Abstract
Purpose
To identify mediators of glioblastoma anti-angiogenic therapy resistance and target these mediators in xenografts.
Experimental Design
We performed microarray analysis comparing bevacizumab-resistant glioblastomas (BRGs) to pre-treatment tumors from the same patients. We established novel xenograft models of anti-angiogenic therapy resistance to target candidate resistance mediator(s).
Results
BRG microarray analysis revealed upregulation versus pre-treatment of receptor tyrosine kinase c-Met, which underwent further investigation because of its prior biologic plausibility as a bevacizumab resistance mediator. BRGs exhibited increased hypoxia versus pre-treatment in a manner correlating with their c-Met upregulation, increased c-Met phosphorylation, and increased phosphorylation of c-Met-activated focal adhesion kinase (FAK) and STAT3. We developed two novel xenograft models of anti-angiogenic therapy resistance. In the first model, serial bevacizumab treatment of an initially responsive xenograft generated a xenograft with acquired bevacizumab resistance, which exhibited upregulated c-Met expression versus pre-treatment. In the second model, a BRG-derived xenograft maintained refractoriness to the MRI tumor vasculature alterations and survival-promoting effects of bevacizumab. Growth of this BRG-derived xenograft was inhibited by a c-Met inhibitor. Transducing these xenograft cells with c-Met shRNA inhibited their invasion and survival in hypoxia, disrupted their mesenchymal morphology, and converted them from bevacizumab-resistant to bevacizumab-responsive. Engineering bevacizumab-responsive cells to express constitutively active c-Met caused these cells to form bevacizumab-resistant xenografts.
Conclusion
These findings support the role of c-Met in survival in hypoxia and invasion, features associated with anti-angiogenic therapy resistance; and growth and therapeutic resistance of xenografts resistant to anti-angiogenic therapy. Therapeutically targeting c-Met could prevent or overcome anti-angiogenic therapy resistance.
Keywords: bevacizumab, resistance, glioblastoma, invasion, c-Met
INTRODUCTION
Vascular endothelial growth factor (VEGF) contributes to the abnormal vascularity of glioblastomas, which promotes their growth and recurrence following standard treatment. Encouraging results from clinical trials of humanized mouse anti-human VEGF antibody bevacizumab in glioblastoma patients (1, 2) led to the 2009 FDA approval of bevacizumab for recurrent glioblastoma treatment.
However, as with other cancers (3), the response to anti-angiogenic therapy in glioblastoma is often transient, with 40–60% rates of radiographic progression after initial tumor regression (1, 2). Glioblastomas developing anti-angiogenic therapy resistance respond poorly to available treatments, with median overall survival below 6 months after acquired bevacizumab resistance (4).
Preclinical studies suggest that tumor cells become resistant to anti-angiogenic therapy through transcriptional upregulation of compensatory pathways increasing production of alternative pro-angiogenic stimuli, recruitment of marrow-derived cells, or invasion, allowing tumor cells to survive and potentially proliferate while the anti-angiogenic target remains inhibited (5). This paradigm represents a departure from resistance to traditional DNA damaging chemotherapy, which typically involves gene mutations. Because transcriptional changes occur more readily than mutations (5), these changes may occur to some extent in all tumors receiving anti-angiogenic therapy, with tumors exhibiting the greatest changes becoming resistant.
To identify mediators of anti-angiogenic therapy resistance, we performed a comprehensive gene expression analysis comparing bevacizumab-resistant glioblastomas (BRGs) to their paired primary tumors, allowing us to identify gene expression changes occurring as patients’ tumors progressed from bevacizumab-responsive to resistant. We subsequently established two novel xenograft models of anti-angiogenic therapy resistance. We targeted a possible mediator of anti-angiogenic therapy resistance identified in our gene expression profile analysis through pharmacologic and genetic disruption in one of our new xenograft models.
MATERIALS AND METHODS
Approvals
This study was approved by the UCSF Committee on Human Research (CHR). Animal procedures were approved by the UCSF Institutional Animal Care and Use Committee (IACUC).
RNA analysis
RNA was extracted from paraffin blocks with the RecoverAll Total Nucleic Acid Isolation Kit (Ambion, Inc., Austin, TX), using 60–120 μm thick slices. Microarray analysis utilized the whole-Genome DASL Assay with HumanRef-8 BeadChips (Illumina, Inc.; San Diego, CA), a platform for FFPE-extracted samples. Microarray data was deposited in ArrayExpress (accession no. E-MTAB-1380). Microarray details, PCR reactions, and primer sequences are in Supplementary Methods.
Immunohistochemistry
Immunohistochemistry and fluorescent in situ hybridization (FISH) details are in Supplementary Methods. c-Met immunostaining was subjectively scored 1–5 by two independent observers blinded to treatment group, and was quantified using ImageJ (NIH; Bethesda, MD). CA9; c-Met; and phosphorylated FAK, STAT3, and c-Met immunostaining were quantified in ImageJ using the measure RGB feature (6) or the immunoRatio macro to measure tissue percentage staining positive.
Cells
HUVEC and U87 were obtained from and authenticated by American Type Culture Collection (ATCC) and passed less than 6 months. HMEC cells were provided by the lab of Gabriele Bergers (UCSF) and were not authenticated. Cell culture, transductions, scratch and brain slice assays, Western blotting, and flow cytometry are described in Supplementary Methods.
Animal work
Animal protocols are in Supplementary Methods. VEGF blockade used 5 mg/kg B20-4.1.1, an antibody targeting mouse and human VEGF (Genentech; South San Francisco, CA) (7) or 10 mg/kg bevacizumab intraperitoneally twice weekly, with multiple xenografts showing identical response to B20-4.1.1 or bevacizumab at these doses.
MRI
MRI protocol is in Supplementary Methods. Tumor permeability (PS) and fractional blood volume (fBV) histograms were averaged for each treatment group.
Statistics
Kaplan-Meier analysis compared treatment group survivals. Wilcoxon rank sum test analyzed nonparametric comparisons. Fisher’s test measured associations between discrete variables. Spearman’s rank correlation determined correlation between continuous variables. Box and whiskers plot outliers were outside 1.5 times interquartile range. P<0.05 was statistically significant.
RESULTS
Patient characteristics
Of 234 bevacizumab-treated GBM patients from 2006–2010 at our institution, 22 were identified who developed radiographic progression after initial response requiring surgery with pre-treatment tissue available (Supplementary Table S1). BRG resection occurred an average of 32 days after last bevacizumab dose (range 14–49 days).
Microarray analysis of BRGs versus pre-treatment paired specimens
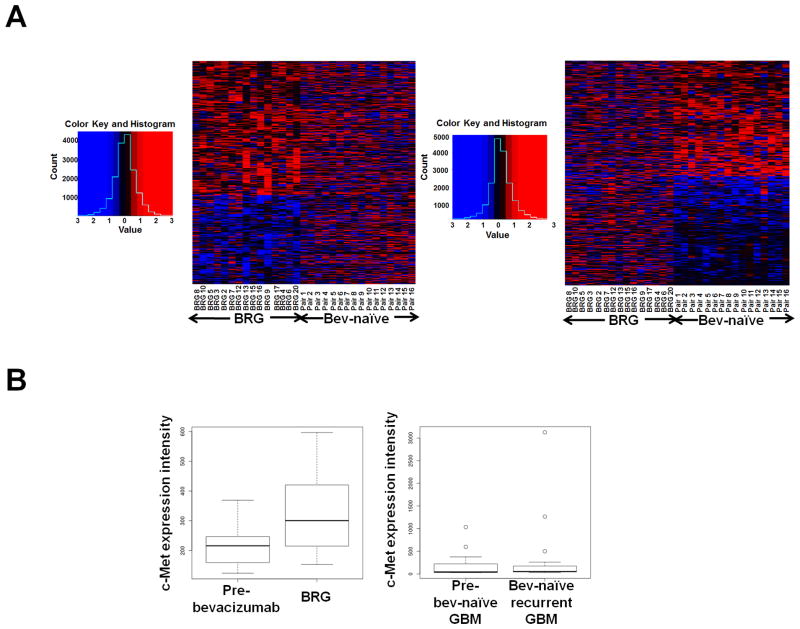
Of over 24,000 transcripts analyzed by microarrays in the 15 BRGs and their pre-treatment GBMs with sufficient RNA, there were 25 upregulated and 14 downregulated genes in BRGs versus their paired pre-treatment specimens with raw P<0.005 (Figure 1A; Supplementary Tables S2–S3). When comparing published microarray data (8) of 16 bevacizumab-naïve GBMs and their recurrences with similar age at recurrence as our BRGs (P=0.1), of the top 1000 genes whose transcription was altered in bevacizumab-naïve recurrent GBMs (raw P ≤ 0.04; Supplementary Table S4), only 33 were also altered with raw P<0.05 in BRGs, suggesting that most BRG transcriptional alterations could be unique to bevacizumab resistance. Receptor tyrosine kinase c-Met, the fifth most upregulated BRG gene (Figure 1B; Supplementary Table S3), was further investigated because its roles in invasion (9) and VEGF-independent angiogenesis (10) offered prior biological plausibility as a mediator of anti-angiogenic therapy resistance. While c-Met intensity in microarray data increased nearly 40% in the 15 BRGs versus pre-treatment, c-Met intensity did not change in the 16 bevacizumab-naïve recurrent GBMs versus their earlier GBMs (Figure 1B). Real time RT-PCR verified c-Met upregulation in BRGs versus pre-treatment and verified microarray data showing unaltered BRG transcription of other factors with prior plausibility as mediators of anti-angiogenic therapy resistance: VEGF, VEGF receptor-2 (VEGFR2), and basic fibroblast growth factor (bFGF) (Supplementary Figure S1). C-Met ligand hepatocyte growth factor (HGF) expression was unaltered in BRG microarray analysis (Supplementary Table S2). Fluorescent in situ hybridization (FISH) revealed unchanged degree of c-Met gene amplification or polysomy 7 in paired pre-treatment GBMs and BRGs (Supplementary Figure S2; Supplementary Table S5) and no c-Met gene amplification in cultured BRG cells (Supplementary Table S5), confirming that c-Met upregulation occurred at the transcription level.
Figure 1. Transcriptional analysis of BRGs and bevacizumab-naïve recurrent GBMs versus paired earlier GBMs from the same patients.
(A) Microarray analysis of 15 BRGs and their paired pre-treatment GBMs, along with 16 bevacizumab-naïve recurrent GBMs and their paired earlier GBMs produced heatmaps in which the 1000 most differentially expressed genes in BRGs (left) or bevacizumab-naïve recurrent GBMs (right) did not exhibit similar alteration patterns in the opposite group. Red/blue=increased/decreased gene expression. (B) Box and whisker plot summarizing microarray c-Met expression changes. Upon recurrence, BRGs exhibited nearly 40% c-Met upregulation, while bevacizumab-naïve recurrent GBMs exhibited unchanged c-Met. Plots show lower extreme (horizontal line below box), lower quartile (box base), median (box center), upper quartile (box roof), upper extreme (horizontal line above box), and outliers (dots outside extremes) for BRGs and their paired “pre-bevacizumab” GBMs (left), along with bevacizumab-naïve (“bev-naïve”) recurrent GBMs and their paired “pre-bev-naïve” GBMs (right).
To determine whether upregulated c-Met expression in BRGs was part of a transition towards expressing genes like c-Met associated with a mesenchymal phenotype, we measured changes in the Pearson correlation coefficients of tumor gene signatures relative to the centroids of three GBM subtypes (mesenchymal, proneural, and proliferative) (8). While published microarray data (8) from 16 paired bevacizumab-naïve GBMs and their recurrences analyzed above showed that these GBMs gained similarity to the mesenchymal centroid, our 15 BRGs analyzed by microarray did not gain similarity to any centroid (Supplementary Figure S3). When dividing BRGs into two types we previously described (11), enhancing BRGs gained more similarity to the mesenchymal centroid than bevacizumab-naïve recurrent GBMs, although this difference was insignificant (P>0.05), while non-enhancing BRGs gained more similarity to the proneural centroid.
C-Met protein expression, downstream effectors, and regulators in BRGs
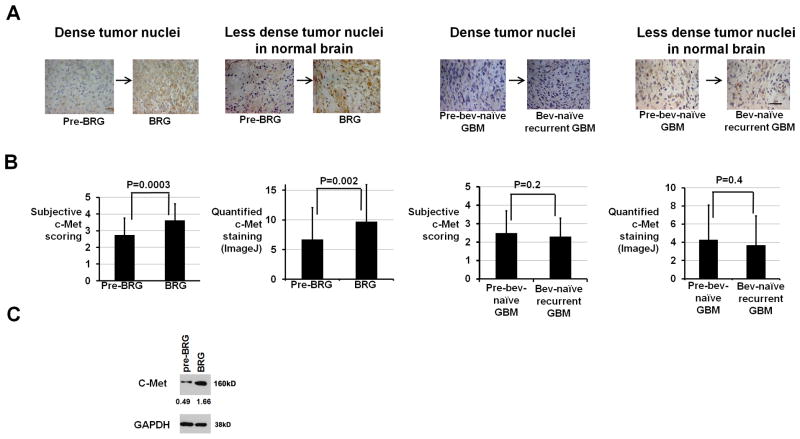
Having confirmed 6.4-fold transcriptional upregulation of c-Met by real time RT-PCR in our 22 BRGs (Supplementary Figure S1), we analyzed c-Met protein expression in BRGs (Figure 2A). Versus pre-treatment, BRGs exhibited 35% increased subjective c-Met staining score (P=0.006; Figure 2B) and 48% increased automated quantification of c-Met staining (P=0.008; Figure 2B), with 16 of 22 BRGs exhibiting increased C-Met staining by both scoring methods (Supplementary Table S1). Western blot revealed 3.4-fold increased c-Met expression in a BRG versus pre-treatment (Figure 2C). Of 2 BRG types we recently described (11), c-Met expression increased comparably in non-enhancing versus enhancing BRGs (P=0.8, data not shown).
Figure 2. C-Met upregulation in BRGs versus pre-treatment.
(A) Shown are representative c-Met immunostainings from areas of dense tumor nuclei or less dense tumor nuclei in normal brain (630x; scale bar=10 μm). (B) Both subjective and automated ImageJ scoring revealed increased c-Met staining after bevacizumab resistance (P=0.0003–0.002) with unchanged c-Met staining after bevacizumab-naïve recurrence (P=0.2–0.4). (C) Western blot revealed 3.4-fold increased c-Met expression in a BRG versus the pre-treatment GBM from the same patient. Numbers represent band densities normalized to GAPDH from the same sample.
We then verified that the increased c-Met expression in BRGs versus pre-treatment caused increased c-Met activity. Phosphorylated c-Met immunostaining nearly doubled in BRGs versus pre-treatment (P<0.05; Supplementary Figure S4). Expression of phosphorylated c-Met downstream effectors focal adhesion kinase (FAK) (12) and STAT3 (13) increased by over 40% and 100%, respectively, in BRGs versus pre-treatment (P=0.01–0.03; Supplementary Figures S5–S6).
We investigated hypoxia, a c-Met transcription regulator (14), after bevacizumab resistance by immunostaining for hypoxia marker CA9. CA9 expression increased 43% after bevacizumab resistance versus before (P<0.05; Supplementary Figure S7), with the magnitude of CA9 increase correlating with c-Met changes (Spearman rs=0.6; P<0.05). Unlike BRGs, a control group of 22 bevacizumab-naïve recurrent glioblastomas and paired initial tumors from our institution with similar average age at recurrence to our BRGs (P=0.3) exhibited unchanged CA9 (P=0.9), c-Met (P=0.2–0.4), or phosphorylated STAT3 (P=0.9) staining upon recurrence (Figures 2A–B; Supplementary Table S6; Supplementary Figures S6–S7), suggesting that the increased hypoxia, c-Met upregulation, and phosphorylation of downstream effectors of c-Met in BRGs could be specific to bevacizumab resistance.
We then investigated if increased c-Met expression in BRGs originated from endothelial cells. Hypoxia and bevacizumab lowered endothelial c-Met expression in two endothelial cell lines and in endothelial cells isolated from a BRG (Supplementary Figure S8A). Immunostaining BRGs revealed no endothelial and c-Met costaining before or after bevacizumab resistance (Supplementary Figure S8B). These findings support GBM cells, not endothelial cells, as the source of c-Met upregulation in BRGs.
Generating a xenograft model of acquired bevacizumab resistance
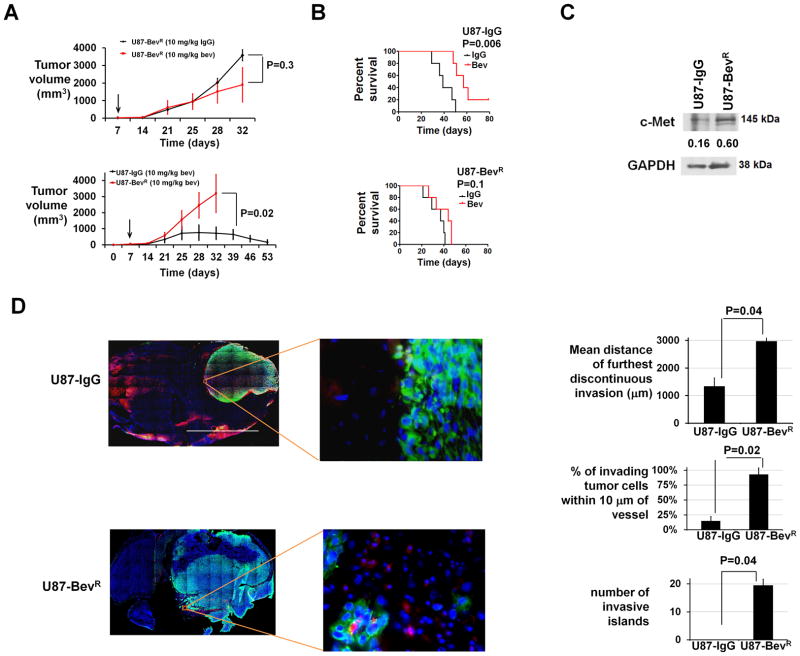
We established a xenograft model of acquired anti-angiogenic therapy resistance by treating subcutaneous U87 glioma cell line-derived xenografts with bevacizumab. The least responsive xenograft, was reimplanted and treated with bevacizumab, and the process was repeated 3 times, producing a U87-BevR xenograft. A control U87-IgG xenograft was generated in parallel with IgG treatment. U87-BevR exhibited no response to bevacizumab versus IgG treatment (P=0.3; Figure 3A), and subcutaneous U87-IgG tumors regressed with bevacizumab while U87BevR grew exponentially during bevacizumab treatment (P=0.02; Figure 3A). Intracranial U87-BevR xenografts were unresponsive to bevacizumab (P=0.1), while intracranial U87-IgG xenografts responded to bevacizumab (P=0.006) (Figure 3B). U87-BevR cells exhibited nearly 4-fold more c-Met protein expression as U87-IgG cells (Figure 3C). Hypoxia increased HGF expression in U87-IgG and U87-BevR cells 3.5- and 4.5-fold, respectively (Supplementary Figure S9). Intracranial U87-BevR xenografts exhibited greater discontinuous invasion, percent of invading tumor cells 10 μm from a vessel, and invasive islands than U87-IgG (P<0.05; Figure 3D).
Figure 3. Xenograft model of acquired anti-angiogenic therapy resistance.
(A) Subcutaneous U87-BevR was unresponsive to bevacizumab versus IgG (P=0.3; upper left), and subcutaneous U87-IgG tumors regressed during bevacizumab treatment while U87-BevR grew exponentially during bevacizumab treatment (P<0.05; lower left). Arrows indicate time bevacizumab treatment began. (B) U87-IgG and U87-BevR xenografts were responsive (P=0.006) and resistant (P=0.1), respectively, to bevacizumab in the orthotopic intracranial environment. (C) Western blot revealed 4-fold more c-Met in U87-BevR than U87-IgG cells. Numbers represent band densities normalized to GAPDH from the same sample. (D) Staining intracranial U87-IgG and U87-BevR xenografts for collagen IV and human vimentin revealed further discontinuous invasion, higher percentage of invasive cells 10 μm from a vessel, and more invasive islands in U87-BevR than U87-IgG (P<0.05). 2x/400x; scale bar=2000/20 μm
Generating a xenograft model of intrinsic bevacizumab resistance
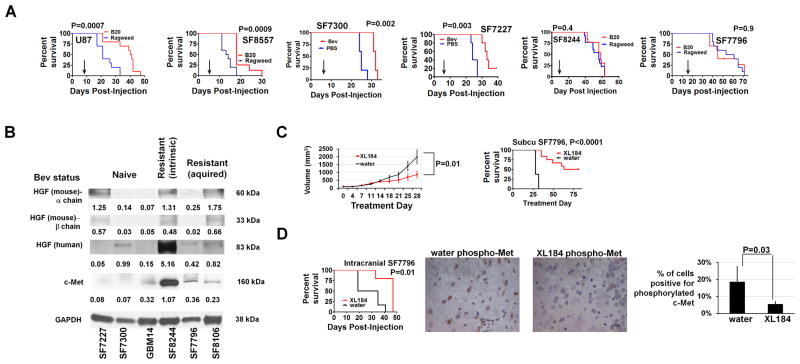
We implanted fresh patient GBM tissue subcutaneously into immunodeficient mice, establishing xenografts from two GBMs which were resected because they acquired bevacizumab resistance after initial responsiveness (SF7796 and SF8106) and one GBM which proved after tissue acquisition to be intrinsically bevacizumab-resistant without radiographic response (SF8244). These xenografts were serially passaged subcutaneously in vivo and formed tumors intracranially. Histologically, SF8106 and SF7796 xenografts exhibited greater distance of white matter invasion (P=0.04) than xenografts from bevacizumab-naïve GBMs (Supplementary Figures S10–13). While the percentage of invasive cells 10 μm from vessels, a marker of perivascular invasion, and islands of 3 or more cells clustered together invading away from the primary mass were higher in BRG-derived xenografts than in most xenografts from bevacizumab-naïve GBMs, these tendencies were insignificant (P=0.1). SF8244, derived from a GBM with intrinsic bevacizumab resistance, exhibited discontinuous and perivascular invasion, albeit less than SF7796 and SF8106.
To determine whether these xenografts maintained the resistance or response to anti-angiogenic therapy found in their patient tumors, we treated xenografts with B20-4.1.1 or bevacizumab. Unlike intracranial U87 cell line-derived xenografts and intracranial SF8557 and SF7300 xenografts established from bevacizumab-naïve GBMs, which responded to VEGF blockade (P=0.0007 U87; P=0.0009 SF8557; P=0.002 SF7300), mice with intracranial SF8244 and SF7796 xenografts exhibited unaltered survival after B20-4.1.1 treatment (P=0.4–0.9) (Figure 4A). While intracranial U87 xenografts exhibited over two-thirds less vascular permeability (PS; P=0.03) and fractional blood volume (fBV, P=0.001) on DCE-MRI after B20-4.1.1 treatment, intracranial SF7796 xenografts exhibited unchanged PS (P=0.4) or fBV (P=0.3) after B20-4.1.1 treatment (Supplementary Figure S14). Thus, BRG-derived xenografts maintained the resistance to VEGF blockade seen in the human tumors they derived from, including resistance to radiological and survival-promoting effects of VEGF blockade.
Figure 4. Xenograft model of intrinsic anti-angiogenic therapy resistance exhibits growth suppression with pharmacologic c-Met blockade.
(A) While immunodeficient mice with intracranial U87, SF8557, SF7300, or SF7227 xenografts exhibited prolonged survival with B20-4.1.1 or bevacizumab versus ragweed control antibody or PBS treatment (n=10/group U87 and SF8557, n=5/group SF7300 and SF7227; P=0.0007 U87, P=0.0009 SF8557, P=0.002 SF7300, P=0.003 SF7227), B20-4.1.1 did not affect survival of mice with intracranial xenografts derived from a GBM with intrinsic (SF8244) or acquired (SF7796) bevacizumab resistance (n=10/group; P=0.4–0.9). Arrows indicate time of treatment initiation. (B) Western blot showing c-Met, human HGF, and mouse HGF expression of 3 xenografts from bevacizumab-naïve GBMs (SF7227, SF7300, and GBM14) and from GBMs with intrinsic (SF8244) or acquired (SF7796 and SF8106) bevacizumab resistance. Numbers represent band densities normalized to GAPDH from the same sample. (C) C-met inhibitor XL184 prevented volumetric growth of subcutaneous SF7796 xenografts after 4 treatment weeks (P=0.01; upper left; n=5/group) and prolonged survival (P<0.0001; upper right). (D) XL184 prolonged survival of mice with intracranial SF7796 xenografts (n=6/group; P=0.01) (lower left). Immunohistochemistry (lower right) revealed reduced phosphorylated c-Met in XL184-treated versus water-treated intracranial SF7796 xenografts (P=0.03). 630x; scale bar=10 μm.
Treating a bevacizumab-resistant glioblastoma xenograft with a c-Met inhibitor
Western blot revealed that combined c-Met and human HGF expression distinguished xenografts derived from GBMs with intrinsic (SF8244) or acquired (SF8106 and SF7796) resistance versus those derived from bevacizumab-naïve GBMs (Figure 4B). We then investigated whether growth of bevacizumab-resistant xenograft SF7796 could be attenuated by XL184, a tyrosine kinase inhibitor targeting c-Met. Treating SF7796 subcutaneous tumors with XL184 for 28 days reduced tumor volume nearly three-fold (P=0.01; Figure 4C). XL184 prolonged the median survivals of mice with subcutaneous SF7796 tumors from 28 to 72 days (P<0.0001; Figure 4C) and intracranial SF7796 tumors from 27 to 47 days (P=0.01; Figure 4D), with nearly four-fold less phosphorylated c-Met staining after XL184 treatment of intracranial SF7796 (P=0.03) (Figure 4D).
Transducing a bevacizumab-resistant xenograft with c-Met shRNA reduces survival in hypoxia and invasiveness and causes bevacizumab responsiveness
We engineered SF7796 cells to express three shRNAs targeting c-Met, each diminishing c-Met protein expression by over 65% versus control shRNAs, with the shRNA diminishing c-Met protein expression the most causing 91% and 55% loss of c-Met mRNA and protein, respectively, relative to control shRNA (Supplementary Figure S15). We also engineered U87-BevR cells to express two shRNAs targeting c-Met, each diminishing c-Met protein expression versus cells transduced with control shRNA and parental U87-BevR cells (Supplementary Figure S16A).
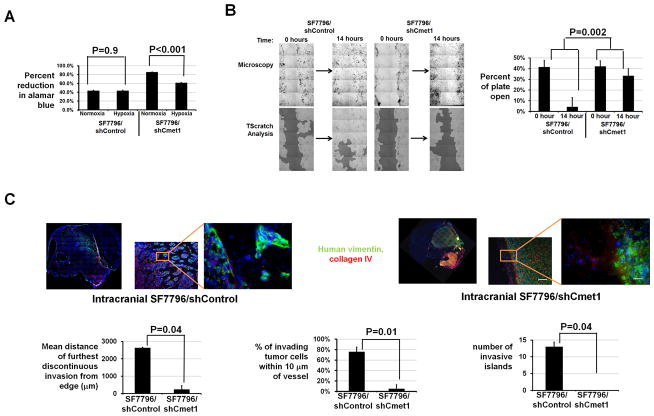
To determine if c-Met upregulation in BRGs promotes survival in the increased hypoxia we identified in BRGs, we investigated the impact of c-Met knockdown on the survival of cells derived from a bevacizumab-resistant xenograft in hypoxia. SF7796/shCmet1 cells grew slightly faster in culture and exhibited 28% fewer cells after 48 hours in hypoxia versus normoxia (P<0.001), while cultured SF7796/shControl cells exhibited unaltered cell numbers after 48 hours in hypoxia (P=0.9; Figure 5A).
Figure 5. Effects of c-Met shRNA on survival in hypoxia, in vitro migration, and in vivo invasion of cells from bevacizumab-resistant xenografts.
(A) Percent Alomar blue reduction, indicating cell survival, was less in SF7796/shCmet1 in hypoxia versus normoxia (P<0.001), but unchanged in SF7796/shControl in hypoxia versus normoxia (P=0.9). (B) After 14 hours, scratches were 91% filled with SF7796/shControl cells, while only 21% scratch reduction occurred with SF7796/shCmet1 cells (P=0.002). (C) Intracranial SF7796/shCmet1-derived xenografts (n=3) exhibited reduced invasive distance from continuous tumor edge (P=0.04), percentage of invading tumor cells 10 μm from a vessel (P=0.01), and invasive islands (P=0.04) than intracranial SF7796/shControl tumors (n=3). Red=collagen IV (vessels); blue=Hoechst nuclear stain; green=human vimentin. 2x/100x/400x, scale bar=2000, 60/20μm.
Because anti-angiogenic therapy resistance can be associated with increased tumor cell invasiveness (15), the impact of c-Met knockdown on the morphology and invasiveness of cells derived from a bevacizumab-resistant xenograft was investigated. SF7796/shCmet1 cells exhibited 40% reduced inverse shape factor, a unitless parameter measuring a cell’s dendricity (16, 17), versus SF7796/shControl cells (P<0.05; Supplementary Figure S17). SF7796/shCmet1 cells were less migratory than SF7796/shControl cells, as scratch assays revealed 91% versus 21% scratch reduction at 14 hours with SF7796/shControl and SF7796/shCmet1 cells, respectively (P=0.002) (Figure 5B). In brain slice invasion assays, SF7796/shControl exhibited more invasive multiple cell islands (P=0.03) and single cells (P=0.04) after 3 days than SF7796/shCmet1 (Supplementary Figure S18). Similarly, the mean fluorescence intensity outside of the core collection of cells after 96 hours was higher after injecting U87-BevR/shControl cells into brain slices than after injecting either U87-BevR/shCmet2 or U87-BevR/shCmet3 cells into brain slices (P<0.001; Supplementary Figure S16B). Thus, c-Met knockdown altered 2 features, survival in hypoxia and invasiveness, associated with anti-angiogenic therapy resistance.
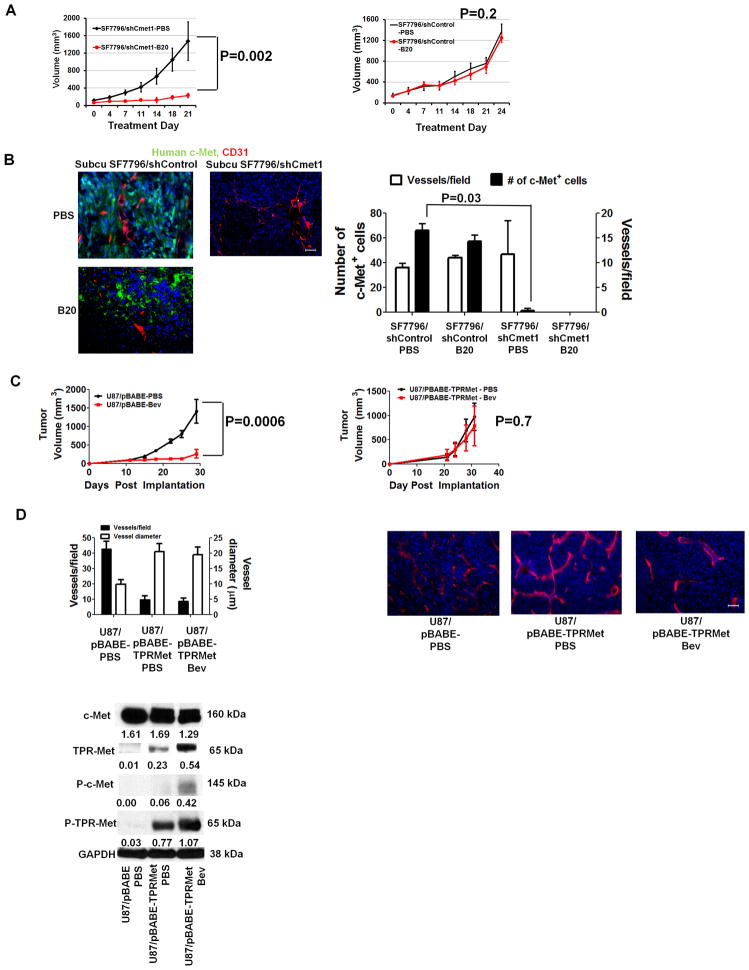
We then established xenografts from SF7796/shCmet1 and SF7796/shControl cells. Intracranial SF7796/shCmet1 xenografts exhibited sharper borders with shorter discontinuous invasion from continuous tumor edge; fewer percentage of invasive cells 10 μm from a vessel; and fewer invasive islands (P<0.05) than intracranial SF7796/shControl xenografts (Figure 5C, Supplementary Figure S19). Serial volumetric analysis of subcutaneous SF7796/shCmet1 and SF7796/shControl xenografts revealed that the latter maintained the B20-4.1.1 resistance of parental SF7796 cells (P=0.2), while SF7796/shCmet1 xenografts exhibited B20-4.1.1 responsiveness (P=0.002) (Figure 6A), suggesting c-Met necessity for xenograft resistance to VEGF blockade. There were similar vessel densities in B20- versus PBS-treated SF7796/shControl xenografts (P=0.8; Figure 6B). C-met staining was reduced in PBS-treated SF7796/shCmet1 xenografts versus PBS-treated SF7796/shControl xenografts (P=0.03), while B20-treated SF7796/shCMet1 xenografts regressed and could not be analyzed (Figure 6B).
Figure 6. Effects of genetic c-Met alteration on xenograft responsiveness to VEGF blockade.
(A) SF7796/shCmet1-derived subcutaneous tumors exhibited B20-4.1.1 responsiveness (n=5/group; P=0.002), while SF7796/shControl-derived subcutaneous tumors exhibited B20-4.1.1 resistance (n=5/group; P=0.2). (B) Immunofluorescence revealed unaltered vessel density in PBS- versus B20-treated subcutaneous SF7796/shControl (P=0.8) and fewer c-Met-positive cells in PBS-treated subcutaneous SF7796/shCmet1 versus PBS-treated subcutaneous SF7796/shControl (P=0.03). B20-4.1.1-treated subcutaneous SF7796/shCmet1 xenografts regressed and could not be analyzed. 200x, scale bar=30 μm. (C) Treating U87/pBABE-Puro-Tpr-met subcutaneous xenografts with bevacizumab (Bev) caused no response versus PBS (n=5/group; P=0.7), while bevacizumab caused U87/pBABE-Puro subcutaneous xenograft regression (n=5/group; P=0.001). Error bars represent standard errors of the mean. (D) Immunohistochemistry revealed that U87/pBABE-Puro-Tpr-met xenografts exhibited fewer (P=0.04), larger (P=0.03) vessels than U87/pBABE-Puro xenografts, with bevacizumab not altering the U87/pBABE-Puro-Tpr-met vascular pattern (P=0.8–0.9). Bevacizumab-treated U87/pBABE-Puro xenografts regressed and could not be analyzed. 200x, scale bar=30 μm. Western blot identified Tpr-met and phosphorylated Tpr-met (P-Tpr-met) in U87/pBABE-Puro-Tpr-met xenografts, but not in U87/pBABE-Puro xenografts. Western blot numbers represent band densities normalized to GAPDH from the same sample.
Transduction of a bevacizumab-responsive xenograft with constitutively active c-Met causes bevacizumab resistance
We engineered U87 cells to express Tpr-met, a cytoplasmic fusion protein escaping lysosomal degradation and causing constitutive c-Met activation (18). Transduction generated U87/pBABE-Puro and U87/pBABE-Puro-Tpr-met cells, with only the former exhibiting phosphorylated Tpr-Met (Supplementary Figure S20). Treating U87/pBABE-Puro-Tpr-met subcutaneous xenografts with bevacizumab caused no response versus PBS treatment (P=0.7), consistent with resistance, while bevacizumab caused U87/pBABE-Puro subcutaneous xenograft regression (P<0.001) (Figure 6C). Immunohistochemistry revealed fewer but larger vessels in U87/pBABE-Puro-Tpr-met xenografts versus U87/pBABE-Puro xenografts (P<0.05), with bevacizumab not altering the U87/pBABE-Puro-Tpr-met xenografts vascular pattern (Figure 6D). Western blot revealed Tpr-met and phosphorylated Tpr-met in U87/pBABE-Puro-Tpr-met xenografts, but not in U87/pBABE-Puro xenografts (Figure 6D). Bevacizumab increased Tpr-Met and phosphorylated Tpr-Met levels in U87/pBABE-Puro-Tpr-met xenografts (Figure 6D).
DISCUSSION
While much heralded, the arrival of angiogenesis inhibitors has been associated with mostly transient responses followed by renewed tumor growth. We used microarray analysis and novel murine models of glioblastoma anti-angiogenic therapy resistance to provide evidence for the role of increased c-Met expression in acquired anti-angiogenic therapy resistance.
While the Bonferroni adjustment to reduce microarray false positives caused significant raw P values to no longer be below 0.05 (Supplementary Table S2), the Bonferroni correction is not crucial for studies like this using microarray data to launch further studies into specific genes with significant raw P values and prior plausibility as candidates (19, 20). C-Met fulfilled these criteria as the fifth most upregulated gene of 24,000 analyzed and because of its roles in invasion (9) and VEGF-independent angiogenesis (10), features associated with angiogenesis inhibitor resistance (5).
Our finding of upregulated c-Met in BRGs versus their paired pre-treatment specimens appeared unique to bevacizumab resistance, as c-Met was not upregulated in bevacizumab-naïve recurrent GBMs. Discrepancies between our findings and a study which noted increased c-Met expression in all recurrent GBMs (21) may reflect that study analyzing c-Met expression as a dichotomous covariate rather than the dual use of subjective and automated scoring in our study.
To functionally examine this observed c-Met upregulation, we established the first two glioblastoma xenograft models of anti-angiogenic therapy resistance. Our first xenograft modeled acquired anti-angiogenic therapy resistance and was established by serially treating cell line-derived xenografts with bevacizumab until they became resistant, generating a stably resistant xenograft line. Like the 22 BRGs we analyzed, this resistant xenograft line exhibited increased c-Met expression compared to its parental sensitive xenograft. Our second xenograft modeled intrinsic anti-angiogenic therapy resistance and was established by implanting BRG tissue into mice, a technique recapitulating GBM biology (22–25). Resulting xenografts maintained the refractoriness to VEGF blockade found in the BRG and exhibited more invasiveness than xenografts from bevacizumab-naïve GBMs. Maintenance of anti-angiogenic therapy resistance in BRG-derived xenografts could reflect persistent resistance-mediating factors from the BRG or invasiveness of the BRG-derived xenograft allowing tumors to grow by vessel cooption whereby neovascularization is unnecessary (22). While our U87-derived model allows valuable comparisons between paired bevacizumab-resistant and bevacizumab-responsive cells derived from the same cell line, the origin of these cells from a several decade old cell line that likely carries alterations from passage in culture is a disadvantage versus our second model which was derived directly from fresh patient specimens.
Further work will need to clarify how c-Met upregulation compensates for VEGF blockade. One possibility is c-Met decreasing apoptosis during VEGF blockade, as shown after EGFR inhibition (26). Our observation of reduced survival in hypoxia of cells expressing c-Met shRNA supports this mechanism. A second possibility is c-Met signaling to VEGFR independent of VEGF, as demonstrated with c-Met and EGFR (27). A third possibility is that c-Met-induced invasion allows escape from hypoxic areas, particularly if the invasion is perivascular (28) and brings tumor cells closer to vessels remaining after anti-angiogenic therapy, as identified in our BRGs and a VEGF-knockout glioma (29). Our finding of bevacizumab resistance in cells expressing cytoplasmic constitutively active c-Met suggests that c-Met upregulation compensates for VEGF blockade even with cytoplasmic c-Met signaling.
The role for c-Met in invasiveness shown in other studies (9) and ours could reflect 3 c-Met effects. First, invasion could reflect chemotaxis to HGF (9). While c-Met-mediated invasion occurs in response to paracrine or autocrine HGF (30), human HGF expression by bevacizumab-resistant xenografts and the inability of mouse HGF to bind human c-Met suggests that autocrine c-Met signaling, which comprises most GBM c-Met signaling (31), contributes to anti-angiogenic therapy resistance. Second, c-Met-induced FAK phosphorylation, which we confirmed in BRGs versus pre-treatment, could promote invasion (32). Third, c-Met-induced altered cell morphology, suggested by our finding that c-Met knockdown reduced inverse shape factor, a marker of non-spherical morphology, could promote invasiveness. C-Met-induced morphologic changes could be caused by STAT3 phosphorylation (33), which we confirmed to be increased in BRGs versus pre-treatment.
While further work will need to identify regulators of the upregulated c-Met we identified in BRGs, our finding that hypoxia and c-Met expression in BRGs both increased versus pre-treatment in correlated fashion suggests that hypoxia caused by anti-angiogenic therapy (34) plays a role. Hypoxia-driven c-Met transcription could be mediated by HIF-1α (14) or hypoxia-induced NF-κB (35), which promotes c-Met transcription (36).
While bevacizumab-naïve GBMs gained similarity to the mesenchymal centroid more than other GBM subtypes, BRGs did not gain similarity to any centroid. While c-Met is more commonly expressed by mesenchymal GBMs than other subtypes, c-Met is not one of the genes defining subtype centroids (8). Given the nearly ubiquitous nature of c-Met upregulation in our BRGs, a mesenchymal transition may occur more frequently after bevacizumab resistance than after bevacizumab-naïve recurrences, as others have suggested (37), but this transition may not be captured by genes defining GBM subtypes and may involve another combination of genes including c-Met, a hypothesis requiring further study.
While we investigated BRG transcriptional changes, anti-angiogenic therapy resistance could also reflect altered translation or post-translational modifications, as suggested by the demonstration that VEGF suppresses c-Met signaling through a VEGFR2/c-Met complex (38), which could mean that bevacizumab-induced VEGF depletion could eliminate basal inhibition of c-Met signaling, which would supplement transcriptional c-Met upregulation we observed in BRGs.
Our findings suggest that c-Met targeting using XL184 or other c-Met inhibitors (39) may be effective as monotherapy for tumors resistant to anti-angiogenic therapy, as we investigated. Alternatively, because genetic c-Met knockdown restored sensitivity to VEGF blockade, c-Met could be targeted alongside VEGF blockade to reduce anti-angiogenic therapy resistance, similar to a report combining a VEGFR inhibitor with XL184 (40). Our findings validate c-Met as a therapeutic target in tumors resistant to anti-angiogenic therapy and provide insight into mechanisms of anti-angiogenic therapy resistance, a problem that has unfortunately limited efficacy of these therapies.
Supplementary Material
STATEMENT OF TRANSLATIONAL RELEVANCE.
The anticancer efficacy of angiogenesis inhibitors like VEGF neutralizing antibody bevacizumab has been hindered by lack of enduring responses. We performed microarray analysis of glioblastomas that acquired resistance to bevacizumab, and chose c-Met, an upregulated gene expressing a receptor tyrosine kinase, for further investigation because of its roles in invasion and VEGF-independent angiogenesis. Elevated hypoxia occurred after bevacizumab resistance in a manner correlating with increased c-Met expression. Growth of a novel xenograft model of anti-angiogenic therapy resistance was inhibited by a c-Met inhibitor. Transducing these xenograft cells with c-Met shRNA inhibited their invasion and survival in hypoxia, and converted them from bevacizumab-resistant to responsive. Transducing bevacizumab-responsive GBM cells to express constitutively active c-Met rendered them bevacizumab-resistant. These findings suggest that c-Met upregulation can mediate anti-angiogenic therapy resistance and that c-Met disruption warrants investigation as a means of overcoming anti-angiogenic therapy resistance, allowing these treatments to fulfill their promise.
Acknowledgments
Work was supported by funding to MKA’s laboratory from the ABTA, the James S. McDonnell Foundation, ACS, the NIH (5K02NS64167-2), and the UCSF Brain Tumor SPORE CA097257. A.J. is an HHMI fellow. Jonathan Levy and Alexander Jackson in Roger Nicoll’s lab assisted in brain slice assays. B20-4.1.1 was provided by Genentech. XL184 was provided by Exelixis.
Footnotes
Conflicts of interest: W.S.C. is co-Founder, CEO, and shareholder of OncoSynergy. M.K.A. is a Scientific Advisory Board Member and shareholder of OncoSynergy.
References
- 1.Vredenburgh JJ, Desjardins A, Herndon JE, 2nd, Marcello J, Reardon DA, Quinn JA, et al. Bevacizumab plus irinotecan in recurrent glioblastoma multiforme. J Clin Oncol. 2007;25:4722–9. doi: 10.1200/JCO.2007.12.2440. [DOI] [PubMed] [Google Scholar]
- 2.Vredenburgh JJ, Desjardins A, Herndon JE, 2nd, Dowell JM, Reardon DA, Quinn JA, et al. Phase II trial of bevacizumab and irinotecan in recurrent malignant glioma. Clin Cancer Res. 2007;13:1253–9. doi: 10.1158/1078-0432.CCR-06-2309. [DOI] [PubMed] [Google Scholar]
- 3.Jain RK, Duda DG, Clark JW, Loeffler JS. Lessons from phase III clinical trials on anti-VEGF therapy for cancer. Nat Clin Pract Oncol. 2006;3:24–40. doi: 10.1038/ncponc0403. [DOI] [PubMed] [Google Scholar]
- 4.Clark AJ, Lamborn KR, Butowski NA, Chang SM, Prados MD, Clarke JL, et al. Neurosurgical management and prognosis of patients with glioblastoma that progress during bevacizumab treatment. Neurosurgery. 2011 doi: 10.1227/NEU.0b013e3182314f9d. [DOI] [PubMed] [Google Scholar]
- 5.Bergers G, Hanahan D. Modes of resistance to anti-angiogenic therapy. Nat Rev Cancer. 2008;8:592–603. doi: 10.1038/nrc2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nair A, Thevenot P, Dey J, Shen J, Sun MW, Yang J, et al. Novel polymeric scaffolds using protein microbubbles as porogen and growth factor carriers. Tissue Eng Part C Methods. 2010;16:23–32. doi: 10.1089/ten.tec.2009.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang WC, Wu X, Peale FV, Lee CV, Meng YG, Gutierrez J, et al. Cross-species vascular endothelial growth factor (VEGF)-blocking antibodies completely inhibit the growth of human tumor xenografts and measure the contribution of stromal VEGF. J Biol Chem. 2006;281:951–61. doi: 10.1074/jbc.M508199200. [DOI] [PubMed] [Google Scholar]
- 8.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–73. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 9.Eckerich C, Zapf S, Fillbrandt R, Loges S, Westphal M, Lamszus K. Hypoxia can induce c-Met expression in glioma cells and enhance SF/HGF-induced cell migration. Int J Cancer. 2007;121:276–83. doi: 10.1002/ijc.22679. [DOI] [PubMed] [Google Scholar]
- 10.Shojaei F, Lee JH, Simmons BH, Wong A, Esparza CO, Plumlee PA, et al. HGF/c-Met acts as an alternative angiogenic pathway in sunitinib-resistant tumors. Cancer Res. 2010;70:10090–100. doi: 10.1158/0008-5472.CAN-10-0489. [DOI] [PubMed] [Google Scholar]
- 11.De Lay M, Jahangiri A, Carbonell WS, Hu YL, Tsao S, Tom MW, et al. Microarray analysis verifies two distinct phenotypes of glioblastomas resistant to anti-angiogenic therapy. Clin Cancer Res. 2012;18:2930–42. doi: 10.1158/1078-0432.CCR-11-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maulik G, Kijima T, Ma PC, Ghosh SK, Lin J, Shapiro GI, et al. Modulation of the c-Met/hepatocyte growth factor pathway in small cell lung cancer. Clin Cancer Res. 2002;8:620–7. [PubMed] [Google Scholar]
- 13.Giannoni P, Scaglione S, Quarto R, Narcisi R, Parodi M, Balleari E, et al. An interaction between hepatocyte growth factor and its receptor (c-MET) prolongs the survival of chronic lymphocytic leukemic cells through STAT3 phosphorylation: a potential role of mesenchymal cells in the disease. Haematologica. 2011;96:1015–23. doi: 10.3324/haematol.2010.029736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hara S, Nakashiro K, Klosek SK, Ishikawa T, Shintani S, Hamakawa H. Hypoxia enhances c-Met/HGF receptor expression and signaling by activating HIF-1alpha in human salivary gland cancer cells. Oral Oncol. 2006;42:593–8. doi: 10.1016/j.oraloncology.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 15.Paez-Ribes M, Allen E, Hudock J, Takeda T, Okuyama H, Vinals F, et al. Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer Cell. 2009;15:220–31. doi: 10.1016/j.ccr.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heffron DS, Mandell JW. Opposing roles of ERK and p38 MAP kinases in FGF2-induced astroglial process extension. Mol Cell Neurosci. 2005;28:779–90. doi: 10.1016/j.mcn.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 17.Levi-Schaffer F, Slovik D, Armetti L, Pickholtz D, Touitou E. Activation and inhibition of mast cells degranulation affect their morphometric parameters. Life Sci. 2000;66:PL283–90. doi: 10.1016/s0024-3205(00)00538-5. [DOI] [PubMed] [Google Scholar]
- 18.Mak HH, Peschard P, Lin T, Naujokas MA, Zuo D, Park M. Oncogenic activation of the Met receptor tyrosine kinase fusion protein, Tpr-Met, involves exclusion from the endocytic degradative pathway. Oncogene. 2007;26:7213–21. doi: 10.1038/sj.onc.1210522. [DOI] [PubMed] [Google Scholar]
- 19.Leung YF, Cavalieri D. Fundamentals of cDNA microarray data analysis. Trends Genet. 2003;19:649–59. doi: 10.1016/j.tig.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 20.Chen JJ, Wang SJ, Tsai CA, Lin CJ. Selection of differentially expressed genes in microarray data analysis. Pharmacogenomics J. 2007;7:212–20. doi: 10.1038/sj.tpj.6500412. [DOI] [PubMed] [Google Scholar]
- 21.Liu W, Fu Y, Xu S, Ding F, Zhao G, Zhang K, et al. c-Met expression is associated with time to recurrence in patients with glioblastoma multiforme. J Clin Neurosci. 2011;18:119–21. doi: 10.1016/j.jocn.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 22.Martens T, Laabs Y, Gunther HS, Kemming D, Zhu Z, Witte L, et al. Inhibition of glioblastoma growth in a highly invasive nude mouse model can be achieved by targeting epidermal growth factor receptor but not vascular endothelial growth factor receptor-2. Clin Cancer Res. 2008;14:5447–58. doi: 10.1158/1078-0432.CCR-08-0147. [DOI] [PubMed] [Google Scholar]
- 23.Engebraaten O, Hjortland GO, Hirschberg H, Fodstad O. Growth of precultured human glioma specimens in nude rat brain. J Neurosurg. 1999;90:125–32. doi: 10.3171/jns.1999.90.1.0125. [DOI] [PubMed] [Google Scholar]
- 24.Horten BC, Basler GA, Shapiro WR. Xenograft of human malignant glial tumors into brains of nude mice. A histopatholgical study. J Neuropathol Exp Neurol. 1981;40:493–511. doi: 10.1097/00005072-198109000-00002. [DOI] [PubMed] [Google Scholar]
- 25.Giannini C, Sarkaria JN, Saito A, Uhm JH, Galanis E, Carlson BL, et al. Patient tumor EGFR and PDGFRA gene amplifications retained in an invasive intracranial xenograft model of glioblastoma multiforme. Neuro-oncol. 2005;7:164–76. doi: 10.1215/S1152851704000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jun HJ, Acquaviva J, Chi D, Lessard J, Zhu H, Woolfenden S, et al. Acquired MET expression confers resistance to EGFR inhibition in a mouse model of glioblastoma multiforme. Oncogene. 2011 doi: 10.1038/onc.2011.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dulak AM, Gubish CT, Stabile LP, Henry C, Siegfried JM. HGF-independent potentiation of EGFR action by c-Met. Oncogene. 2011;30:3625–35. doi: 10.1038/onc.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubenstein JL, Kim J, Ozawa T, Zhang M, Westphal M, Deen DF, et al. Anti-VEGF antibody treatment of glioblastoma prolongs survival but results in increased vascular cooption. Neoplasia. 2000;2:306–14. doi: 10.1038/sj.neo.7900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du R, Lu KV, Petritsch C, Liu P, Ganss R, Passegue E, et al. HIF1alpha induces the recruitment of bone marrow-derived vascular modulatory cells to regulate tumor angiogenesis and invasion. Cancer Cell. 2008;13:206–20. doi: 10.1016/j.ccr.2008.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su W, Gutmann DH, Perry A, Abounader R, Laterra J, Sherman LS. CD44-independent hepatocyte growth factor/c-Met autocrine loop promotes malignant peripheral nerve sheath tumor cell invasion in vitro. Glia. 2004;45:297–306. doi: 10.1002/glia.10340. [DOI] [PubMed] [Google Scholar]
- 31.Xie Q, Bradley R, Kang L, Koeman J, Ascierto ML, Worschech A, et al. Hepatocyte growth factor (HGF) autocrine activation predicts sensitivity to MET inhibition in glioblastoma. Proc Natl Acad Sci U S A. 2011;109:570–5. doi: 10.1073/pnas.1119059109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agochiya M, Brunton VG, Owens DW, Parkinson EK, Paraskeva C, Keith WN, et al. Increased dosage and amplification of the focal adhesion kinase gene in human cancer cells. Oncogene. 1999;18:5646–53. doi: 10.1038/sj.onc.1202957. [DOI] [PubMed] [Google Scholar]
- 33.Boccaccio C, Ando M, Tamagnone L, Bardelli A, Michieli P, Battistini C, et al. Induction of epithelial tubules by growth factor HGF depends on the STAT pathway. Nature. 1998;391:285–8. doi: 10.1038/34657. [DOI] [PubMed] [Google Scholar]
- 34.Keunen O, Johansson M, Oudin A, Sanzey M, Rahim SA, Fack F, et al. Anti-VEGF treatment reduces blood supply and increases tumor cell invasion in glioblastoma. Proc Natl Acad Sci U S A. 2011;108:3749–54. doi: 10.1073/pnas.1014480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Culver C, Sundqvist A, Mudie S, Melvin A, Xirodimas D, Rocha S. Mechanism of hypoxia-induced NF-kappaB. Molecular and cellular biology. 2010;30:4901–21. doi: 10.1128/MCB.00409-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Bacco F, Luraghi P, Medico E, Reato G, Girolami F, Perera T, et al. Induction of MET by ionizing radiation and its role in radioresistance and invasive growth of cancer. J Natl Cancer Inst. 2011;103:645–61. doi: 10.1093/jnci/djr093. [DOI] [PubMed] [Google Scholar]
- 37.Carbone C, Moccia T, Zhu C, Paradiso G, Budillon A, Chiao PJ, et al. Anti-VEGF treatment-resistant pancreatic cancers secrete proinflammatory factors that contribute to malignant progression by inducing an EMT cell phenotype. Clin Cancer Res. 2011;17:5822–32. doi: 10.1158/1078-0432.CCR-11-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu KV, Chang JP, Parachoniak CA, Pandika MM, Aghi MK, Meyronet D, et al. VEGF Inhibits Tumor Cell Invasion and Mesenchymal Transition through a MET/VEGFR2 Complex. Cancer Cell. 2012;22:21–35. doi: 10.1016/j.ccr.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Comoglio PM, Giordano S, Trusolino L. Drug development of MET inhibitors: targeting oncogene addiction and expedience. Nat Rev Drug Discov. 2008;7:504–16. doi: 10.1038/nrd2530. [DOI] [PubMed] [Google Scholar]
- 40.You WK, Sennino B, Williamson CW, Falcon B, Hashizume H, Yao LC, et al. VEGF and c-Met blockade amplify angiogenesis inhibition in pancreatic islet cancer. Cancer Res. 2011;71:4758–68. doi: 10.1158/0008-5472.CAN-10-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.