Abstract
Growth and remodelling impact the network topology of complex systems, yet a general theory explaining how new links arise between existing nodes has been lacking, and little is known about the topological properties that facilitate link-prediction. Here we investigate the extent to which the connectivity evolution of a network might be predicted by mere topological features. We show how a link/community-based strategy triggers substantial prediction improvements because it accounts for the singular topology of several real networks organised in multiple local communities - a tendency here named local-community-paradigm (LCP). We observe that LCP networks are mainly formed by weak interactions and characterise heterogeneous and dynamic systems that use self-organisation as a major adaptation strategy. These systems seem designed for global delivery of information and processing via multiple local modules. Conversely, non-LCP networks have steady architectures formed by strong interactions, and seem designed for systems in which information/energy storage is crucial.
Digging into the properties of complex networks is fundamental for the definition of general paradigms of complex systems, in which complexity is distinctively generated by the topological integration of many interacting parts. The Erdős–Rényi (ER) model, was proposed in 1959 to analyse an idealised random network, as characterised by a fixed number of nodes and by a uniform probability that two nodes are randomly connected1. Nearly four decades later, in 1998, Watts and Strogatz (WS) deepened our insight into the relationship between random processes and the rise of topological properties in real networks2. The WS model (also named small-world network) untangled how regular networks - with a fixed number of nodes and edges - react if an increasing number of edges are randomly rewired with uniform probability2. A WS network presents significantly higher clustering and comparable mean geodesic distance than a random graph of the same size and mean node-degree2. In particular, the mean geodesic distance in a WS network is ‘small’ relative to the network size - at most a logarithmic function of the total number of nodes in the network3 - and we now know that several real networks follow this paradigm.
Only one year later, while investigating the topological evolution of real networks, Barabási and Albert (BA) shed new light on complex network processes4. Voicing the assumption of an open network, the BA model (consistent with a previous study of de Solla Price5 on scientific citation networks) explained how the scale-invariance of many real networks originates from a specific growth process, in which a new node tends with higher probability to be linked to those nodes (network hubs) that already have a large number of neighbours. This paradigm, named preferential attachment (PA)4, together with the underlying idea of popularity6 is just one facet of node attractiveness in growing networks, another important aspect being similarity6.
However, topological evolution when exclusively new links are added to the network has yet to be congruously formalised, and it connects with a practical and front-line issue, namely the link-prediction problem7. Many applications have to predict new links in large and sparse complex networks merely with the knowledge of network topology, and new solutions could impact both science and engineering positively. Meanwhile, link-prediction reflects the extent to which the evolution of a network might be modelled on the basis of topological features intrinsic to the network itself7,8.
Here, we inspect three central questions that stem naturally from the state of the art: 1) how can the prediction of new links on the exclusive basis of network topology (topological link-prediction) be improved? 2) is it possible to define a network paradigm that enables the prediction of new links in many real networks, on the assumption that their topologies are shaped in accordance with a single, general link-growing process (as postulated in the paradigm)? 3) is it possible to conjecture any connection between a topological network paradigm and a class of physical systems?
Results
A local community approach to link-prediction
A myriad of complicated techniques for topological link-prediction with two or more parameters to tune have been proposed7,9,10, even inspired by concepts that originate from statistical mechanics and theory of disordered systems. Nevertheless, such elegant techniques are at the moment mere proof of concepts rather than concrete methods to apply on real problems. Apart from the problem to tune the parameters in an unsupervised framework, the greatest obstacle is their prohibitive computational time, which in practice relegates their application to toy networks of very small dimensions (few hundreds of nodes), in comparison to the giant networks used in real problems. For these reasons, in order to answer the first question, we preferred to focus our attention on efficient and parameter-free Node-Neighbourhood-based approaches7, which are commonly employed in both research and application, and whose design is inspired by the main patterns characterising the topology of complex networks7,11. Node-Neighbourhood-based approaches assign a likelihood score to each pair of non-connected nodes (candidate-links), and then produce a ranked list in decreasing order to ‘advocate’ candidate interactions. The common neighbours (CN) index11 is the progenitor of these methods and follows the natural intuition that the likelihood that two nodes x and y interact increases if their sets of first-node-neighbours 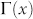 and
and 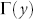 overlap substantially:
overlap substantially: 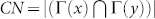 . The other indices are often a variation or generalisation of CN7: Jaccard's index (JC) is a normalisation of CN12, Adamic & Adar (AA)13 and Resource Allocation (RA)14 give more importance to CNs with low degree, while Preferential Attachment (PA)11 is the degree product of nodes x and y (for the formulae see Table 1 and for details see Supplementary Information (SI) section IIIA). In contrast to the existing methodologies, which are focused on groups of common nodes and their node neighbours, we here embrace a strategic shift from nodes to links that represents a new way to treat complex networks15. In particular, Ahn et al. reconceived communities as groups of links rather than as mere groups of nodes, and proposed a link-based approach for graph-partition that outperforms node-based techniques15. However, the potential of the link/community viewpoint is still largely unexplored, and we sensed that this strategy might also be successful for the design of novel link-prediction indices; our interest is to introduce a new philosophy in the formulation of parameter-free/neighbourhood-based indices, advocating a shift in perspective from nodes to links, and in particular from nodes to community links.
. The other indices are often a variation or generalisation of CN7: Jaccard's index (JC) is a normalisation of CN12, Adamic & Adar (AA)13 and Resource Allocation (RA)14 give more importance to CNs with low degree, while Preferential Attachment (PA)11 is the degree product of nodes x and y (for the formulae see Table 1 and for details see Supplementary Information (SI) section IIIA). In contrast to the existing methodologies, which are focused on groups of common nodes and their node neighbours, we here embrace a strategic shift from nodes to links that represents a new way to treat complex networks15. In particular, Ahn et al. reconceived communities as groups of links rather than as mere groups of nodes, and proposed a link-based approach for graph-partition that outperforms node-based techniques15. However, the potential of the link/community viewpoint is still largely unexplored, and we sensed that this strategy might also be successful for the design of novel link-prediction indices; our interest is to introduce a new philosophy in the formulation of parameter-free/neighbourhood-based indices, advocating a shift in perspective from nodes to links, and in particular from nodes to community links.
Table 1. Table of formulae for the classical and CAR-based neighbourhood techniques.
| Type | Name of the Index | Formulation |
|---|---|---|
| Classical | Common Neighbours (CN) | 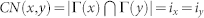 |
| Preferential Attachment (PA) | 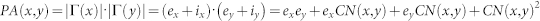 |
|
| Adamic & Adar (AA) | 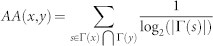 |
|
| Resource Allocation (RA) | 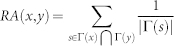 |
|
| Jaccard (JC) | 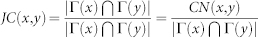 |
|
| CAR-based | CAR | 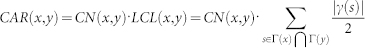 |
| CPA | 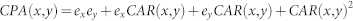 |
|
| CAA | 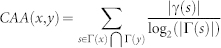 |
|
| CRA | 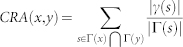 |
|
| CJC | 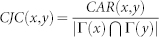 |
x and y are network nodes; s is a common neighbour node of x and y; 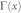 refers to the set of neighbours of x;
refers to the set of neighbours of x; 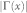 refers to the cardinality of set
refers to the cardinality of set 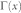 , which is equivalent to the degree of x; γ(s) refers to the sub-set of neighbours of s that are also common neighbours of x and y, thus γ(s) is the local community degree of s; ex refers to the external degree of x, computed considering the neighbours of x that are not common neighbours of x and y; ix refers to the internal degree of x that is equivalent to the number of common neighbours shared by x and y. For the Matlab code to compute all the indices in the table see SI, section IIID.
, which is equivalent to the degree of x; γ(s) refers to the sub-set of neighbours of s that are also common neighbours of x and y, thus γ(s) is the local community degree of s; ex refers to the external degree of x, computed considering the neighbours of x that are not common neighbours of x and y; ix refers to the internal degree of x that is equivalent to the number of common neighbours shared by x and y. For the Matlab code to compute all the indices in the table see SI, section IIID.
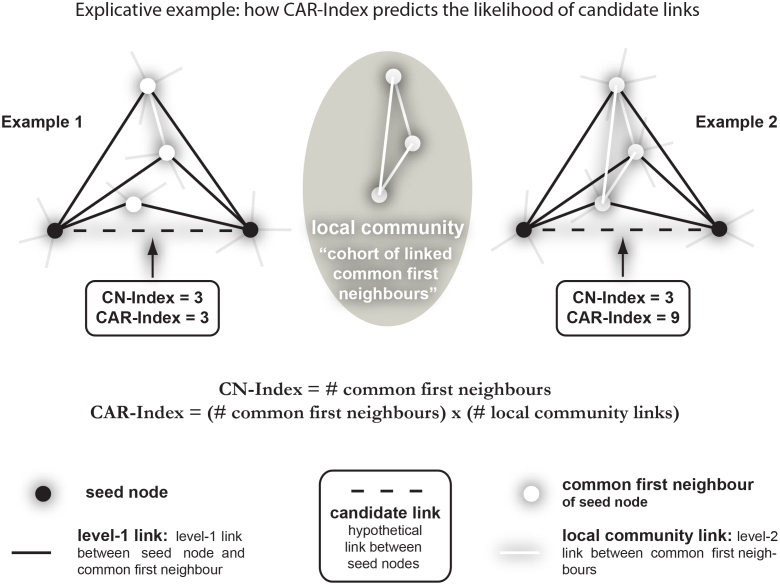
The CAR index (see Fig. 1 for definition and examples, and SI section IIID) stems from the fusion of the old node-based and new link-based perspectives. CAR suggests that two nodes are more likely to link together if their common-first-neighbours are members of a strongly inner-linked cohort, named a local-community (LC), and the LC's internal links are called local-community-links (LCL, see Fig. 1). A consequence of this formulation is that, in respect to CN, CAR offers more discriminative resolution between candidate-links characterised by the same number of common-first-neighbours (Fig. 1), and this boosting in resolution is clearly derived by the use of the link/community perspective, which is introduced adopting LCL in the CAR formula (for details see Fig. 1 and SI section IIID). To demonstrate that the formulation of CAR is not a banal trick, but the precise introduction of a link/community strategy in designing neighbourhood-based indices, we propose for each of the above mentioned classical methods a CAR-based variant. If LCL is seen as an index enhancer, it can be plugged into PA, AA, RA and JC indices so that these techniques also shift to the link/community perspective and in the rest of the article we will extensively prove the value of this idea. Table 1 provides the formula of the considered classical indices and their respective CAR-based variations, for details on their derivation please refer to SI section IIID, where we also provide a demonstration, which, under the assumption of very sparse networks, proves that CAR and CAA give the same ranking, evidence that we confirm also experimentally by simulations on real networks (SI section IIID and Fig. S2).
Figure 1. CAR index.
When a link or a node directly interacts with a seed node, it belongs to the first-level-neighbourhood and conveys first-level topological information. Conversely, a link or a node that interacts with the first-level-neighbourhood conveys second-level information. Second-level information is valuable and its use can significantly improve topological link-prediction, but unfortunately it is also very noisy, and for this reason difficult to integrate with the first-level information. CAR is designed to capture and filter meaningful second-level information by exploiting common-first-neighbours. The topological information conveyed by the internal links between common-first-neighbours is valuable second-level information. Indeed, the more the common-first-neighbours reciprocally interact, the more they represent a local-community, which in turn encompasses the two seed nodes and thus increases their candidate-link likelihood. Here we introduce the idea that the likelihood of a candidate-link is a function of both the number of common-first-neighbours (as in the CN index) and of the number of links between them (local-community links), as expressed in the formula of CAR. The two explicative examples clarify how CAR increases discriminative resolution between candidate-links with the same number of common-first-neighbours.
Link-prediction in brain connectomes
To test the performance of CAR's indices against the classical indices, we propose an innovative benchmark problem that originates from the neurosciences. The notion of connectome16 suggests that the brain circuitry can be outlined as a network of neurons connected by links, which are synapses. Several neuroscientific studies have demonstrated that certain forms of learning consist of synaptic modifications17, while the number of neurons remains basically unaltered18. A first model of this process was proposed in 1949 by Hebb and subsequently used in Hopfield's model of associative memory18. The Hebbian learning theory assumes that different engrams (memory traces) are memorised by the differing neurons' cohorts that are co-activated within a given synaptic network. It is also termed cell-assembly hypothesis18, because these neuron-assemblies are shaped during engram formation by a re-tuning of the strengths of all the adjacent synapses extant in the network. Recent studies18, however, demonstrated that learning new motor or sensory tasks is associated with the development of new and non-overlapping sets of persistent synapses. Ziv et al. commented on these findings by suggesting that synapse-assemblies, rather than cell-assemblies, might be viewed as the elementary entities of stored memories18, which in turn amounts to a link/community reinterpretation of learning and memory processes in neuroplasticity. We thought to test this synapse-assemblies hypothesis in a computational framework, where the formation of new synapses during learning might also be influenced by the local-synapse-communities extant in the network. Consequently, we proposed to use the topological prediction of new links in a brain-connectome to model the part of the growth and remodelling process that is conditioned by the connectome topology during synaptic formation.
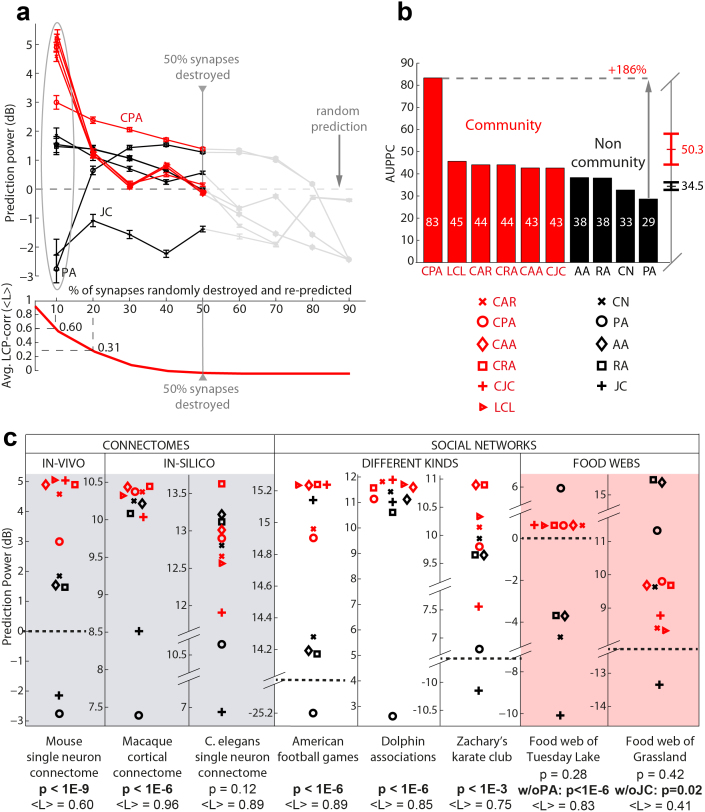
Figs. 2a and 2b show the prediction power of CAR's indices versus that of the classical node-neighbourhood-based indices. We considered the first and (to date) only available in-vivo single neuron connectome (obtained by means of in vivo two-photon calcium imaging in combination with large-scale electron microscopy) that reports mouse primary visual cortex (layers 1, 2/3 and upper 4) synaptic connections between neurons19. The evaluation was performed by destroying uniformly at random a certain percentage of synapses and by re-predicting them with the indices' ranking (details are in the Methods section and in the SI, section IV). CAR's indices proved to be the best methods and performed significantly better (p-value < 0.05) than the classical node-based models such as CN in the destruction and re-prediction of up to 50% of the original connectome synapses (Fig. 2a–b). Interestingly, the 50% deletion level seems to be a critical value. Synaptic deletion beyond this limit induced a strong reduction in performance in all indices; nevertheless CPA (which is the CAR-based variant of PA) and PA were notably the only indices whose prediction remained stable in the given condition - with CPA performing also as the best index overall. This finding suggests that for synaptic deletion greater than 50%, a significant quantity of connectome nodes is also deleted causing network disconnection into different components and network clustering coefficient reduction. The consequence is a general drop in performance for all the non-PA based indices20 that, as a result, assign a score of zero to candidate links whose seed nodes are located in different components. On the contrary PA formulation is based on the product of the seed nodes’ degrees only, thus its performance is less affected. By definition, this result also corroborates the reliability of the analysed connectome and of the experimental approach used to detect it. Additionally, the result confirms that the design of our computational experiment was correct, and illustrates, as expected, that initial link-growth is dominated by a self-organising process of node-preferential-attachment4.
Figure 2. Random deletion followed by topological link re-prediction in brain connectomes, social and ecological networks.
(a) In-vivo single neuron connectome: mouse visual cortex neuro-synaptic connections. Sets of links (synapses) from the mouse connectome were progressively removed uniformly at random. Each prediction technique assigned a score to the missing synapses and this score was used to sort a list of candidate synapses. Upon removal of n links from the network, the same number was taken from the top of the candidate list; by comparing the candidate set with the removed set, we assessed the technique's precision. Since this process was repeated 1000 times for each sparsification level (ranging from 10% to 90% of removed synapses), in practice mean precision and standard error were considered at each stage. To assess deviation from the mean random-predictor performance, the Prediction-Power was computed in decibel (dB):  ; thus, considering the different levels of sparsification, a Prediction-Power-Curve was generated and the area under this curve (AUPPC) was adopted as a comprehensive measure of performance (see Methods section and SI, section IV). Notice that the progressive removal of links from the original topology made the average LCP-corr drop down to the point where it is almost 0. (b) Since deletion of more than 50% synapses caused node isolation and the disappearance of local communities, the AUPPC for the plots in panel (a) was computed considering only half of the experimental range (10% to 50%). CAR-based indices provided an average AUPPC of more than 50, and the performance of the best approach (CPA), represented a 186% improvement over the lowest performing technique (PA). JC was not reported. (c) The performance of CAR-based and classical predictors was also studied over 8 different networks when 10% of the links were removed 1000 times uniformly at random (which ensures community preservation and a fair benchmarking, see average LCP-corr values <L> below each plot). The difference in performance between the CAR-based (in red) and the classical techniques (in black) was statistically significant every time the studied network exhibited a LCP structure (see p-values, below each plot computed as a permutation test with 1000 sampling realizations).
; thus, considering the different levels of sparsification, a Prediction-Power-Curve was generated and the area under this curve (AUPPC) was adopted as a comprehensive measure of performance (see Methods section and SI, section IV). Notice that the progressive removal of links from the original topology made the average LCP-corr drop down to the point where it is almost 0. (b) Since deletion of more than 50% synapses caused node isolation and the disappearance of local communities, the AUPPC for the plots in panel (a) was computed considering only half of the experimental range (10% to 50%). CAR-based indices provided an average AUPPC of more than 50, and the performance of the best approach (CPA), represented a 186% improvement over the lowest performing technique (PA). JC was not reported. (c) The performance of CAR-based and classical predictors was also studied over 8 different networks when 10% of the links were removed 1000 times uniformly at random (which ensures community preservation and a fair benchmarking, see average LCP-corr values <L> below each plot). The difference in performance between the CAR-based (in red) and the classical techniques (in black) was statistically significant every time the studied network exhibited a LCP structure (see p-values, below each plot computed as a permutation test with 1000 sampling realizations).
Furthermore, we considered two different in-silico connectomes, the Macaque cortical connectome and the C. elegans frontal ganglia connectome, which previous studies had assembled in order to merge partial information obtained from disparate literature and database sources21. Here, in particular, we concentrated our attention on the predictors' performances when random deletion of only 10% of links is applied, to ensure that the community structure (if present) is still preserved, and in order to objectively compare the proposed community-based indices versus the classical ones (details on the procedures used for these and all the comparisons of Fig. 2c are in the Methods section and in the SI, section IV). Although CAR's indices performed largely better than random predictors in these two simulations too (Fig. 2c), their superiority over the classical indices was diminished but still statistically significant in the Macaque connectome, and comparable to the classical methods in the C. elegans connectome. This finding may be due to: i) the unreliability and paucity of the two different connectomes, which do not directly derive from in-vivo investigations; ii) the absence of sufficient topological information that is generated by the neuroplasticity process of learning. In particular, the Macaque cortical connectome (aka corticocortical network) is incomplete and differs in anatomical scale: the links are long-range-projections between cortical areas and subareas within one hemisphere of the primate brain21. Although the C. elegans frontal ganglia connectome is reconstructed at the synaptic level, in-silico merging causes it to be similarly unreliable. Significantly, it has recently been emphasised how published wiring diagrams for C. elegans are neither accurate nor complete and self-consistent22. Nevertheless, the process of synaptic formation (or, to be precise, at least that part of the process that depends on connectome topology) between existing neurons seems to be more accurately predicted by our local-community-based models than by PA and the other classical neighbourhood-based approaches, and this is the first substantial result we have obtained for link-prediction in computational network biology. A straightforward implication of this finding might open up new avenues in computational learning models, in which the Hebbian theory might be complemented by an epitopological learning theory, whereby engram formation stems from the growth of additional synapses within local-communities of pre-existing synapses. A second important implication is that our local-community-based interpretation of synaptic learning could also be a computational evidence to support the recently proposed unifying hypothesis on the autistic brain called Intense World Syndrome (IWS), which sustains that the core pathology of the autistic brain is hyper-reactivity and hyper-plasticity of local neuronal circuits23. Markram et al. advocate that such excessive neuronal processing and plasticity in circumscribed circuits lead to excessive neuronal learning: hyper-perception, hyper-attention, and hyper-memory, which may lie at the heart of most autistic symptoms23. Thus, we speculate that our local-community-based interpretation of synaptic learning might be also adopted as part of a computational model to simulate some of the basic mechanisms that are assumed by the IWS to explain how the autistic person is an individual with far above average capabilities (due to enhanced perception, attention and memory)23.
Link-prediction in social and ecological networks
The CAR's indices were in general significantly better than classical ones also when applied to diverse kinds of complex networks such as the human/animal social networks (Fig. 2c). Only the food webs represented a case apart. For the food web of Tuesday Lake (excluding the performance of PA) the CAR-based indices were the only ones that performed better than random, and in general significantly better than the classical indices. For the food web of Grassland species (excluding the performance of JC) we registered a paradoxical behaviour, since it is the only network where the performance of classical methods is significantly better than CAR-based indices (the topological motivation of this result will be further investigated in the next section). There is, nevertheless, an explanation for these opposite behaviours based on the structure, characteristics and members of these two ecosystems. While the Tuesday Lake is regarded as one of the most complete and organised webs in the field of ecology24,25, the Grassland web is considered highly unsaturated26. Moreover, the Grassland web has a linear structure that in ecology is known as the cascade model: a serial trophic organisation, where the bigger organism eats the smaller. In contrast, the Tuesday Lake web has a pyramidal organisation that differs from the cascade model in the distribution of links between basal, intermediate, and top communities of species25. Thus, the Grassland web has a highly sparse and almost linear connectivity, which makes it hard for local communities to emerge; on the contrary, the Tuesday Lake web has a more densely connected and organised structure, which is actually related with the presence of local communities at different levels of the pyramidal organization.
As a further investigation, we compared CAR's indices with two sophisticated statistical inference techniques: the Hierarchical Random Graph27 and the Stochastic Block Model28 - see SI section IIIC for details. The comparison followed the same procedure showed in Fig. 2c, and we did not detect any remarkable increase in performance of these advanced algorithms in comparison to the family of CAR-based community indices (see SI, Fig. S1). Details on the methods and results of this last comparison are in SI, section IV.
Link-prediction in protein interactomes
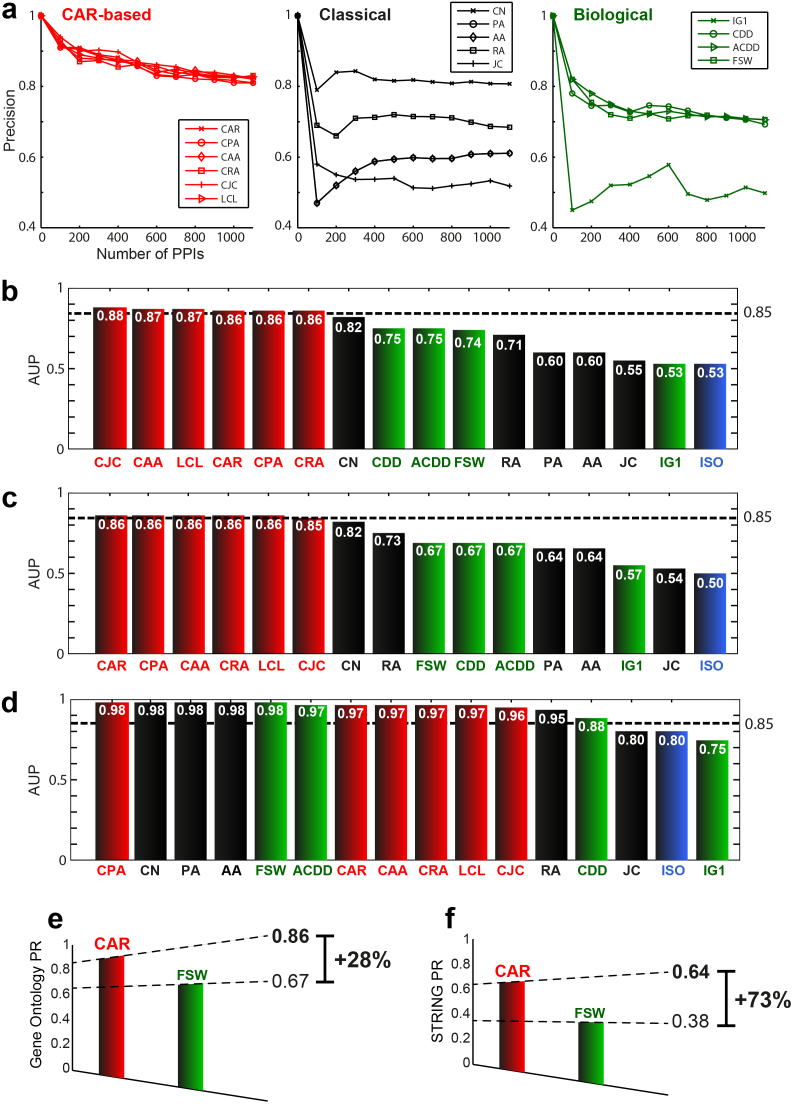
A second aspect of the link-prediction problem regards the inference of missing interactions from an observed network7. In varying disciplines, a network is constructed on the basis of experiments, and for at least two reasons some links might not be observable: i) by nature, the links are not directly detectable; ii) the experiments and/or the execution time are very expensive. The problem of observability affects systems biology, a discipline in which the topological prediction of novel interactions in protein networks (interactomes) is particularly useful, especially and specifically when other types of information, such as biological prior knowledge, are not available29. This problem differs from the original formulation (link-prediction in network evolution over time) discussed above, and the class of link-prediction indices (named bio-inspired indices, SI, section IIIB) currently employed in systems biology derives from methods invented to infer similar attributes between adjacent protein nodes29: the Interaction Generality (IG1) Index30 originates from phenomenological evidences in experimental protein interaction detection; the Czekanowski-Dice Dissimilarity (CDD, as well as its adjusted version ACDD)31,32 and the Functional Similarity Weight (FSW)33 stem from methods for protein functional prediction in interactomes; and ISOMAP (ISO)29,34 is based on high-dimensional properties and embedding of protein networks. The strategy used for evaluation differs too29, and a candidate protein interaction is judged to be correctly predicted if the association between the two linked proteins has relevance in the Gene Ontology (GO) categories (Methods sections and SI, section V). Typically, a level of precision (on the basis of significant association in GO) is recursively evaluated for sets of increasing size of the best first-ranked candidate interactions (Methods section and SI, section V). A precision curve is obtained, and the area under this curve (AUP) is a measure of performance. Yeast networks are the preferred benchmark, because of the large amount of information available for yeast in terms of both detected interactions and GO associations15,29. We accordingly re-analysed three different, independently produced, yeast networks that had been used in previous studies for performance testing (SI, sections V; and Table S3 in section IX). CAR and CAR-based indices not only substantially outperformed the other methods in the three networks (they are the only indices to attain AUP always equal or higher than 0.85, Fig. 3b–d), but also proved to be the most efficient, in that it simultaneously provided consistent robustness (Fig. 3e,f and SI Table S1) and the lowest maximum-computational time (SI Table S1).
Figure 3. Topological link-prediction in protein interactomes.
(a) Precision curve of each technique by ascertaining whether the top-ranked candidate links had meaningful association in Gene Ontology (GO). For clarity, only CAR-based, classical and bio-inspired prediction techniques are shown; the rest of the results and plots appear in SI, section V. (b–d) The Area Under the Precision curve (AUP) summarises the general performance for each technique in the three analysed networks: Network1 (Ben−Hur & Noble, 2005), Network2 (Chen et al., 2006), Network3 (Chen et al., 2006). (e) Gene Ontology (GO) performance robustness (PR) is computed as the minimum AUP among the networks and used for comparison between CAR and FSW. (f) In-silico STRING validation: the first 100 best interactions for each method are tested in STRING. The minimum number of verified-interactions among the three networks represents the minimum precision, which is a measure of performance robustness (PR) for comparison - with regard to STRING - between CAR and FSW.
We besides investigated the extent to which the topological link-prediction could be practically useful. To this end, we proposed an in-silico validation (SI, section VI), and tested the 100 best ranked interactions, both for CAR and for FSW (reference method for the bio-inspired indices), with the STRING database35. Considered to be the most complete and reliable database of protein-protein interactions (PPIs), STRING integrates multiple sources of information and provides a confidence score for each interaction35. Once again, CAR confirmed the benefit of using a link/community strategy, and performed impressively during this validation test, both in each single network evaluation (SI, section VI, Fig. S4a-c) and in the general evaluation of robustness (Fig. 3f).
Summarizing, we observe that in general using the CAR-trick to convert the classical methods to the new link-community perspective, generates in several complex networks a significant increase in link prediction performance, suggesting that a new family of community-based link predictors is here proposed. In addition, we notice that whenever the community structure of the network (if present) is preserved, it can be exploited to achieve more accurate predictions using the new CAR-based family of link predictors.
The local community paradigm (LCP) in real networks
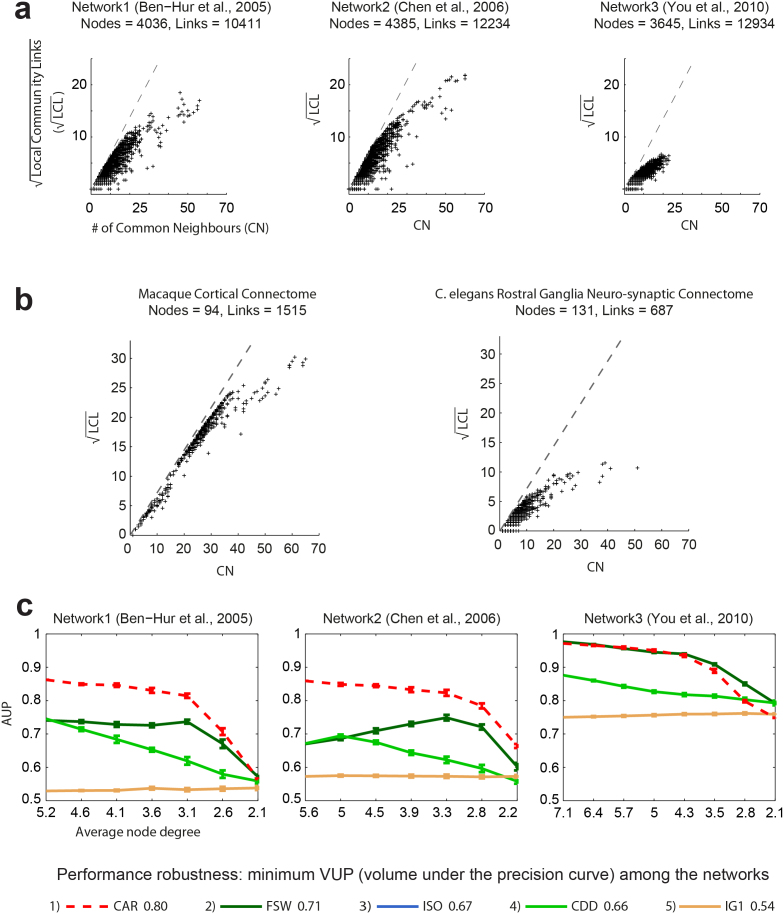
A necessary condition for the application of CAR's indices is that, during its growth, the network in question evolves in accordance with a general process, one of whose distinctive features is the development of diverse, overlapping and hierarchically organised local-communities15. We defined this form of topological self-organisation as a local-community-paradigm (LCP), and we therefore propose the LCP-decomposition-plot (LCP-DP) to visualise and investigate the effect of LCP on network topology. The LCP-DP is a form of network-decomposition because each real link in the network is plotted in a bi-dimensional space according both to its number of CNs (reported on the x-axis) and to the respective number of LCLs (number of links between the common-first-neighbours, reported on the y-axis). More specifically, given that the number of LCL is a squared function of the CNs (SI, section VIII), we found it more convenient to report the square root of LCL on the y-axis, so as to linearize the visualisation. The result of this decomposition is a plot that offers a link-based visualisation of the analysed network and provides information on the presence and size of its local-communities.
Fig. 4a shows the LCP-DP for the three previously employed yeast PPI networks. Surprisingly, we discovered that Network3 differs from the other two, whose LCP-DP patterns resemble each other (Fig 4a). In particular, the maximal local-community size for Network3 is around 20 CNs, while for the other two networks the maximal local-community size is about 60 CNs. Considering that all the networks have a comparable number of nodes and links, and that the maximal size of local-communities in Network3 is 1/3 of that in the other networks, we can conclude that Network3 is more strongly characterised by small-local-communities. The fact that multiple small-local-communities in a large and sparse network most probably do not overlap with each other explains why all the indices (CAR-based, classical and bio-inspired) performed in a comparable manner and with better results in Network3 (Fig. 3d) than in the other two networks (Fig. 3b–c), where instead the CAR-based performed significantly better than the others. Following a similar rationale, we observe that the C. elegans connectome (Fig. 4b, left panel), although larger (more nodes) and sparser (less links) than the Macaque connectome (Fig. 4b, right panel), is more strongly characterised by small-local-communities (Fig. 4b, left panel), and this explains why the performances of the CAR-based and classical indices were not significantly different, and were overall higher in this connectome than in the others (almost all the indices were between 12.5 and 13.5 dB, Fig. 2c). From the topological similarity between Network3 and the C. elegans connetome, evidenced by using the LCP-DP, we can now infer that a very sparse and clustered network topology, characterised by the presence of multiple small-local-communities that do not overlap with each other, improves link prediction in general and minimise the difference between CAR-based and classical indices. Conversely, in the extreme and opposite case of very densely connected networks, the occurrence of large and indistinct communities - which most likely overlap reciprocally and envelop the small-local-communities - erases the community distinction that is fundamental for the efficiency of CAR and of prediction methods in general. This is easily demonstrable because the more a network tends to the ideal case of a fully-connected network, the more it converges towards a unique and large single-community. On the other hand, in Fig. 4c we clearly demonstrate that when we randomly sparsify the networks in question (sparsification is a procedure that enables the performance testing in relation to unpredictable random variations in the original network topologies) CAR, whenever the network community structure is preserved, is more efficient and robust than the other indices in prediction of candidate protein interactions (for details on this computational experiment refer to SI, section VII).
Figure 4. LCP-DP, LCP-correlation and sparsification-experiment on PPIs.
(a) Using the LCP-DP for investigation of the network topology and for visualization of the LCP-correlation in protein interactomes. (b) Using the LCP-DP for investigation of the network topology and for visualization of the LCP-correlation in brain connectomes. (c) Testing the prediction robustness of CAR and the other indices during PPI network sparsification by random link deletion. For each point 10 network realizations are computed, and the average AUP with standard-error for each index (computed with the same procedure as that used for Fig. 3b-d) is reported. The random deletion adopted progressive levels (10%, 20%, and so forth, until network connectivity was lost) of the links in the original network. By varying the level of sparsification, we produced a curve of average-AUP values, so that the area under this curve became a volume. On the basis of normalised AUP, we were able to compute what we called ‘volume’ under the precision curve (VUP). VUP is an advanced performance measure in the sense that it accounts both for random variations in the original network topology and for differing levels of sparsification. Isomap (ISO), which is the only embedding-based method, is not plotted for reasons of display clarity, but its VUP is reported.
A second discovery, which emerges from the LCP-DP, is the strong correlation between the two variables CN and LCL, which we call the LCP-correlation (LCP-corr), and which might be interpreted as a typical feature of LCP networks (Fig. 4a–b). More specifically, the LCP-correlation is defined as the Pearson-correlation coefficient between the variables CN and LCL as plotted in the LCP-DP (computation details are in SI, section VIII). Looking at the curve of average LCP-correlation values - computed for the Mouse connectome configurations, in the randomly destroyed synapses experiment of Fig. 2a – we observe that the LCP-correlation decreases significantly between 10% (LCP-corr = 0.60) and 20% (LCP-corr = 0.31) of removed synapses, and this is a confirmation that the choice to focus our attention on the comparison of the predictors when 10% only of the links are randomly removed, was a correct procedure to preserve the community structure. In fact the average LCP-correlation values of the 8 partially destroyed complex networks adopted in Fig. 2c are always higher (except for the Grassland web) than LCP-corr = 0.5. On the other hand, also the examples provided in Fig. 4a (Network1 has LCP-corr = 0.92; Network2 has LCP-corr = 0.95; Network3 has LCP-corr = 0.90) might lead one to suspect that large LCP-correlation coefficients are invariably associated with the occurrence of LCP, and small LCP-correlation coefficients with LCP's non-occurrence. Therefore, to investigate the extent to which this hypothesis is generally valid, we considered a total of 45 networks from differing fields (15 biological, 10 social, 10 atomic, 1 power grid, 9 roadmaps - see SI, section IX).
As evidenced in Fig. 5, where we juxtapose a few paradigmatic examples, the scenery is more intriguing than we had expected: we found that the region of LCP-correlations between 0.8 and 0.4 represents a threshold (a sort of intermediate region) that distinguishes networks that are characterised by LCP from those that are not. We want to acknowledge that also Lancichinetti et al.36 went close to the formalisation of the LCP paradigm; and the observation of the dichotomy between LCP and non-LCP networks. In fact, they provided a systematic empirical analysis of the statistical properties of communities in diverse types of large complex networks36, and evidenced that the mesoscopic organisation of networks of the same category is extraordinarily similar, concluding that: although the community size distributions are always wide, certain categories of networks consist mainly of tree-like communities, while others have denser modules.
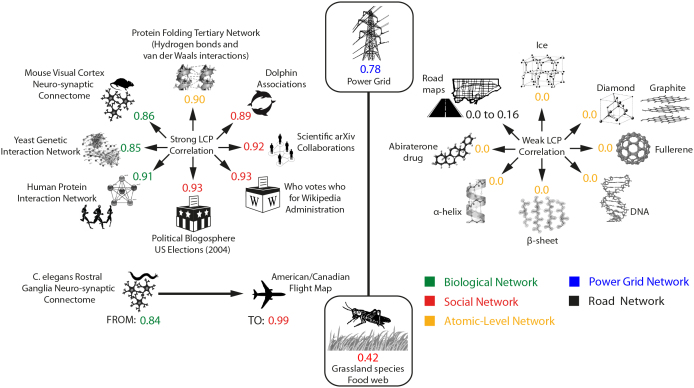
Figure 5. LCP in real networks.
Examples of LCP networks (left panel) and non-LCP networks (right panel). The LCP networks we found range from LCP-corr 0.84 (C. elegans rostral ganglia neuro-synaptic connectome) to 0.99 (American/Canadian flight map). The networks in the center (Power Grid and Grassland species food web) are borderline cases. The adjacency matrices of all the networks for which we computed LCP-corr are provided at the link in SI, section IX. For licence information on the individual elements of this figure please see SI, section X.
If we investigate the networks at the extremity of the intermediate region, we find that the power grid network is a borderline case, one that can be considered neither as LCP nor as non-LCP, and this finding has strong significance. It has been proved that power grid networks, which are human-designed and propagate electricity rather than information, are neither scale-free nor clustered37, that they have homogenous topology and that they are easy to control38. The other interesting borderline case is the Grassland species food web that with LCP-correlation of 0.42 is closer to non-LCP networks, and this can motivate its paradoxical behaviour detected in Fig. 2c (after 10% of random link removal, LCP-corr = 0.41) and discussed above.
On the other hand, the upper-bound case is represented by the air transportation network, which shows a 0.99 LCP-corr (Fig. 5 left side, American/Canadian flight map). We mined the literature to find an explanation for the contradiction between the result obtained for the air transportation and for the road transportation networks (Fig. 5 right side, LCP-corr ranges from 0 to 0.16). Guimera et al.39 provided a possible answer, which we shall now cite: ≪ …We find that the worldwide air transportation network is a scale-free small-world network. In contrast to the prediction of scale-free network models, however, we find that the most connected cities are not necessarily the most central, resulting in anomalous values of the centrality. We demonstrate that these anomalies arise because of the multicommunity structure of the network. We identify the communities in the air transportation network and show that the community structure cannot be explained solely based on geographical constraints and that geopolitical considerations have to be taken into account. …≫. Although Guimera et al. did not formalise any paradigm, we recognise an ante litteram discovery of the LCP in their intuition of the need for a new explanation (besides the scale-free and the small-world paradigms) to characterise the topology of air transportation networks more precisely. Meanwhile, the social (geopolitical) interpretation of the flight networks clarifies the mismatch with the road networks, which are shaped more by geographic constraints, and further justifies our choice to allocate the flight map among the social networks.
The conclusions of Guimera et al. - and the need to formalise them in a paradigm such as LCP - appear clearer if interpreted in the light shed by two theories recently posited, one by Boguñá et al.37 on the navigability of complex networks, and the other by Liu et al.38 on the controllability of the same. Boguñá et al. illustrated the greedy-routing of information in a network through an example of passenger air-travel. They showed how greedy behaviour takes a passenger from a small airport to larger hub airports, which significantly reduces the distance to the destination (zoom-out coarse-grained search), and how hubs are thus crucial for global delivery. Once a hub near the destination is reached, hubs are not needed anymore because a less-connected neighbouring airport can take the passenger to the desired city (zoom-in fined-grained search). Therefore, some particular non-hub nodes centred in local modules are fundamental for the local processing of the general function implemented on the network. On the other hand, Liu et al. found that low-degree nodes (and, counter-intuitively, not hubs) play the most important part in the full controllability of complex networks, and this finding fits with Boguñá's, because hub nodes may be viewed as collectors and distributors of information: if one of the near-destination hubs is unavailable, another one can compensate. They act as intermediary relaxing points that avoid bottlenecks and direct data to more important nodes (driver or processor nodes) that accomplish a dedicated function within local modules, where they naturally assume a position of centrality. Similar conclusions are discussed by Lancichinetti et al. as well36.
We also introduced the analysis of atomic-level networks. On the right side of Fig. 5, we showed that many organic molecules (their crystals, reticula and lattices) and secondary-biological structures have a network topology that is non-LCP. In similar vein, we investigated the network topology in the tertiary-biological structures of proteins: said structures are generated by various classes of non-covalent interactions that occur between protein residues and determine the typical protein folding conformation. Residue interaction networks (RINs) have recently been used to describe the protein three-dimensional structure as a graph, where nodes represent residues and edges physico-chemical interactions, e.g. hydrogen bonds or van der Waals contacts40. Various topological properties have been calculated over RINs and have been correlated with differing aspects of protein structure and function40. We found that both the hydrogen bond network of human glutathione peroxidase 4 (GPX4, LCP-corr = 0.90) and the van der Waals contact network of human triosephosphateisomerase (TIM barrel, LCP-corr = 0.88) - which were the only RINs available to use40 - are LCP (Fig. 5 right side, and SI, section IX). Consequently, we envisage that this finding might be extended beyond the examples here quoted, to confirm that the LCP-state detected within tridimensional-protein conformations could be a generic property of some tertiary-biological structures.
The LCP in idealised networks
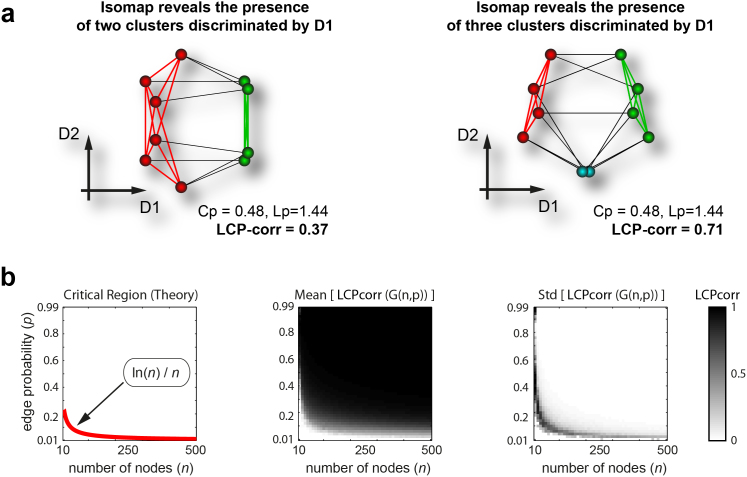
LCP accounts for community-based structure in the topology of complex networks, and extends present knowledge to a degree that, along with the small-world and preferential-attachment paradigms, may enhance our understanding of systems of interacting units, their evolution and self-organisation. The need for, and value of, such a novel paradigm are further investigated in the following examples, which deal with referential idealised models. The networks in Fig. 6a are two diverse random regular graphs (random graphs for which each node has the same degree) with the same number of nodes. The BA paradigm (which is based on power-law node distribution) does not detect any difference between these two networks, since the degree distribution of a random regular graph corresponds to a single value that is also the fixed node degree. Even the WS paradigm (small-world) does not detect any difference, since the two networks have identical clustering coefficients and characteristic path lengths. This is further proof that, as clarified in the original article2, the small-world paradigm is inapplicable to the detection of topological changes that emerge exclusively at a local structural level in the network. Boguñá et al.37 showed, firstly, that behind each network there is a hidden metric space that is closely related to the network topology and, secondly, that global mapping may be inferred from local distances. In a way, the same principle is exploited by Isomap41, a landmark algorithm for embedding that was mainly designed to visualise the hidden structure of a dataset or of a network in a bi-dimensional space. Surprisingly, the embedding in two dimensions by Isomap suggests substantial divergence between the hidden metric spaces of the two random regular networks, as well as a likewise substantial difference in the local topology (Fig. 6a). The paradigm we propose is the only one that clearly identifies this topological difference within the hidden metric space of the two networks (Fig. 6a). The LCP-correlation is 0.37 for the network on the left, which consists of only two clusters, while for the second network, which consists of three clusters, the LCP-correlation is, as expected, higher (LCP-corr = 0.71, Fig. 6a on the right side).
Figure 6. LCP in idealised networks.
(a) Isomap embedding of two random regular graphs (Nodes = 10, Degree = 5) with equal clustering coefficients (Cp) and characteristic path lengths (Lp) but different LCP-correlation. Neither the BA nor the WS paradigm were able to explain the topological difference between these two random regular graphs, which share the same node numbers and the same node degree. The paradigm we propose is the only one that, consistent with Isomap embedding, clearly highlights the community organisation diversity present in the hidden metric spaces of the two networks. (b) Testing the LCP-correlation for differing Erdős–Rényi random graphs G(n,p) by varying the number of nodes n (from 10 to 500, step 10) and the edge probability p (from 0.01 to 0.99, step 0.02). For each combination (n,p) the LCP-correlation was evaluated 100 times for calculation of mean and standard deviation. The theoretical critical region computed by Erdős and Rényi in their model (plot on the left) is strikingly detectable both by the mean (plot in the centre) and by the standard-deviation (plot on the right) LCP-correlation.
Idealised networks are very useful to test the generality of a hypothesis in different configurations, which in turn are easily generated artificially through a known model and controlled by certain parameters2. Such networks are particularly crucial to investigations into the behaviour of a given measurement around a critical region that hosts a transition between differing macroscopical states of the system. We simulated differing ER networks by varying the two parameters used in the model G(n, p), namely network size (number of nodes n) and edge probability (each edge is included in the graph with probability p independent from every other edge). We performed 100 realizations for each combination of model parameters and plotted in Fig. 6b the mean (displayed in the centre) and the standard deviation (displayed on the right) of the LCP-correlation values computed for the 100 network realizations. In their original 1960 paper1, Erdős and Rényi mathematically described the behaviour of G(n, p) at various values of p. One of their main conclusions was that 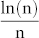 represents a critical threshold for the connectedness of G(n, p). In particular, they proved that: i) If
represents a critical threshold for the connectedness of G(n, p). In particular, they proved that: i) If 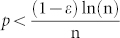 then a graph in G(n, p) will be disconnected because it certainly contains several isolated vertices (which implies the loss of local network communities); ii) if
then a graph in G(n, p) will be disconnected because it certainly contains several isolated vertices (which implies the loss of local network communities); ii) if 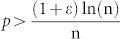 then a graph in G(n, p) will be most likely connected (which implies the preservation of local network communities). Interestingly, we discovered that the theoretical transition region (Fig. 6b, plot on the left) is accurately detected and visualised by the LCP-correlation (Fig. 6b, in the centre and on the right) in our experiments. From the mean of the LCP-correlation values, as plotted in the centre of Fig. 6b, we discovered that the majority of all possible generable ER models strongly follow the LCP (black area above the critical region). The necessarily small percentage of non-LCP models appear in the white area under the critical region. This result confirms that under the critical region, several isolated vertices appear in the graph, while above the critical region a topology characterised by several community structures begins to emerge. Meanwhile, prompted by standard deviation analysis (Fig. 6b on the right) we learn that the generation of graphs with intermediate values for LCP-correlation occurs very rarely and is only possible when it coincides with the instability expressed in the critical transition region (dark region in the plot); in the other zones (white part of the plot), standard deviation is close to zero and the LCP-correlation value is very stable.
then a graph in G(n, p) will be most likely connected (which implies the preservation of local network communities). Interestingly, we discovered that the theoretical transition region (Fig. 6b, plot on the left) is accurately detected and visualised by the LCP-correlation (Fig. 6b, in the centre and on the right) in our experiments. From the mean of the LCP-correlation values, as plotted in the centre of Fig. 6b, we discovered that the majority of all possible generable ER models strongly follow the LCP (black area above the critical region). The necessarily small percentage of non-LCP models appear in the white area under the critical region. This result confirms that under the critical region, several isolated vertices appear in the graph, while above the critical region a topology characterised by several community structures begins to emerge. Meanwhile, prompted by standard deviation analysis (Fig. 6b on the right) we learn that the generation of graphs with intermediate values for LCP-correlation occurs very rarely and is only possible when it coincides with the instability expressed in the critical transition region (dark region in the plot); in the other zones (white part of the plot), standard deviation is close to zero and the LCP-correlation value is very stable.
In conclusion, this simulation suggests that the generation of: i) ER networks with LCP-corr higher than 0.80 is very frequent; ii) ER networks with LCP-corr close to 0 is not frequent; iii) ER networks with intermediate LCP-corr is very infrequent. These findings are in line with the ones we obtained for real networks, where the identification of LCP networks was very easy (Fig. 5b, left side), both at diverse physical levels and in differing domains (biological, social, atomic), while the identification of non-LCP networks was difficult and their occurrence was almost exclusively detected at the atomic level (Fig. 5b, right side). Interestingly, except for the power grid (which showed 0.78 LCP-corr, a borderline value) and few others, real networks with intermediate levels of LCP-correlation were not identified.
Taken together, these discoveries are important steps towards the answer to the second question formalised in the introduction.
Discussion
The third question posed in the introduction conjectures a sort of generalised systemic parallelism between the occurrence of certain topological properties (as formalised in a paradigm) and the relevance of some physical properties. In effect, LCP networks (Fig. 5 left side) are related to dynamic and heterogeneous systems that are characterised by weak interactions42 (relatively expensive or relatively strong) that in turn facilitate network evolution and remodelling; these are typical features of social and biological systems as well as, at the atomic level, of tertiary protein structures. According to Liu et al.38, it should be more difficult to achieve full control of these systems by manipulating a few network driver-nodes. In contrast, non-LCP networks (Fig. 5 right side) characterise steady and homogeneous systems that are assembled through strong (often quite expensive) interactions, difficult to erase. Given their homogenous structure, such systems should be easier to control38. This argument is particularly valid for the power grid, which is not densely connected (network density is proven to increase controllability), but whose topology is homogeneous enough to be easily controllable38 - a required specification in human-engineered networks.
The LCP architecture facilitates not only the rapid delivery of information across the various network modules, but also the local processing. On the other hand, the non-LCP architecture is more useful for processes where: i) the storage of information (or energy) is at least as important as its delivery; ii) the cost of creating new interactions is excessive; iii) the creation of a redundant and densely connected system is strategically inadvisable. An emblematic example is the road networks, for which the money and time costs of creating additional roads are very high, and in which a community of strongly connected and crowded links resembles an impractical labyrinth.
While the small-world-paradigm treated the main effect of remodelling in real networks, and the scale-free-paradigm offered an innovative view of network growth in terms of node-preferential-attachment, the LCP is a first attempt to advance a link/community-based-interpretation of the epitopological learning component that appears in many cognitive, social and evolutionary processes.
Methods
Link removal and re-prediction in connectomes, social and ecological networks
A specific amount of synapses, equal to 10% of the total in the mouse connectome, was destroyed uniformly at random. This process was repeated 1000 times, and we generated 1000 diverse and random sparsified connectome topologies with 10% less synapses with respect to the original connectome. Following the same procedure we generated 1000 diverse and random sparsified topologies for each sparsification level, ranging the amount of removed synapses from 10% to 90% of the original, according to the following progressive levels of sparsification: 10%, 20%, 30%, …, 90% (see Fig. 2a). The link-prediction techniques (see Table 1), along with a random predictor, were applied to the diverse connectome topologies, and a list of candidate interactions sorted by likelihood score was obtained for each sparsified connectome configuration. The proportion of top-ranked candidate interactions that at each sparsification level matched the removed synapses - which is a measure of precision - was used to assess the performance in prediction. Since this process was repeated 1000 times for each sparsification level, in practise mean precision and standard error were considered for each stage. To characterise the deviation of each predictor from randomness, we transformed the indices' mean precisions at each sparsification level into decibels as: 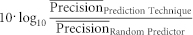 taking the mean performance of the random predictor as a reference. This transformed measure was named prediction power. The predictive power, measured at different sparsification levels, generated a prediction power curve (see Fig. 2a), and the area under this curve (AUPPC) summarised the power of each predictor. Since deletion of more than 50% synapses caused node isolation and the disappearance of local communities, to have a fair comparison, the AUPPC in Fig. 2B was computed considering only up to 50% of links removed in the Mouse Connectome. More details on this part are given in SI, section IV.
taking the mean performance of the random predictor as a reference. This transformed measure was named prediction power. The predictive power, measured at different sparsification levels, generated a prediction power curve (see Fig. 2a), and the area under this curve (AUPPC) summarised the power of each predictor. Since deletion of more than 50% synapses caused node isolation and the disappearance of local communities, to have a fair comparison, the AUPPC in Fig. 2B was computed considering only up to 50% of links removed in the Mouse Connectome. More details on this part are given in SI, section IV.
To further test the link-prediction power of classical and CAR-based indices, we inspected their performance over 8 different networks (see Fig. 2C). Their prediction power was tested when only 10% of the network links were destroyed, in order to be sure that the community structure of the analysed networks was preserved, and thus to guarantee a fair comparison between all different predictors. Finally, we performed a permutation test with 1000 resampling realizations, to assess whether the mean prediction power of the CAR-based indices was significantly different and possibly higher than the mean prediction power of the classical indices; a p-value threshold of 0.05 was considered statistically significant (see SI, section IV).
Link prediction in protein interactomes
The classical (see Table 1; and SI, section IIIA), the bio-inspired (see SI, section IIIB) and the CAR-based indices (see Table 1; and SI, section IIID) were applied to three different yeast protein interactomes. The proteins involved in the predicted candidate interactions were annotated according to their Gene Ontology (GO) terms, and their GO semantic similarities were measured over the three different gene ontologies (see SI, section V). A level of precision (on the basis of significant associations in GO; see SI, section V) was recursively evaluated for increasing-size sets of the best first-ranked candidate interactions. A precision curve was obtained, and the area under this curve (AUP) was considered as measure of performance.
In-silico validation of the best protein interaction candidates
Considering CAR and FSW as reference methods, we took the top-ranked 100 candidate interactions from each of the two techniques' list and intersected them with the entire STRING Database version 9.0, which was queried in February 2012. We reported: 1) how many protein pairs per 100 were validated for each network; 2) the average STRING confidence and its standard deviation; 3) the average GO confidence and its standard deviation. In addition, we reported the Robustness of each of these indicators across the networks (see SI, section VI).
Network sparsification in protein interactomes
This experiment is similar to the one performed for the connectome. However, instead of the prediction power we measured the AUP at each sparsification level, and since protein networks are significantly larger than the previous considered networks, 10 (as in Liu et al.38) different sparsified network topologies were generated instead of 1000. Another difference is that the simulation stops whenever network connectivity is lost, instead of when 90% of original links are removed, because embedding techniques such as ISOMAP can be applied only to networks with a single connected component.
LCP-decomposition-plot and LCP-correlation
A point in the LCP-decomposition-plot (LCP-DP) corresponds to a link from the network, and its coordinates are specified by the number of shared neighbours between the interacting nodes (CN, on the x-axis), and the squared root of the number of local community links (LCL, on the y-axis) between the CN (see SI, section VIII). The LCP-correlation (LCP-corr) quantifies the linear dependency between CN and LCL, and is based on their Pearson correlation coefficient (see SI, section VIII).
LCP in idealised networks
We simulated differing Erdős-Rényi networks by varying the two parameters used in the model G(n, p), namely the number of nodes n and edge probability p. We performed 100 realizations for each combination of model parameters and plotted the mean LCP-corr and its standard deviation in Fig. 6.
Additional Methods' information and any associated references are given in the Supplementary Information that is available on the online version of the paper.
Supplementary Material
Supplementary Information
Acknowledgments
We thank all members of the Integrative-Systems-Biology Lab at KAUST for their support; Prof. Vladimir Bajic, Dr Taewoo Ryu, and Loqmane Seridi for inspiring the sparsification experiments in protein networks; Prof. Franco Maria Montevecchi for the useful hints; Dr Joshua Vogelstein for providing the mouse connectome; Ewa Aurelia Miendlarzewska, María Contreras, and Anthony Hoang for their reviews; William Cooke for language revision.
Author Contributions C.V.C. and T.R. envisioned the study. C.V.C. invented the CAR and LCP theory, the CAR-index and the CAR-based variants of the classical indices, the LCP-DP and the LCP-correlation, and their applications. C.V.C. conceived the theoretical demonstrations provided in supplementary information, and G.A.L. inspected their correctness. C.V.C. and G.A.L. designed the experiments, algorithms, codes and performed the computational analysis. C.V.C., G.A.L. and TR analysed the results. C.V.C. wrote the article with input and corrections from G.A.L. and T.R., while G.A.L. wrote the SI with input and corrections from C.V.C. and T.R. T.R. led, directed and supervised the project.
07/21/2015
A correction has been published and is appended to both the HTML and PDF versions of this paper. The error has not been fixed in the paper.
References
- Erdős P. & Rényi A. On the evolution of random graphs. Publications of the Mathematical Institute of the Hungarian Academy of Sciences 5, 17–61 (1960). [Google Scholar]
- Watts D. J. & Strogatz S. H. Collective dynamics of 'small-world' networks. Nature 393, 440–442 (1998). [DOI] [PubMed] [Google Scholar]
- Newman M. E. J. Models of the small world. J Stat Phys 101, 819–841 (2000). [Google Scholar]
- Barabasi A. L. & Albert R. Emergence of scaling in random networks. Science 286, 509–512 (1999). [DOI] [PubMed] [Google Scholar]
- Price D. D. S. A General Theory of Bibliometric and Other Cumulative Advantage Processes. J Am Soc Inf Sci Tec 27, 292–306 (1976). [Google Scholar]
- Papadopoulos F., Kitsak M., Serrano M. A., Boguna M. & Krioukov D. Popularity versus similarity in growing networks. Nature 489, 537–540 (2012). [DOI] [PubMed] [Google Scholar]
- Liben-Nowell D. & Kleinberg J. The link-prediction problem for social networks. J Am Soc Inf Sci Tec 58, 1019–1031 (2007). [Google Scholar]
- Wang W. Q., Zhang Q. M. & Zhou T. Evaluating network models: A likelihood analysis. Epl-Europhys Lett 98 (2012). [Google Scholar]
- Lu L. Y. & Zhou T. Link prediction in complex networks: A survey. Physica A 390, 1150–1170 (2011). [Google Scholar]
- Getoor L. & Diehl C. P. Link Mining: A Survey. ACM SIGKDD Explorations Newsletter (2005). [Google Scholar]
- Newman M. E. J. Clustering and preferential attachment in growing networks. Phys Rev E 64 (2001). [DOI] [PubMed] [Google Scholar]
- Jaccard P. Distribution de la flore alpine dans le bassin des Dranses et dans quelques régions voisines. Bulletin de la Société Vaudoise des Sciences Naturelles 37, 241–272 (1901). [Google Scholar]
- Adamic L. A. & Adar E. Friends and neighbors on the Web. Social Networks 25, 211–230 (2003). [Google Scholar]
- Zhou T., Lu L. Y. & Zhang Y. C. Predicting missing links via local information. Eur Phys J B 71, 623–630 (2009). [Google Scholar]
- Ahn Y. Y., Bagrow J. P. & Lehmann S. Link communities reveal multiscale complexity in networks. Nature 466, 761–U711 (2010). [DOI] [PubMed] [Google Scholar]
- Sporns O., Tononi G. & Kotter R. The human connectome: A structural description of the human brain. Plos Comput Biol 1, 245–251 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti V. et al. Protein fingerprints of cultured CA3-CA1 hippocampal neurons: comparative analysis of the distribution of synaptosomal and cytosolic proteins. BMC Neurosci 9, 36 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv N. E. & Ahissar E. NEUROSCIENCE New tricks and old spines. Nature 462, 859–861 (2009). [DOI] [PubMed] [Google Scholar]
- Bock D. D. et al. Network anatomy and in vivo physiology of visual cortical neurons. Nature 471, 177–U159 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X., Zhao J. C. & Xu K. Link prediction in complex networks: a clustering perspective. Eur Phys J B 85 (2012). [Google Scholar]
- Kaiser M. & Hilgetag C. C. Nonoptimal component placement, but short processing paths, due to long-distance projections in neural systems. Plos Comput Biol 2, 805–815 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney L. R., Chen B. L., Paniagua E., Hall D. H. & Chklovskii D. B. Structural Properties of the Caenorhabditis elegans Neuronal Network. Plos Comput Biol 7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markram H., Rinaldi T. & Markram K. The intense world syndrome--an alternative hypothesis for autism. Front Neurosci 1, 77–96 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. E., Schittler D. N., Raffaelli D. G. & Reuman D. C. Food webs are more than the sum of their tritrophic parts. P Natl Acad Sci USA 106, 22335–22340 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiqi L. Food Webs: From Connectivity to Energetics. (Elsevir Academic Press, 2005).
- Tscharntke T., Vidal S. & Hawkins B. A. Parasitoids of grass-feeding chalcid wasps: a comparison of German and British communities. Oecologia 129, 445–451 (2001). [DOI] [PubMed] [Google Scholar]
- Clauset A., Moore C. & Newman M. E. J. Hierarchical structure and the prediction of missing links in networks. Nature 453, 98–101 (2008). [DOI] [PubMed] [Google Scholar]
- Guimera R. & Sales-Pardo M. Missing and spurious interactions and the reconstruction of complex networks. P Natl Acad Sci USA 106, 22073–22078 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z.-H., Lei Y.-K., Gui J., Huang D.-S. & Zhou X. Using manifold embedding for assessing and predicting protein interactions from high-throughput experimental data. Bioinformatics (Oxford, England) 26, 2744–2751 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito R., Suzuki H. & Hayashizaki Y. Interaction generality, a measurement to assess the reliability of a protein-protein interaction. Nucleic acids research 30, 1163–1168 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun C. et al. Functional classification of proteins for the prediction of cellular function from a protein-protein interaction network. Genome biology 5, R6–R6 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Wong L. & Chua H. N. Complex discovery from weighted PPI networks. Bioinformatics 25, 1891–1897 (2009). [DOI] [PubMed] [Google Scholar]
- Chua H. N., Sung W.-K. & Wong L. Exploiting indirect neighbours and topological weight to predict protein function from protein-protein interactions. Bioinformatics (Oxford, England) 22, 1623–1630 (2006). [DOI] [PubMed] [Google Scholar]
- Kuchaiev O., Rasajski M., Higham D. J. & Przulj N. Geometric de-noising of protein-protein interaction networks. Plos Comput Biol 5, e1000454 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D. et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic acids research 39, D561–568 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancichinetti A., Kivela M., Saramaki J. & Fortunato S. Characterizing the Community Structure of Complex Networks. Plos One 5 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguñá M., Krioukov D. & Claffy K. C. Navigability of complex networks. Nature Physics 5, 74–80 (2008). [Google Scholar]
- Liu Y. Y., Slotine J. J. & Barabasi A. L. Controllability of complex networks. Nature 473, 167–173 (2011). [DOI] [PubMed] [Google Scholar]
- Guimera R., Mossa S., Turtschi A. & Amaral L. A. N. The worldwide air transportation network: Anomalous centrality, community structure, and cities' global roles. P Natl Acad Sci USA 102, 7794–7799 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A. J. M. et al. RING: networking interacting residues, evolutionary information and energetics in protein structures. Bioinformatics 27, 2003–2005 (2011). [DOI] [PubMed] [Google Scholar]
- Tenenbaum J. B., de Silva V. & Langford J. C. A global geometric framework for nonlinear dimensionality reduction. Science (New York, N. Y.) 290, 2319–2323 (2000). [DOI] [PubMed] [Google Scholar]
- Csermely P. Weak links : the universal key to the stability of networks and complex systems. (Springer, Berlin; London; 2009).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information