Abstract
Proper ras1 function is required for normal sexual function in the yeast Schizosaccharomyces pombe. We have found a gene in S. pombe, sar1, that encodes a product capable of regulating ras1 function. sar1 is a member of an expanding family of RAS GTPase-activating proteins (GAPs) that includes mammalian GAP, the yeast Saccharomyces cerevisiae IRA proteins, and the product of the human neurofibromatosis locus, NF1 sar1, like these other proteins, can complement the loss of IRA function in S. cerevisiae. Computer analysis shows that the highest degree of sequence conservation is restricted to a very small number of diagnostic residues represented by the motif Phe-Leu-Arg-X-X-X-Pro-Ala-X-X-X-Pro. We find no evidence that sar1 is required for the effector function of ras1.
Full text
PDF



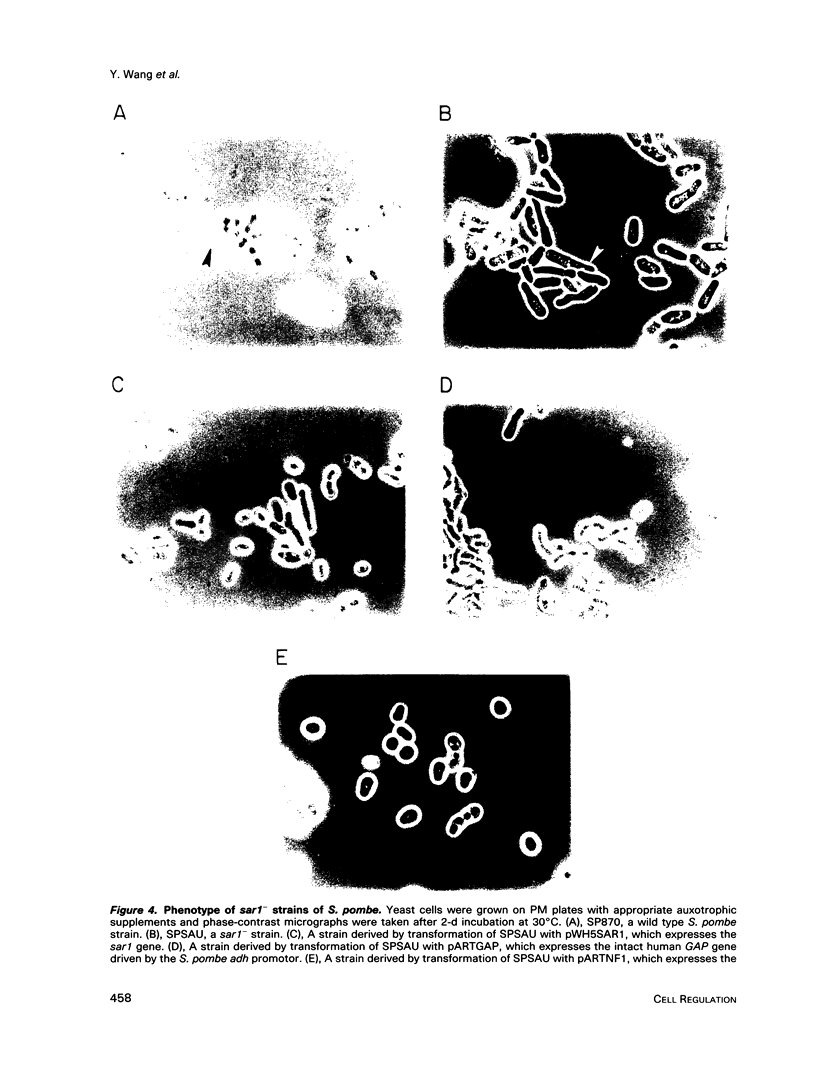

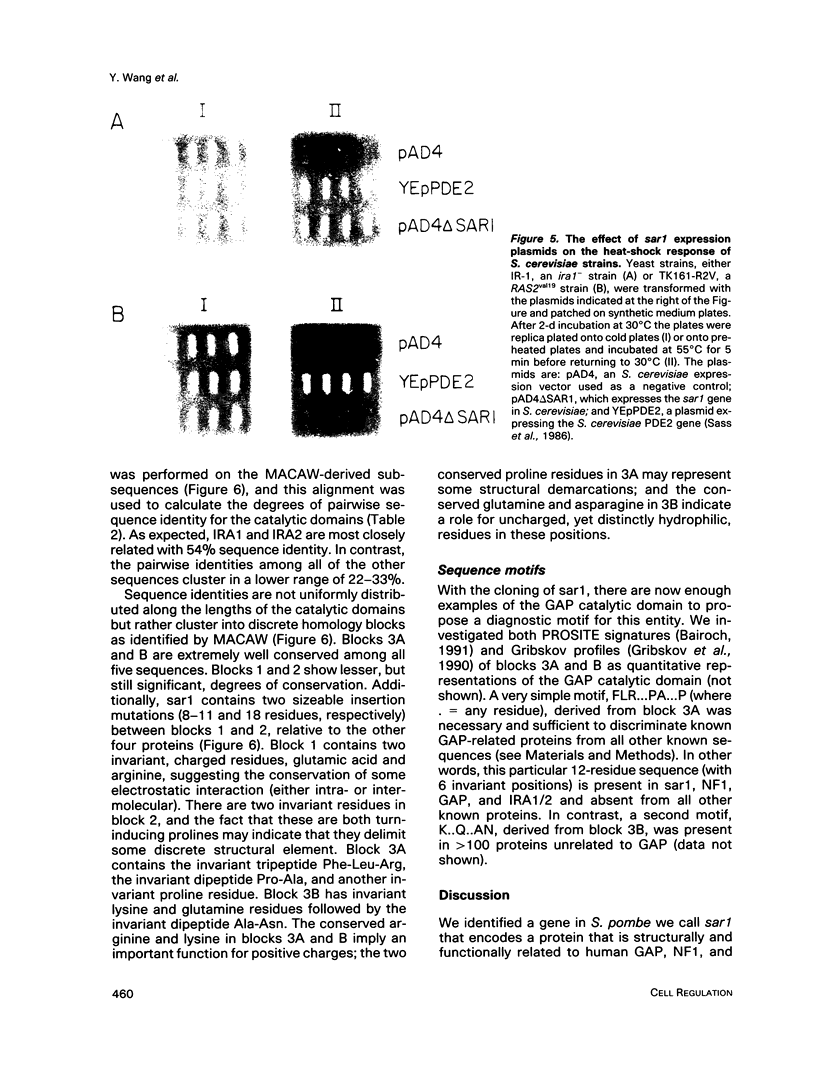





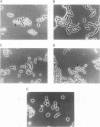
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adari H., Lowy D. R., Willumsen B. M., Der C. J., McCormick F. Guanosine triphosphatase activating protein (GAP) interacts with the p21 ras effector binding domain. Science. 1988 Apr 22;240(4851):518–521. doi: 10.1126/science.2833817. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ballester R., Marchuk D., Boguski M., Saulino A., Letcher R., Wigler M., Collins F. The NF1 locus encodes a protein functionally related to mammalian GAP and yeast IRA proteins. Cell. 1990 Nov 16;63(4):851–859. doi: 10.1016/0092-8674(90)90151-4. [DOI] [PubMed] [Google Scholar]
- Ballester R., Michaeli T., Ferguson K., Xu H. P., McCormick F., Wigler M. Genetic analysis of mammalian GAP expressed in yeast. Cell. 1989 Nov 17;59(4):681–686. doi: 10.1016/0092-8674(89)90014-7. [DOI] [PubMed] [Google Scholar]
- Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990 Nov 8;348(6297):125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991 Jan 10;349(6305):117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- Buchberg A. M., Cleveland L. S., Jenkins N. A., Copeland N. G. Sequence homology shared by neurofibromatosis type-1 gene and IRA-1 and IRA-2 negative regulators of the RAS cyclic AMP pathway. Nature. 1990 Sep 20;347(6290):291–294. doi: 10.1038/347291a0. [DOI] [PubMed] [Google Scholar]
- Calés C., Hancock J. F., Marshall C. J., Hall A. The cytoplasmic protein GAP is implicated as the target for regulation by the ras gene product. Nature. 1988 Apr 7;332(6164):548–551. doi: 10.1038/332548a0. [DOI] [PubMed] [Google Scholar]
- Field J., Broek D., Kataoka T., Wigler M. Guanine nucleotide activation of, and competition between, RAS proteins from Saccharomyces cerevisiae. Mol Cell Biol. 1987 Jun;7(6):2128–2133. doi: 10.1128/mcb.7.6.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui Y., Kaziro Y. Molecular cloning and sequence analysis of a ras gene from Schizosaccharomyces pombe. EMBO J. 1985 Mar;4(3):687–691. doi: 10.1002/j.1460-2075.1985.tb03684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui Y., Kozasa T., Kaziro Y., Takeda T., Yamamoto M. Role of a ras homolog in the life cycle of Schizosaccharomyces pombe. Cell. 1986 Jan 31;44(2):329–336. doi: 10.1016/0092-8674(86)90767-1. [DOI] [PubMed] [Google Scholar]
- Gibbs J. B., Schaber M. D., Allard W. J., Sigal I. S., Scolnick E. M. Purification of ras GTPase activating protein from bovine brain. Proc Natl Acad Sci U S A. 1988 Jul;85(14):5026–5030. doi: 10.1073/pnas.85.14.5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribskov M., Lüthy R., Eisenberg D. Profile analysis. Methods Enzymol. 1990;183:146–159. doi: 10.1016/0076-6879(90)83011-w. [DOI] [PubMed] [Google Scholar]
- Hughes D. A., Fukui Y., Yamamoto M. Homologous activators of ras in fission and budding yeast. Nature. 1990 Mar 22;344(6264):355–357. doi: 10.1038/344355a0. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S., Altschul S. F. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2264–2268. doi: 10.1073/pnas.87.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman D. J., Altschul S. F., Kececioglu J. D. A tool for multiple sequence alignment. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4412–4415. doi: 10.1073/pnas.86.12.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G. A., Viskochil D., Bollag G., McCabe P. C., Crosier W. J., Haubruck H., Conroy L., Clark R., O'Connell P., Cawthon R. M. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell. 1990 Nov 16;63(4):843–849. doi: 10.1016/0092-8674(90)90150-d. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass P., Field J., Nikawa J., Toda T., Wigler M. Cloning and characterization of the high-affinity cAMP phosphodiesterase of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9303–9307. doi: 10.1073/pnas.83.24.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler G. D., Altschul S. F., Lipman D. J. A workbench for multiple alignment construction and analysis. Proteins. 1991;9(3):180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Matsumoto K., Toh-E A. IRA1, an inhibitory regulator of the RAS-cyclic AMP pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Feb;9(2):757–768. doi: 10.1128/mcb.9.2.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Nakafuku M., Satoh T., Marshall M. S., Gibbs J. B., Matsumoto K., Kaziro Y., Toh-e A. S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell. 1990 Mar 9;60(5):803–807. doi: 10.1016/0092-8674(90)90094-u. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Nakafuku M., Tamanoi F., Kaziro Y., Matsumoto K., Toh-e A. IRA2, a second gene of Saccharomyces cerevisiae that encodes a protein with a domain homologous to mammalian ras GTPase-activating protein. Mol Cell Biol. 1990 Aug;10(8):4303–4313. doi: 10.1128/mcb.10.8.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trahey M., McCormick F. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science. 1987 Oct 23;238(4826):542–545. doi: 10.1126/science.2821624. [DOI] [PubMed] [Google Scholar]
- Wigler M., Field J., Powers S., Broek D., Toda T., Cameron S., Nikawa J., Michaeli T., Colicelli J., Ferguson K. Studies of RAS function in the yeast Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 1988;53(Pt 2):649–655. doi: 10.1101/sqb.1988.053.01.074. [DOI] [PubMed] [Google Scholar]
- Yatani A., Okabe K., Polakis P., Halenbeck R., McCormick F., Brown A. M. ras p21 and GAP inhibit coupling of muscarinic receptors to atrial K+ channels. Cell. 1990 Jun 1;61(5):769–776. doi: 10.1016/0092-8674(90)90187-j. [DOI] [PubMed] [Google Scholar]
- Zhang K., DeClue J. E., Vass W. C., Papageorge A. G., McCormick F., Lowy D. R. Suppression of c-ras transformation by GTPase-activating protein. Nature. 1990 Aug 23;346(6286):754–756. doi: 10.1038/346754a0. [DOI] [PubMed] [Google Scholar]