Abstract
Despite the great potential of small interfering RNA (siRNA) as a therapeutic agent, progress in this area has been hampered by a lack of efficient biocompatible transfection agents. Recently, cationic shell-crosslinked knedel-like nanoparticles (cSCKs) were found to possess lower cytotoxicity and better transfection ability for phosphorothioate ODNs and plasmid DNA than the commonly used cationic lipid-based agent Lipofectamine. To determine the usefulness of cSCKs for siRNA transfection, a small library of cSCKs with varying percentage of primary and tertiary amines was assessed for its ability to bind to siRNA, inhibit siRNA degradation in human serum, and to transfect HeLa and mouse macrophage cell lines. The silencing efficiency in HeLa cells was greatest with the cSCK with 100% primary amines (pa100) as determined by their viability following transfection with cytotoxic and non-cytotoxic siRNAs. cSCK-pa100 showed greater silencing efficiency than Lipofectamine 2000 in the HeLa cells, as well in 293T and human bronchial epithelial (HEK) cells, but was comparable in human bronchial epithelial (BEAS-2B) cells and human mammary epithelial (MCF10a) cells. cSCK-pa100 also showed greater silencing of iNOS expression than Lipofectamine 2000 in a mouse macrophage cell line, and provided greater protection from serum degradation, demonstrating its potential usefulness as an siRNA transfection agent. The siRNA silencing of iNOS at lower concentrations of siRNA could be enhanced by complexation with the fusogenic GALA peptide, which was shown to enhance endosomal escape following uptake.
Introduction
Small interfering RNAs (siRNA) have found extensive application for the suppression of gene expression, and have great promise as therapeutic agents (KURRECK, 2009; Lares et al., 2010; Wang et al., 2010; Watts and Corey, 2010). siRNAs are not membrane permeable, however, and require auxiliary agents for efficient transport to the cytosol where the RNA targets reside. Though viral carrier systems are the most efficient, they often have associated undesirable side effects (Couto and High, 2010; Liu and Berkhout, 2011). To circumvent this problem, various types of synthetic, non-viral delivery systems have been developed such as cationic liposomes, lipoplexes, and polyplexes (Whitehead et al., 2009; David et al., 2010; Wang et al., 2010; Gao et al., 2011). We have recently shown that cationic shell-crosslinked knedel-like nanoparticles (cSCKs), containing primary amines in the shell, form electrostatic complexes with negatively charged plasmid DNA and antisense phosphorothioate 2′-OMe oligonucleotides (ps-MeON) and efficiently transfect them into cells (Zhang et al., 2009b). The cSCKs could also be used to deliver neutral peptide nucleic acids with very high efficiency, through electrostatic complexation with a duplex formed with a partially complementary oligodeoxynucleotide, or through conjugation via a bioreductively cleavable linker (Zhang et al., 2009b). Shell-crosslinked nanoparticles are an attractive and versatile platform (Nystrom and Wooley, 2011) for the development of nucleic acid delivery agents because their synthetic design enables their size and shape (Zhang et al., 2008a), charge and buffering capacity (Zhang et al., 2010; Shrestha et al., 2012), degradability (Li et al., 2008), stealth character (Sun et al., 2008), and ligand presentation (Nystrom and Wooley, 2008; Zhang et al., 2008b) to be easily tailored for a particular target.
The ability of cSCKs to efficiently transfect cells with nucleic acids can be attributed to their ability to facilitate 3 key steps in the transfection process (Zhang et al., 2009a). First, cSCKs can bind to nucleic acids through electrostatic interactions between protonated amines in their shell with negatively charged nucleic acids or hybrids. Second, cSCKs are readily endocytosed through electrostatic interactions between the protonated amines in the shell and the negatively charged membrane surface, followed by vesicle formation induced by the spherical but flexible structure of the cSCK. Third, cSCKs facilitate release of the nucleic acid payload from the endosome into the cytoplasm, presumably by rupturing or destabilizing the endosome through the proton sponge effect (Boussif et al., 1995). In this mechanism, unprotonated amines in the shell neutralize protons during endosomal acidification, thereby driving additional protons, counterions, and water into the endosome that ultimately cause its rupture through an increase in osmotic pressure.
To determine whether the transfection efficiency could be further optimized for a particular nucleic acid payload by manipulating the proton and phosphate binding affinity of the shell, a small library of cSCKs containing differing proportions of primary (pa) and tertiary (ta) amines was also synthesized (Fig. 1) (Zhang et al., 2010). The cationic cSCK-pa25-ta75 was found to be best for transfecting an enhanced green fluorescent protein reporter plasmid DNA into human cervical cancer (HeLa) cells at an N/P [cSCK or cationic micelle (cM) nitrogen to siRNA phosphate] ratio of 20, while cSCK-pa50-ta50, at an N/P ratio 10, was best for delivery of a splice correcting ps-MeON. It was also found that increasing the proportion of tertiary amines relative to primary amines reduced the cytotoxicity. Herein we report that cSCK-pa100 showed the highest silencing efficiency among the various cSCKs in HeLa cells, and was better than Lipofectamine 2000 in various other human cell lines, and in knocking down inducible nitric oxide synthase (iNOS) in a macrophage cell line. We also demonstrate the ability of the optimal cSCK to protect siRNA from degradation in human serum, and the ability of added fusogenic glutamic acid-alanine-leucine-alanine repeat (GALA) peptide to enhance knockdown of iNOS mRNA expression, through enhancing endosomal escape rather than cell entry.
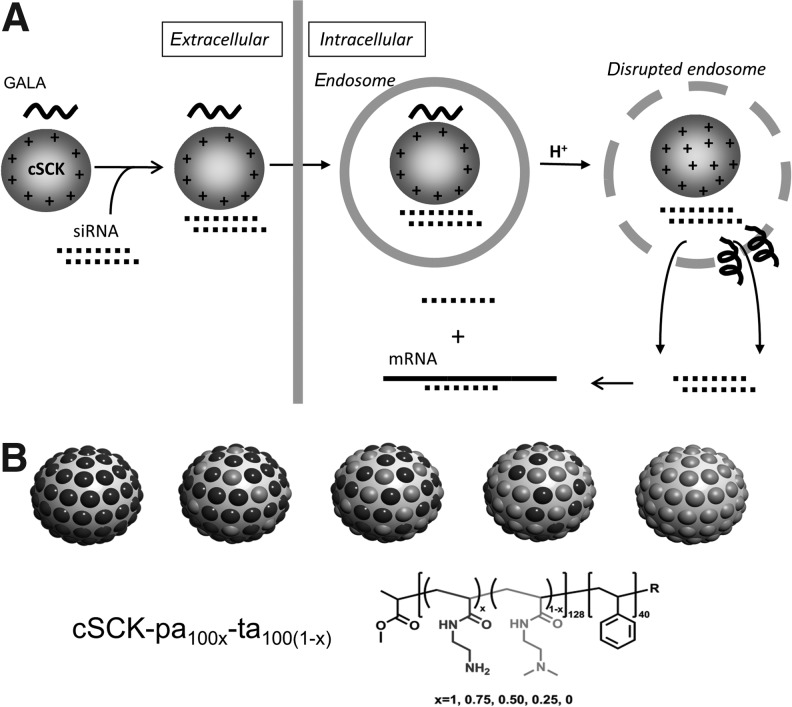
FIG. 1.
Cationic shell crosslinked nanoparticle-mediated delivery of siRNA. (A) Scheme showing various key steps in the delivery of small interfering RNA (siRNA) by a cationic shell crosslinked nanoparticle (cSCK). In the first step siRNA is assembled electrostatically with or without the fusogenic glutamic acid-alanine-leucine-alanine repeat (GALA) peptide by binding to positively charged protonated amines. In the second step, the cationic particle interacts with the cell surface and is endocytosed. In the third step, the remaining unprotonated amines neutralize protons during endosomal acidification (proton sponge effect) to increase osmotic pressure and disrupt the endosome, facilitating siRNA release. At the same time, carboxylate residues on the GALA peptide become protonated causing its release from the cSCK and inducing a conformational change that further destabilizes the endosome, further enhancing release of siRNA. (B) cSCK structure with differing ratios of primary:tertiary amines in the shell.
Materials and Methods
Materials
All solvents and chemicals were purchased from Sigma-Aldrich and used without further purification unless otherwise indicated. The HeLa, MCF-10A (human mammary epithelial), and RAW264.7 (mouse monocyte-macrophage) cell lines were obtained from the American Type Culture Collection. The human embryonic kidney cell line (HEK), 293T (a variant of human embryonic kidney cell line), and human bronchial epithelial cell line (BEAS-2B) were gifts from Dr. Steven Brody. Lipofectamine™ 2000 was obtained from Invitrogen Co., Polyfect® from Qiagen Inc., and ExGen500 from Fermentas, Inc. AllStars Hs Cell Death Control siRNA and AllStars Negative Control siRNA were purchased from Qiagen Inc. fluorescein amidite (FAM)-labeled glyceraldehyde 3-phosphate dehydrogenase (GAPDH) siRNA, and the iNOS481 siRNA (GGCCACAUCGGAUUUCACUtt)•(AGUGAAAUCCGAUGUGGCCtt) were from Applied Biosystems/Ambion, Inc. The iNOS481 siRNA with 3′-Cy5-labeled antisense siRNA was from IDT. The iNOS antibody, NOS2(c-11) was from Santa Cruz Biotechnology Inc. Steady-Glo® Luciferase Assay reagent, CellTiter-Glo® Luminescent Cell Viability and CellTiter 96 Non-Radioactive Cell Proliferation Assay Kits were from Promega Co. All cell culture media, TRIzol Reagent, Dextran Cascade Blue (10,000 MW, anionic, lysine fixable) and Lysotracker Blue DND-22 were from Invitrogen, Inc. Turbo DNA-Free Kit, High-Capacity cDNA Reverse Transcription Kit and Power SYBR Green Master Mix were from Ambion. Lipopolysaccharide (LPS) cytochalasin D, chlorpromazine hydrochloride, and methyl-beta-cyclodextrin were from Sigma, and interferon-gamma (γ-IFN) was from Peprotech. The non-fluorescently labeled cSCKs used in the studies without GALA were prepared as previously described (Zhang et al., 2009b ; Zhang et al., 2010) with dynamic light scattering (DLS): (Dh)vol=14±2 nm and a zeta potential of 21.7 mV (pH 5.5). A 1 mg/mL solution of cSCK-pax-ta100-x corresponds to an amine concentration of 6.5 mM.
Gel retardation assays
The iNOS481 siRNA was 5′-labeled by [γ-32P]-ATP with T4 polynucleotide kinase. Serial amounts of cSCKs were mixed with 50 nM radiolabeled siRNA at N/P ratios of 0, 0.5:1, 1:1, 2:1, 4:1, 8:1, 16:1, and 32:1 in phosphate-buffered saline (PBS) buffer for 30 minutes and then mixed with loading buffer (6×), and loaded to 15% native polyacrylamide gel.
Cell culture
cSCK-mediated siRNA transfection was evaluated on HeLa, RAW264.7, HEK, 293T, BEAS-2B, and MCF-10A (human mammary epithelial line) cells. Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS), streptomycin (100 μg/mL), and penicillin (100 units/mL) at 37°C in a humidified atmosphere with 5% CO2.
cSCKs•siRNA silencing efficiency assays
Cells (HeLa, 293T, HEK, BEAS-2B, and MCF-10A) were seeded in a 96-well microtiter plate at a density of 1×104 cells per well and cultured for 24 hours in 100 μL DMEM containing 10% FBS. AllStars Hs Cell Death Control siRNA or AllStars Negative Control siRNA was complexed with cSCKs at predetermined N/P ratios in 20-μL opti-MEM solution and incubated for 15 minutes before use. At the time of the transfection experiment, the medium was replaced with 80 μL of fresh medium, to which the cSCK•siRNA or Lipofectamine2000•siRNA complexes were added for a final siRNA concentration of 100 nM. After 48 hours, cell death was quantified by the CellTiter-Glo® Luminescent Cell Viability Assay using 100 μL of CellTiter-Glo reagent. The contents were mixed and the plate was shaken for 3 minutes at 600 rpm, and then allowed to incubate at room temperature for 10 minutes to stabilize the luminescence signal. Luminescence intensities were recorded on a Luminoskan Ascent® luminometer (Thermo Scientific) with an integration time of 1 second per well. The relative cell viability was calculated by the following equation:
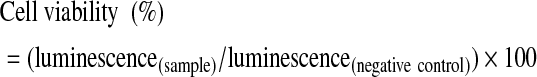 |
(1) |
Where luminescence(negative control) was obtained in the absence of the transfection reagent and luminescence(sample) was obtained in the presence of cSCKs or Lipofectamine 2000. The data was fit to a standard sigmoidal Hill equation to determine the lethal concentration 50 (LC50) for the transfection agents shown below:
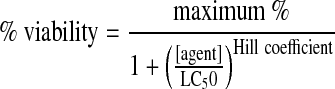 |
(2) |
Confocal microscopy and flow cytometry of cSCK•siRNA complexes
HeLa cells (5×105) were plated in 35-mm MatTek glass bottom microwell dishes (MatTek Co.) 24 hours prior to transfection. FAM-labeled GAPDH siRNA (Ambion) was complexed with cSCKs at predetermined N/P ratios in 500 μL opti-MEM solution and incubated for 15 minutes before use. Prior to transfection, the medium in each dish was replaced with 2.0 mL of fresh DMEM, to which cSCK/siRNA complexes were added for a final siRNA concentration of 100 nM. The plates were then incubated at 37°C for another 4 hours. Each plate was washed 3 times with PBS buffer and viewed under bright field and fluorescent conditions using a Leica TCS SP2 inverted microscope, with excitation by an argon (Ar) laser (488 nm). Cells were also collected by trypsinization, pelleted, and resuspended in 0.5 mL PBS for flow cytometric analysis on an fluorescence-activated cell sorting (FACS)-calibur instrument (Becton Dickinson) equipped with a 488-nm Ar laser. For each sample, 20,000 events were collected by list-mode data that consisted of side scatter, forward scatter, and fluorescence emission centered at 530 nm (FL1). The fluorescence was collected at a logarithmic scale with a 1,024-channel resolution. CellQuest software (Becton Dickinson) was applied for the analyses.
Inhibition of iNOS expression by cSCKs•siRNA complexes
RAW264.7 cells were seeded in a 24-well plate with 1×105 cells per well in 600 μL DMEM with 10% FBS and incubated for 24 hours. iNOS siRNA, si-481 (GGCCACAUCGGAUUUCACUtt)•(AGUGAAAUCCGAUGUGGCCtt) or AllStars Negative Control siRNA was complexed with cSCKs at predetermined N/P ratios or the other transfection agents in 100 μL opti-MEM solution and incubated for 15 minutes before use. At the time of the transfection experiment, the cells were washed 3 times with PBS and replaced with 400 μl DMEM (no FBS), to which the cSCK/siRNA or the control siRNA transfection agent complex were added for a final siRNA concentration of 100 nM. Six hours later, LPS (lipopolysaccharides from Escherichia coli 055:B5, Sigma, 1 μg/mL), IFN-gamma (γ-IFN, mouse, recombinant, E. coli, Calbiochem, 100 ng/mL; LPS), and 60 μL FBS was added with DMEM to make a total of 600 μL and incubated for another 24 hours. An aliquot of the supernatant (50 μL) was removed for the Griess assay (Promega Co.) and quantified by the absorbance at 540 nm. Then the cells were harvested for western blotting analysis. The cells were first lysed in 30 μL lysis buffer [1% sodium dodecyl sulfate (SDS), 1.0 mM sodium ortho-vanadate, 10 mM Tris pH 7.4]. The Lysates were centrifuged at 12,000 rpm for 10 minutes at 4°C. Equal amounts of supernatant (containing about 20 μg protein) were mixed with Tris–glycine SDS sample buffer (2×) and boiled for 5 minutes. The proteins were subjected to 10% SDS-polyacrylamide gel electrophoresis for detection of β-actin and iNOS, respectively. The separated proteins were then transferred onto polyvinylidene difluoride membrane (Amersham Hybond™-P, GE Healthcare). iNOS was detected by using monoclonal anti-iNOS antibody NOS2(C-11) at 1:200 (Santa Cruz Biotechnology, Inc.). β-actin was detected using mouse anti- β-actin monoclonal antibody at 1:20,000 (GenScript Corporation). The secondary antibody was peroxidase-conjugated goat-anti-mouse immunoglobin G (H+L) (Jackson ImmunoResearch Laboratories, Inc.) at a dilution of 1:20,000. Signals were detected by enhanced chemiluminescence (GE Healthcare) according to the manual provided with the product.
Degradation of siRNA by human serum
siRNA481 duplexes (0.766 μM) were incubated alone or in the presence of cSCK (50 μg/mL, N/P=10:1) or Lipofectamine 2000 (13 and 200 μg/mL) for 30 minutes in 25 μL of PBS buffer. The siRNA solutions were then incubated with 25 μL of 100% human serum (Sigma) at 37°C, under 5% CO2. Aliquots of 10 μL were withdrawn at 0, 3, 6, and 24 hours and immediately frozen in 0.5% SDS at −80°C. Samples were subjected to 2% agarose gel electrophoresis and visualized by staining with ethidium bromide. Band intensities were quantified with Quantity One Software and plotted versus time to calculate the half-life of siRNA degradation in human serum.
Synthesis and characterization of peptide GALA
Peptide GALA (WEAALAEALAEALAEHLAEALAEALEALAA) was synthesized continuously on universal support xanthenylamine polyethylene glycol-polystyrene resin with standard solid-phase Fmoc (9-fluorenylmethoxycarbonyl) chemistry on an APEX 369 peptide synthesizer (AAPPTec). After completion of automated synthesis, peptide was cleaved from the solid support with 88% trifluoroacetic acid (TFA), 5% H2O, 5% phenol, and 2% triisopropylsilane for 3 hours and then precipitated with diethyl ether. The peptide was purified by reverse-phase chromatography on a Microsorb-MV 300-5 C-18 column, (250×4.6 mm column, 300 Å pore size, Varian, Inc.) with a method of 0–40% B/40 minutes, 40–100% B/5 minutes, 100–0%, B/3 minutes, where A=0.1% TFA in water and B=0.1% TFA in CH3CN. The fraction containing the pure peptide was evaporated to dryness in a Savant Speedvac, redissolved in 7:3 dimethyl sulfoxide (DMSO)/H2O and characterized by MALDI-TOF (Applied Biosystems) using CHCA as a matrix.
cSCKs for GALA studies
The cSCKs for the GALA studies were prepared from poly(acrylamidoethylamine)160-b-poly(styrene)30 (PAEA160-b-PS30) according to the previously published procedure (Zhang et al., 2009b; Zhang et al., 2010). DLS: (Dh)num=22±6 nm; with a (Dh)vol=33±22 nm; (Dh)int=157±140 nm Zeta potential of 28±2 mV (pH 5.5). These particles were then conjugated to Alexa Fluor 488-NHS ester as follows. To a cSCK solution (3.5 mL, 0.85 mg/mL) was added sodium carbonate (20 μL of 1.0 M aqueous solution) to adjust the pH to ca. 8.0. Alexa Fluor 488 (0.1 mg, 0.1 μM) was added from a stock solution (1 mg/0.5 mL) in DMSO to tether ca. 1 dye per polymer chain and was stirred overnight in darkness at RT. The solution was dialyzed (MWCO ca. 6000-8000 Da) against 150 mM PBS buffer for 3 days in an aluminum-foil wrapped beaker to give a 0.8 mg/mL solution. Dye concentration was quantified by ultraviolet and visible absorption spectroscopy to be 5.10 μM or approximately 0.1 dye per polymer chain. DLS: (Dh)num=23±7 nm and; (Dh)vol=34±20 nm; (Dh)int=120±91 nm. Zeta potential=37±1 mV (pH 5.5).
Effect of GALA on siRNA binding to cSCK
iNOS siRNA481 (GGCCACAUCGGAUUUCACUtt)•(AGUGAAAUCCGAUGUGGCCtt) was 5′-labeled by [γ-32P]-ATP with T4 polynucleotide kinase (Fermentas) by a standard labeling procedure indicated in the instructions. The cSCK (10 μg/mL) were mixed with iNOS siRNA481 (50 nM or 100 nM) and GALA (0, 0.5, 1, and 2 μM) in Opti-MEM 1 for 30 minutes. The complexes were then mixed with loading buffer (6×), and loaded to 10% native polyacrylamide gel.
Quantitative real-time polymerase chain reaction analysis of iNOS silencing by cSCK•siRNA•GALA complexes
RAW 264.7 cells were seeded in 6-well plate at a density of 3×105 cells per well and cultured in 1,200 μL DMEM containing 10% FBS. Twenty-four hours later, cells were activated with LPS and IFN-gamma. After another 24 hours, cSCK (10 μg/mL) was mixed with siRNA481 (0 and 50 nM) and GALA (0, 0.1, 0.2, and 0.3 μM) for 30 min to form electrostatic complexes and transfected to RAW 264.7 cells with fresh culture medium. Eight hours later, cells were washed twice with PBS and total RNA was extracted using TRIzol Reagent (Invitrogen) according to the manufacturer's instruction and quantified spectrophotometrically by Nanophotometer (Implen). Approximately 500 ng of total RNA was treated with Turbo DNA-free kit (Ambion) and reverse transcribed with high-capacity cDNA reverse transcription kit (Ambion) according to standard protocols supplied by the manufacturer. cDNA synthesis was performed for 10 minues at 25°C and 2 hours at 37°C, followed by heat inactivation for 5 minutes at 85°C. The real-time polymerase chain reaction (RT-PCR) assay was performed using the Applied Biosystems StepOne Plus Real-Time PCR System (Applied Biosystems) with 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. PCR reactions were carried out in 25 μL of reaction mixture consisting of 12.5 μL Power SYBR Green Master Mix (2*) (Ambion), 200 nM primers, cDNA, and water. The primers used to amplify iNOS were d(TGGTGGTGACAAGCACATTT) and d(AAGGCCAAACACAGCATACC), and to amplify GAPDH were d(TGGAGAAACCTGCCAAGTATG) and d(GTTGAAGTCGCAGGAGACAAC). The threshold cycle (CT) was calculated by the instrument's software (StepOne). The expression of each mRNA was calculated by the comparative CT method (ΔΔCT). The average of 4 determinations was calculated.
Cytotoxicity assay of cSCK•siRNA•GALA complexes
The cytotoxicities of cSCK•siRNA•GALA complexes were examined by CellTiter 96 Non-Radioactive Cell proliferation Assay (Promega). RAW 264.7 cells were seeded in a 96-well plate at a density of 2×104 cells per well and cultured in 100 μL DMEM containing 10% FBS. One day prior to transfection, cells were activated with LPS and IFN-gamma to induce iNOS mRNA. Twenty-four hours later, the medium was replaced with 100 μL fresh culture medium containing complexes consisting of 10 μg/mL cSCKs, iNOS siRNA481 (0, 50, and 100 nM), and various concentrations of GALA (0, 0.3, 0.6, and 1.0 μM). After another 24 hours, 15 μL of dye solution was added to each well and incubated at 37°C in a humidified CO2 incubator. Four hours later, 100 μL of stop solution was added to each well and incubated overnight. The absorbance of colored formazan product was recorded at 570 nm using a microplate reader (Molecular Devices). An average of 3 determinations was made.
Cell uptake of cSCK•siRNA•GALA complexes by flow cytometry
RAW 264.7 cells were plated in 6-well plates at a density of 3×105 cells per well and cultured in 1 mL DMEM containing 10% FBS. Twenty-four hours prior to transfection, RAW 264.7 cells were activated by LPS and IFN-gamma. At the time of transfection, complexes of AlexaFluor488-cSCK (10 μg/mL), Cy5-siRNA (50 nM), and GALA (0, 0.2, 0.5, 0.8, and 1 μM) were added to RAW264.7 cells, and dishes were returned to the cell culture incubator. On the next day, cells were washed 3 times with PBS, collected by trypsinization, pelleted, and resuspended in 0.5 mL PBS for flow cytometric analysis on an FACS-calibur instrument (Becton Dickinson) equipped with a 488-nm argon laser and a 633 nm helium-neon laser. For each sample, 10,000 events were collected and the fluorescence from 2 channels was detected on a logarithmic scale respectively. The data were processed by FlowJo software.
Effect of inhibitors on cSCK uptake in RAW 264.7 cell line
RAW 264.7 cells were plated in 6-well plates at a density of 3×105 cells per well and cultured in 1 mL DMEM containing 10% FBS. Twenty-four hours prior to transfection, RAW 264.7 cells were activated by LPS and IFN-gamma and then transfected with AlexaFluor488-cSCK-pa100 (10 μg/mL). One hour prior to transfection, cells were treated with cytochalasin D (average of 2–50 μM), chlorpromazine (20 μM) and methyl-beta-cyclodextrin (20 μM) at maximal concentrations shown not to be significantly cytotoxic. Cells were then incubated for an additional hour and washed 3 times with PBS, collected by trypsinization, pelleted, and resuspended in 0.5 mL PBS for flow cytometric analysis on an FACS-calibur instrument (Becton Dickinson) equipped with a 488 nm Ar laser. For each sample, 10,000 events were collected and the fluorescence was detected on a logarithmic scale respectively. The data were processed by FlowJo software.
Confocal fluorescent microscopy of siRNA and cSCK non-colocalization with tracker dyes
RAW 264.7 cells were plated in 35-mm MatTek glass-bottom microwell dishes (MatTek) at a density of 2.5×104 cells per well and cultured in 150 μL DMEM containing 10% FBS. RAW 264.7 cells were activated by LPS and IFN-gamma 24 hours prior to transfection. Then complexes of AlexaFluor488-cSCK (10 μg/mL), Cy5-siRNA (50 nM) and GALA (0 and 0.2 μM) were added to RAW264.7 cells and incubated for 24 hours at 37°C and examined by confocal microscopy on a Nikon A1 Confocal Microscope. In one experiment, 1 μL of Hoechst 33342 (10 μg/mL) was added to each dish 1 hour before confocal microscopy to stain the nucleus. In another experiment, Lysotracker-Blue (50 nM) was added to each dish 2 hours before confocal microscopy. In a third experiment, Dextran-Cascade Blue (100 μg/mL) was added together with the cSCK, siRNA, and GALA to the RAW264.7 cells and incubated for 8 hours before confocal microscopy. In each experiment, the dishes were washed 3 times with PBS buffer before confocal microscopy. The degree of non-colocalization between the siRNA and cSCK with each other or the tracker dye was determined by Image J Green and Red Puncta Colocalization Macro developed by Ruben K. Dagda and Daniel J. Swiwarski from University of Pittsburgh.
Results and Discussion
Synthesis and characterization of the cSCK nanoparticles
The cationic cSCKs were prepared as previously described by the self-assembly of amphiphilic block copolymers in water, followed stabilization through crosslinking of the amine in the shell layer (Zhang et al., 2009b; Zhang et al., 2010). The amphiphilic block copolymer, poly(acrylamidoethylamine)128-b-polystyrene40 (PAEA128-b-PS40), was obtained by amidation of a common precursor for SCK construction, poly(acrylic acid)-b-polystyrene, by reaction with mono-Boc-protected 1,2-ethylene diamine and subsequent deprotection under acidic conditions. The amine composition was varied by coamidation with various proportions of 2-dimethylaminoethylamine. This chemical transformation afforded an amphiphilic block copolymer bearing large quantities of primary (pa) and/or tertiary (ta) amines, which after micellar assembly were crosslinked to give cSCK-pa100x-ta100(1-x) (Fig. 1). Because the micelle containing 100% tertiary amine could not be crosslinked due to the lack of primary amines, it was used as an uncrosslinked cationic micellar nanoparticle, cM-ta100. The nanoparticles were 9–11 nm in diameter, as observed by transmission electron microscopy, and 14–18 nm in hydrodynamic diameter, as measured by dynamic light scattering. All particles were positively charged at pH 5.5 or pH 7.0, as determined by zeta potential measurements.
cSCK binding affinity for siRNA
The binding affinities of the cSCKs and cM for siRNA were measured by a gel retardation assay with 50 nM 5′-32P-labeled siRNA (iNOS siRNA481 GGCCACAUCGGAUUUCACUtt•AGUGAAAUCCGAUGUGGCCtt) at increasing N/P ratios (Fig. 2). While complete binding of the siRNA could be achieved at an N/P ratio of 4 with the primary amine modified cSCK-pa100 (100% primary amine), a ratio of 8 was required for cSCK-pa50-ta50 (50% primary amine and 50% tertiary amine), and an N/P ratio of 32 for the tertiary amine micelle cM-ta100. The increasing N/P ratio for binding with increasing tertiary amine content may be the result of steric hindrance to phosphate binding caused by the methyl groups on the tertiary amines.
FIG. 2.
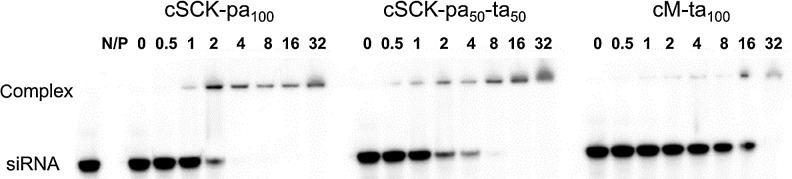
Electromobility shift assay of siRNA binding to cSCK at different N/P [cSCK or cationic micelle (cM) nitrogen to siRNA phosphate] ratios. 32P-labeled inducible nitric oxide synthetase (iNOS)481 siRNA (50 nM) was incubated with poly(acrylamidoethylamine)128-b-poly(styrene)40 (PAEA128-b-PS40) cSCKs at N/P ratios of 0, 0.5, 1, 2, 4, 8, 16, and 32 in phosphate-buffered saline (PBS) buffer for 30 minutes and electrophoresed on a 15% native polyamide gel. The concentration of cSCK was 0.31 μg/mL for an N/P of 1.
Silencing efficiency and cytotoxicity of cSCK•siRNA complexes
To rapidly and quantitatively compare the silencing efficiences of the cSCK•siRNA complexes the viabilities of cells transfected with cytotoxic and non-cytotoxic siRNAs were compared (Fig. 3). The non-toxic siRNA was used to determine the intrinsic cytotoxicity of the cSCK•siRNA complex and the cytotoxic siRNA was used to determine the silencing efficiency of the cSCK•siRNA complex from the increase in cytotoxicity caused by the cytotoxic siRNA. The optimal concentration of cSCK for siRNA delivery would therefore be one that showed the maximal difference between the viability of the cells transfected with the cytotoxic and non-cytotoxic siRNAs. By using this general method, optimal conditions for siRNA silencing could be determined for any cell line, without the need for cell lines bearing a specific reporter gene, or the use of RT-PCR and western assays. The cell viability was assayed with the CellTiter-Glo™ Luminescent assay for measuring cellular ATP concentration after treatment with AllStars Hs Cell Death Control siRNA and AllStars Negative Control siRNAs. The AllStars Hs Cell Death Control siRNA is a commercially available mixture of highly potent siRNAs targeting ubiquitously expressed human genes that are essential for cell survival. Knockdown of these essential genes results in cell death. AllStars Negative Control siRNA has no significant homology to any known mRNA sequence for mammalian genes and is used as a negative control to assay non-specific effects of siRNA transfection.
FIG. 3.
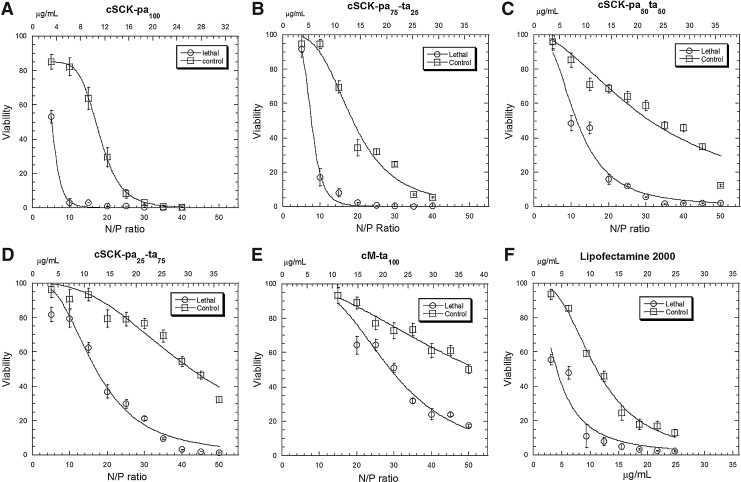
Cytotoxicity assay for siRNA silencing efficiency. HeLa cells were transfected with 100 nM AllStars Hs Cell Death Control siRNA or AllStars Negative Control siRNA in the presence of PAEA128-b-PS40 cSCKs at different N/P ratios, or Lipofectamine 2000 at the same μg/mL as for cSCK-pa100. After 48 hours, the relative cell viability was quantified using CellTiter-Glo® Luminescent Cell Viability Assay. The concentration of the cSCKs for N/P=1 were: cSCK-pa100 0.62 μg/mL (A); cSCK-pa75-ta25 0.72 μg/mL (B); cSCK-pa50-ta50 0.72 μg/mL (C); cSCK-pa25-ta75 0.72 μg/mL (D); and cM-ta100 0.74 μg/mL (E). The concentrations of Lipofectamine 2000 (F) and Polyfect (not shown) were 3.1, 6.2, 9.3, 12.4, 15.5, 18.6, 21.7, and 24.8 μg/mL, respectively, which were equivalent to the concentrations used in panel A for cSCK-pa100.
To determine the optimal N/P ratio for siRNA silencing with a given type of cSCK, we tested cell viability for a range of cSCK/siRNA ratios from N/P of 5 to 40 or 50 (corresponding to about 3.5–35 μg/mL) relative to non-transfected cells. For preparations with cSCK-pa100, increasing the N/P ratio from 5 to 10 decreased the viability of the cells transfected with the cytotoxic siRNA from about 50% to 5%, while the viability of the cells transfected with the non-cytotoxic siRNA only decreased from about 85% to 80% compared to non-transfected cells. Since the viability of the cells appears to have plateaued at 85% at the lowest concentration of cSCK, this value may actually represent 100% viability for this set of experiments and was normalized to this value. When the N/P ratio was changed from 10 to 20, the viability with the cytotoxic siRNA approached 0%, whereas the viability with the non-cytotoxic siRNA decreased from 80% to 30%, presumably due to the inherent cytotoxicity of the cSCK bearing a non-targeted siRNA. Thus, for preparations with cSCK-pa100, an N/P ratio of 10 would appear to afford the optimal combination of siRNA transfection efficiency and minimal nanoparticle cytotoxicity. To get a more quantitative assessment of the transfection efficiency, the survival curves were fit to a Hill equation (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/nat) from which the silencing efficiency at a given cSCK concentration could be plotted as the difference between cell survival with the non-toxic and the toxic siRNAs (Fig. 4A). From this plot we can readily see that the maximum silencing efficiency of 93% was at 6 μg/mL of cSCK-pa100 corresponding to an N/P ratio of 9.7.
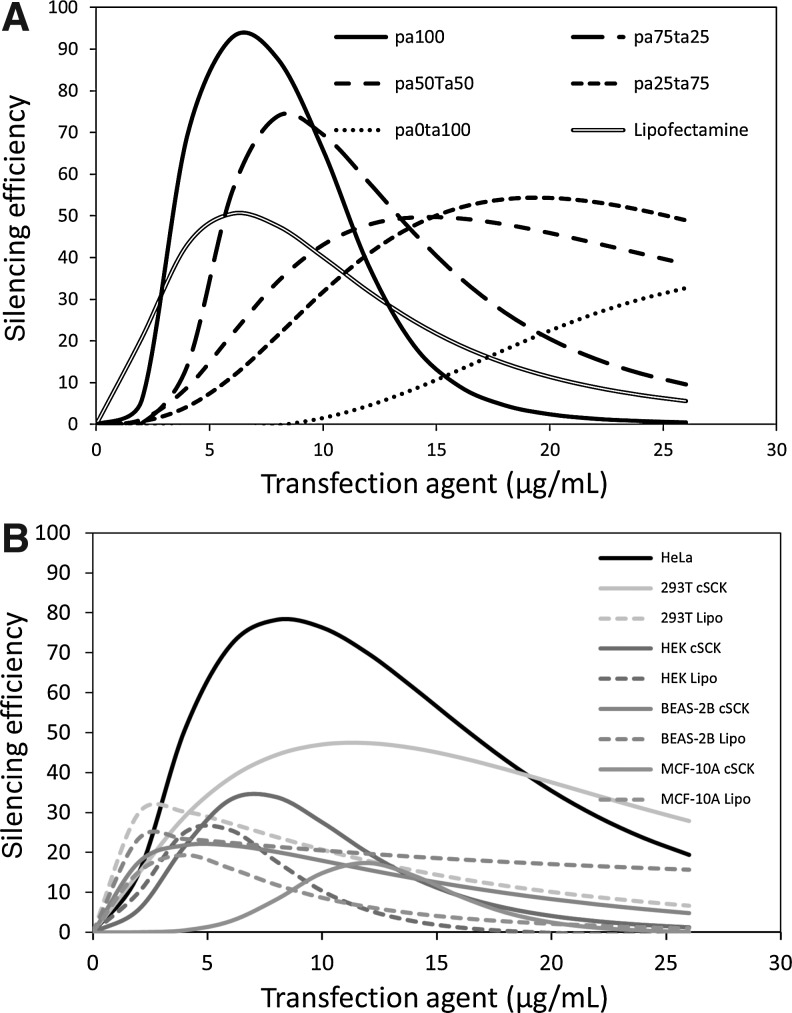
FIG. 4.
Silencing efficiency of the cSCK nanoparticles compared with Lipofectamine 2000. The silencing efficiency was calculated as the difference between the survival rate (%) of cells transfected with the AllStars Negative Control siRNA, and the survival rate (%) of the AllStars Cell Death Control siRNA using the lethal concentration 50 (LC50) and Hill slope coefficients derived from the raw data. (A) The silencing efficiency for PAEA128-b-PS40 derived cSCKs with different shell compositions in HeLa derived from the data in Supplementary Table S1, and (B) for silencing in different cell lines with PAEA160-b-PS30 cSCK-pa100 using data in Supplementary Table S2 (Supplementary Data are available online at www.liebertpub.com/nat).
The optimal nanoparticle concentration and N/P ratios for maximum silencing efficiency of the other cSCKs were determined in a similar way (Fig. 4A). For cSCK-pa75-ta25, 8 μg/mL or an N/P ratio of 11 gave a maximum silencing efficiency of 74%, resulting from 11% viability with the cytotoxic siRNA and 85% viability with the non-cytotoxic siRNA (Fig. 3B). For cSCK-pa50-ta50, a higher concentration of 15 μg/mL or an N/P ratio of 20.8 gave maximum silencing efficiency of 50%, resulting from 18% viability with the cytotoxic siRNA and 67% viability with the non-cytotoxic siRNA (Fig. 3C). For cSCK-pa25-ta75, 19 μg/mL or an N/P ratio of 26.4 gave a maximum silencing efficiency of 54% resulting from about 23% viability with the cytotoxic siRNA, and 76% viability with the non-toxic siRNA (Fig. 3D). For cM-ta100 35 μg/mL or an N/P ratio of 47 was optimal, showing about 18% viability with the cytotoxic siRNA, and about 55% viability with the non-toxic siRNA (Fig. 3E). Though increasing the tertiary amine concentration of the cSCK was effective at decreasing its background toxicity, it also decreased the transfection efficiency of the cSCK, making these combinations less desirable for siRNA transfection than the parent cSCK-pa100.
In comparison with cSCK-pa100, which had an optimal silencing efficiency of 93% at 6 μg/mL, Lipofectamine 2000 only had a 51% silencing efficiency at its optimal concentration of 6 μg/mL (Fig. 4A). So while cSCK-pa100 and Lipofectamine 2000 have the same LC50 value on a gm/mL basis (Supplementary Table S1), cSCK-pa100 appears to be much more efficient at intracellular delivery of the siRNA. This feature is reflected in the much larger Hill coefficient (equation 1, section 2.4) for toxic siRNA delivery for cSCK-pa100 (5.2) than for Lipofectamine 2000 (1.8) (Supplementary Table S1). Polyfect did not exhibit any ability to transfect siRNA, showing approximately the same cytotoxicity with the lethal and control siRNAs at all concentrations (data not shown). While cSCK-pa100 showed the highest silencing efficiency, its window of effectiveness was slightly narrower than cSCK-pa75-ta25 due to its higher toxicity. For in vivo applications where cells may be exposed to a greater range of nanoparticle concentrations, a less efficient but less toxic nanoparticle may be more desirable.
The silencing efficiency of cSCK-pa100 was also later examined in a series of other human cell lines in comparison to Lipofectamine 2000 (Supplementary Fig. S1; Fig. 4B). In these experiments a new preparation of cSCK that was used for the fusogenic GALA experiments to be described later, which had a slightly different composition (PAEA160-b-PS30 vs. PAEA128-b-PS40). The silencing efficiency of this preparation was first determined in HeLa cells by the same cell viability method described above with toxic and control siRNAs (Supplementary Fig. S1; Supplementary Table S2), and found to be quite similar to the first preparation, though it showed a somewhat lower peak efficiency of about 78% at a slightly higher 8 μg/mL (Fig. 4B) compared to 93% and 6 μg/mL for the original cSCK. The silencing efficiency was found to be less in HEK cells (human embryonic kidney cells, 34% at 8 μg/mL) and 293T cells (a variant of HEK cells, 47% at 12 μg/mL), but still better than Lipofectamine 2000. The silencing efficiency was less, however, and about comparable to Lipofectamine 2000 in BEAS-2B (human bronchial epithelial cell line) and MCF-10a (human mammary epithelial cell line).
Confocal and flow cytometry assays of cSCK•siRNA transfection efficiency
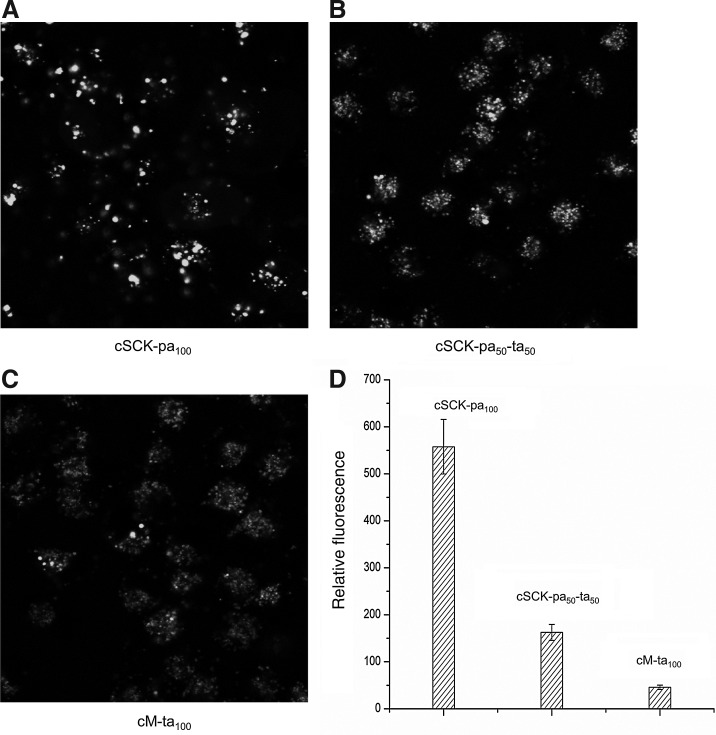
The PAEA128-b-PS40 cSCKs were also evaluated for their ability to transfect a fluorescently labeled siRNA into HeLa cells by confocal microscopy and by flow cytometry (Fig. 5). Fluorescently-labeled GAPDH siRNA (GAPDH-F) was mixed with cSCK-pa100 (N/P=10), cSCK-pa50-ta50 (N/P=10), and cM-ta100 (N/P=20), and then incubated with HeLa cells. After 4 hours the cells were observed by confocal microscopy (Fig. 5A–C) and quantified by flow cytometry (Fig. 5D). Confocal microscopy showed that the siRNAs were mainly trapped in endosomes, indicating that endocytosis was the principal uptake mechanism. Quantitative flow cytometry further indicated that the HeLa cells took up 3 times more GAPDH siRNA in the presence of cSCK-pa100 than in the presence of cSCK-pa50-ta50, and 10 times more than cM-ta100. These results suggest that the greater transfection efficiency of cSCK-pa100 is due to better uptake, rather than by some other mechanism, such as endosomal release or release of the siRNA from the cSCK.
FIG. 5.
Fluorescence confocal microscopy and flow cytometry analysis of siRNA delivery by PAEA128-b-PS40 cSCKs. Fluorescently labeled GAPDH siRNA (100 nM) was complexed with cSCK-pa100, cSCK-pa50-ta50, and cM-ta100 at N/P ratios of 10, 10, and 20 and incubated with HeLa cells for 4 h and then examined by fluorescence confocal microscopy (A–C) and flow cytometry (D).
Inhibition of iNOS expression with cSCK•siRNA complexes
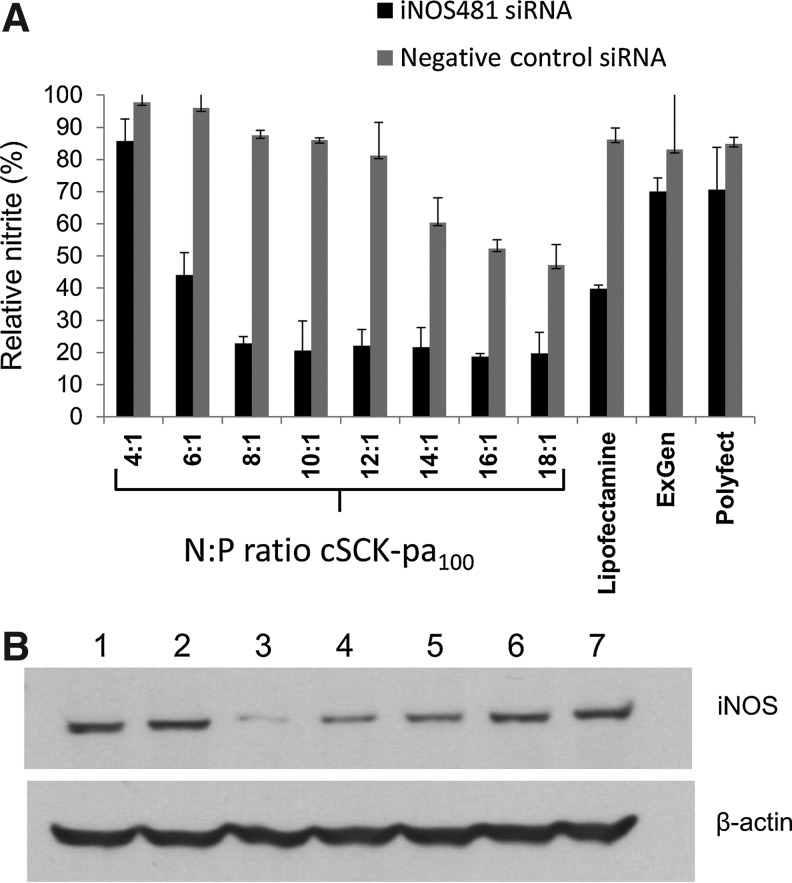
To test the ability of the cSCK•siRNA system to knock down the expression of a specific gene, we investigated the inhibition of iNOS expression (Fig. 6). iNOS is induced in acute lung injury (ALI) and at high levels further aggravates the condition through cell damage caused by the nitric oxide (NO) that it produces (MEHTA, 2005). Inhibition of iNOS has been shown to enhance cell survival and thereby improve the outcome of ALI (Hosogi et al., 2008). Because iNOS induction in acute lung injury has been mainly ascribed to alveolar macrophages (Altmann et al., 2012), we chose the RAW264.7 mouse macrophage cell line for transfection experiments which is known to greatly upregulate iNOS expression in response to LPS/γ-IFN (Noda and Amano, 1997). For this study, we chose to investigate only the parent PAEA128-b-PS40 cSCK-pa100 because it was one of the better studied of the cSCKs (Zhang et al., 2009b; Zhang et al., 2010) and showed the best siRNA silencing efficiency with the toxic siRNA assay described above. We used the iNOS481 siRNA (GGCCACAUCGGAUUUCACUtt•AGUGAAAUCCGAUGUGGCCtt) that we had previously determined to be very efficient in knocking down iNOS when transfected by Lipofectamine 2000 (Fang et al., 2010) and the non-lethal Allstar negative siRNA as a control. The iNOS and control siRNAs (100 nM) were combined with the cSCK-pa100 at different N/P ratios and incubated with mouse peritoneal macrophage RAW264.7 cells in comparison to Lipofectamine 2000, ExGen500, and Polyfect, which serve as positive transfection agent controls. Following transfection, the RAW264.7 cells were activated with LPS and γ-IFN. After an additional 24 hours, a portion of the supernatant (50 μL) was assayed for the production of nitrite by the Griess reagent, which could be quantified by the absorbance at 540 nm (Fig. 6A). The cells were then harvested for western blotting analysis of iNOS protein expression. The greatest inhibition of NO production with the minimum of background toxicity occurred at N/P ratios 8 and 10, corresponding to 5 and 6.2 μg/mL of cSCK. At these concentrations, 77% and 79% inhibition was observed, respectively, with 12% to 14% non-specific inhibition likely due to cSCK toxicity, corresponding to silencing efficiencies of 65% at both concentrations. Curve fitting also gave a maximum efficiency of 65% at 6 μg/mL (Supplementary Fig. S2). The cSCK was better than Lipofectamine 2000, which only achieved about 60% inhibition at the same level of toxicity as for the cSCK (14%) corresponding to a net silencing efficiency of 46%, while ExGen500 and Polyfect showed very poor transfection efficiency. The Griess results were also confirmed by western blotting (Fig. 6B).
FIG. 6.
cSCK-pa100 mediated silencing of iNOS expression by siRNA. Anti-iNOS siRNA481 (100 nM) was transfected into mouse monocyte-macrophage RAW264.7 cells using PAEA128-b-PS40 cSCK-pa100 at different N/P ratios (for N/P=1, cSCK 0.62 μg/mL), Lipofectamine 2000 (1.7 μg/mL), ExGen 500 (18.2 μM), or Polyfect (6.8 μg/mL). After 6 hours iNOS expression was induced by treatment with LPS (lipopolysaccharides from Escherichia coli 055:B5) and γ-IFN (gamma interferon). After 24 hours the supernatant was analyzed for NO production by the Griess assay, and the cells were analyzed for iNOS production by western blotting. (A) Griess assay; (B) western blotting, lane 1, cSCK (6.2 μg/mL), no siRNA; lane 2, 100 nM AllStars Negative Control siRNA with cSCK (6.2 μg/mL); lane 3, 100 nM iNOS siRNA481 with cSCK (6.2 μg/mL); lane 4, 100 nM iNOS siRNA481 with Lipofectamine 2000 (1.7 μg/mL); lane 5, 100 nM iNOS siRNA481 with Polyfect (6.8 μg/mL); lane 6, 100 nM iNOS siRNA481 with ExGen 500 (18.2 μM); lane 7, 100 nM iNOS siRNA481 alone, without any transfection agent.
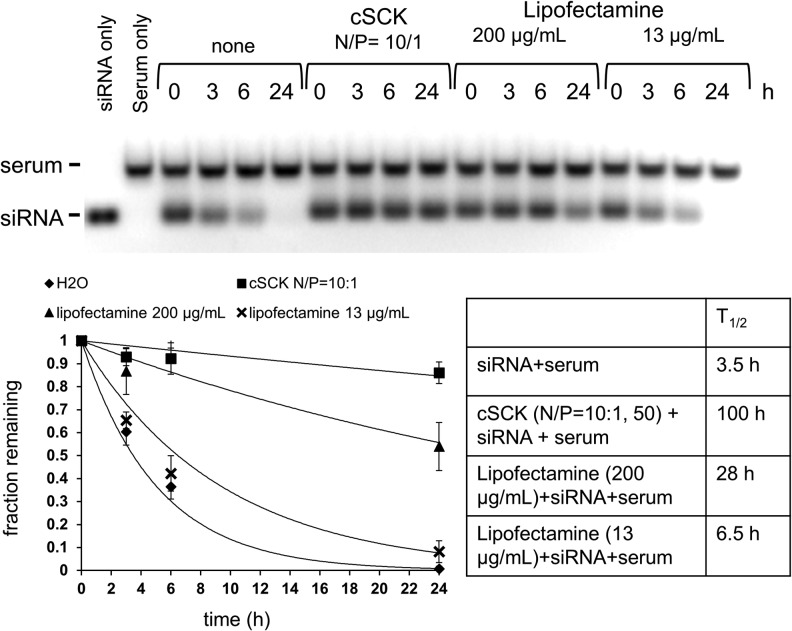
Protection of siRNA from degradation by human serum by cSCK
In addition to being able to transport an siRNA into a cell, a transfection agent must also be able to protect an siRNA from degradation during its transit to cell. To determine the ability of the PAEA128-b-PS40 cSCK-pa100 to protect siRNAs from degradation by nucleases present in human serum we made use of unlabeled siRNA and monitored its degradation by agarose gel electrophoresis with ethidium bromide staining as previously described (Bartlett and Davis 2007a, 2007b). As shown in Fig. 7, siRNAs in the presence of 50% human serum were degraded rapidly with a half-life of 3.5 hours. However, siRNAs complexed with cSCK were degraded rather slowly, with a half-life of 100 hours. In contrast, complexes of siRNA with 13 μg/mL and 200 μg/mL of Lipofectamine 2000 had half-lives of 6.5 and 28 hours respectively. The greater protection afforded by the cSCK is probably due to better sequestration of the siRNA within the cationic shell of the nanoparticle than can be afforded by the much thinner cationic layer on the surface of a cationic lipid.
FIG. 7.
Protection of siRNA degradation by cSCK-pa100. siRNA481 (0.77 μM final concentration) alone or complexed with PAEA128-b-PS40 cSCK-pa100 (50 μg/mL, N/P=10:1) or Lipofectamine 2000 (13 and 200 μg/mL) was incubated in 50% human serum (Sigma) at 37°C, under 5% CO2 for the times shown, electrophoresed on a 2% agarose gel, and visualized by staining with ethidium bromide.
Enhancement of cSCK•siRNA knockdown of iNOS mRNA by the GALA peptide
While the cSCK nanoparticles appear to facilitate endosomal escape of nucleic acid cargo by the proton sponge effect, the exact efficiency of this process is not known, and it was of interest to see if this critical step could be further enhanced with fusogenic peptides (Nakase et al., 2010; Varkouhi et al., 2011). Among the various fusogenic peptides shown to enhance endosomal escape, the GALA peptide (Li et al., 2004) was chosen for study because it has a net negative charge, which would enable it to be delivered together with the siRNA in an electrostatic complex with the cationic SCK nanoparticles. The GALA peptide is a 30 amino acid synthetic amphiphilic peptide (WEAALAEALAEALAEHLAEALAEALEALAA) with a repeating Glu-Ala-Leu-Ala, which also contains a His and a Trp as spectroscopic probes. The design of the sequence was based on a mutant sequence of haemagglutinin subunit (HA-2) from influenza virus, which mediates endosomal escape (Subbarao et al., 1987). The GALA peptide undergoes a pH-dependent conformational change from a random coil to an alpha-helix when the pH drops from 7.0 to 5.0, leading to the interaction with lipid bilayers and the destabilization of endosomal membrane (Parente et al., 1990a; Parente et al., 1990b; Fattal et al., 1994). GALA has been shown to improve their intracellular delivery of DNA, ODNs and siRNA with cationic liposomes, dendrimers, and polylysine (Futaki et al., 2005; Yamada et al., 2005; Sasaki et al., 2008; Hatakeyama et al., 2009; Kobayashi et al., 2009; Wang et al., 2009; Ukawa et al., 2010; Akita et al., 2011a, 2011b; Khalil et al., 2011; Nakase et al., 2011). Although there are several mechanisms proposed for membrane perturbation induced by GALA-based gene delivery systems (Simoes et al., 1999; Sasaki et al., 2008), the actual mechanism is still not yet well understood.
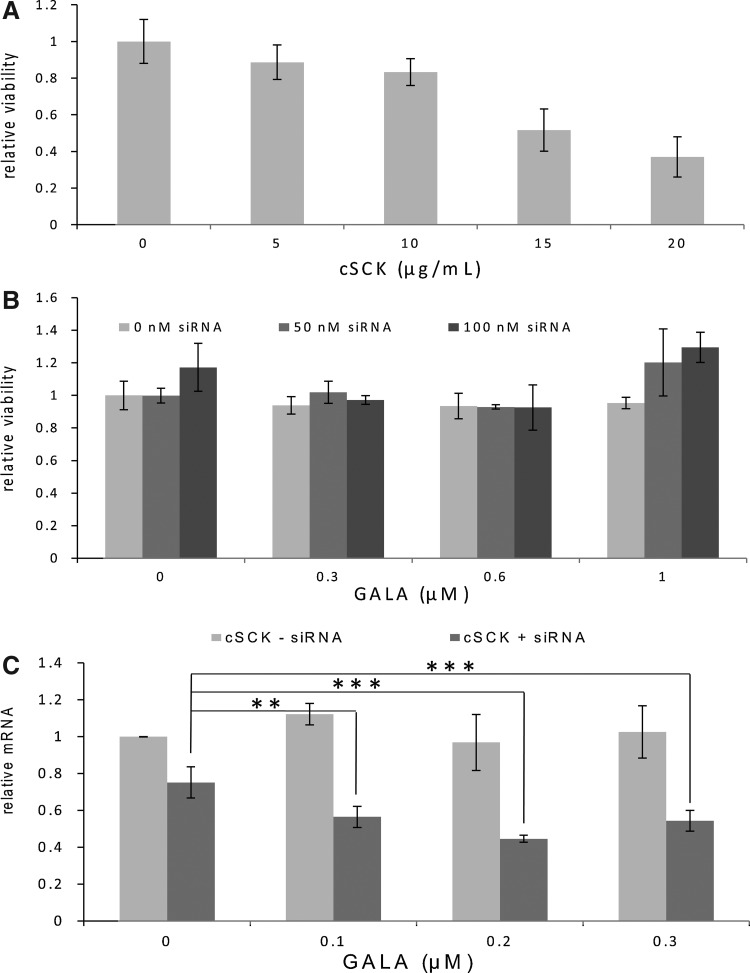
To test whether fusogenic peptide GALA could enhance iNOS mRNA knockdown by cSCK•siRNA, we used quantitative RT-PCR to directly monitor mRNA levels in RAW 264.7 cells. For all the following studies, we used the PAEA160-b-PS30 cSCK-pa100. These particles behaved similarly to the original particles in that they quantitatively bound both 50 or 100 nM siRNA at 10 μg/mL cSCK, and at this cSCK concentration siRNA binding was unaffected by the addition of 0.5 to 2.0 μM GALA (Supplementary Fig. S3). As shown in Fig. 8C, addition of GALA (0, 0.1, 0.2, and 0.3 μM) to 10 μg/mL of cSCK in the absence of siRNA did not cause any knockdown of iNOS mRNA, indicating that fusogenic peptide GALA by itself does not have any direct effect. While addition of varying concentrations of GALA did not significantly affect knockdown of iNOS mRNA with 10 μg/mL cSCK and 100 nM siRNA, a significant effect was observed when the concentration of the siRNA was further decreased to 50 nM. At this concentration of siRNA only 25% knockdown of iNOS mRNA was observed in the absence of GALA, but addition of 0.2 μM GALA significantly increased the knockdown to 55% (P<0.001). The difference in observed knockdown was not due to differences in cytotoxic effects from the GALA in the presence of the siRNA as ascertained from independent MTT assays of the complexes (Fig. 8B). Thus, GALA could be used to lower the concentration of siRNA needed to achieve knockdown.
FIG. 8.
iNOS mRNA silencing by cSCK•siRNA•GALA nanocomplex in RAW264.7 cells. (A) cytoxicity of PAEA160-b-PS30 cSCK-pa100 after 24 h in RAW cells that had been induced for 24 hours with LPS/γ-IFN as determined by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. (B) Cytotoxicity of cSCK•GALA complexes in induced RAW cells in the absence or presence of different siRNA concentrations as determined by the MTT assay. (C) Knockdown efficiency in RAW cells with cSCK (10 μg/mL)•siRNA (50 nM)•GALA (0, 0.1, 0.2, 0.3 μM) nanocomplexes in induced RAW cells. iNOS mRNA level was measured by real-time polymerase chain reaction after 8 hours. The P value for the observed difference in expression with and without GALA was <0.01.
Mechanism of GALA enhancement of iNOS knockdown by cSCK•siRNA
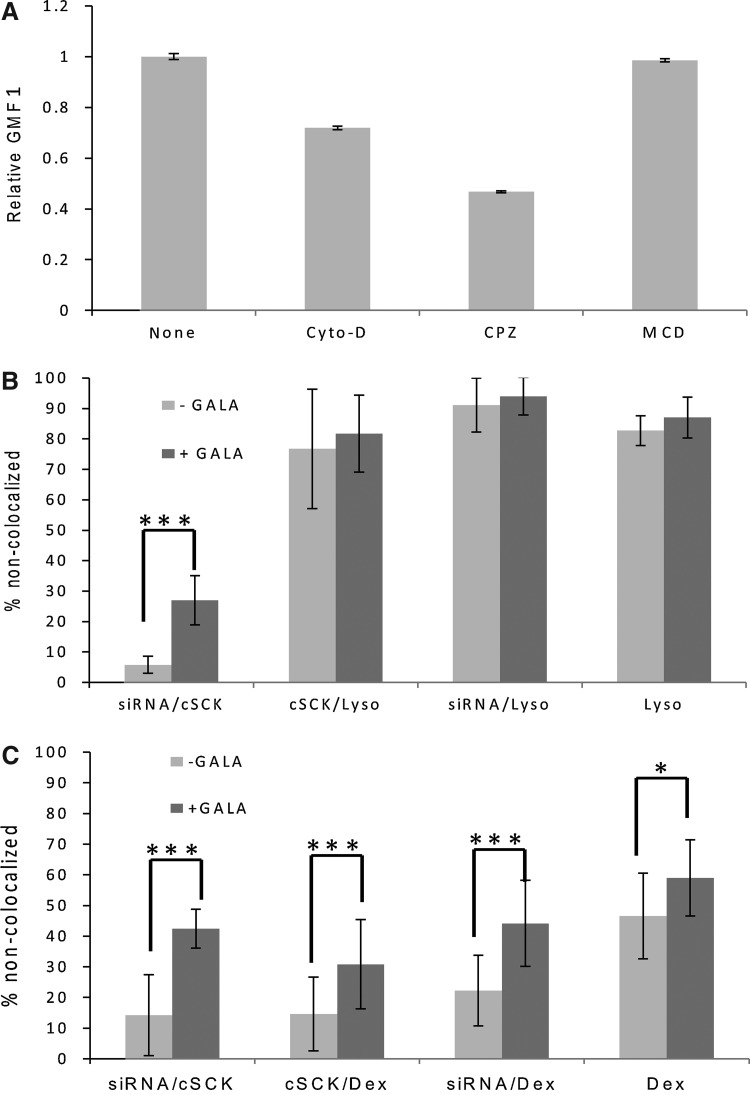
To determine whether GALA might be enhancing siRNA silencing by enhancing nanoparticle uptake and/or endosomal escape, we first carried out a study of nanoparticle uptake. To quantitatively measure the cellular uptake of cSCK•siRNA•GALA in the RAW 264.7 cell line, we used flow cytometry to monitor the uptake of a dual fluorescence-labeled nanocomplex of Alexafluor 488-labeled cSCK, Cy5-labeled siRNA in the presence and absence of the GALA peptide. Geometric mean fluorescence intensities of the AlexaFluor 488 and Cy5 channels show that with an increase of fusogenic peptide GALA, the relative uptake of cSCK (Supplementary Fig. S4A) and siRNA (Supplementary Fig. S4B) were both slightly decreased but not significantly different. The reduction in uptake efficiency, if any, could be explained by a net reduction in the positive charge of the cSCK upon addition of the negatively charged GALA, which reduces endocytosis efficiency. The observation that GALA may at most reduce uptake efficiency suggests that the enhanced mRNA silencing observed upon addition of GALA must more likely be due to enhanced endosomal escape or some other process following uptake.
To determine what process GALA might be affecting, we examined the non-colocalization of the siRNA and nanoparticle by confocal microscopy using a concentration of the nanocomplex that showed a significant effect of GALA on RNA knockdown. Thus, iNOS-induced RAW 264.7 cells were incubated with 10 μg/mL AlexaFluor488-cSCK with 50 nM Cy5-siRNA with and without 0.2 μM GALA for 24 hours and then examined at 250×magnification (Supplementary Fig. S5). We found both nanocomplexes could be efficiently uptaken by RAW 264.7 cells, but in the absence of GALA, the siRNA was almost completely colocalized with the cSCK (Supplementary Fig. S5A) showing only 3.3±2.7 % that was non-colocalized (n=10 images). In the presence of the fusogenic peptide GALA, 20±9 % of the siRNA was non-colocalized with the cSCK (n=8 images, P=0.000) (Supplementary Fig. S5B), suggesting that GALA must be enhancing separation of the siRNA from the cSCK, possibly by enhancing escape of the siRNA from the endosomes in preference to the cSCK.
Effect of inhibitors on cellular uptake of cSCKs in RAW 264.7 cells
To gain further insight into the uptake and trafficking of the nanocomplexes in the cells, the effects of known endocytosis inhibitors (Lundin et al., 2008; Vercauteren et al., 2010) on cSCK uptake by the RAW cells were investigated (Fig. 9). Cytochalasin D is known to inhibit phagocytosis and macropinocytosis by inhibiting F-actin elongation. Chlorpromazine is an inhibitor of clathrin-mediated endocytosis, since it disrupts the assembly of adaptor protein 2 and clathrin, which is essential for the formation of clathrin-coated vesicles. Methyl-β-cyclodextrin is an inhibitor of caveolin-dependent endocytosis as it disrupts lipid rafts by binding and removing cholesterol from membranes. The effect of these inhibitors on cSCK uptake and cell viability were examined over a range of concentrations (Supplementary Fig. S6) and the effect on cSCK uptake summarized in Fig. 9A. Entry of cSCK into RAW 264.7 cells was most greatly reduced (about 50% at 20 μM; Supplementary Fig. S6B) by chlorpromazine which inhibits clathrin-dependent endocytosis. Entry was also inhibited, but to a lesser extent by cytochalasin D (average of 30 % for 2–50 μM; Supplementary Fig. S6A), suggesting that macropinocytosis or phagocytosis might also play a role in cSCK internalization in the RAW cells. It would appear that caveolin-mediated endocytosis is not involved, since methyl-β-cyclodextrin had no effect at any concentration (Supplementary Fig. S6C).
FIG. 9.
Endocytosis inhibition and tracking studies. (A) Inhibition experiments. iNOS-induced RAW 264.7 cells were incubated with cytochalasin D (Cyto-D) (average of 2–50 μM), chlorpromazine (CPZ) (20 μM) and methyl-beta-cyclodextrin, (MCD) (20 μM) for 1 hour and then incubated with PAEA160-b-PS30 AlexaFluor488-cSCKpa100 (10 μg/mL) for another hour, and cSCK uptake (n=3) was determined by flow cytometry. (B) Degree of non-colocalization of siRNAs and cSCKs with Lysotracker in the presence and absence of GALA. Induced RAW264.7 cells were incubated with siRNA (50 nM)•AlexaFluor488-cSCKpa100 (10 μg/mL)-Cy5•GALA (0, 0.2 μM) complexes for 22 hours, after which Lysotracker-Blue (50 nM) was added. Two hours later the cells were analyzed by confocal microscopic imaging. The degree of non-colocalization of siRNA with cSCK (siRNA/cSCK), siRNA with Lysotracker (siRNA/Lyso), cSCK with Lysotracker (cSCK/Lyso), and Lysotracker with siRNA and cSCK (Lyso) was determined by Image J Green and Red Puncta Colocalization Macro (n=7 images without GALA, and n=10 with GALA with 2–3 cells/image). (C) Degree of non-colocalization of siRNAs and cSCKs with dextran-cascade blue. AlexaFluor488-cSCK-pa100 (10 μg/mL), Cy5-siRNA (50 nM), and GALA (0, 0.2 μM) with dextran-cascade blue (100 μg/mL) were transfected to RAW264.7 cells for 8 hours followed by confocal microscopic imaging. The degree of non-colocalization of siRNA with cSCK (siRNA/cSCK), siRNA with dextran (siRNA/Dex), cSCK with dextran (cSCK/Dex) and dextran with siRNA and cSCK (Dex) was determined as above (n=10 images without GALA and n=16 with GALA with 2–3 cells/image).
Colocalization with endosome trackers
Additional insight into the mechanism of uptake and trafficking was obtained with the use of endosome trackers. One standard endosomal tracker is Lysotracker, which accumulates in acidic vesicles, such as late endosomes and lysosomes. Most interestingly, there was little colocalization of either the cSCK or the siRNA with this dye in the absence or presence of GALA (Supplementary Fig. S7; Fig. 9B), suggesting that the cSCK and siRNA were escaping the endosomal pathway prior to entering late endosome or lysosomes, or that the cSCK was preventing acidification due to its proton sponge properties. Addition of GALA, however, significantly enhanced the separation of the siRNA from the cSCK from 5.8±2.8% to 26.9±8.1% (P<0.001). The colocalization of the siRNA and nanoparticle with dextran-cascade blue, a tracker of fluid phase uptake, which was indicated by sensitivity to cytochalasin D was then studied (Supplementary Fig. S8; Fig. 9C). Cells were incubated with the nanocomplex and dextran-cascade blue and observed by confocal microscopy after 8 hours. In the absence of GALA, 22±12% of the siRNA was non-colocalized with dextran-labeled endosomes and increased significantly (P<0.001) to 44±14% in the presence of GALA. Likewise, 15±12% of the cSCK was non-colocalized with the Dextran in the absence of GALA, but increased significantly (P<0.001) to 31±15% in the presence of GALA. About 50% of the Dextran was non-colocalized with the siRNA and cSCK. These results indicate that a substantial fraction of the nanocomplexes may be entering by macropinocytosis or phagocytosis, and that GALA may in fact be enhancing endosomal escape of both siRNAs and cSCKs. The behavior of the cSCKs with respect to Lysotracker and Dextran is very similar to what has been previously reported for uptake of a DNA lipoplex in Chinese hamster ovary cells (Zhang et al., 2011) and polyplexes in human hepatocellular liver carcinoma cells (Goncalves et al., 2004).
Conclusion
Of a series of cationic shell-crosslinked nanoparticles, cSCKs, with varying percentages of primary to tertiary amines, the parent cSCK-pa100, with only primary amines showed the best siRNA silencing efficiency in HeLa cells, and was much better than Lipofectamine 2000 and Polyfect. The better silencing efficiency of cSCK-pa100 in HeLa cells could be attributed to better cell uptake, indicating an important role for primary amines in this process. Silencing efficiency for cSKC-pa100 was not as good in other human cell lines, indicating that cSCK nanoparticles may have to be tailored for each specific cell type. As indicated in the introduction, the synthetic design of cSCKs makes them ideal for such an optimization process, as it is quite straightforward to manipulate their size and shape, charge density and buffering capacity, stealth and targeting properties. The other important advantage of cSCK-pa100 for eventual in vivo use was its ability to greatly inhibit the degradation of siRNA by human serum, which it does so much more effectively than Lipofectamine 2000, presumably by sequestering the siRNA deeply within the shell and thereby encumbering access to the siRNA by serum enzymes. cSCK-pa100 was also very good for siRNA knockdown of iNOS in a mouse macrophage cell line, which would be of great interest for treatment of acute lung injury. The high charge density of the cSCK also enables co-delivery of the fusogenic GALA peptide which was able to enhance siRNA silencing at lower siRNA concentrations by facilitating endosomal escape. The studies as a whole demonstrate the potential of shell crosslinked nanoparticles as a tunable delivery platform for siRNA and auxiliary peptides. Ongoing studies are focusing on other agents for facilitating endosomal escape, such as the incorporation of histamine into the shell (Shrestha et al., 2012), as well as the development of a biodegradable version of the cSCK for silencing iNOS induction in acute lung injury animal models.
Supplementary Material
Acknowledgments
We thank Dianne Duncan of the Washington University Biology Department Imaging Facility for assisting the image quantification analysis. This work was supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health as a Program of Excellence in Nanotechnology (HHSN268201000046C).
Author Disclosure Statement
No competing financial interests exist.
References
- AKITA H. KOGURE K. MORIGUCHI R. NAKAMURA Y. HIGASHI T. NAKAMURA T. SERADA S. FUJIMOTO M. NAKA T. FUTAKI S., et al. Reprint of: Nanoparticles for ex vivo siRNA delivery to dendritic cells for cancer vaccines: programmed endosomal escape and dissociation. J. Control Release. 2011a;149:58–64. doi: 10.1016/j.jconrel.2010.08.023. [DOI] [PubMed] [Google Scholar]
- AKITA H. MASUDA T. NISHIO T. NIIKURA K. IJIRO K. HARASHIMA H. Improving in vivo hepatic transfection activity by controlling intracellular trafficking: the function of GALA and maltotriose. Mol. Pharm. 2011b;8:1436–1442. doi: 10.1021/mp200189s. [DOI] [PubMed] [Google Scholar]
- ALTMANN C. ANDRES-HERNANDO A. MCMAHAN R.H. AHUJA N. HE Z. RIVARD C.J. EDELSTEIN C.L. BARTHEL L. JANSSEN W.J. FAUBEL S. Macrophages mediate lung inflammation in a mouse model of ischemic acute kidney injury. Am. J. Physiol. Renal Physiol. 2012;302:F421–432. doi: 10.1152/ajprenal.00559.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARTLETT D.W. DAVIS M.E. Effect of siRNA nuclease stability on the in vitro and in vivo kinetics of siRNA-mediated gene silencing. Biotechnology and bioengineering. 2007a;97:909–21. doi: 10.1002/bit.21285. [DOI] [PubMed] [Google Scholar]
- BARTLETT D.W. DAVIS M.E. Physicochemical and biological characterization of targeted, nucleic acid-containing nanoparticles. Bioconjug. Chem. 2007b;18:456–468. doi: 10.1021/bc0603539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOUSSIF O. LEZOUALC'H F. ZANTA M.A. MERGNY M.D. SCHERMAN D. DEMENEIX B. BEHR J.P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. U.S.A. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COUTO L.B. HIGH K.A. Viral vector-mediated RNA interference. Curr. Opin. Pharmacol. 2010;10:534–542. doi: 10.1016/j.coph.2010.06.007. [DOI] [PubMed] [Google Scholar]
- DAVID S. PITARD B. BENOIT J.P. PASSIRANI C. Non-viral nanosystems for systemic siRNA delivery. Pharmacol. Res. 2010;62:100–114. doi: 10.1016/j.phrs.2009.11.013. [DOI] [PubMed] [Google Scholar]
- FANG H. SHEN Y. TAYLOR J.S. Native mRNA antisense-accessible sites library for the selection of antisense oligonucleotides, PNAs, and siRNAs. RNA. 2010;16:1429–1435. doi: 10.1261/rna.1940610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FATTAL E. NIR S. PARENTE R.A. SZOKA F.C., JR Pore-forming peptides induce rapid phospholipid flip-flop in membranes. Biochemistry. 1994;33:6721–6731. doi: 10.1021/bi00187a044. [DOI] [PubMed] [Google Scholar]
- FUTAKI S. MASUI Y. NAKASE I. SUGIURA Y. NAKAMURA T. KOGURE K. HARASHIMA H. Unique features of a pH-sensitive fusogenic peptide that improves the transfection efficiency of cationic liposomes. J. Gene Med. 2005;7:1450–1458. doi: 10.1002/jgm.796. [DOI] [PubMed] [Google Scholar]
- GAO Y. LIU X.L. LI X.R. Research progress on siRNA delivery with nonviral carriers. International journal of nanomedicine. 2011;6:1017–1025. doi: 10.2147/IJN.S17040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GONCALVES C. MENNESSON E. FUCHS R. GORVEL J.-P. MIDOUX P. PICHON C. Macropinocytosis of polyplexes and recycling of plasmid via the clathrin-dependent pathway impair the transfection efficiency of human hepatocarcinoma cells. Mol. Ther. 2004;10:373–385. doi: 10.1016/j.ymthe.2004.05.023. [DOI] [PubMed] [Google Scholar]
- HATAKEYAMA H. ITO E. AKITA H. OISHI M. NAGASAKI Y. FUTAKI S. HARASHIMA H. A pH-sensitive fusogenic peptide facilitates endosomal escape and greatly enhances the gene silencing of siRNA-containing nanoparticles in vitro and in vivo. J. Control. Release. 2009;139:127–132. doi: 10.1016/j.jconrel.2009.06.008. [DOI] [PubMed] [Google Scholar]
- HOSOGI S. IWASAKI Y. YAMADA T. KOMATANI-TAMIYA N. HIRAMATSU A. KOHNO Y. UEDA M. ARIMOTO T. MARUNAKA Y. Effect of inducible nitric oxide synthase on apoptosis in Candida-induced acute lung injury. Biomed. Res. 2008;29:257–266. doi: 10.2220/biomedres.29.257. [DOI] [PubMed] [Google Scholar]
- KHALIL I.A. HAYASHI Y. MIZUNO R. HARASHIMA H. Octaarginine- and pH sensitive fusogenic peptide-modified nanoparticles for liver gene delivery. J. Control. Release. 2011;156:374–380. doi: 10.1016/j.jconrel.2011.08.012. [DOI] [PubMed] [Google Scholar]
- KOBAYASHI S. NAKASE I. KAWABATA N. YU H.H. PUJALS S. IMANISHI M. GIRALT E. FUTAKI S. Cytosolic targeting of macromolecules using a pH-dependent fusogenic peptide in combination with cationic liposomes. Bioconjug. Chem. 2009;20:953–959. doi: 10.1021/bc800530v. [DOI] [PubMed] [Google Scholar]
- KURRECK J. RNA interference: from basic research to therapeutic applications. Angew. Chem. Int. Ed. Engl. 2009;48:1378–1398. doi: 10.1002/anie.200802092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LARES M.R. ROSSI J.J. OUELLET D.L. RNAi and small interfering RNAs in human disease therapeutic applications. Trends Biotechnol. 2010;28:570–579. doi: 10.1016/j.tibtech.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI W. NICOL F. SZOKA F.C., JR GALA: a designed synthetic pH-responsive amphipathic peptide with applications in drug and gene delivery. Adv. Drug Deliv. Rev. 2004;56:967–985. doi: 10.1016/j.addr.2003.10.041. [DOI] [PubMed] [Google Scholar]
- LI Y. DU W. SUN G. WOOLEY K.L. pH-Responsive shell cross-linked nanoparticles with hydrolytically labile cross-links. Macromolecules. 2008;41:6605–6607. [Google Scholar]
- LIU Y.P. BERKHOUT B. miRNA cassettes in viral vectors: problems and solutions. Biochimi. Biophys. Acta. 2011;1809:732–745. doi: 10.1016/j.bbagrm.2011.05.014. [DOI] [PubMed] [Google Scholar]
- LUNDIN P. JOHANSSON H. GUTERSTAM P. HOLM T. HANSEN M. LANGEL U. EL Andaloussi S. Distinct uptake routes of cell-penetrating peptide conjugates. Bioconjug. Chem. 2008;19:2535–2542. doi: 10.1021/bc800212j. [DOI] [PubMed] [Google Scholar]
- MEHTA S. The effects of nitric oxide in acute lung injury. Vascul. Pharmacol. 2005;43:390–403. doi: 10.1016/j.vph.2005.08.013. [DOI] [PubMed] [Google Scholar]
- NAKASE I. KOBAYASHI S. FUTAKI S. Endosome-disruptive peptides for improving cytosolic delivery of bioactive macromolecules. Biopolymers. 2010;94:763–770. doi: 10.1002/bip.21487. [DOI] [PubMed] [Google Scholar]
- NAKASE I. KOGURE K. HARASHIMA H. FUTAKI S. Application of a fusiogenic peptide GALA for intracellular delivery. Methods Mol. Biol. 2011;683:525–533. doi: 10.1007/978-1-60761-919-2_37. [DOI] [PubMed] [Google Scholar]
- NODA T. AMANO F. Differences in nitric oxide synthase activity in a macrophage-like cell line, RAW264.7 cells, treated with lipopolysaccharide (LPS) in the presence or absence of interferon-gamma (IFN-gamma): possible heterogeneity of iNOS activity. J. Biochem. 1997;121:38–46. doi: 10.1093/oxfordjournals.jbchem.a021566. [DOI] [PubMed] [Google Scholar]
- NYSTROM A.M. WOOLEY K.L. Thiol-functionalized shell crosslinked knedel-like (SCK) nanoparticles: a versatile entry for their conjugation with biomacromolecules. Tetrahedron. 2008;64:8543–8552. doi: 10.1016/j.tet.2008.04.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NYSTROM A.M. WOOLEY K.L. The importance of chemistry in creating well-defined nanoscopic embedded therapeutics: devices capable of the dual functions of imaging and therapy. Acc. Chem. Res. 2011;44:969–978. doi: 10.1021/ar200097k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARENTE R.A. NADASDI L. SUBBARAO N.K. SZOKA F.C., JR Association of a pH-sensitive peptide with membrane vesicles: role of amino acid sequence. Biochemistry. 1990a;29:8713–8719. doi: 10.1021/bi00489a030. [DOI] [PubMed] [Google Scholar]
- PARENTE R.A. NIR S. SZOKA F.C., JR Mechanism of leakage of phospholipid vesicle contents induced by the peptide GALA. Biochemistry. 1990b;29:8720–8728. doi: 10.1021/bi00489a031. [DOI] [PubMed] [Google Scholar]
- SASAKI K. KOGURE K. CHAKI S. NAKAMURA Y. MORIGUCHI R. HAMADA H. DANEV R. NAGAYAMA K. FUTAKI S. HARASHIMA H. An artificial virus-like nano carrier system: enhanced endosomal escape of nanoparticles via synergistic action of pH-sensitive fusogenic peptide derivatives. Anal. Bioanal. Chem. 2008;391:2717–2727. doi: 10.1007/s00216-008-2012-1. [DOI] [PubMed] [Google Scholar]
- SHRESTHA R. ELSABAHY M. FLOREZ-MALAVER S. SAMARAJEEWA S. WOOLEY K.L. Endosomal escape and siRNA delivery with cationic shell crosslinked knedel-like nanoparticles with tunable buffering capacities. Biomaterials. 2012;33:8557–8568. doi: 10.1016/j.biomaterials.2012.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIMOES S. SLEPUSHKIN V. PIRES P. GASPAR R. DE LIMA M.P. DUZGUNES N. Mechanisms of gene transfer mediated by lipoplexes associated with targeting ligands or pH-sensitive peptides. Gene Ther. 1999;6:1798–1807. doi: 10.1038/sj.gt.3301015. [DOI] [PubMed] [Google Scholar]
- SUBBARAO N.K. PARENTE R.A. SZOKA F.C., JR. NADASDI L. PONGRACZ K. pH-dependent bilayer destabilization by an amphipathic peptide. Biochemistry. 1987;26:2964–2972. doi: 10.1021/bi00385a002. [DOI] [PubMed] [Google Scholar]
- SUN G. HAGOOLY A. XU J. NYSTROM A.M. LI Z. ROSSIN R. MOORE D.A. WOOLEY K.L. WELCH M.J. Facile, efficient approach to accomplish tunable chemistries and variable biodistributions for shell cross-linked nanoparticles. Biomacromolecules. 2008;9:1997–2006. doi: 10.1021/bm800246x. [DOI] [PubMed] [Google Scholar]
- UKAWA M. AKITA H. MASUDA T. HAYASHI Y. KONNO T. ISHIHARA K. HARASHIMA H. 2-Methacryloyloxyethyl phosphorylcholine polymer (MPC)-coating improves the transfection activity of GALA-modified lipid nanoparticles by assisting the cellular uptake and intracellular dissociation of plasmid DNA in primary hepatocytes. Biomaterials. 2010;31:6355–6362. doi: 10.1016/j.biomaterials.2010.04.031. [DOI] [PubMed] [Google Scholar]
- VARKOUHI A.K. SCHOLTE M. STORM G. HAISMA H.J. Endosomal escape pathways for delivery of biologicals. J. Control Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- VERCAUTEREN D. VANDENBROUCKE R.E. JONES A.T. REJMAN J. DEMEESTER J. DE SMEDT S.C. SANDERS N.N. BRAECKMANS K. The use of inhibitors to study endocytic pathways of gene carriers: optimization and pitfalls. Mol. Ther. 2010;18:561–569. doi: 10.1038/mt.2009.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG J. LU Z. WIENTJES M.G. AU J.L. Delivery of siRNA therapeutics: barriers and carriers. AAPS J. 2010;12:492–503. doi: 10.1208/s12248-010-9210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG Y. MANGIPUDI S.S. CANINE B.F. HATEFI A. A designer biomimetic vector with a chimeric architecture for targeted gene transfer. J. Control Release. 2009;137:46–53. doi: 10.1016/j.jconrel.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WATTS J.K. COREY D.R. Clinical status of duplex RNA. Bioorg. Med. Chem. Lett. 2010;20:3203–3207. doi: 10.1016/j.bmcl.2010.03.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHITEHEAD K.A. LANGER R. ANDERSON D.G. Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAMADA Y. SHINOHARA Y. KAKUDO T. CHAKI S. FUTAKI S. KAMIYA H. HARASHIMA H. Mitochondrial delivery of mastoparan with transferrin liposomes equipped with a pH-sensitive fusogenic peptide for selective cancer therapy. Int. J. Pharm. 2005;303:1–7. doi: 10.1016/j.ijpharm.2005.06.009. [DOI] [PubMed] [Google Scholar]
- ZHANG K. FANG H. CHEN Z. TAYLOR J.S. WOOLEY K.L. Shape effects of nanoparticles conjugated with cell-penetrating peptides (HIV Tat PTD) on CHO cell uptake. Bioconjug. Chem. 2008;19:1880–1887. doi: 10.1021/bc800160b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG K. FANG H. SHEN G. TAYLOR J.S. WOOLEY K.L. Well-defined cationic shell crosslinked nanoparticles for efficient delivery of DNA or peptide nucleic acids. Proc. Am. Thorac. Soc. 2009a;6:450–457. doi: 10.1513/pats.200902-010AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG K. FANG H. WANG Z. LI Z. TAYLOR J.S. WOOLEY K.L. Structure-activity relationships of cationic shell-crosslinked knedel-like nanoparticles: shell composition and transfection efficiency/cytotoxicity. Biomaterials. 2010;31:1805–1813. doi: 10.1016/j.biomaterials.2009.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG K. FANG H. WANG Z. TAYLOR J.S. WOOLEY K.L. Cationic shell-crosslinked knedel-like nanoparticles for highly efficient gene and oligonucleotide transfection of mammalian cells. Biomaterials. 2009b;30:968–977. doi: 10.1016/j.biomaterials.2008.10.057. [DOI] [PubMed] [Google Scholar]
- ZHANG K. ROSSIN R. HAGOOLY A. CHEN Z. WELCH M.J. WOOLEY K.L. Folate-mediated cell uptake of shell-crosslinked spheres and cylinders. J. Polym. Sci. A Polym. Chem. 2008b;46:7578–7583. doi: 10.1002/pola.23020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG X.-X. ALLEN P.G. GRINSTAFF M. Macropinocytosis Is the major pathway responsible for DNA transfection in CHO cells by a charge-reversal amphiphile. Mol. Pharm. 2011;8:758–766. doi: 10.1021/mp100366h. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.