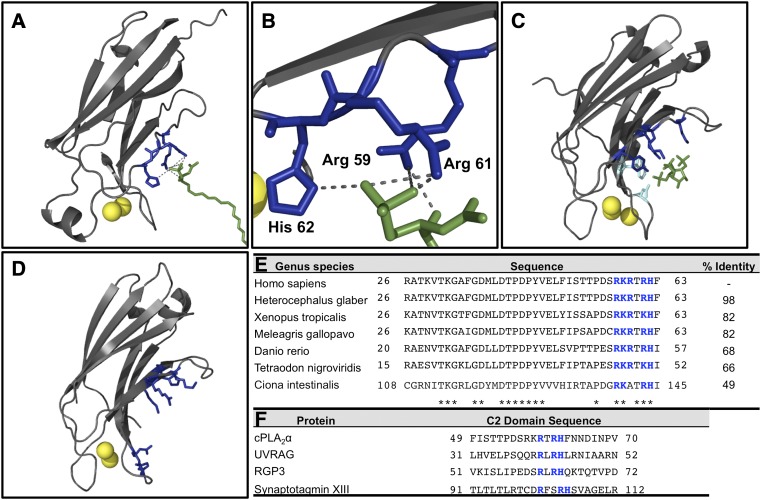
The cPLA2α C2 domain sequence was defined as residues 1–130 for the searching parameters. Homology among other organisms was investigated using a nonredundant protein-protein basic local alignment search tool (BLAST) with no excluded organisms on the default search parameters. Percent identity was calculated for the proteins compared with residues 1–130 of the query search by BLAST. The range of organisms was selected based on their diversity and is not inclusive of all organisms containing the basic sequence highlighted. To locate other C2 domains containing the RxRH motif, the conserved domain database engine on National Center for Biotechnology Information was utilized to aggregate C2 domain sequences (cd00030). At this point, the search was not limited to any particular organism. Once a positive hit was found, the Homo sapiens analog was found using BLAST. Sequences also found in Homo sapiens containing the RxRH motif were input into Fig. 8F. The search parameters did not include all C2 domains, but rather are limited to the subset included in the CD model under cd00030 that contained a reasonable sequence in Homo sapiens as of July 2012.
Fig. 8.
C2 domains bind different anionic lipids through their β-groove cationic residues. A: A model of cPLA2α-C2 with a C1P molecule docked into the C1P binding site. The cPLA2α-C2 crystal structure is depicted in gray, Ca2+ in yellow, cationic C1P binding site in blue, and C1P in green; the model displays the cPLA2α-C2 binding C1P. B: Higher magnification shows more clearly the specific interactions between Arg59, Arg61, and His62 and C1P in the C2 domain of cPLA2α. C: The PKCα C2 domain is shown (3GPE) coordinated to Ins(1,4,5)P3 with highlighted PI(4,5)P2 binding residues. Basic residues are shown in dark blue, and hydrophobic residues are shown in teal. D: The rabphilin 3A C2 domain (2K3H) is shown with purported PI(4,5)P2 binding residues in dark blue. E: The RxRH motif is conserved in cPLA2 C2 domains from diverse organisms. Asterisks indicate identical residues, and the β-groove residues are bolded in blue. F: The RxRH motif is found in the β-groove of other C2 domains, including UVRAG, which binds C1P.
RESULTS
cPLA2α-C2 domain C1P binding specificity is determined by Arg59, Arg61, and His62
The preferential binding of cPLA2α to zwitterionic lipids such as PC is well established (10, 45), as is the increased binding affinity for C1P-containing vesicles over PC membranes (19, 24, 31). To quantify the strength and selectivity of binding to these lipids, the equilibrium binding constant (Kd) for WT cPLA2α-C2 and single point mutations was determined using SPR for POPC or POPC-C1P (97:3) vesicles, which are tethered intact on a L1 sensor chip. The WT protein bound to C1P vesicles ten times stronger than to POPC vesicles alone (Fig. 1D, E), whereas R57A and K58A displayed small reductions (1.5- and 2-fold, respectively) in binding C1P-containing vesicles (Fig. 1D, E), suggesting a nonspecific or weak electrostatic role in C1P binding. Other mutations of the cationic β-groove, R59A, R61A, and H62A, displayed a 6.7- to 8.7-fold reduction in binding affinity for C1P-containing vesicles, signifying their role in determining C1P specificity. Previously, all single, double, and triple mutations in the full-length enzyme retained PC binding, Ca2+ binding, and activation properties that were non-C1P dependent (24, 31). Similarly, mutations of the C2 domain prepared in this study retained WT binding properties to POPC vesicles (Fig. 1D, E), underscoring the specific role of Arg59, Arg61, and His62 in C1P binding.
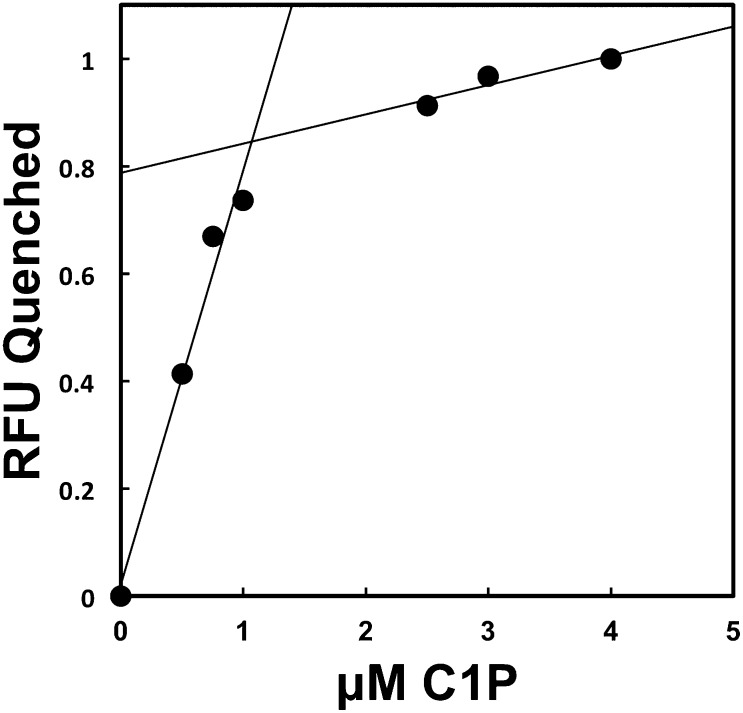
To calculate the stoichiometric ratio of C1P to cPLA2α-C2, tryptophan fluorescence was used, inasmuch as cPLA2α-C2 has an endogenous tryptophan near the basic patch that is quenched as the molar ratio of C1P increases in concentration. As vesicles containing POPC-POPE-C1P (50:40:10) were titrated into a fixed concentration of cPLA2α-C2, quenching was measured until no further change was detectable. The stoichiometry of binding was calculated as previously described by fitting the rise and saturation phases of the fluorescence data (37). The intersection of the lines gave a ratio of 1:1.16 ± 0.13, which is approximately 1 C1P to 1 cPLA2α-C2 (Fig. 2). These data were further confirmed using vesicles containing 5% C1P, which yielded a ratio of 1.02 ± 0.14 (data not shown). Spatially, this result is logical, considering the size and overall topology of the cationic patch. In addition, MD simulations also suggest the binding of one C1P by the cationic patch (see below).
Fig. 2.
Determination of the C1P binding stoichiometry of cPLA2α-C2. Titration of POPC-POPE-C1P (50:40:10) vesicles into 1 μM cPLA2α-C2 as measured by tryptophan relative fluorescence (RFU) quenched. Through fitting the rise and saturation portions of the data with linear regression, a stoichiometric ratio of 1:1.16 ± 0.13 (cPLA2α-C2-C1P) was determined in duplicate.
Arg59, Arg61, and His62 are important for cellular translocation of cPLA2α
WT cPLA2α and full-length mutant proteins expressing EGFP were transfected into A549 cells and subsequently treated with A23187 ionophore to increase cytoplasmic Ca2+ and induce cPLA2α translocation to cellular membranes (Fig. 3A). To confirm the in vitro SPR results, single residues located in the basic patch of cPLA2α were mutated along with other basic residues that have not been shown to bind to C1P. Cellular translocation responses to a cytoplasmic Ca2+ increase were analyzed for WT and mutants using confocal microscopy. Although mutants R57A and K58A (Fig. 3A, B) show a slightly decreased ability to translocate to membranes, mutations of distant basic residues R26A and K29A (Fig. 3A) did not have a statistically significant effect (Fig. 3B). Quantitative analysis of the membrane translocation of WT and mutants demonstrated that Arg59, Arg61, and His62 significantly decreased cellular translocation, suggesting that C1P binding plays a critical role in the protein's ability to translocate to intracellular membranes.
C2 orientation of binding with and without C1P
To assess the importance of Ca2+ in docking the cPLA2α-C2 domain to membranes and to discern the effect of C1P on this binding, we performed MD simulations with and without Ca2+ and C1P. In this study, the starting orientation of the protein with respect to the membrane in both simulation sets was the same as the experimentally validated binding orientation with POPC (44). Ca2+ ions were also kept intact as they were solved in the crystal structure (46). In situations where a single Ca2+ was used in the simulations, Ca2+ ion 2 was removed.
The simulations resulted in both predicted and other potentially important mechanistic results. When POPC and Ca2+ were present, the domain formed a stable complex with the bilayer, with only a small deviation from the binding orientation (Fig. 4B). This was in agreement with previous experimental (5, 9, 51) and computational (52) studies. In fact, when the Ca2+ ions were removed from the system, the domain showed very little affinity for the bilayer and drifted away by 5 Å in the first 4 ns, further validating the accuracy of protein-lipid interface in this model system. As shown in Fig. 4B, the domain then remained away from the bilayer for the remainder of the simulation time. When C1P was introduced into the bilayer, in the presence of Ca2+, the domain formed a steadfast protein-membrane complex (Fig. 4C). Interestingly, the domain showed even less deviation after 6 ns from the binding orientation than with just POPC, suggesting that the domain forms a more-stable complex (i.e., higher membrane affinity), which further supports the SPR results, cellular studies, and previous findings (19, 24, 28, 31). In addition, C1P further strengthened the binding of the C2 domain in the absence of one Ca2+ ion, demonstrating its role in promoting C2 domain docking to zwitterionic membranes.
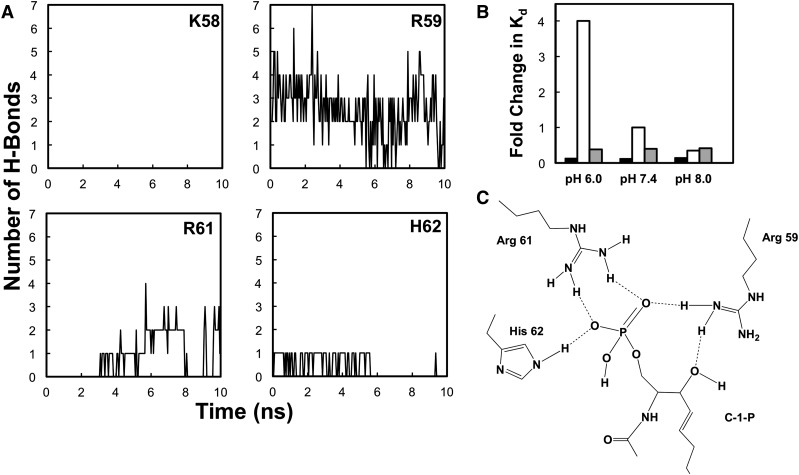
Hydrogen bonding between β-groove basic residues and C1P
A further examination of the interface between the cPLA2α-C2 and the bilayer revealed the presence of multiple hydrogen bonds between cationic residues and the C1P headgroup (Fig. 5A, C). These cationic residues are located in the cationic patch shown in Fig. 3C. Interestingly, the H-bonding is most significant for Arg59, which H-bonds with the phosphomonoester headgroup and the backbone hydroxyl of C1P (Fig. 5C). Additionally, Arg61 and His62 form significant H-bonds with the phosphomonoester headgroup of C1P. The number of hydrogen bonds between C1P and individual cationic residues in the C1P binding site was counted along the simulation time. We found that on average, the domain forms three to four hydrogen bonds with the C1P headgroup over the simulation time. Of these H-bonds, Arg59 and Arg61 have the highest contribution, with at least two and one H-bond(s) during the majority of the simulation time, respectively. His62 also forms at least one H-bond, whereas Arg57 (not shown) or Lys58 does not form any detectable H-bonds with C1P, owing to their spatial distance from the C1P headgroup. Thus, nonspecific electrostatic interactions may be the main contribution of Arg57 and Lys58 to C1P binding, which is supported by a small reduction in binding affinity and cellular membrane translocation for these mutations.
Fig. 5.
MD simulations demonstrate the selectivity of basic residues in H-bonding with the C1P headgroup. A: Predicted number of H-bonds contributed by each residue in the basic patch during the MD simulation. B: SPR binding results for POPC (black bars), POPC-C1P (97:3) (white bars), or POPC-PI(4,5)P2 (97:3) (gray bars) plotted as relative Kd value, with the Kd value for binding POPC-C1P vesicles at pH 7.4 set to 1. Each Kd value was determined in triplicate at each pH value to calculate and average and a standard deviation. C: The predicted H-bonds from the MD simulation depict a C1P binding site that is supported by in vitro C1P binding and cellular translocation data.
An experimental analysis of cPLA2α-C2 and C1P interactions
To experimentally validate the MD output for the C1P binding site, a C1P analog devoid of the hydroxyl group on the sphingosine backbone (Fig. 6A) was compared with C1P in vitro for C2 domain binding and in A549 cells (Fig. 6) for cPLA2α translocation. Identical SPR surfaces with C1P or N-hexadecanoyl-3-deoxy-sphingosine-1-phosphate (deoxy-C1P) were used to test the binding response of the C2 domain. The C2 domain demonstrated lower saturation equilibrium values in the SPR studies, with a 24% reduction in Req values for the deoxy-C1P surface compared with the C1P-containing vesicles (Fig. 6B). Additionally, the Req values for the deoxy-C1P surface were comparable to Req values for the C2 domain to POPC vesicles alone, suggesting that the backbone hydroxyl group is an important determinant of C2 domain C1P binding. A549 cells were treated with exogenous lipid through a dodecane-ethanol vehicle to test the effects of each lipid on cPLA2α translocation (38). Although membrane translocation was lower than in cells treated with the Ca2+ ionophore A23187, the translocation was observed in ∼17% of cells when C1P was added to A549 cells but was not detectable in cells treated with vehicle alone or deoxy-C1P (Fig. 6C, D). In the presence of A23187, which induces maximal translocation of cPLA2α owing to higher cytoplasmic Ca2+ concentrations, treatment with deoxy-C1P displayed a statistically significant reduction in cPLA2α translocation, suggesting that deoxy-C1P may act as a dominant negative by masking the endogenous C1P present in A549 cells (Fig. 6E, F). Treatment with additional C1P had no statistical effect in the presence of A23187, as shown by 83% translocation, because C1P binding is probably saturated at the higher calcium concentration created by the ionophore (Fig. 6E, F). These results provide experimental evidence regarding the importance of the sphingosine backbone hydroxyl group of C1P as an interaction determinant of the C2 domain (Fig. 5C).
Because His62 was shown to hydrogen bond to C1P in the MD simulations, we sought to experimentally assess the pH dependency of this interaction compared with PC and PI(4,5)P2. Previously, His protonation at acidic pH was shown to enhance the affinity of a number of phosphoinositide binding domains for phosphoinositides (53). Using SPR, Kd values at pH 6.0, 7.4, and 8.0 were determined for POPC, POPC-C1P (97:3), or POPC-PI(4,5)P2 (97:3) vesicles and demonstrate a strong relationship between pH and affinity for C1P-containing vesicles (Fig. 5B). The interaction is 4-fold stronger at pH 6.0 than at pH 7.4 and is further reduced at pH 8.0. In contrast, binding affinity to POPC or POPC-PI(4,5)P2 vesicles was not influenced by pH. The pH dependence of selective binding provided additional experimental evidence to validate the importance of His62 in the cPLA2α-C1P interaction.
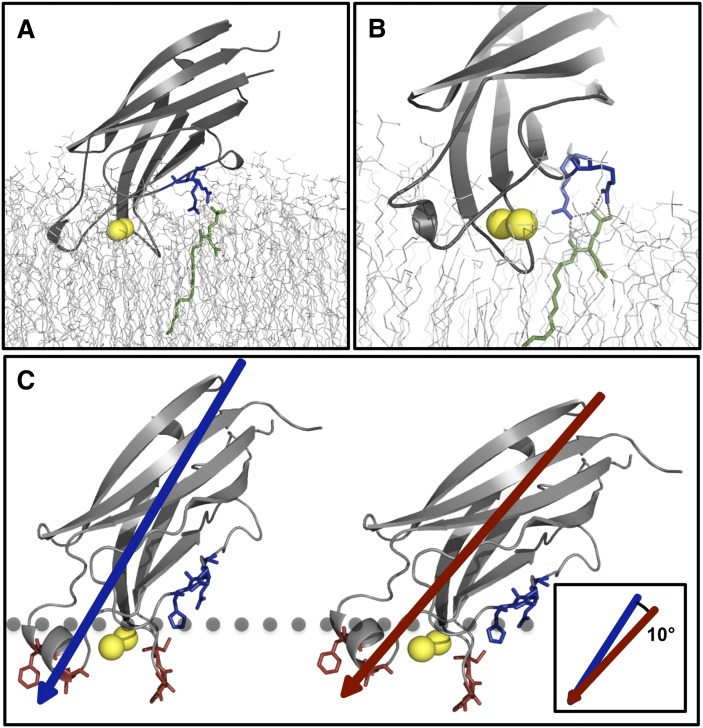
The C2 domain tilts toward the membrane to bind C1P
To further characterize the interfacial membrane interaction that the cPLA2α-C2 domain forms in the presence of C1P, we also examined the binding orientation in the absence and presence of C1P. Recently, electron paramagnetic resonance (44, 54, 55) and X-ray reflectivity (45) studies have elucidated the cPLA2α-C2 domain orientation in the presence of POPC lipids; however, the orientation when C1P is present in the membrane is still unknown. We found that starting with the same orientation (44) as that with only POPC, the domain tilts by ∼10° toward the bilayer over the course of the simulation in the presence of C1P (Fig. 7). This tilting facilitates formation of stable H-bonds between cationic residues and the C1P headgroup. This observation is further supported by the fact that the C1P headgroup is much smaller in size than the POPC headgroup and is thought to be more deeply buried, and at physiological pH, the C1P acyl chains have been found to tilt away from the surface normal of the monolayer (56). Thus, Arg59, Arg61, and His62 may further facilitate formation of hydrogen bonds by extending their side chains toward the bilayer (for instance Arg59 in Fig. 7B), which has previously been shown for Lys and Arg residues in peptides that bind PA (57).
Fig. 7.
The C2 domain rotates upon membrane docking to bind C1P embedded in the membrane. A: The structural output from the cPLA2α-C2 MD simulation in a POPC membrane docked to a C1P molecule. B: Arg59 and Arg61 shown in blue coordinating the C1P headgroup during the simulation. C: cPLA2α-C2 is shown with hydrophobic membrane-penetrating residues (red), basic C1P binding β-groove residues (blue), and Ca2+ ions (yellow); the phosphorus atoms in the headgroups of the membrane lipids are represented by the gray dotted line. The shift in cPLA2α-C2 required to bind to the phosphomonoester C1P is depicted, which results in a shift about the z-axis by ∼10° to reach the phosphate headgroup.
DISCUSSION
cPLA2α-C2 has previously been shown to deeply penetrate PC membranes in a Ca2+- dependent manner (10, 44, 45). Ca2+ has been shown to act as an electrostatic switch, which reduces the negative charge surrounding calcium-binding loops and significantly reduces the desolvation penalty (58) associated with membrane insertion. This allows the C2 domain to deeply insert into zwitterionic membranes and also to drive cellular localization (7, 9, 59). Most phospholipids contain a larger headgroup than C1P through the attachment of another molecule, meaning that phosphomonoester lipids (i.e., C1P and PA) are probably spatially embedded deeper into the membrane interface. This is an important point in C1P binding, because although C1P does not induce C2 domain membrane penetration (28), the current study demonstrates that membrane penetration is a prerequisite for the β-groove to contact the C1P headgroup. Therefore, for peripheral proteins such as the C2 domain to bind to C1P specifically, they must penetrate deeply into the membrane. It has been reported that C1P has the ability to impact both the activity and the association of cPLA2α (18, 24, 31), but much less is known about the molecular basis of C1P recognition by the C2 domain. In this study, we have characterized the molecular mechanism of C1P recognition by the C2 domain of cPLA2α using in vitro biophysical assays, cellular translocation studies, and MD simulations.
It is established that the C2 domain of cPLA2α binds specifically to membranes in a Ca2+- dependent manner, whereas the catalytic domain binds to membranes independent of Ca2+, albeit weakly (6, 25). This functionality allows the C2 domain to act as a Ca2+ sensor in cells. Because the translocation of cPLA2α was significantly reduced when C1P binding residues where mutated, we show that the cellular membrane binding capabilities of cPLA2α-C2 are not only Ca2+ dependent but also appear to rely on the domain's ability to specifically interact with C1P (Fig. 3A, B). The increased affinity for C1P greatly affects the protein's ability to stably localize to internal membranes, which is presumed to have a direct impact on the catalytic domain's ability to produce AA.
At first sight, C1P and PA look structurally very similar (Fig. 6A), but note that there is a hydroxyl group on the sphingosine backbone capable of hydrogen bonding in C1P that is not present in the glycerophospholipid PA. Through the production of single mutations, the experimental data demonstrate that Arg59, Arg61, and His62 are essential for in vitro affinity for C1P (Fig. 1D, E). The MD simulations confirm C1P's contribution to membrane binding and provide further insight regarding the importance of Arg59, Arg61, and His62 to C1P binding (Figs. 4–6). With this experimental and computational data, we modeled the binding site for the cPLA2α-C2 C1P interaction (Fig. 5). Notice that Arg59 coordinates the hydroxyl group that is specific for C1P over PA in addition to the headgroup phosphate. This interaction was further validated using deoxy-C1P, a synthetic C1P derivative lacking the hydroxyl group on the sphingosine backbone (Fig. 6). The additional interaction with the backbone hydroxyl group supports the specificity of the C2 domain for recognition of C1P over PA as demonstrated in earlier studies (19), inasmuch as neither the C2 domain nor cPLA2α exhibits enhanced affinity or cellular translocation in response to deoxy-C1P. In addition, binding studies conducted at varying pH levels demonstrate that the C1P interaction is pH dependent, supporting the importance of His62 protonation in the C1P headgroup coordination (Fig. 5B). Perhaps this result is not surprising, inasmuch as His protonation has been shown to play a key role in phosphoinositide binding (53); but to the best of our knowledge, this is the first evidence that sphingolipid binding can be regulated by pH. The higher affinity of the C2 domain for C1P in an acidic environment could have important physiological implications, because cellular pH can vary during inflammation (60) or cancer (61) with a more-profound acidic pH. Because cPLA2α has been implicated in inflammatory diseases and cancers (15, 16), the pH dependence of cPLA2α translocation and activation at acidic pH warrants further investigation.
Now that we have shown the origin of C2 domain C1P binding, it is important to ask how this domain is able to locate a membrane-embedded C1P molecule. Previous studies have demonstrated that owing to its small phosphomonester headgroup, C1P would be slightly buried and also tilted in a membrane at physiological pH (56). Because cPLA2α-C2 binds to PC membranes in a Ca2+- dependent manner, we hypothesize that this docking serves to allow the protein to sample the membrane for C1P. Upon locating a C1P, the domain locks down though a 10° shift toward the membrane (Fig. 7C). This shift increases the number of interactions with C1P and also enhances hydrophophobic interactions with the membrane for the adjacent calcium-binding loop. Because the C2 domain binds to membranes with increased affinity, it orients the catalytic domain close to its corresponding substrate and also elongates the membrane residence time of the enzyme (19, 28). Our results confirm the previous reports that cPLA2α-C2 binds with ∼10-fold higher affinity to membranes containing C1P and support the notion that C1P would negligibly increase or induce membrane penetration (28) but would, rather, maximize hydrophobic contacts when bound to C1P. This also supports a mechanism in which docking to PC drives the binding whereas C1P serves as a coincidence detector to maximize cellular localization to sites enriched with C1P.
A large number of peripheral proteins and lipid binding domains act as coincidence detectors (62, 63). These proteins contain either multiple lipid binding sites or the ability to recognize a membrane physical property such as charge or curvature in addition to a key target lipid. The role of coincidence detection in C2 domains was first suggested by Fukuda and coworkers (64), who demonstrated the ability of the C2B domain of synaptotagmin II and IV to bind inositol polyphosphates. Subsequently, several C2 domains were identified as binding phosphoinositides through their β-groove cationic residues (Fig. 8C, D) in both Ca2+-dependent and -independent modes (65). Binding to PI(4,5)P2 is the most-common ascribed function of this patch in C2 domains, but binding can often be nonselective for anionic lipids, inasmuch as the cationic patch is exposed, highly charged, and contains a less-defined pocket than selective PI-binding domains (53). In contrast, the C2 domain of cPLA2α is selective for C1P over other anionic lipids (19), which is supported by in vitro and cellular studies but also a distinct β-groove structure (Fig. 8A). The C2 domain of cPLA2α lacks consensus PI binding residues in both presence and position (Fig. 8A and supplementary Fig. I) among C2 domains. The C1P binding site is localized to β strand 3 in the C2 domain and is sequential in nature, whereas PI binding sites in PKCα (Fig. 8C, and supplementary Fig. I) and other C2 domains (Fig. 8D, and supplementary Fig. I) are composed of charged and aromatic residues from β strands 3 and 4 as well as the adjacent calcium binding loop (Figs. 8C, D, and supplementary Fig. I).
The current study has identified the conserved sequence RxRH in the cPLA2α-C2 domain as the C1P binding site. These residues are essential for C1P binding both in vitro and in a cellular system. Additionally, adjacent basic residues Arg57 and Lys58 preceding the RxRH motif play a role in anionic C1P binding, albeit weakly and more suggestive of nonspecific electrostatic effects (34). Amino acid sequence analysis of C2 domains indicates that the RxRH sequence is found in a similar position in at least three other C2 domains (Fig. 8F). This includes the C2 domains of UV resistance-associated gene (UVRAG, alternatively called p63) and regulator of G-protein signaling 3 (RGP3) (Fig. 8F). Recently, the structure of RGP3 was solved (PDB ID: 3FBK), harboring a similar site for the RxRH motif on the interfacial binding surface of the β-groove (see supplementary Fig. II). Interestingly, preliminary studies with UVRAG indicate its ability to bind C1P (Stahelin, R. V. and Chalfant, C. E., unpublished observations), suggesting that a subset of C2 domains may bind C1P. Recently, TACE has been shown to bind C1P specifically over PA and sphingonsine-1-phosphate (30). Although the origin of TACE C1P specificity is unknown, TACE harbors several conserved cationic patches (RxRH-like sequences) (30). Further work will be needed to characterize proteins such as TACE and will aid in the full characterization of the molecular variability in the cationic cluster of selective C1P binding proteins. To date, the origin of specificity has not been shown for proteins or cationic peptides that bind PA (66, 67); however, in many cases, binding to C1P has not been tested. In closing, this study should provide a framework to discover and characterize new C1P binding proteins to better understand binding to the small phosphomonoester headgroup.
Supplementary Material
Footnotes
Abbreviations:
- AA
- arachidonic acid
- BCA
- bicinchoninic acid
- BLAST
- basic local alignment search tool
- C1P
- ceramide-1-phosphate
- cPLA2α
- group IVA cytosolic phospholipase A2
- deoxy-C1P
- N-hexadecanoyl-3-deoxy-sphingosine-1-phosphate
- MD
- molecular dynamics
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPE
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
- RGP3
- regulator of G-protein signaling 3
- SPR
- surface plasmon resonance
- TACE
- TNF-α-converting enzyme
- UVRAG
- UV resistance-associated gene
- WT
- wild type
This work was supported by grants from the American Heart Association (SDG0735350N and GRNT12080254) to R.V.S.; NIH/HL-072925, NIH/CA-154314, VA Merit Award BX001792, VA Research Career Scientist Award, and a U.S.–Israel Binational Science Foundation/BSF#2011380 to C.E.C. K.E.W. is supported by an American Heart Association Predoctoral Fellowship (AHA 11PRE7640028) and a NIH CBBI Training Fellowship (T32GM075762). This work was also supported by the Indiana University School of Medicine-South Bend Imaging and Flow Cytometry Core Facility (R.V.S).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two figures.
REFERENCES
- 1.Clark J. D., Schievella A. R., Nalefski E. A., Lin L. L. 1995. Cytosolic phospholipase A2. J. Lipid Mediat. Cell Signal. 12: 83–117 [DOI] [PubMed] [Google Scholar]
- 2.Dessen A., Tang J., Schmidt H., Stahl M., Clark J. D., Seehra J., Somers W. S. 1999. Crystal structure of human cytosolic phospholipase A2 reveals a novel topology and catalytic mechanism. Cell. 97: 349–360 [DOI] [PubMed] [Google Scholar]
- 3.Leslie C. C. 1997. Properties and regulation of cytosolic phospholipase A2. J. Biol. Chem. 272: 16709–16712 [DOI] [PubMed] [Google Scholar]
- 4.Leslie C. C., Gangelhoff T. A., Gelb M. H. 2010. Localization and function of cytosolic phospholipase A2 at the Golgi. Biochimie. 92: 620–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corbin J. A., Evans J. H., Landgraf K. E., Falke J. J. 2007. Mechanism of specific membrane targeting by C2 domains: localized pools of target lipids enhance Ca2+ affinity. Biochemistry. 46: 4322–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans J. H., Leslie C. C. 2004. The cytosolic phospholipase A2 catalytic domain modulates association and residence time at the Golgi membranes. J. Biol. Chem. 279: 6005–6016 [DOI] [PubMed] [Google Scholar]
- 7.Evans J. H., Spencer D. M., Zweifach A., Leslie C. C. 2001. Intracelluar calcium signals regulating cytosolic phospholipase A2 translocation to internal membranes. J. Biol. Chem. 276: 30150–30160 [DOI] [PubMed] [Google Scholar]
- 8.Nalefski E. A., Falke J. J. 1998. Location of the membrane-docking face on the Ca2+-activated C2 domain of cytosolic phospholipase A2. Biochemistry. 37: 17642–17650 [DOI] [PubMed] [Google Scholar]
- 9.Stahelin R. V., Rafter J. D., Das S., Cho W. 2003. The molecular basis of differential subcellular localization of C2 domains of protein kinase C-alpha and group IVA cytosolic phospholipase A2. J. Biol. Chem. 278: 12452–12460 [DOI] [PubMed] [Google Scholar]
- 10.Bittova L., Sumandea M., Cho W. 1999. A structure-function study of the C2 domain of cytosolic phospholipase A2. Identification of essential calcium ligands and hydrophobic membrane binding residues. J. Biol. Chem. 274: 9665–9672 [DOI] [PubMed] [Google Scholar]
- 11.Stahelin R. V., Cho W. 2001. Roles of calcium ions in the membrane binding of C2 domains. Biochem. J. 359: 679–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimizu T., Ohto T., Kita Y. 2006. Cytosolic phospholipase A2: biochemical properties and physiological roles. IUBMB Life. 58: 328–333 [DOI] [PubMed] [Google Scholar]
- 13.Kerkelä R., Boucher M., Zaka R., Gao E., Harris D., Piuhola J., Song J., Serpi R., Woulfe K. C., Cheung J. Y., et al. 2011. Cytosolic phospholipase A(2)α protects against ischemia/reperfusion injury in the heart. Clin. Transl. Sci. 4: 236–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hewson C. A., Patel S., Calzetta L., Campwala H., Havard S., Luscombe E., Clarke P. A., Peachell P. T., Matera M. G., Cazzola M., et al. 2012. Preclinical evaluation of an inhibitor of cytosolic phospholipase A2α for the treatment of asthma. J. Pharmacol. Exp. Ther. 340: 656–665 [DOI] [PubMed] [Google Scholar]
- 15.Tai N., Kuwabara K., Kobayashi M., Yamada K., Ono T., Seno K., Gahara Y., Ishizaki J., Hori Y. 2010. Cytosolic phospholipase A2 alpha inhibitor, pyrroxyphene, displays anti-arthritic and anti-bone destructive action in a murine arthritis model. Inflamm. Res. 59: 53–62 [DOI] [PubMed] [Google Scholar]
- 16.Sundarraj S., Kannan S., Thangam R., Gunasekaran P. 2012. Effects of the inhibition of cytosolic phospholipase A(2)α in non-small cell lung cancer cells. J. Cancer Res. Clin. Oncol. 138: 827–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gentile M. T., Reccia M. G., Sorrentino P. P., Vitale E., Sorrentino G., Puca A. A., Colucci-D'Amato L. 2012. Role of cytosolic calcium-dependent phospholipase A2 in Alzheimer's disease pathogenesis. Mol. Neurobiol. 45: 596–604 [DOI] [PubMed] [Google Scholar]
- 18.Pettus B. J., Bielawska A., Subramanian P., Wijesinghe D. S., Maceyka M., Leslie C. C., Evans J. H., Freiberg J., Roddy P., Hannun Y. A., et al. 2004. Ceramide 1-phosphate is a direct activator of cytosolic phospholipase A2. J. Biol. Chem. 279: 11320–11326 [DOI] [PubMed] [Google Scholar]
- 19.Subramanian P., Stahelin R. V., Szulc Z., Bielawska A., Cho W., Chalfant C. E. 2005. Ceramide 1-phosphate acts as a positive allosteric activator of group IVA cytosolic phospholipase A2 alpha and enhances the interaction of the enzyme with phosphatidylcholine. J. Biol. Chem. 280: 17601–17607 [DOI] [PubMed] [Google Scholar]
- 20.Balsinde J., Balboa M. A., Li W. H., Llopis J., Dennis E. A. 2000. Cellular regulation of cytosolic group IV phospholipase A2 by phosphatidylinositol bisphosphate levels. J. Immunol. 164: 5398–5402 [DOI] [PubMed] [Google Scholar]
- 21.Casas J., Gijon M. A., Vigo A. G., Crespo M. S., Balsinde J., Balboa M. A. 2006. Phosphatidylinositol 4,5-bisphosphate anchors cytosolic group IVA phospholipase A2 to perinuclear membranes and decreases its calcium requirement for translocation in live cells. Mol. Biol. Cell. 17: 155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leslie C. C., Channon J. Y. 1990. Anionic phospholipids stimulate arachidonyl-hydrolyzing phospholipase A2 from macrophages and reduce the calcium requirement for activity. Biochim. Biophys. Acta. 1045: 261–270 [DOI] [PubMed] [Google Scholar]
- 23.Mosior M., Six D. A., Dennis E. A. 1998. Group IV cytosolic phospholipase A2 binds with high affinity and specificity to phosphatidylinositol 4,5-bisphosphate resulting in dramatic increases in activity. J. Biol. Chem. 273: 2184–2191 [DOI] [PubMed] [Google Scholar]
- 24.Stahelin R. V., Subramanian P., Vora M., Cho W., Chalfant C. E. 2007. Ceramide-1-phosphate binds group IVA cytosolic phospholipase a2 via a novel site in the C2 domain. J. Biol. Chem. 282: 20467–20474 [DOI] [PubMed] [Google Scholar]
- 25.Das S., Cho W. 2002. Roles of catalytic domain residues in interfacial binding and activation of group IV cytosolic phospholipase A2. J. Biol. Chem. 277: 23838–23846 [DOI] [PubMed] [Google Scholar]
- 26.Tucker D. E., Ghosh M., Ghomashchi F., Loper R., Suram S., John B. S., Girotti M., Bollinger J. G., Gelb M. H., Leslie C. C. 2009. Role of phosphorylation and basic residues in the catalytic domain of cytosolic phospholipase A2alpha in regulating interfacial kinetics and binding and cellular function. J. Biol. Chem. 284: 9596–9611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Falke J. J. 2012. Lipid targeting domain with dual-membrane specificity that expands the diversity of intracellular targeting reactions. Proc. Natl. Acad. Sci. USA. 109: 1816–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian P., Vora M., Gentile L. B., Stahelin R. V., Chalfant C. E. 2007. Anionic lipids activate group IVA cytosolic phospholipase A2 via distinct and separate mechanisms. J. Lipid Res. 48: 2701–2708 [DOI] [PubMed] [Google Scholar]
- 29.Kooijman E. E., Sot J., Montes L. R., Alonso A., Gericke A., de Kruijff B., Kumar S., Goni F. M. 2008. Membrane organization and ionization behavior of the minor but crucial lipid ceramide-1-phosphate. Biophys. J. 94: 4320–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamour N. F., Wijesinghe D. S., Mietla J. A., Ward K. E., Stahelin R. V., Chalfant C. E. 2011. Ceramide kinase regulates the production of tumor necrosis factor α (TNFα) via inhibition of TNF-α converting enzyme. J. Biol. Chem. 286: 42808–42817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamour N. F., Subramanian P., Wijesinghe D. S., Stahelin R. V., Bonventre J. V., Chalfant C. E. 2009. Ceramide 1-phosphate is required for the translocation of group IVA cytosolic phospholipase A2 and prostaglandin synthesis. J. Biol. Chem. 284: 26897–26907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kates M. 1986. Techniques of Lipidology. Burdon R.H., van Knippenberg P.H., editors. 2nd edition Elsevier Science Publishers B.V., Amsterdam: 114–115 [Google Scholar]
- 33.Bittova L., Stahelin R. V., Cho W. 2001. Roles of ionic residues of the C1 domain in protein kinase C-alpha activation and the origin of phosphatidylserine specificity. J. Biol. Chem. 276: 4218–4226 [DOI] [PubMed] [Google Scholar]
- 34.Stahelin R. V., Cho W. 2001. Differential roles of ionic, aliphatic, and aromatic residues in membrane-protein interactions: a surface plasmon resonance study on phospholipases A2. Biochemistry. 40: 4672–4678 [DOI] [PubMed] [Google Scholar]
- 35.Anderluh G., Besenicar M., Kladnik A., Lakey J. H., Macek P. 2005. Properties of nonfused liposomes immobilized on an L1 Biacore chip and their permeabilization by a eukaryotic pore-forming toxin. Anal. Biochem. 344: 43–52 [DOI] [PubMed] [Google Scholar]
- 36.Stahelin R. V., Long F., Diraviyam K., Bruzik K. S., Murray D., Cho W. 2002. Phosphatidylinositol 3-phosphate induces the membrane penetration of the FYVE domains of Vps27p and Hrs. J. Biol. Chem. 277: 26379–26388 [DOI] [PubMed] [Google Scholar]
- 37.Lai C. L., Landgraf K. E., Voth G. A., Falke J. J. 2010. Membrane docking geometry and target lipid stoichiometry of membrane-bound PKCα C2 domain: a combined molecular dynamics and experimental study. J. Mol. Biol. 402: 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wijesinghe D. S., Subramanian P., Lamour N. F., Gentile L. B., Granado M. H., Bielawska A., Szulc Z., Gomez-Munoz A., Chalfant C. E. 2009. Chain length specificity for activation of cPLA2alpha by C1P: use of the dodecane delivery system to determine lipid-specific effects. J. Lipid Res. 50: 1986–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Humphrey W., Dalke A., Schulten K. 1996. VMD: visual molecular dynamics. J. Mol. Graphics. 14: 33–38, 27–28 [DOI] [PubMed] [Google Scholar]
- 40.Mihailescu D., Smith J. C. 2000. Atomic detail peptide-membrane interactions: molecular dynamics simulation of gramicidin S in a DMPC bilayer. Biophys. J. 79: 1718–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feller S. E., Zhang Y. H., Pastor R. W., Brooks B. R. 1995. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 103: 4613–4621 [Google Scholar]
- 42.Darden T., York D., Pedersen L. 1993. Particle mesh Ewald: An N•log(N) method for Ewald sums in large systems. J. Chem. Phys. 98: 10089–10092 [Google Scholar]
- 43.Essmann U., Perera L., Berkowitz M. L., Darden T., Lee H., Pedersen L. G. 1995. A smooth particle mesh Ewald method. J. Chem. Phys. 103: 8577–8593 [Google Scholar]
- 44.Frazier A. A., Wisner M. A., Malmberg N. J., Victor K. G., Fanucci G. E., Nalefski E. A., Falke J. J., Cafiso D. S. 2002. Membrane orientation and position of the C2 domain from cPLA2 by site-directed spin labeling. Biochemistry. 41: 6282–6292 [DOI] [PubMed] [Google Scholar]
- 45.Málková S., Long F., Stahelin R. V., Pingali S. V., Murray D., Cho W., Schlossman M. L. 2005. X-ray reflectivity studies of cPLA2{alpha}-C2 domains adsorbed onto Langmuir monolayers of SOPC. Biophys. J. 89: 1861–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perisic O., Fong S., Lynch D. E., Bycroft M., Williams R. L. 1998. Crystal structure of a calcium-phospholipid binding domain from cytosolic phospholipase A2. J. Biol. Chem. 273: 1596–1604 [DOI] [PubMed] [Google Scholar]
- 47.Wang J., Wang W., Kollman P. A., Case D. A. 2006. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 25: 247–260 [DOI] [PubMed] [Google Scholar]
- 48.Case D. A., Cheatham T. E., III, Darden T., Gohlke H., Luo R., Merz K. M., Jr, Onufriev A., Simmerling C., Wang B., Woods R. J. 2005. The Amber biomolecular simulation programs. J. Comput. Chem. 26: 1668–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang J., Wolf R. M., Caldwell J. W., Kollman P. A., Case D. A. 2004. Development and testing of a general amber force field. J. Comput. Chem. 25: 1157–1174 [DOI] [PubMed] [Google Scholar]
- 50.Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kale L., Schulten K. 2005. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26: 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nalefski E. A., Wisner M. A., Chen J. Z., Sprang S. R., Fukuda M., Mikoshiba K., Falke J. J. 2001. C2 domains from different Ca2+ signaling pathways display functional and mechanistic diversity. Biochemistry. 40: 3089–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jaud S., Tobias D. J., Falke J. J., White S. H. 2007. Self-induced docking site of a deeply embedded peripheral membrane protein. Biophys. J. 92: 517–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kutateladze T. G. 2010. Translation of the phosphoinositide code by PI effectors. Nat. Chem. Biol. 6: 507–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malmberg N. J., Falke J. J. 2005. Use of EPR power saturation to analyze the membrane-docking geometries of peripheral proteins: applications to C2 domains. Annu. Rev. Biophys. Biomol. Struct. 34: 71–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Malmberg N. J., Van Buskirk D. R., Falke J. J. 2003. Membrane-docking loops of the cPLA2 C2 domain: detailed structural analysis of the protein-membrane interface via site-directed spin-labelling. Biochemistry. 42: 13227–13240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kooijman E. E., Vaknin D., Bu W., Joshi L., Kang S. W., Gericke A., Mann E. K., Kumar S. 2009. Structure of ceramide-1-phosphate at the air-water solution interface in the absence and presence of Ca2+. Biophys. J. 96: 2204–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kooijman E. E., Tieleman D. P., Testerink C., Munnik T., Rijkers D. T., Burger K. N., de Kruijff B. 2007. An electrostatic/hydrogen bond switch as the basis for the specific interaction of phosphatidic acid with proteins. J. Biol. Chem. 282: 11356–11364 [DOI] [PubMed] [Google Scholar]
- 58.Murray D., Honig B. 2002. Electrostatic control of the membrane targeting of C2 domains. Mol. Cell. 9: 145–154 [DOI] [PubMed] [Google Scholar]
- 59.Evans J. H., Gerber S. H., Murray D., Leslie C. C. 2004. The calcium binding loops of the cytosolic phospholipase A2 C2 domain specify targeting to Golgi and ER in live cells. Mol. Biol. Cell. 15: 371–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Punnia-Moorthy A. 1987. Evaluation of pH changes in inflammation of the subcutaneous air pouch lining in the rat, induced by carrageenan, dextran, and Staphylococcus aureus. J. Oral Pathol. 16: 36–44 [DOI] [PubMed] [Google Scholar]
- 61.Gerweck L. E., Seetharaman K. 1996. Cellular pH gradient in tumor versus normal tissue: potential exploitation for the treatment of cancer. Cancer Res. 56: 1194–1198 [PubMed] [Google Scholar]
- 62.Moravcevic K., Oxley C. L., Lemmon M. A. 2012. Conditional peripheral membrane proteins: facing up to limited specificity. Structure. 20: 15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scott J. L., Musselman C. A., Adu-Gyamfi E., Kutateladze T. G., Stahelin R. V. 2012. Emerging methodologies to investigate lipid-protein interactions. Integr. Biol. (Camb). 4: 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fukuda M., Aruga J., Niinobe M., Aimoto S., Mikoshiba K. 1994. Inositol-1,3,4,5-tetrakisphosphate binding to C2B domain of IP4BP/synaptotagmin II. J. Biol. Chem. 269: 29206–29211 [PubMed] [Google Scholar]
- 65.Cho W., Stahelin R. V. 2006. Membrane binding and subcellular targeting of C2 domains. Biochim. Biophys. Acta. 1761: 838–849 [DOI] [PubMed] [Google Scholar]
- 66.Kooijman E. E., Burger K. N. 2009. Biophysics and function of phosphatidic acid: a molecular perspective. Biochim. Biophys. Acta. 1791: 881–888 [DOI] [PubMed] [Google Scholar]
- 67.Stace C. L., Ktistakis N. T. 2006. Phosphatidic acid- and phosphatidylserine-binding proteins. Biochim. Biophys. Acta. 1761: 913–926 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.