Abstract
The upregulation or mutation of C-MYC has been observed in gastric, colon, breast, and lung tumors and in Burkitt’s lymphoma. However, little is known about the role C-MYC plays in gastric adenocarcinoma. In the present study, we intended to investigate the influence of C-MYC on the growth, proliferation, apoptosis, invasion, and cell cycle of the gastric cancer cell line SGC7901 and the gastric cell line HFE145. C-MYC cDNA was subcloned into a constitutive vector PCDNA3.1 followed by transfection in normal gastric cell line HFE145 by using liposome. Then stable transfectants were selected and appraised. Specific inhibition of C-MYC was achieved using a vector-based siRNA system which was transfected in gastric cancer cell line SGC7901. The apoptosis and cell cycles of these clones were analyzed by using flow cytometric assay. The growth and proliferation were analyzed by cell growth curves and colony-forming assay, respectively. The invasion of these clones was analyzed by using cell migration assay. The C-MYC stable expression clones (HFE-Myc) and C-MYC RNAi cells (SGC-MR) were detected and compared with their control groups, respectively. HFE-Myc grew faster than HFE145 and HFE-PC (HFE145 transfected with PCDNA3.1 vector). SGC-MR1, 2 grew slower than SGC7901 and SGC-MS1, 2 (SGC7901 transfected with scrambled control duplexes). The cell counts of HFE-Myc in the third, fourth, fifth, sixth, and seventh days were significantly more than those of control groups (P < 0.05). Those of SGC-MR1, 2 in the fourth, fifth, sixth, and seventh days were significantly fewer than those of control groups (P < 0.05). Cell cycle analysis showed that proportions of HFE-Myc and SGC-MR cells in G0–G1 and G2–M were different significantly with their control groups, respectively (P < 0.05). The apoptosis rate of HFE-Myc was significantly higher than those of control groups (P < 0.05). Results of colony-forming assay showed that the colony formation rate of HFE-Myc was higher than those of control groups; otherwise, the rate of SGC-MR was lower than those of their control groups (P < 0.05). The results of cell migration assay showed that there were no significant differences between experimental groups and control groups (P > 0.05). In conclusion, C-MYC can promote the growth and proliferation of normal gastric cells, and knockdown of C-MYC can restrain the growth and proliferation of gastric cancer cells. It can induce cell apoptosis and help tumor cell maintain malignant phenotype. But it can have not a detectable influence on the ability of invasion of gastric cancer cells.
Keywords: Gastric cancer, C-MYC gene, Cell cycle, RNAi
Introduction
Globally, gastric cancer is the second most common malignancy. Although the incidence of the disease has been declining for the past few decades, each year, roughly 798,000 people are diagnosed with gastric cancer worldwide (9.9% of total cancer cases), and 628,000 people die from the disease (12.1% of cancer deaths) [1]. In China, gastric cancer is the most common carcinoma. A variety of factors can affect the complex multi-factorial and multistage process of gastric carcinogenesis [2–4]. Some researchers have reported that C-MYC gene can play a very important role in gastric carcinogenesis. C-MYC proto-oncogene is one of the most frequently activated oncogenes, and is estimated to be involved in 20% of all human cancers, affecting about 100,000 US cancer deaths per year [5, 6]. The cancer deaths concerned with this gene in China is still not clear. It is therefore critical that the functions of C-MYC in gastric cancer and other cancers are well delineated.
C-MYC gene is an important member of MYC proto-oncogene family. The oncogene MYC family encodes the transcription factor proteins N-myc, c-myc, and L-myc which are implicated in the regulation of cell proliferation and differentiation [7, 8], and apoptosis [9, 10]. N-myc, c-myc, and L-myc are also widely expressed in developing and adult tissues and organs, which are also involved in neoplasia. The C-MYC oncogene (located at chromosomal band 8q24) encodes a transcriptional factor that regulates a variety of genes related to proliferation, differentiation, and apoptosis [11]. The C-MYC protein is a transcription factor which regulates a large series of downstream genes.
Some studies have shown an association between C-MYC deregulation and gastric cancer. C-MYC overexpression has been described in over 40% of GC [12]. Calcagno et al. [13] reported that C-MYC protein was expressed in all cases of both intestinal- and diffuse-type gastric adenocarcinoma samples of individuals from Northern Brazil. Kozma et al. [14] and Yang et al. [15] reported that higher C-MYC expression was associated with the presence of metastasis. Onoda et al. [16] also found that C-MYC mRNA levels were higher in metastatic than in primary lesions. Han et al. [17] described that patients with high levels of C-MYC expression had poor disease-free survival. Therefore, its expression may represent an aggressive phenotype of GC. The gene overexpression has also been seen in early GC when tumor invasion is confined to the mucosa or submucosa regardless of the presence of lymph node metastasis [17–21]. Xu et al. [22] noticed that C-MYC protein expression increased progressively from chronic active gastritis, gastric ulcer, and mild nonclassic proliferation to progressive GC. Lan et al. [23] found that C-MYC expression was higher in GC than in chronic gastritis, intestinal metaplasia, and dysplasia. In one word, the results of these researches above mentioned have suggested that C-MYC could enhance the canceration of gastric epithelial cells.
To date, most of these studies have been carried out on human gastric cancer tissues while the relationship between C-MYC and parameters of tumor cell proliferation has not been studied much or well characterized. Therefore, we have chosen to study C-MYC function in tumor cells, using the gene transfection and the RNAi technique that allows specific upregulation and downregulation of C-MYC gene expression to determine what role, if any, this protein has in contributing to the biological activity of transformed cells. We have used a human gastric cancer cell line SGC7901 and a human normal gastric cell line HFE145 to analyze the role of C-MYC. We report that C-MYC expression is crucial for tumor cell growth, survival, and the maintenance of tumor cell parameters that may contribute to malignant potential.
Materials and methods
Materials
C-MYC monoclonal antibody was purchased from Abcam Company (Cambridge, MA, USA), Rabbit antimouse HPR (1:1,000; Dako, Copenhagen, Denmark) was used to recognize corresponding proteins. PCDNA3.1 vector was preserved by our laboratory; common cell culture plates were purchased from Orange Company (Belgium). Transwell cell culture plates were purchased from Castar Company, and Watrigel gel was purchased from BD Company (USA). AnexinV-FITC apoptosis detection kit was purchased from Biosea Biotechnology Co., Ltd (Beijing, China). All the primers used in this research were synthesized by Shanghai Boya Biotechnology Co., Ltd (Shanghai, China).
Cell lines and culture
The human gastric adenocarcinoma cell line (SGC7901) was obtained from Shanghai Cell Research Institute of Chinese Scientific Academy, stored and transfer cultured in our laboratory. The human gastric cell line (HFE145) was preserved by our department. DMEM containing 10% calf serum, 100 IU/mL penicillin, and 100 IU/mL streptomycin were used as conventional culture medium. The culture procedures were taken under 37°C, 5% CO2, and saturation humidity.
Investigation of C-MYC expression in SGC7901 and HFE145 cell line
The expressions of C-MYC in SGC7901 and HFE145 were detected to determine whether the cell lines could use in the research. RT-PCR and immunocytochemical assay were performed to detect the expression of C-MYC in cells, and the results showed that there was a high expression level of C-MYC in SGC7901 cell, but HFE145 was a C-MYC-defective cell strain. The two cell lines were all suitable for transfection or RNAi experiments, respectively.
Transfections
Human gastric HFE145 cells were transfected with the cDNA of C-MYC gene. For the stable transfections, the cDNA was inserted into the pcDNA3.1 expression vector (Invitrogen, San Diego, CA, USA) between the EcoRI and BamHI sites. The orientation of the insert was confirmed by restriction digestion and DNA sequencing. The pcDNA3.1 vector inserted with C-MYC cDNA was renamed as pcDNA3/MYC which was transfected into HFE145 cells using a Lipofectamine2000 liposome transfection kit (Boehringer Mannheim, Indianapolis, IN, USA). The pcDNA3.1 empty vector transfection group and the blank control group (only liposome was added, and there was no vector DNA transfected) were established. Putative transfectants were then selected by antibiotic resistance in cell culture medium containing 800 μg/ml G418. After 6 weeks of culture in the presence of G4l8, the remaining cells were isolated with cloning cylinders and transferred into 24-well dishes. 17 and 12 clones were, respectively, obtained in the pcDNA3/Myc vector transfection group (HFE-Myc) and pcDNA3.1 empty vector transfection group (HFE-PC). These selected clones were taken for identification and frozen for future use.
C-MYC siRNA synthesis and manipulation
The nucleic acid sequence of C-MYC was obtained from Genbank. OligoEngine software was used to select two targeted fragments for RNAi as follows: 1′-CAGCGAGGATATCTGGAAGAA; 2′-CTCCACACATCAGCACAACTA. These 21 nucleotide DNA sequences were corresponding to C-MYC (Genebank V00568) coding nucleotides.
To control for the specificity of the RNAi effect, an independent DNA duplex was designed and produced, respectively, with 1-point mutations compared with the C-MYC siRNA duplex; this served as the control for the transfection procedure. Scrambled control sequence was as follows: (1) CAGCTAGGATATCTGGAAGAA; (2) CTC CACACATCAGAACAACTA. A BLAST search of the C-MYC siRNA against the Genbank _ EMBL _ DDBJ _ PDB sequences was performed, and the results showed that only one gene, human mRNA encoding the c-myc oncogene (C-MYC), is targeted. BLAST searches of the control siRNA against the same sequences showed no other human gene to be targeted. According to the selected targeted fragments, two C-MYC siRNA duplexes and two scrambled control duplexes were synthesized as follows:
The C-MYC siRNA duplexes:
S1
cmy-sh-1F: GATCCGCGAGGATATCTGGAAGAATTCAAGAGATTCTTCCAGATATCCTCGCTGA
cmy-sh-1R: AGCTTCAGCGAGGATATCTGGAAGAATCTCTTGAATTCTTCCAGATATCCTCGCG
S2
cmy-sh-2F: GATCCCCACACATCAGCACAACTATTCAAGAGATAGTTGTGCTGATGTGTGGAGA
cmy-sh-2R: AGCTTCTCCACACATCAGCACAACTATCTCTTGAATAGTTGTGCTGATGTGTGGG
Scrambled control duplexes:
S1s
cmy-sh-1F: GATCCGCGAGGATATCTGGAAGAATTCAAGAGATTCTTCCAGATATCCTAGCTGA
cmy-sh-1R: AGCTTCAGCTAGGATATCTGGAAGAATCTCTTGAATTCTTCCAGATATCCTCGCG
S2s
cmy-sh-2F: GATCCCCACACATCAGCACAACTATTCAAGAGATAGTTGTTCTGATGTGTGGAGA
cmy-sh-2R: AGCTTCTCCACACATCAGAACAACTATCTCTTGAATAGTTGTGCTGATGTGTGGG
All the DNA fragments were chemically synthesized, purified, and annealed by Dharmacon Research (Dharmacon Research, Lafayette, CO, USA) and manipulated according to the manufacturer’s instructions. These duplex DNA fragments were inserted into pSliencer™ 4.1-CMV neo vector (Ambion, Austin, Texas, USA), and then vector-based siRNAs were amplified and verified by DNA sequencing.
Cell culture and RNA transfection
SGC7901 cells were grown in DMEM medium (Invitrogen, Paisley, UK) supplemented with 10% fetal calf serum (FCS) without antibiotics. Twenty-four hours before transfection, 1 × 106 cells were seeded into a T75 flask in 15 ml DMEM medium with 10% FCS. Lipofectamine2000 liposome transfection kit (Boehringer Mannheim, Indianapolis, IN, USA) was used to transfect short interference RNAs (siRNAs) into cells according to the procedures recommended by the manufacturer. An amount of 1 nmol siRNA duplex was used for each T75 flask. In cases of apparent overconfluence, assessed by simple visual examination, cells were split on the second day after the transfection, but the cells were maintained in various media containing the vector-based siRNA duplexes at the appropriate concentration. All assays were carried out on day 5 after transfection, except the growth curve study, which was initiated on day 2 after the transfection and continued over the subsequent 7 days. Two groups of SGC7901 cell transfected with two siRNA duplexes were renamed as SGC-MR1 and SGC-MR2. Other two groups of SGC7901 cell transfected with scrambled control duplexes were renamed as SGC-MS1 and SGC-MS2, respectively. On the fifth day after transfection, the effect of siRNA on the gene expression was analyzed.
Analysis of transfectants
The RT-PCR and Western blotting analysis were, respectively, performed to detect the mRNA and protein of C-MYC in each sample, and immunocytochemical analysis was used to detect the expression of C-MYC protein in the HFE-Myc and HFE-PC cells in situ. In immunocytochemical analysis, human colorectal cancer cell line SW480 was immunocytochemical detected as positive control [24], and HFE-Myc cells were immunostained using PBS instead of C-MYC antibody as negative control.
Cell growth curves
All of 17 HFE-Myc cell clones, 12 HFE-PC cell clones, and untreated HFE145 cell were used. The cells of each clone were inoculated into 24-well culture plate at the concentration of 5 × 104/ml. After the cells completely plating, they were washed once with PBS, and then trypsinized in 0.5 ml of Trypsin/EDTA and counted in triplicates at 1–7 days using a cell counter (Beckman Coulter, Inc., Fullerton, CA, USA). The mean values of all of 17 HFE-Myc cell clones and 12 HFE-PC cell clones on different time were calculated, and growth curves were plotted.
Two days after the siRNA transfection, cells of SGC-MR1, 2, SGC-MS1, 2 and untreated SGC7901 groups were trypsinized and seeded at 5 × 104 per well in 6-well plates. Subsequently, cells were trypsinized and counted each day after plating.
Colony formation assay
Each clone of HFE-Myc, HFE-PC, and untreated HFE145 groups was detected. 1,000 cells of each clone were, respectively, seeded in a 9-cm cell culture dish. After 18 days’ culture in DMEM containing fetal calf serum, cell colonies were then fixed and stained with 0.5% methylene blue (Sigma, Poole, Dorset, UK) in ethanol. All colonies visible by eye with more than 50 cells were counted separately for each sample, and their clone formation rates were evaluated (clone formation rate = number of clones in each dish/1,000). Three reduplicate dishes were used from each clone.
On the fourth day after transfection, 1,000 cells from each sample of SGC-MR1, 2, SGC-MS1, 2 and untreated SGC7901 groups were seeded in 6-well plates and kept growing in 4 ml DMEM with 10% FCS until 14 days after transfection. Cell colonies were then fixed and stained with 0.5% methylene blue in ethanol. All colonies visible by eye were counted separately for each sample, and their clone formation rates were evaluated.
Analysis of cell cycle and apoptosis
All cell clones of HFE-Myc, HFE-PC, and untreated HFE145 groups and all cell samples of SGC-MR1, 2, SGC-MS1, 2 and untreated SGC7901 groups were detected by flow cytometry for analysis of cell cycle and apoptosis. When the cells covered 70% of the area of cell culture plates, serum-free culture medium was used for synchronization. After 24-h continuous culture, the cells were harvested and fixed by 100% ethanol, and then prepared for single cell suspensions. After DNA staining, the cell cycles of the samples were measured on a FACS Calibur cytometer. The analysis software was CellQuest. In cell apoptosis analysis, after synchronization and 24-h continuous culture, the cells were harvested and fixed, PI and AnexinV-FITC double staining was performed, and flow cytometry was used to detect the apoptosis of cells. Three replicate tests on every sample were performed in each group; the average values of these groups were calculated, respectively, and comparison between these groups was conducted.
Cell migration assay
Cell migration assays were performed using FCS-coated polycarbonate filters (8 μm pore size; Transwell, Becton–Dickinson, Franklin Lakes, NJ, USA). The membrane undersurface was coated with 200 μl FCS for 1 h at 37°C and blocked with 200 μl migration buffer (0.5% BSA in DMEM) for 30 min at 37°C. The lower chamber was filled with 500 μl of migration buffer, following which cells were plated in the upper chamber of four wells per treatment at a density of 1 × 105 in 100 μl of migration buffer and incubated at 37°C for 4 h. Following incubation, cells in the upper compartment were trypsinized and counted by the CASY 1 counter (Sharfe System, Reutlingen, Germany). Cells that had migrated to the lower surface of the filter were also trypsinized and counted. The migration rate was obtained by dividing the cell number in the lower chamber by the sum of the cell number found in both the lower chamber and the upper chamber.
Results
Expression of C-MYC gene in SGC7901 and HFE145 cell lines
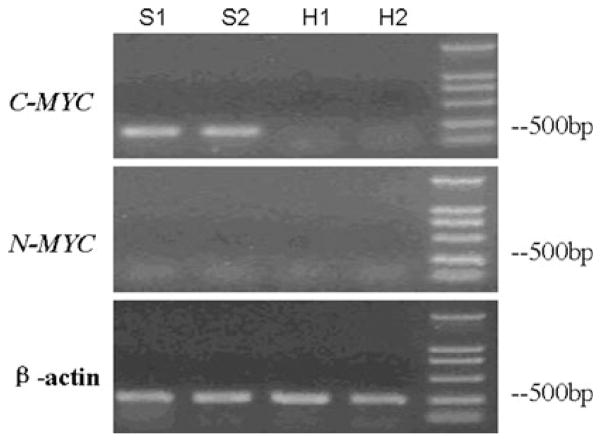
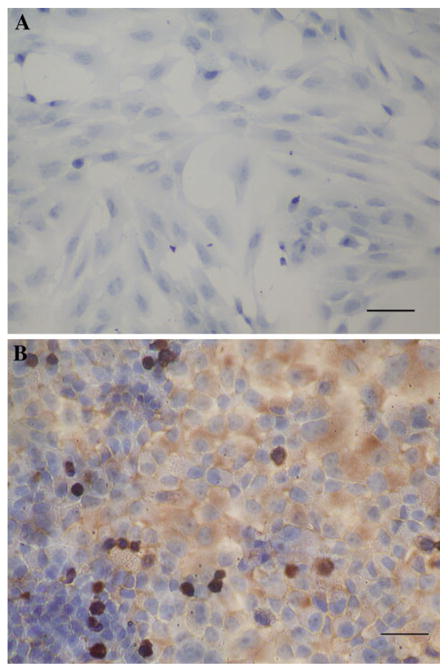
The expression of C-MYC gene in gastric adenocarcinoma cell strain SGC7901 and gastric cell strain HFE145 were detected by RT-PCR and immunocytochemical analysis. All the results in HFE145 cell strain were negative, which indicated that there was no detectable expression of C-MYC gene in untreated HFE145 cells. On the other hand, the results in SGC7901 cell strain were positive. The results of immunocytochemical assay showed that the expression of C-MYC gene in SGC7901 cells was mainly distributed in nucleus and cytoplasm, and there was no obvious positive signal in cell membrane (Figs. 1, 2).
Fig. 1.
The results of RT-PCR for C-MYC in SGC7901 cell and HFE145 cell. S1, S2 were the results of RT-PCR for C-MYC, N-MYC, and β-actin control in SGC7901 cell. H1 and H2 were the results of RT-PCR for C-MYC, N-MYC, and β-actin control in HFE145 cell. The results showed that there was C-MYC expression in SGC7901 cell but no detectable expression of C-MYC gene in HFE145 cell. On the other hand, there was no detectable expression of N-MYC gene in SGC7901 or HFE145 cell
Fig. 2.
The immunocytochemistry result of C-MYC in HFE145 and SGC7901 cell lines (×200). a There was no positive signal in HFE145 cell. The results showed that there was no detectable expression of C-MYC gene in HFE145 cell. Bar 20 μm. b The brown positive signals were mainly distributed in cytoplasm and nucleus. The results showed that there was expression of C-MYC gene in SGC7901 cell line. Bar 25 μm). (Color figure online)
Expression of C-MYC gene in transfectants
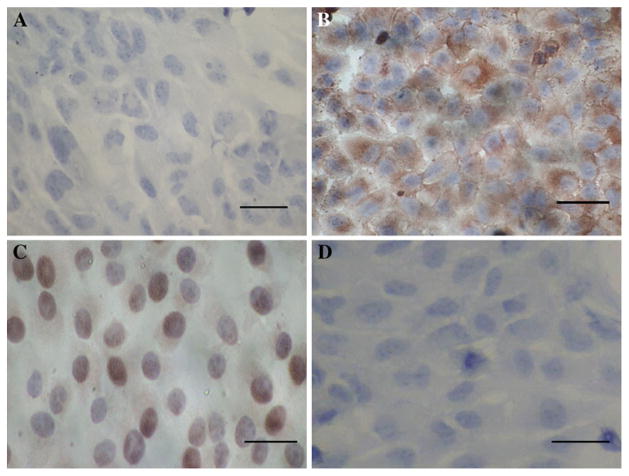
The results of RT-PCR, western blotting, and immunocytochemical analysis showed that the expression of C-MYC gene significantly increased in HFE-Myc cells when compared with the untreated HFE145 cells or HFE-PC cells, respectively. On the other hand, the results of immunocytochemical assay showed that the expression of C-MYC gene in HFE-Myc cells was mainly distributed in cytoplasm and nucleus (Figs. 3, 4, 5). The results of RT-PCR and western blotting showed that C-MYC siRNA can efficiently and specifically decrease endogenous C-MYC expression. As shown in Figs. 6 and 7, only the C-MYC siRNA caused the downregulation of C-MYC protein, while C-MYC protein levels in the scrambled siRNA and untreated control samples showed no significant change. These results indicate that only specific siRNA caused complementary mRNA degradation and thus the decrease of the corresponding protein (Figs. 6, 7).
Fig. 3.
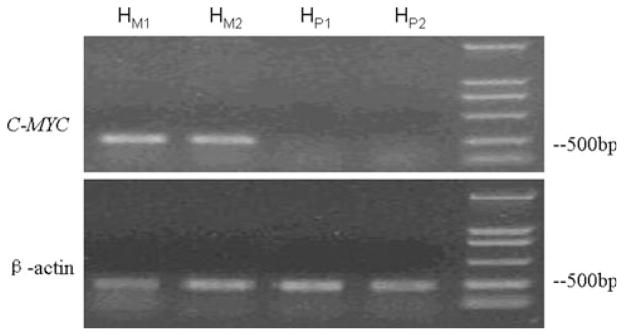
The RT-PCR results of C-MYC in HFE-Myc and HFE-PC cells. HM1 and HM2 were the results of RT-PCR for C-MYC and β-actin control in HFE-Myc cell. HP1 and HP2 were the results of RT-PCR for C-MYC and β-actin control in HFE-PC cell. The results showed that there were expressions of C-MYC gene in HFE-Myc cell but no expression in HFE-PC cell
Fig. 4.
The immunocytochemistry results of C-MYC in HFE-PC and HFE-Myc cells (×400). a There was no positive signal in HFE -PC cell. Bar 20 μm. b The brown positive signals were mainly distributed in cytoplasm of HFE-Myc cells. The results showed that there was expression of C-MYC gene in HFE-Myc cell line. Bar 20 μm. c The brown positive signals were mainly distributed in nucleus and cytoplasm of positive control cells. Bar 20 μm. d There was no positive signal in HFE-Myc cells which were immunostained using PBS instead of C-MYC antibody as negative control. Bar 20 μm. (Color figure online)
Fig. 5.
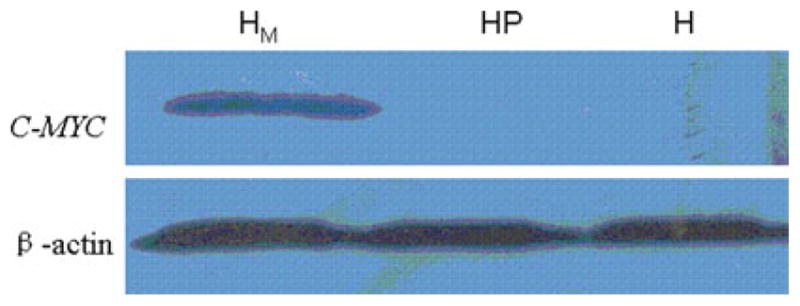
The western blot results of C-MYC in HFE-Myc, HFE-PC, and HFE145 cells. HM was the result of western blot for C-MYC and β-actin control in HFE-Myc cell, HP was the result of western blot for C-MYC and β-actin control in HFE-PC cell, and H was that in HFE145 cell. The results showed that there was expression of C-MYC gene in HFE-Myc cell but no expression in HFE-PC and HFE145 cell
Fig. 6.
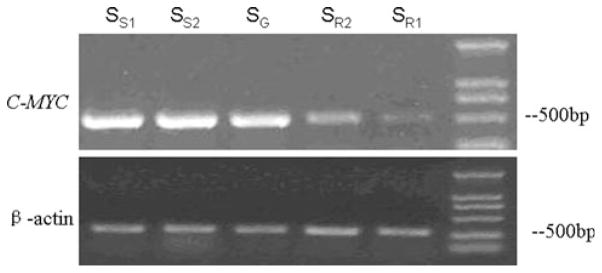
The RT-PCR results of C-MYC in SGC-MR1, SGC-MR2, SGC-MS1, SGC-MS2, and untreated SGC7901 cells. SS1 and SS2 were the results of RT-PCR for C-MYC and β-actin control in SGC-MS1 and SGC-MS2 cells, respectively; SR1 and SR2 were the results of RT-PCR for C-MYC and β-actin control in SGC-MR1 and SGC-MR2 cells, respectively. SG was that in untreated SGC7901 cell. The results showed that the expression level of C-MYC gene in SGC-MR1 and SGC-MR2 cells was decreased when compared with the control groups
Fig. 7.
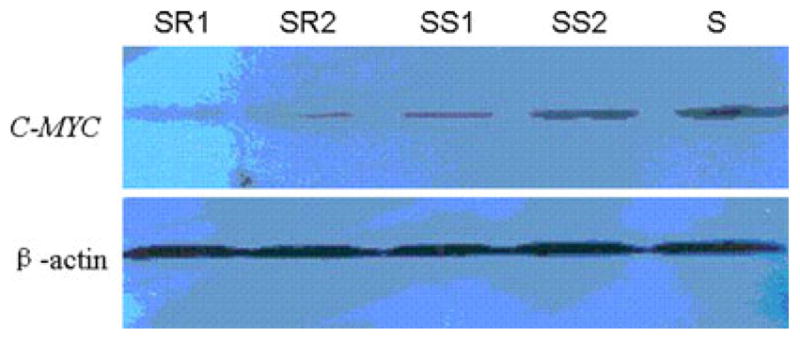
The western blot results of C-MYC in SGC-MR1, SGC-MR2, SGC-MS1, SGC-MS2, and untreated SGC7901 cells. SR1 and SR2 were the results of western blot for C-MYC and β-actin control in SGC-MR1 and SGC-MR2 cells, respectively; SS1 and SS2 were the results of western blot for C-MYC and β-actin control in SGC-MS1 and SGC-MS2 cells, respectively. S was that in untreated SGC7901 cell. The results showed that the expression level of C-MYC gene in SGC-MR1 and SGC-MR2 cells was decreased when compared with the control groups
The influence of C-MYC gene on cell growth
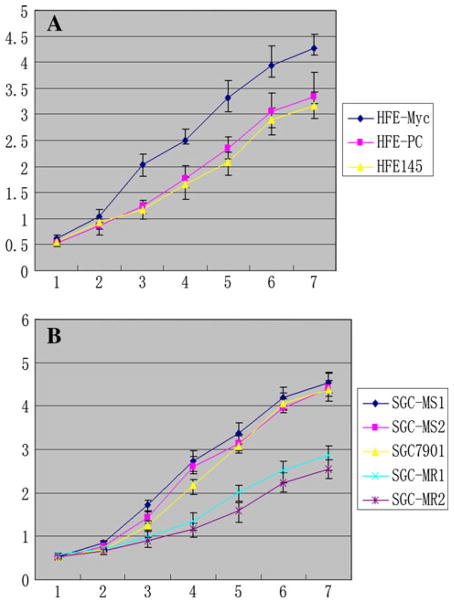
The results of cell growth curve assay showed that HFE-Myc cells grew significantly faster than HFE-PC and untreated HFE145 cells, respectively (P < 0.05), and there was no significant difference between the control groups. On days 3,4, 5, 6, and 7 after inoculation, the average cell counts of HFE-Myc group were 2.04 × 105, 2.51 × 105, 3.32 × 105, 3.93 × 105, and 4.28 × 105, respectively, which were significantly more than those of the two control groups (P < 0.05) (Fig. 8a).
Fig. 8.
The growth curves of C-MYC-transfected and C-MYC-RNAi cell lines. a The growth curves of HFE-Myc, HFE-PC, and HFE145 groups. b The growth curves of SGC-MR1, 2, SGC-MS1, 2, and untreated SGC7901 groups. The unit of vertical axis was ×105; horizontal axis was the number of days
The growth curves of SGC-MR1, SGC-MR2, SGC-MS1, SGC-MS2, and untreated SGC7901 (Fig. 8b) groups showed that treatment with C-MYC siRNA, but not with scrambled siRNA, could restrain cell growth over a period of 7 days. On days 4, 5, 6 and 7 after inoculation, the average cell counts of SGC-MR1 group were 1.35 × 105, 2.02 × 105, 2.51 × 105, and 2.86 × 105, respectively, which resembled those of SGC-MR2 group and were significantly fewer than those of the control groups (P < 0.05).
The influence of C-MYC gene on colony formation
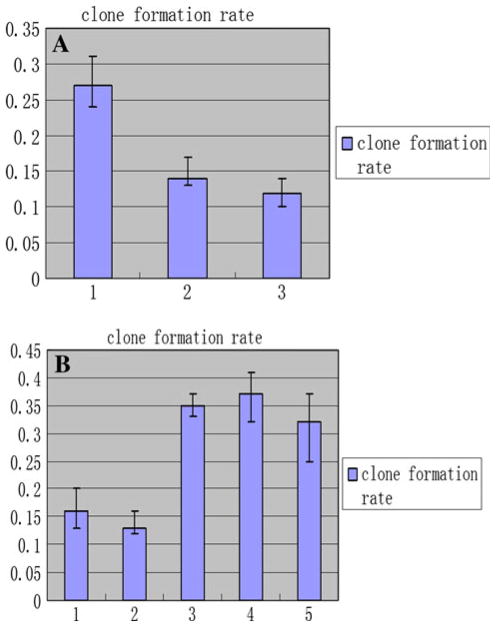
The clone formation rates of the HFE-Myc (0.27 ± 0.04) were significantly higher than that of their control groups, respectively (P < 0.05). There was no significant difference between these control groups (P > 0.05) (Fig. 9a). It is apparent that transfection with C-MYC gene increased the capacity of these cells to establish colonies to a highly significant degree.
Fig. 9.
The results of colony formation assay for C-MYC-transfected and C-MYC-RNAi cell lines. a The results of colony formation assay of HFE-Myc, HFE-PC and HFE145 cells. 1 the colony formation rate of HFE-Myc cell, 2 that of HFE-PC cell, 3 that of HFE145 cell line. b The results of colony formation assay of SGC-MR1, SGC-MR2, SGC-MS1, SGC-MS2, and untreated SGC7901 cell lines. 1, 2 the colony formation rate of SGC-MR1, SGC-MR2 cells, respectively; 3, 4 that of SGC-MS1, SGC-MS2 cells, respectively; 5 that of SGC7901 cell line
An example of data from a colony-forming assay for SGC-MR cells is shown in Fig. 10b which was significantly lower than that of their control groups, respectively (P < 0.05) (Fig. 9b). It is apparent that transfection with C-MYC siRNA decreased the capacity of these tumor cells to establish colonies.
Fig. 10.
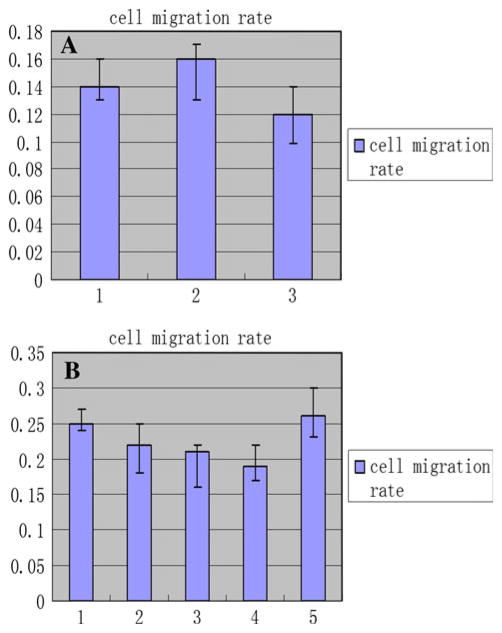
The results of cell migration assay for C-MYC-transfected and C-MYC-RNAi cell lines. a The results of cell migration assay of HFE-Myc, HFE-PC, and untreated HFE145 cell lines. 1 the cell migration rate of HFE-Myc cell line, 2 that of HFE-PC cell line, 3 that of untreated HFE145 cell line. b The results of cell migration assay of SGC-MR1, SGC-MR2, SGC-MS1, SGC-MS2, and untreated SGC7901 cell lines. 1, 2 the cell migration rate of SGC-MR1, SGC-MR2 cells, respectively; 3, 4 that of SGC-MS1, SGC-MS2 cells, respectively; 5 that of SGC7901 cell line
C-MYC is important for cell cycle progression
The results of the effect of C-MYC on individual experiments measuring cell cycle progression were summarized in Tables 1 and 2. It was clear that upregulation or downregulation of C-MYC gene expression led to consistent changes in the number of cells in G0/G1, S, and G2/M phase.
Table 1.
The different cell cycle of HFE-Myc, HFE-PC, and HFE145 groups
| Group | Clone number | n | G0–G1 (%) | G2–M (%) | S (%) |
|---|---|---|---|---|---|
| HFE-Myc | 17 | 3 | 64.66 ± 7.43* | 25.66 ± 3.79* | 10.45 ± 4.18* |
| HFE-PC | 12 | 3 | 43.84 ± 5.10 | 6.38 ± 1.16 | 41.18 ± 6.43 |
| HFE145 | 1 | 3 | 46.68 ± 8.35 | 5.71 ± 1.25 | 38.32 ± 7.04 |
P < 0.05, compared with the group of HFE145
Table 2.
The different cell cycle of SGC-MR1, SGC-MR2, SGC-MS1, SGC-MS2, and untreated SGC7901 groups
| n | G0–G1 (%) | G2–M (%) | S (%) | |
|---|---|---|---|---|
| SGC-MR1 | 3 | 25.57 ± 3.77* | 3.75 ± 0.40* | 71.03 ± 7.98* |
| SGC-MR2 | 3 | 34.09 ± 5.39 | 4.85 ± 2.32* | 61.04 ± 6.21* |
| SGC-MS1 | 3 | 37.98 ± 6.11 | 15.37 ± 2.62 | 48.83 ± 10.35 |
| SGC-MS2 | 3 | 44.35 ± 7.21 | 13.55 ± 3.16 | 43.52 ± 6.36 |
| SGC7901 | 3 | 42.46 ± 6.15 | 16.78 ± 2.46 | 41.02 ± 6.65 |
P < 0.05, compared with the group of SGC7901
The influence of C-MYC gene on cell apoptosis
The apoptosis assay results showed that the average apoptosis rates of all cell clones in HFE-Myc, HFE-PC and untreated HFE145 groups were 5.81 ± 0.71, 2.96 ± 0.56, and 2.63 ± 0.35%, respectively, and there was statistical significant difference between HFE-Myc and their control groups (P < 0.05). The results of apoptosis assay in SGC-MR1, SGC-MR2, SGC-MS1, SGC-MS2, and untreated SGC7901 groups were 2.63 ± 0.24, 2.44 ± 0.16, 2.25 ± 0.32, 2.56 ± 0.27, and 1.99 ± 0.37%, respectively, and there was no statistical significant difference between them (P > 0.05). It appeared that upregulation of C-MYC gene expression had an important influence on cell apoptosis.
Role of C-MYC in cancer cell migration
Because individual cell migration is an important characteristic of invasive tumor cells or others, we examined the effects of C-MYC modulation on migration. The migration rates of HFE-Myc, HFE-PC, untreated HFE145 cells, and C-MYC siRNA transfected cells were showed in Fig. 10. Unfortunately, we were unable to observe statistically significant migration differences in the experimental groups and their control groups (P > 0.05).
Statistical analysis
In present research, the data were expressed as mean ± SD. One-way ANOVA test and Student’s t test were performed. Obtained data were analyzed using SPSS 11.0 (SPSS, USA) statistical software package; Statistical significance was defined as P < 0.05.
Discussion
Gastric cancer remains a common disease worldwide, with a poor prognosis and low survival rates. Its molecular controls are poorly understood, and the involvement of a number of different cell death or proliferation-modifying genes has been proposed. Among these genes, dysregulation of the transcription factor C-MYC is characteristic of gastric adenocarcinoma as well as other malignancies. C-MYC affects diverse roles in regulating critical biological functions including cell-cycle regulation, apoptosis, metabolism, cellular differentiation, cell adhesion, promotion of neovascularization, and others [5, 25–27].
In the present study, cell growth and proliferation was significantly increased in C-MYC-transfected normal gastric cells and significantly decreased in C-MYC-RNAi gastric cancer cells. The results of the present study provide more support for an oncogenic (proliferative) role in gastric cancer and concur with those of Langlois et al. [28], who also recorded c-myc expression in association with higher mitosis. C-MYC is also well known for the direct and indirect regulation of cellular genes involved in transcription and the mitotic program as well as mediating nuclear events associated with growth factor stimulation [29–31]. In conclusion, the molecular and biologic factors that control the balance between cell proliferation and apoptosis in development and progression of gastric cancers are complex, but the changes of C-MYC gene expression have an effective influence on growth and proliferation of gastric cancer cell.
The results of cell cycle assay suggested that upregulation of C-MYC expression could increase the cells in G2/M stage significantly; otherwise, downregulation of C-MYC expression decreased the proportion of cell in G2/M stage. Our results showed that upregulation of C-MYC expression could reduce the number of cells in S-phase while downregulation of C-MYC expression could increase it. Kristin et al. [32] has conformed that c-Myc overexpression in primary human fibroblasts markedly accelerates S-phase while c-Myc deficient fibroblasts exhibit a prolonged S-phase by detecting the duration of S-phase of mammalian cells when the expression of C-MYC increased and decreased. According to their results, we speculated that lesser cells in S-phase could be found because more cells came into G2/M from S-phase for a shorter duration of S-phase. On the other hand, more cells in S-phase also could be detected for a longer duration of S-phase of c-Myc-deficient cell. These results provided more support for C-MYC’s role of promoting cell growth and proliferation. Many investigators have uncovered target genes that encode proteins that regulate the cell cycle. The genes that consistently emerge and contain E-boxes bound by c-Myc in ChIP assays are cyclins D1 and D2, CDK4, and cyclin B1 [33–35]. Cyclin A, cyclin B, and cdk4 were also identified as MYC targets in Drosophila [36]. It is notable that in vivo genetic studies have implicated a role for CDK4 in C-MYC-mediated transformation [37]. A role for D-type cyclins is more controversial. However, it has been shown that fibroblasts lacking cyclins D1–D3 fail to be transformed by c-Myc, suggesting an important role for D-type cyclins in C-MYC-mediated transformation [38]. C-MYC represses the CDK inhibitors p21 and p15INK4A through an interaction with the Miz-1 protein at the core promoter [39–41]. In essence, C-MYC directly regulates genes involved in cell cycle regulation.
By cell apoptosis assay, we found that upregulation of C-MYC expression could increase the cell apoptosis rate in gastric cell line. It is seemed that C-MYC could induce gastric cell apoptosis, but the results of many researches implied that the influence of C-MYC on cell apoptosis was very complex, and there were contradictory conclusions in their reports. Other in vitro experiments show that stable transfection of C-MYC and subsequent protein activation in low C-MYC-expressing melanoma cell lines is able to sensitize these cells to apoptosis under multiple types of stress [42]. The deregulated expression of C-MYC also induces genes that contribute to apoptosis under nutrient or growth factor deprivation; however, C-MYC target genes involved in apoptosis remain to be fully elucidated. A number of excellent reviews are available on C-MYC and apoptosis [10, 43, 44]. The downregulation of C-MYC expression by specific RNAi could have no detectable influence on gastric cancer cell apoptosis. The reason could be that the change of cell apoptosis had not been displayed in the short experimental time. The further research must be needed to confirm the result.
Our results showed that upregulation or downregulation of C-MYC expression could have no influence on cellular migration, so the gene could not impact invasion of gastric cancer cell. The mechanism by which C-MYC impacts cellular migration in gastric cancer is entirely unknown at present. The change of genes that encode cytoskeletal and cell adhesion proteins by C-MYC may contribute to neoplastic transformation and invasion of susceptible cell lines [45, 46]. The altered expression of cytoskeletal genes also influences cellular morphology or invasion ability. For example, C-MYC null fibroblasts possess many actin stress fibers and focal adhesions as compared to null fibroblasts reconstituted with C-MYC [47]. A proteomics approach identified the cytoskeletal proteins actin and cdc42 as downregulated in C-MYC reconstituted fibroblasts. These changes suggest that C-MYC plays a role in enhancing fibroblast motility. These researches suggested that C-MYC could promote cancer cell invasion, but it affects individual gastric cancer cell migration and such possibilities currently are under investigation.
In general, the positive immunohistochemical staining of C-MYC as a transcription factor can be detected in nucleus. But in our research, the positive staining was located in cytoplasm and nucleus. In fact, the results of many researches have suggested that nuclear and cytoplasmic C-MYC staining was common in many human tissue or cell. Geisler et al. [48] reported that C-MYC cytoplasmic staining was present in 75.2% of the tumors, and nuclear staining was present in 66.9% in 121 patients suffering with endometrial carcinoma. Ruzinova et al. [49] also found that nuclear and cytoplasmic C-MYC staining was common in B-cell Lymphomas, and a primarily nuclear or mixed nuclear and cytoplasmic staining pattern for C-MYC could be highly predictive of a C-MYC translocation in the disease. It is well known that C-MYC protein is synthesized in cytoplasm. The synthesis of of C-MYC protein could increase uncommonly in some tumor cells or the cells transfected with C-MYC gene, and C-MYC protein could accumulate in cytoplasm, so cytoplasmic c-myc staining should be possible.
Conclusion
The results of the present investigation demonstrated that overexpression of C-MYC gene can influence some biological characteristics of normal gastric cell. C-MYC can promote the growth and proliferation of these cells and help tumor cell maintain malignant phenotype. But it can have a negative influence on the ability of invasion of gastric cancer cells. Specific knockdown of C-MYC can reduce gastric cancer cell growth and proliferation, so the gene could be a potential target of gene therapy.
Acknowledgments
The authors wish to thank Drs. Haili Huang and Gangshi Wang, and Nurse Weidi You, Weihua Wang et al., for handling patient contacts. We wish to thank the Forth Military Medical University of PLA for providing means for the current investigation.
Footnotes
Conflict of interest statement The authors declare no competing interest.
Contributor Information
Lin Zhang, Email: stepinghuns@yahoo.com.cn, Department of Gastroenterology, The 309 Hospital of PLA, Beijing 100091, People’s Republic of China.
Yanhong Hou, Department of Gastroenterology, The 309 Hospital of PLA, Beijing 100091, People’s Republic of China.
Hassan Ashktorab, Medicine and Cancer Center, Howard University Cancer Center, Washington, DC, USA.
Liucun Gao, Department of Gastroenterology, The 309 Hospital of PLA, Beijing 100091, People’s Republic of China.
Yanjie Xu, Department of Gastroenterology, The 309 Hospital of PLA, Beijing 100091, People’s Republic of China.
Kai Wu, Department of Gastroenterology, The 309 Hospital of PLA, Beijing 100091, People’s Republic of China.
Junshan Zhai, Department of Gastroenterology, The 309 Hospital of PLA, Beijing 100091, People’s Republic of China.
Lei Zhang, Department of Gastroenterology, The 309 Hospital of PLA, Beijing 100091, People’s Republic of China.
References
- 1.Parkin DM, Poisani P, Ferlay Global cancer statistics. CA Cancer J Clin. 1999;49(1):33–64. doi: 10.3322/canjclin.49.1.33. [DOI] [PubMed] [Google Scholar]
- 2.Correa P. Human gastric carcinogenesis: a multistep and multifactorial process—First American Cancer society award lecture on cancer epidemiology and prevention. Cancer Res. 1992;52(24):6735–6740. [PubMed] [Google Scholar]
- 3.Gonzalez CA, Sala N, Capella G. Genetic susceptibility and gastric cancer risk. Int J Cancer. 2002;100(3):249–260. doi: 10.1002/ijc.10466. [DOI] [PubMed] [Google Scholar]
- 4.Xue FB, Xu YY, Wan Y, et al. Association of H. pylori infection with gastric carcinoma: a meta analysis. World J Gastroenterol. 2001;7(6):801–804. doi: 10.3748/wjg.v7.i6.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18(19):3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 6.Dang CV. C-MYC target genes involved in cell growth, apoptosis, an metabolism. Mol Cell Biol. 1999;19(1):1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole MD, McMahon SB. The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene. 1999;18(19):2916–2924. doi: 10.1038/sj.onc.1202748. [DOI] [PubMed] [Google Scholar]
- 8.Obaya AJ, Mateyak MK, Sedivy JM. Mysterious liaisons: the relationship between C-MYC and the cell cycle. Oncogene. 1999;18(19):2934–2941. doi: 10.1038/sj.onc.1202749. [DOI] [PubMed] [Google Scholar]
- 9.Hoffman B, Liebermann DA. The proto-oncogene c-myc and apoptosis. Oncogene. 1998;17(25):3351–3357. doi: 10.1038/sj.onc.1202592. [DOI] [PubMed] [Google Scholar]
- 10.Prendergast GC. Mechanisms of apoptosis by C-Myc. Oncogene. 1999;18(19):2967–2987. doi: 10.1038/sj.onc.1202727. [DOI] [PubMed] [Google Scholar]
- 11.Dang CV, Resar LM, Emison E, et al. Function of the C-MYC oncogenic transcription factor. Exp Cell Res. 1999;253(1):63–77. doi: 10.1006/excr.1999.4686. [DOI] [PubMed] [Google Scholar]
- 12.Milne AN, Sitarz R, Carvalho R, et al. Early onset gastric cancer: on the road to unraveling gastric carcinogenesis. Curr Mol Med. 2007;7(1):15–28. doi: 10.2174/156652407779940503. [DOI] [PubMed] [Google Scholar]
- 13.Calcagno DQ, Leal MF, Seabra AD, et al. Interrelationship between chromosome 8 aneuploidy, C-MYC amplification and increased expression in individuals from northern Brazil with gastric adenocarcinoma. World J Gastroenterol. 2006;12(38):6207–6211. doi: 10.3748/wjg.v12.i38.6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozma L, Kiss I, Hajdu J, et al. C-myc amplification and cluster analysis in human gastric carcinoma. Anticancer Res. 2001;21(1B):707–710. [PubMed] [Google Scholar]
- 15.Yang GF, Deng CS, Xiong YY, et al. Expression of nuclear factor-kappa B and target genes in gastric precancerous lesions and adenocarcinoma: association with Helicobactor pylori cagA (+) infection. World J Gastroenterol. 2004;10(4):491–496. doi: 10.3748/wjg.v10.i4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Onoda N, Maeda K, Chung YS, et al. Overexpression of c-myc messenger RNA in primary and metastatic lesions of carcinoma of the stomach. J Am Coll Surg. 1996;182(1):55–59. [PubMed] [Google Scholar]
- 17.Han S, Kim HY, Park K, et al. c-Myc expression is related with cell proliferation and associated with poor clinical outcome in human gastric cancer. J Korean Med Sci. 1999;14(5):526–530. doi: 10.3346/jkms.1999.14.5.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakata B, Onoda N, Chung YS, et al. Correlation between malignancy of gastric cancer and c-myc DNA amplification or overexpression of c-myc protein. Gan To Kagaku Ryoho. 1995;22(Suppl 2):176–179. [PubMed] [Google Scholar]
- 19.Sanz-Ortega J, Steinberg SM, Moro E, et al. Comparative study of tumor angiogenesis and immunohistochemistry for p53, c-ErbB2, c-myc and EGFr as prognostic factors in gastric cancer. Histol Histopathol. 2000;15(2):455–462. doi: 10.14670/HH-15.455. [DOI] [PubMed] [Google Scholar]
- 20.Ishii HH, Gobe GC, Pan W, et al. Apoptosis and cell proliferation in the development of gastric carcinomas: associations with c-myc and p53 protein expression. J Gastroenterol Hepatol. 2002;17(9):966–972. doi: 10.1046/j.1440-1746.2002.02805.x. [DOI] [PubMed] [Google Scholar]
- 21.Costa Raiol LC, Figueira Silva EC, Mendes da Fonseca D, et al. Interrelationship between MYC gene numerical aberrations and protein expression in individuals from northern Brazil with early gastric adenocarcinoma. Cancer Genet Cytogenet. 2008;181(1):31–35. doi: 10.1016/j.cancergencyto.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 22.Xu AG, Li SG, Liu JH, et al. Function of apoptosis and expression of the proteins Bcl-2, p53 and C-myc in the development of gastric cancer. World J Gastroenterol. 2001;7(3):403–406. doi: 10.3748/wjg.v7.i3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lan J, Xiong YY, Lin YX, et al. Helicobacter pylori infection generated gastric cancer through p53-Rb tumor-suppressor system mutation and telomerase reactivation. World J Gastroenterol. 2003;9(1):54–58. doi: 10.3748/wjg.v9.i1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hashimoto K, Nakagawa Y, Morikawa H, et al. Co-overexpression of DEAD box protein rck/p54 and c-myc protein in human colorectal adenomas and the relevance of their expression in cultured cell lines. Carcinogenesis. 2001;22(12):1965–1970. doi: 10.1093/carcin/22.12.1965. [DOI] [PubMed] [Google Scholar]
- 25.Henriksson M, Luscher B. Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv Cancer Res. 1996;68:109–182. doi: 10.1016/s0065-230x(08)60353-x. [DOI] [PubMed] [Google Scholar]
- 26.Oster SK, Ho CS, Soucie EL, et al. The myc oncogene: MarvelouslY Complex. Adv Cancer Res. 2002;84:81–154. doi: 10.1016/s0065-230x(02)84004-0. [DOI] [PubMed] [Google Scholar]
- 27.Evan GI, Wyllie AH, Gilbert CS, et al. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69(1):119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 28.Langlois NE, Lamb J, Eremin O, et al. Apoptosis in colorectal carcinoma occurring in patients aged 45 years and under: relationship to prognosis, mitosis, and immunohistochemical demonstration of p53, c-myc and bcl-2 protein products. J Pathol. 1997;182(4):392–397. doi: 10.1002/(SICI)1096-9896(199708)182:4<392::AID-PATH874>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 29.Xiangming C, Natsugoe S, Takao S, et al. Preserved Smad4 expression in the transforming growth factor beta signaling pathway is a favorable prognostic factor in patients with advanced gastric cancer. Clin Cancer Res. 2001;7(2):277–282. [PubMed] [Google Scholar]
- 30.Vaux DL, Cory S, Adams JM. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature. 1988;335(6189):440–442. doi: 10.1038/335440a0. [DOI] [PubMed] [Google Scholar]
- 31.Fernandez PC, Frank SR, Wang L, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17(9):1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson K, Asawachaicharn N, Denise A. c-Myc accelerates S-phase and requires WRN to avoid replication stress. PLoS One. 2009;4(6):e5951. doi: 10.1371/journal.pone.0005951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bouchard C, Dittrich O, Kiermaier A, et al. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev. 2001;15(16):2042–2047. doi: 10.1101/gad.907901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menssen A, Hermeking H. Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc Natl Acad Sci USA. 2002;99(9):6274–6279. doi: 10.1073/pnas.082005599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hermeking H, Rago C, Schuhmacher M, et al. Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci USA. 2000;97(5):2229–2234. doi: 10.1073/pnas.050586197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orian A, van Steensel B, Delrow J, et al. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 2003;17(9):1101–1114. doi: 10.1101/gad.1066903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miliani de Marval PL, Macias E, Rounbehler R, et al. Lack of cyclin-dependent kinase 4 inhibits c-myc tumorigenic activities in epithelial tissues. Mol Cell Biol. 2004;24(17):7538–7547. doi: 10.1128/MCB.24.17.7538-7547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozar K, Ciemerych MA, Rebel VI, et al. Mouse development and cell proliferation in the absence of D-cyclins. Cell. 2004;118(4):477–491. doi: 10.1016/j.cell.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 39.Wu S, Cetinkaya C, Munoz-Alonso MJ, von der Lehr N, et al. Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter. Oncogene. 2003;22(3):351–360. doi: 10.1038/sj.onc.1206145. [DOI] [PubMed] [Google Scholar]
- 40.Seoane J, Pouponnot C, Staller P, et al. TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat Cell Biol. 2001;3(4):400–408. doi: 10.1038/35070086. [DOI] [PubMed] [Google Scholar]
- 41.Claassen GF, Hann SR. A role for transcriptional repression of p21CIP1 by c-Myc in overcoming transforming growth factor beta induced cell-cycle arrest. Proc Natl Acad Sci USA. 2000;97(17):9498–9503. doi: 10.1073/pnas.150006697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peltenburg LT, de Bruin EC, Meersma D, et al. c-Myc is able to sensitize human melanoma cells to diverse apoptotic triggers. Melanoma Res. 2004;14(1):3–12. doi: 10.1097/00008390-200402000-00002. [DOI] [PubMed] [Google Scholar]
- 43.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2(10):764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 44.Nilsson JA, Cleveland JL. Myc pathways provoking cell suicide and cancer. Oncogene. 2003;22(56):9007–9021. doi: 10.1038/sj.onc.1207261. [DOI] [PubMed] [Google Scholar]
- 45.Yang BS, Geddes TJ, Pogulis RJ, et al. Transcriptional suppression of cellular gene expression by c-Myc. Mol Cell Biol. 1991;11(4):2291–2295. doi: 10.1128/mcb.11.4.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang BS, Gilbert JD, Freytag SO, et al. Overexpression of Myc suppresses CCAAT transcription factor/nuclear factor 1-dependent promoters in vivo. Mol Cell Biol. 1993;13(5):3093–3102. doi: 10.1128/mcb.13.5.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shiio Y, Donohoe S, Yi EC, et al. Quantitative proteomic analysis of Myc oncoprotein function. EMBO J. 2002;21(19):5088–5096. doi: 10.1093/emboj/cdf525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geisler JP, Geisler HE, Manahan KJ, et al. Nuclear and cytoplasmic c-myc staining in endometrial carcinoma and their relationship to survival. Int J Gynecol Cancer. 2004;14(1):133–137. doi: 10.1111/j.1048-891x.2004.14027.x. [DOI] [PubMed] [Google Scholar]
- 49.Ruzinova MB, Caron T, Rodig SJ. Altered Subcellular Localization of c-Myc Protein Identifies Aggressive B-cell Lymphomas Harboring a c-MYC Translocation. Am J Surg Pathol. 2010;34(6):882–891. doi: 10.1097/PAS.0b013e3181db83af. [DOI] [PubMed] [Google Scholar]