Abstract
RNA interference technology is becoming an integral tool for target discovery and validation.; With perhaps the exception of only few studies published using arrayed short hairpin RNA (shRNA) libraries, most of the reports have been either against pooled siRNA or shRNA, or arrayed siRNA libraries. For this purpose, we have developed a workflow and performed an arrayed genome-scale shRNA lethality screen against the TRC1 library in HeLa cells. The resulting targets would be a valuable resource of candidates toward a better understanding of cellular homeostasis. Using a high-stringency hit nomination method encompassing criteria of at least three active hairpins per gene and filtered for potential off-target effects (OTEs), referred to as the Bhinder–Djaballah analysis method, we identified 1,252 lethal and 6 rescuer gene candidates, knockdown of which resulted in severe cell death or enhanced growth, respectively. Cross referencing individual hairpins with the TRC1 validated clone database, 239 of the 1,252 candidates were deemed independently validated with at least three validated clones. Through our systematic OTE analysis, we have identified 31 microRNAs (miRNAs) in lethal and 2 in rescuer genes; all having a seed heptamer mimic in the corresponding shRNA hairpins and likely cause of the OTE observed in our screen, perhaps unraveling a previously unknown plausible essentiality of these miRNAs in cellular viability. Taken together, we report on a methodology for performing large-scale arrayed shRNA screens, a comprehensive analysis method to nominate high-confidence hits, and a performance assessment of the TRC1 library highlighting the intracellular inefficiencies of shRNA processing in general.
Introduction
In 2006, Fire and Mello won the Nobel Prize in Physiology or Medicine for their discovery that double-stranded RNA molecules can trigger suppression of gene expression at the mRNA level; a process later coined RNA interference (RNAi). This discovery was accompanied by a vast new world of tiny regulatory RNA molecules that are profoundly changing the way we think about target discovery and gene regulation. Almost a decade later, RNAi as a screening technology platform has opened the door to functional genomics screen approaches to discover, validate current targets, or merely to elucidate gene function within pathways and signaling networks.1 This has allowed scientists to perform up to genome-wide simultaneous gene knockdowns at an industrial scale against arrayed or pooled RNAi libraries, and with a battery of assay readouts ranging from simple cell metabolic measurements to highly sophisticated deep sequencing methods.
Mammalian RNAi screens have been conducted in various fields and largely focused on cancer biology addressing a wide diversity of biological questions. To date, we estimate ∼300 RNAi screens have been reported in the literature ranging from basic research questions to studies of various biological processes, such as stem cell biology and host–pathogen interactions. Surprisingly, only a handful of arrayed short hairpin RNA (shRNA) screens were reported among the 300 list questioning the technical feasibility of performing such screens in terms of libraries, methodology, workflow, and screening data analysis. Among them were the following studies: (1) viability screen for oncogenic KRAS dependency against a TRC (The RNAi Consortium) library covering 1,000 human genes,2 (2) viability screen for genetic vulnerabilities across multiple cancer cell lines against a TRC library covering 1,000 human genes,3 (3) viability screen for mitochondrial dependence upon mTORC2 addiction performed against a TRC1 library covering 14,308 murine genes,4 and (4) viability screen for genetic vulnerabilities in a multiple myeloma cell line against a TRC library covering 1,000 human genes.5 Of note, the first published pooled genome-wide siRNA screen was a synthetic lethal assay in response to treatment with paclitaxel and led to the nomination of 87 hits;6 despite such an endeavor, concerns over hit identification and nomination, for follow-up confirmatory studies, have been recently raised.7–10 This is reflected in some examples, where despite the use of identical RNAi libraries and similar cell lines, gene targets reported in some of these reports were found to be oceans apart, highlighting discordance in screening data output in support of the raised concerns.2,3,5,8,11 Thus, while RNAi screening offers the premise to systematically identify genes involved in and associated with a given biological function or process as a new dimension to our understanding of biology, there are currently some inherent limitations preventing the technology from fulfilling its ultimate promise of providing new tools for medicine to tackle diseases; due, in part, to the current practices of hit nomination lacking off-target effects (OTEs) filtering.1,8,12
RNAi screening workflow can be divided into three critical and essential areas, all relevant to potential high-confidence hit nomination: first, RNAi library choice and cargo delivery to cells; second, assay development, optimization, and readout considerations; and finally, screening data analysis: active identification, hit gene nomination, and most importantly ruling out the different facets of OTEs associated with the technology. We explored the RNAi library choice by opting for a lentiviral-enabled shRNA library arrayed in 384-well microtiter plates; we reasoned that an arrayed format has several advantages as compared to the commonly used pooled approaches in that cargo delivery through viral transduction offers a much more controlled delivery of one hairpin per well, and yielding stable gene silencing even in those challenging cell types, such as primary cells, stem cells, or neurons as opposed to transient knockdown observed for siRNA duplexes. The arrayed as opposed to the pooled approaches eliminate the need and tedious tasks of deconvolution of active shRNA hairpins; and potentially allow for a more robust hit nomination through individual hairpin scoring and OTE filtering at the hairpin sequence level; a task impossible to perform for pooled shRNA libraries.
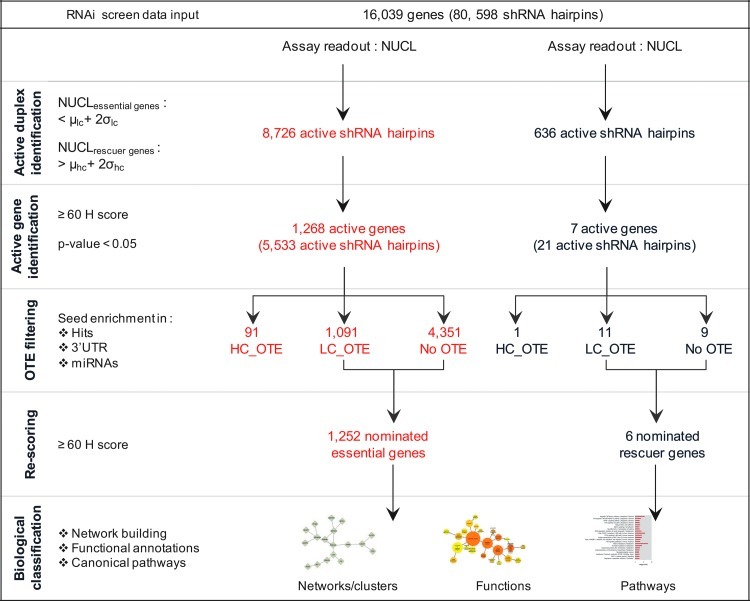
To this end, we performed an arrayed genome-wide shRNA lethality screen against the TRC1 library of 80,598 hairpins targeting 16,039 genes with an average coverage of five hairpins per gene to profile essential genes in HeLa cells; and scored using automated whole-well microscopy and quantification of Hoechst-stained nuclei as the assay readout, based on residual nuclei over a 10-day incubation period. We applied a high-stringency hit nomination method encompassing criteria of at least three active hairpins per gene and filtered for potential OTEs, referred to as the Bhinder–Djaballah analysis (BDA) method,12 and leading to the identification of 1,252 lethal candidates; the knockdown of which resulted in severe growth inhibition, and 6 rescuer candidates; the knockdown of which enhanced cellular growth. In this report, we focus on thoroughly describing the systematic workflow we have built to develop and execute on arrayed shRNA genome-wide lethality screens, followed by the application of the BDA method throughout the data analysis process to filter out all the different facets of OTEs, such as miRNA sequence mimics and/or 3′ UTR seed sequence enrichment, inherent, but mostly ignored problems in RNA screening data analysis; we also discuss the TRC1 library performance and provide a list of 239 gene candidates deemed independently validated as a resource to be used toward fundamental system's biology understanding of gene regulation for cellular homeostasis.
Materials and Methods
Cell Culture and Materials
HeLa cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1 mM glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin, and grown under a humidified atmosphere of 5% CO2–95% air at 37°C as previously described.13 Cell culture supplies were purchased from Life Technologies (Carlsbad, CA) and Sigma-Aldrich (St Louis, MO). Paraformaldehyde was purchased from Electron Microscopy Science (Hatfield, PA).
Liquid Dispensing and Automation System
Several liquid dispensing devices were used throughout this study. Lentiviral particles were plated and transferred using a 384 stainless steel head with disposable low-volume polypropylene tips on a PP-384-M Personal Pipettor (Apricot Designs, Monrovia, CA). The addition of cell suspensions and growth media was performed using the Multidrop 384 (Thermo Fisher Scientific, Waltham, MA). Cell fixation and staining was performed using the ELx405 automated washer (Biotek, Winooski, VT). Assay plates were incubated in the Cytomat 6001 H automated incubator (Thermo Fisher Scientific) under controlled humidity at 37°C and 5% CO2–95% air. The screen was performed on a fully automated linear track robotic platform (Thermo Fisher Scientific) using several integrated peripherals for plate handling, cell incubators, liquid dispensing, and detection systems.
Arrayed Lentiviral Particle shRNA Library
The arrayed TRC1 library is a hairpin-based library comprising of a 21-base stem and a 6-base loop with each hairpin cloned into the pLKO.1 vector harboring a puromycin drug resistance marker.14 The library contains 80,598 shRNA hairpin clones targeting 16,039 human genes with an average coverage of 5 hairpins per gene. Each hairpin oligonucleotides sequence design has a 21-nucleotide (nt) passenger strand, a 6-nt loop, and followed with a 21-nt guide strand (Fig. 1A). The design called for a 4-nt overhang on both the 5′ and 3′ ends, with the following sequences CCGG and TTTT, respectively. A sequence analysis revealed that the 6-nt loop is highly conserved across the whole library and with a sequence motif of CTCGAG found in 99.9% of the shRNA hairpins. High-throughput lentiviral production of each hairpin was outsourced to Sigma-Aldrich, resulting in several copies of the library arrayed in 384-well microtiter plates and in a transduction-ready assay-plate format. The lentiviral particles harboring shRNA hairpins were pre-arrayed as single clones per well into 295 intermediate 384-well microtiter polypropylene plates (ABgene, Rockford, IL), leaving columns 13 and 14 empty for control additions, and with built-in puromycin selection control wells in rows Owells:15–24 and Pwells:15–24. The library is stored at −80°C. The overall lentivirus titer yields were at least 106 TU/mL, a yield sufficient enough to achieve multiplicity of infectivity (MOI) up to five per screening campaign.
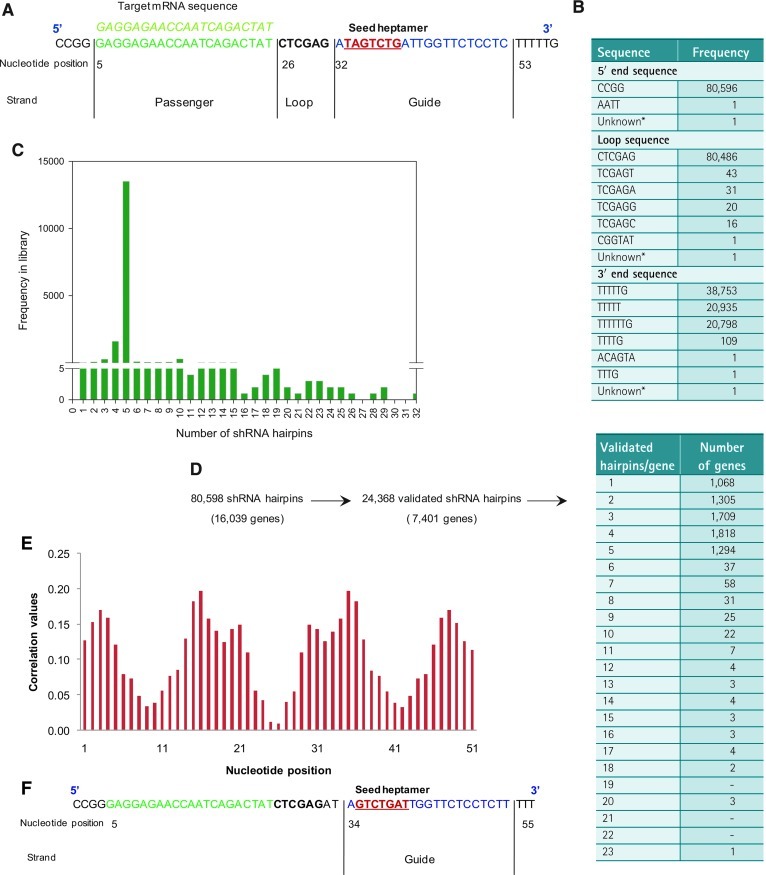
Fig. 1.
shRNA hairpin design, TRC1 library attributes, and 7-mer seed selection for off-target effect (OTE) analysis. (A) A graphical representation of the shRNA hairpin oligonucleotide sequence showing the start position of the seed heptamer as per the ideal assumption method. (B) Hairpin loop, 5′ overhang and 3′ overhang sequence design and representation in the TRC1 library of 80,598 shRNA hairpins. (C) Frequency distribution of number of hairpins per gene in the TRC1 library (D) The TRC1 library validation data obtained from Sigma-Aldrich showing the number of independently validated shRNA hairpins and their respective genes. (E) Hairpin sequence analysis for optimal seed sequence determination using the empirical seed selection (ESS) method. (F) A graphical representation of the shRNA hairpin oligonucleotide sequence showing the start position of the seed heptamer as per the ESS method.
Control Lentiviral Particles
The following lentiviral particles were used as negative transduction controls: a pLKO.1-puro nontargeting shRNA control (SHC002V) harboring a unique shRNA hairpin control designed using an internal scrambled sequence and a pLKO.1-puro empty vector control (SHC001V). As a functional-positive control for transduction efficiency, lentiviral particles harboring shRNA hairpins against PLK1 (TRCN0000121325, TRCN0000121072, and TRCN0000199639) were used. Transduction efficiency was also assessed using lentiviral particles harboring the pLKO.1-puro-CMV-TurboGFP expressing vector (SHC003V).
Assay Development and Optimization
Assay development and optimization were carried out as a matrix format-based design in 384-microtiter plates, encompassing time course experiments for cell growth kinetics, polybrene tolerance, puromycin killing curve, shRNA transduction efficiency assessment, and control shRNA performances. Where indicated, cell seeding densities and dose–response reagent additions were performed as doubling serial dilutions. All transduction experiments were conducted by including an 8-min spin performed on a bench top centrifuge at room temperature to facilitate viral particle integration host cells. This matrix-based design of experiment allows for a rapid assessment of best conditions for the screen as well as taking into account a cell growth time course.
Genome-wide shRNA Screen for Essential Genes
For the arrayed genome-wide screen, the preplated shRNA library arrayed in 295 intermediate plates was first thawed from storage at room temperature, the plates were then spun for 1 min at a speed of 130 g on a bench top centrifuge, after which 4 μL was transferred into the designated assay plate, seeded on the previous day with 500 HeLa cells in 45 μL of growth media supplemented 8 μg/mL polybrene, and achieving a final MOI of 5. The assay plates were then centrifuged for 8 min at a speed of 340 g and transduced assay plates were incubated for 2 days in dedicated Cytomat automated incubators (Thermo Fisher Scientific) under controlled humidity at 37°C and 5% CO2–95% air. At day 3 post-transduction, a virus-containing medium was swapped with 50 μL of fresh growth medium supplemented with 3.2 μg/mL puromycin for selection. Assay plates under puromycin selection were incubated for another 4 days at which time, the selection medium was swapped with 50 μL of fresh growth medium and cells were left to recover for one more day. Cells were then fixed and stained followed by imaging of Hoechst-stained nuclei. The total run time for the assay is 8 days. Of note, each assay plates contained high- and low-control (LC) wells; high-control (HC) wells are those wells in column 14 and contained puromycin-untreated HeLa cells. The LC wells are those wells in rows Owells:15–24 and Pwells:15–24 and contained puromycin-treated HeLa cells; thus, we have 16 HC and 20 LC wells per assay plate screened.
Automated Image Acquisition and Analysis
Images of stained cells in 384-well microplates were acquired using the IN Cell Analyzer 2000 (INCA2000; GE Healthcare, Piscataway, NJ) as previously described.15,16 The INCA2000 is a wide-field automated epifluorescence microscope equipped with a large chip CCD camera allowing for whole-well imaging. The 4× magnifying objective with Plan Apo, chromatic aberration-free infinity (CF160), and 0.20NA was used. Images were acquired using a custom-made polychroic. Images of Hoechst-stained nuclei in the 4′,6-diamidino-2-phenylindole channel were acquired using 350/50 nm excitation and 437/58 nm emission at an exposure time of 100 ms. Images of resulting wells during transduction assessment were acquired for green fluorescent protein (GFP) in the fluorescein isothiocyanate channel using 490/20 nm excitation and 525/36 nm emission at 450 ms. One tile was imaged per well covering 100% of the well, with an acquisition time of 4 s per well and total of 25.6 min per 384-well microtiter plate. Images acquired by the INCA2000 were analyzed with the Developer Toolbox 1.7 software (GE Healthcare) using a custom-developed image analysis protocol as previously described.15,16 Hoechst-stained nuclei were identified after post processing using object-based segmentation on the blue channel. Automated image analysis using our custom-developed protocol allowed us to extract nuclei count used for quantification of remaining cells as a cytotoxicity index referred to as NUCL.
The Z′ Factor and the Signal-to-Background Ratio
The Z′ factor was used to determine the overall assay performance during the screen. The Z′ factor constitutes a dimensionless parameter that ranges from 1 (infinite separation) to <0. The Z′ factor is defined as Z′=1–(3σc++3σc−)/|μc+−μc−|, where σc+, σc−, μc+ and μc− are the standard deviations (σ) and averages (μ) of the high (C+) and low (C−) control wells.17 The Z′ factor between 0.5 and 1 indicates an excellent assay with good separation between controls. The Z′ factor between 0 and 0.5 indicates a marginal assay and <0 signifies a poor assay with overlap between positive and negative controls. The signal-to-background ratio (S/B) is defined as S/B=μc+/μc−, where μc+ and μc− are the averages (μ) of the high (C+) and low (C−) control wells, respectively. An S/B ratio of >3 is considered as a good assay window separation indicator.16,17
Screening Data Analysis Using the BDA Method
Active duplex scoring
The genome-wide screen performance was scored using the cytotoxicity index of residual Hoechst-stained nuclei count per well referred to as NUCL. To identify active shRNA hairpins, which cause severe growth inhibition of HeLa cells, individual shRNA hairpins were first scored for their activity based on a NUCL threshold determined at +2 standard deviations (σ) from the mean of the LC wells. In parallel, the shRNA hairpins that conferred growth advantages were scored for their activity based on a NUCL threshold determined at +2σ from the mean of HC wells. The outliers in the control data were identified and removed using the interquartile range before determining NUCL thresholds. The genes corresponding to the active shRNA hairpins were subjected to a breakpoint analysis as previously described.12
Initial gene hit nomination
The active genes were nominated from the active shRNA hairpin list using a hit rate per gene score (H score) with a high-stringency threshold of ≥60 as previously described.12 The gene coverage of the TRC1 library is inherently heterogeneous:14 we find that 4% of the genes have more than five hairpins (and up to 32 in some cases), 14% of them have less than five hairpins with some covered by only one hairpin, and the remaining 84% of the genes have an exact coverage of five hairpins. Considering the inherent gene coverage heterogeneity of the TRC1 library, our H score evaluation caters to the majority of genes in the TRC1 library. However, for those minority genes with more than five hairpins of coverage, we have also made provisions to the H score analysis whereby a t-test is performed to determine if the performance of the active duplexes was significantly different from the performance of the inactive ones.12 The threshold for the P values was set at <0.05; those genes with at least three active shRNA duplexes that met these criteria were considered and nominated as active genes. The active gene candidates that led to a significant growth inhibition in HeLa cells were classified as “essentials” while those conferring growth advantages were classified as “rescuers” and will be referred to as such for the rest of this report. Statistical analysis for hit selection was performed using Perl scripts and Sigmaplot (SYSTAT, San Jose, CA).
OTE filtering
The overall active shRNA hairpins for both classes were further assessed for OTEs. The OTE activity was determined based on the 7-mer seed sequence in the antisense (guide) strand. The OTE analysis was performed twice based on the two different methods of seed sequence selection: (1) seed sequence starting from nt position 33 as determined by its ideal location on the hairpin oligonucleotide sequence and (2) seed sequence starting from nt position 35 as determined empirically using the empirical seed selection (ESS) method.12 The union of the results from both of the above methods was considered for OTE filtering. OTEs were then scored on the basis of three criteria:
Seed sequence enrichment among active hairpins using the whole library as a background and the threshold was set at a hypergeometric P value of <0.05.
Seed sequence enrichment in the human 3′UTR sequences obtained from the University of California at Santa Cruz (UCSC) genome browser18 from human genome assembly GRCh37/hg19 (genome.ucsc.edu); the threshold was set at >10% enrichment in 3′UTR sequences.12 The 3′UTR sequences <10 nt in length were excluded from the analysis.
Seed sequence enrichment in human miRNA sequences obtained from miRbase release 1819; the threshold was at least a single exact seed sequence. The information on the experimentally validated miRNA targets was obtained from TARBASE.20
The active shRNA hairpins that qualified in all three criteria were deemed as high-confidence OTEs (HC_OTE), and eliminated from further consideration; for those that qualified in at least two criteria were flagged as low-confidence OTEs (LC_OTE) and kept for further analysis as previously described.12
Rescoring and final hit nomination
The active shRNA hairpins selected as HC_OTEs were removed from subsequent analysis. The active gene list was then rescored for the remaining active shRNA hairpins deemed not affected by OTEs. After rescoring, the gene candidates that failed to meet the criterion of an H score of ≥60 were filtered out as false positives due to OTEs. For those genes which had an H score of <60, but ≥4 active duplexes, a t-test was performed to determine if the performance of the active duplexes was significantly different from the performance of the inactive duplexes as previously reported.12 The threshold for P values was set at <0.05 and those genes with ≥4 active duplexes that met these criteria were considered active. The remaining genes constituted the final list of nominated essential and rescuer gene candidates.
Biological classification
Biological classifications were performed to identify enrichments in the following three categories: (1) over-connected gene clusters, (2) functional classes, and (3) canonical pathway associations as previously described.12 Cytoscape's MiMi plug-in was first used to create a master network from the list of nominated genes. The protein–protein interaction databases used to construct the networks in MiMI were BIND, CCSB, DIP, HPRD, KEGG, MDC, MINT, and REACTOME.21 Cytoscape's MCODE plug-in was used to find over-connected gene clusters within the master network.22 BiNGO was used to visualize enriched gene ontology (GO) categories in Cytoscape.23 Functional classes were assigned to the nominated hits based on GO term enrichments and were done using the DAVID Functional Annotation Tool24 (www.david.abcc.ncifcrf.gov). Canonical pathway analysis was done using Gene Go's Metacore software (www.genego.com/metacore.php). The threshold for the statistical level of significance was determined at a P value of <0.05. Biological functions information was also obtained using the PANTHER classification system.25
Comparative Performance Analysis of the Human Versus the Murine TRC1 Libraries
Since Colombi and coworkers reported the only other published genome scale and arrayed shRNA screen against the murine TRC1 library, we sought to compare performances with its human counterpart since their screen readout was cellular viability using the Alamar Blue assay in the murine Pten mutant cell line 6.5. A list of the 800 essential genes from the Colombi screen was obtained for the analysis.4 17,777 human orthologs for mouse genes were obtained from the Mouse Genome Informatics (MGI) database for mammalian orthology in the form of gene symbols and their corresponding Entrez ids.26,27 The overlap analysis was performed only between those genes for which an ortholog was found and a match was determined based on the Entrez gene IDs obtained from MGI data set. A hypergeometric P value was calculated for the number of common genes28 based on the probability of finding an overlap at least as extreme as observed between the two lists being compared; and the two lists were assumed to be derived from the pool of 17,777 known orthologs. The P value threshold for rejecting the null hypothesis (H0) of random overlap was selected at <0.05. The P value was calculated using Perl.
Results
The TRC1 Library Attributes
The TRC1 shRNA hairpin-based library was constructed within the lentivirus plasmid vector pLKO.1-puro with hairpin sequences designed and developed by The RNAi Consortium at the Broad Institute.14 Each hairpin oligonucleotide sequence design has a 21-nt passenger strand, a 6-nt loop, and followed with a 21-nt guide strand (Fig. 1A). The design seems to have a 4–5-nt overhang on either or both the 5′ and 3′ ends, with the following predominant sequence at the 5′ end of CCGG (99.99%), and either TTTTTG (48.08%), TTTT (25.97%), TTTTTTG (25.80%), or TTTTG (0.13%) at the 3′ end of the hairpin oligonucleotide sequence. The predominant loop sequence is CTCGAG with 99.86% abundance (Fig. 1B). The library contains 80,598 hairpin clones targeting 16,039 genes with the following coverage of 12% of those with less than 5 hairpins, 84% with 5 hairpins, and 4% with greater than 5 and up to 32 hairpins per gene (Fig. 1C). The knockdown validation of the TRC1 hairpin clones was provided by Sigma-Aldrich in a joint collaboration with the Broad Institute. The knockdown of a given gene was measured by SYBR green-based real-time quantitative PCR (RT-qPCR) relative to a nontargeting control clone in lentivirus transduced cells. The threshold of acceptance of a validated clone was set at a 50% reduction of the mRNA expression as compared to the control. Based on the knockdown data validation provided by Sigma-Aldrich, 22,368 out of 80,598 clones of the TRC1 library were deemed validated; that is 30% of the library, and translating into 7,401 out of 16,039 genes as independently validated (Fig. 1D). By applying a more stringent validation criterion of at least three validated hairpin clones per gene, only 5,028 genes met the criterion and are deemed high-confidence validated genes in the TRC1 library (Fig. 1D).
Seed Heptamer Selection in shRNA Hairpin Oligonucleotide
Seed was defined as the heptamer nt sequence from nt position two to eight on the guide strand from its 5′ end. Based on the target sequence reverse complementary match, an ideal start location of the seed heptamer was determined at nt position 33 of the shRNA hairpin oligonucletotide (Fig. 1A). We have also used the ESS method to determine the seeds on the oligonucleotide, which empirically determined the seed sequence based on the correlation between the seed, start position, and the screen data output to yield an optimal location of the seed heptamer in a processed shRNA duplex.12 This method generates a list of correlation values associated with each nt position on the shRNA hairpin oligonucleotide and the results can be visualized as a distribution histogram with two peaks representing the most probable start position of the seed heptamer on the oligonucleotide (Fig. 1E). One of the peaks is for the guide strand, while the other one is for the passenger strand and the two peaks occur twice; thus, four in total, due to the sequence symmetry of the oligonucleotide. Applying the ESS method to our screen data output of raw NUCL, the guide strand peak was found at nt position 35, indicative of the start position of the seed heptamer (Fig. 1F), which is two nt positions away from the ideal seed heptamer start position at nt position 33. Taken together, we have generated two lists of the seed heptamers, one selected based on the ideal seed location, while the other selected based on the ESS method and have conducted the OTE filtering analysis separately for these two lists. The results thus obtained were merged to get a final list of HC_OTEs that were removed from subsequent analysis.
Automated Microscopy-Based shRNA Assay Development Workflow
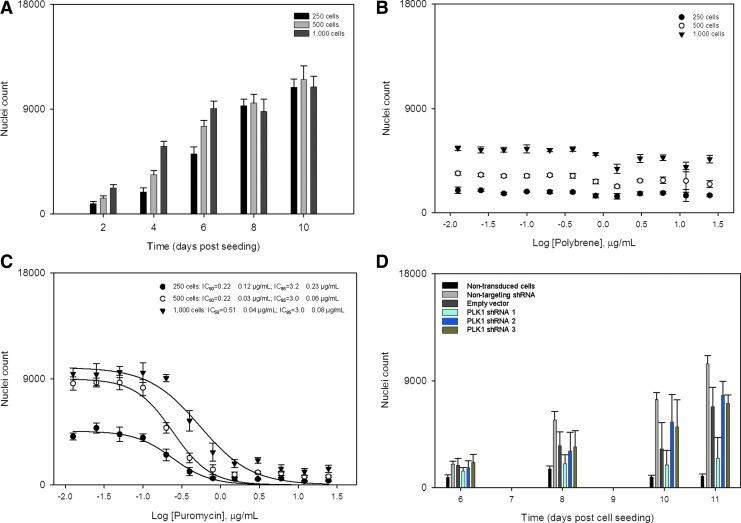
The various assay parameters were optimized in a 384-well format to determine the optimal assay conditions for the screen. HeLa cells were seeded at three densities of 250, 500, and 1,000 cells per well and allowed to grow for up to 10 days while monitoring their growth; linear growth was observed mostly for the 500 cell seeding density (Fig. 2A, Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/adt). The effects of polybrene and puromycin on nuclei count were assessed at the same cell densities and at concentrations ranging from 12 ng/mL to 25 μg/mL for both. Polybrene was well tolerated in the assay across the three cell seeding densities and over 4 days (Fig. 2B). Puromycin, on the other hand, was found to be cytotoxic as expected allowing us to generate a kill curve for each cell density tested over 4 days post puromycin addition (Fig. 2C). Calculated IC50 values for the puromycin kill curve were in the 200–500 ng/mL range; whereas the IC95 values were in the 3 μg/mL range (Fig. 2C). Our optimization efforts led to the following assay conditions for the subsequent studies: a cell seeding density of 500 cells per well in a growth media supplemented with 8 μg/mL polybrene; and a puromycin selection concentration of 3 μg/mL.
Fig. 2.
Design of experiment to optimize experimental assay conditions for the screen. (A) HeLa cell density growth kinetics at three seeding densities of 250, 500, and 1,000 cells per well, (B) dose–response studies of HeLa cells tolerance to polybrene addition, (C) dose–response studies of puromycin kill curve of HeLa cells estimating the IC50 and IC95 values, and (D) control shRNA hairpin transduction experiments using HeLa cell density of 500 cells per well in 8 μg/mL polybrene. Experiments were conducted by seeding HeLa cells as indicated in 384-well microtiter tissue-treated plates and monitoring their growth over a course of up to 10 days as indicated. Cell fixing, imaging, and nuclei quantification were performed as described in Material and Methods.
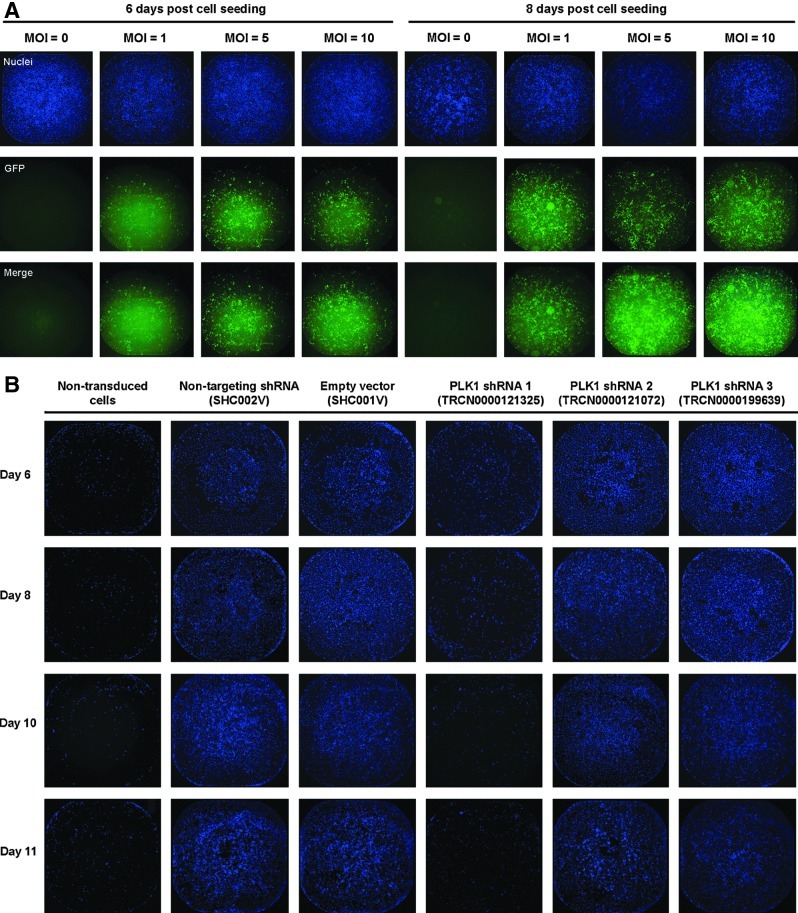
To determine the optimal MOI for use in the screen, we performed transduction experiments using lentiviral particles harboring the pLKO.1-puro-CMV-TurboGFP expressing vector and scoring for GFP fluorescence at three MOI values of 1, 5, and 10. We did not observe much difference in GFP intensities for MOIs 5 and 10 at either 6 or 8 days post cell seeding (Fig. 3A) translating into transduction efficiencies of up to 50%. Finally, we performed a small validation time course of control lentiviral particles transduction for 2 days followed by puromycin selection for up to 7 days at three MOIs of 1, 5, and 10; during which we assessed the optimal time for selection followed by 1 day cell recovery (Supplementary Fig S2). The negative control particles (scrambled and empty pLKO.1 vector) exhibited puromycin resistance at a concentration of 3 μg/mL; whereas the functional positive control particles targeting PLK1 had one shRNA hairpin active (TRCN0000121325) killing cells as expected (Fig. 2D). The two other hairpins targeting PLK1 (TRCN0000121072 and TRCN0000199639) were found to be inactive in our assay. Resulting whole-well images of Hoechst-stained nuclei for the transduction at an MOI of 5 are shown (Fig. 3B). The resulting optimized assay conditions call for seeding 500 cells in 8 μg/mL polybrene for 1 day, transducing the cells at an MOI of 5 for 2 days, puromycin selection at a concentration of 3 μg/mL for 4 days; let cells recover in fresh growth media for 1 day before fixing, staining the nuclei, and image acquisition on the INCA2000 (Table 1).
Fig. 3.
Whole-well images of control lentiviral particle transduction efficiency assessment. (A) HeLa cells were transduced with lentiviral particles harboring the pLKO.1-puro-CMV-TurboGFP expressing vector at three different multiplicity of infectivity (MOI) values of 1, 5, and 10. Resulting images for enhanced GFP production and Hoechst-stained nuclei at either 6 or 8 days post cell seeding are shown. (B) Resulting nuclei-stained images from the control shRNA hairpin transduction and performance at an MOI of 5 and over a time course of up to 11 days post cell seeding and puromycin selection. Cell fixing, imaging, and nuclei quantification were performed as described in Material and Methods. GFP, green fluorescent protein.
Table 1.
Assay Workflow Using the Optimized Conditions and Experimental Steps for the Execution of the Genome-Wide shRNA Lethality Screen in HeLa Cells
| Step | Parameter | Value | Description |
|---|---|---|---|
| 1 | Cell plating | 45 μL | 500 cells per well in growth media containing 8 μg/mL polybrene |
| 2 | Incubation time | 24 h | 37°C, 5% CO2, dedicated Cytomat incubator, controlled humidity |
| 3 | Cell transduction | 4 μL | Arrayed lentiviral particle shRNA library |
| 4 | Assay plate spin | 8 min | Spin assay plate on bench top centrifuge at 340 g |
| 5 | Incubation time | 48 h | 37°C, 5% CO2, dedicated Cytomat incubator, controlled humidity |
| 6 | Puromycin selection | 50 μL | Growth media containing 3 μg/mL puromycin |
| 7 | Incubation time | 96 h | 37°C, 5% CO2, dedicated Cytomat incubator, controlled humidity |
| 8 | Cell recovery | 50 μL | Growth media |
| 9 | Incubation time | 24 h | 37°C, 5% CO2, dedicated Cytomat incubator, controlled humidity |
| 10 | Fix | 50 μL | Wash twice with 1×PBS, 4% paraformaldehyde (v/v) for 20 min |
| 11 | Nuclei staining | 50 μL | 10 μM Hoechst solution containing 0.05% Triton X-100 (v/v) |
| 12 | Image acquisition | 360 nm/450 nm (ex/em) | INCA2000 automated microscope |
| 13 | Image analysis | Multiparametric analysis using Developer Toolbox 1.7 software |
Step Notes
1. Dispense cells into assay plates with Multidrop 384.
3. Transfer lentiviral particles on the PP-384-M Personal Pipettor using a custom 384 head; 30 s per plate.
6,8,10. Aspirate on the ELx405 automated washer and dispense with Multidrop 384; 1 min per plate.
11. Dispense into assay plates with Multidrop 384.
12. 4 s per well with a total imaging time of 25.6 min per plate.
13. Analysis of Hoechst-stained nuclei, 8 min per plate.
Genome-Wide shRNA Hairpin Lethality Screen Performance
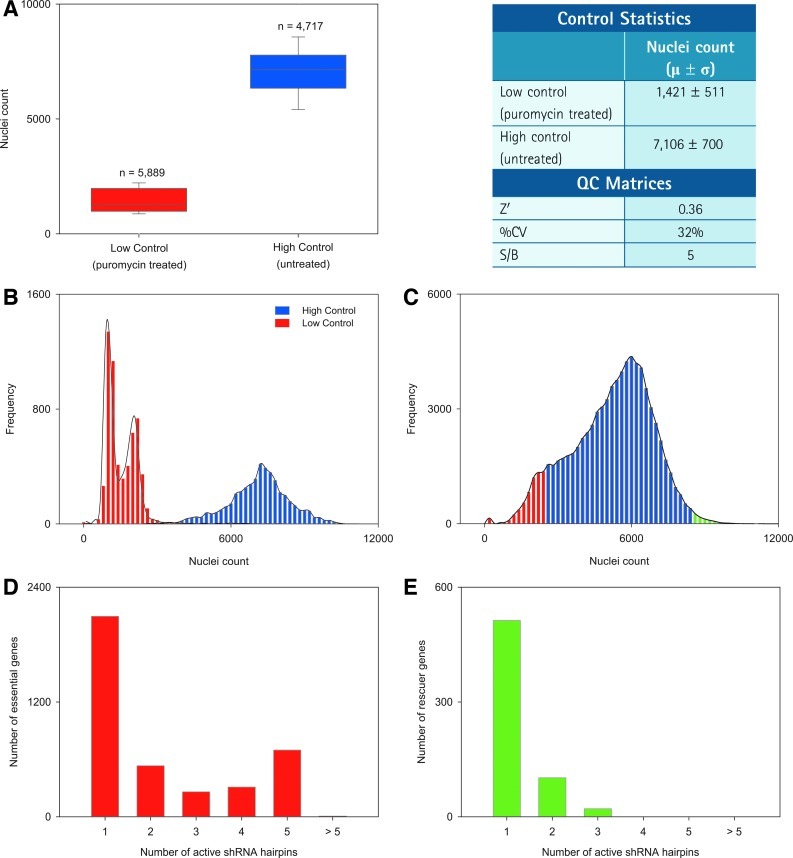
Using the optimized conditions and established workflow (Table 1), we have successfully executed on an arrayed genome-wide lethality screen against the TRC1 lentiviral library in HeLa cells to identify essential genes, the knockdown of which would result in severe growth inhibition. In addition, our image-based screening allows us to also score for rescuer genes, the knockdown of which would enhance cellular growth. The TRC1 library contains 80,598 shRNA hairpins targeting 16,039 genes and arrayed in 295 384-well microtiter plates with columns 13 and 14 left empty for puromycin-untreated HC wells and rows Owells:15–24 and Pwells:15–24 left empty for puromycin-treated LC wells, both in the absence of lentiviral-enabled shRNA transduction. The puromycin treatment was employed as the best way to define the screen boundaries, independently of lentiviral-enabled shRNA transduction, which is heterogeneous in nature. To monitor the assay's performance throughout the screen, a box plot analysis of HC and LC wells shows a good NUCL separation from an average of 1,421 for puromycin-treated wells to an average of 7,106 counts for the untreated wells (Fig. 4A), and translating into a S/B ratio of 5, which lies well within the accepted range of >3 and further ascertains the quality of the screen for the control separation window. An average Z′ value of 0.36 was obtained for the screen indicative of a marginal cell-based assay and to some extent confirming sensitivity to the puromycin selection treatment. To further assess the overall performance of the controls, a frequency distribution plot was generated showing a wider distribution in the HC wells reflective of cellular growth heterogeneity (Fig. 4B), and what appears to be a bimodal distribution in the LC wells, perhaps, reflective of differential sensitivities of the HeLa cells to puromycin (Fig. 4B); a similar plot was generated for each of the 80,598 individual shRNA hairpins in the TRC1 library showing a bimodal distribution as would be expected since not all hairpins would induce cytotoxicity effects on the HeLa cells (Fig. 4C). The hairpins that lead to severe growth inhibition in HeLa cells, therefore essential for viability, would lie to the left-hand side of the histogram (colored in red); while those which confer potential growth advantage to HeLa cells therefore rescuers, would lie to the right-hand side of the histogram (colored in green).
Fig. 4.
Assessment of the assay performance during screening. (A) Box plot and statistical analysis of control wells nuclei count showing a good signal separation. (B) Histogram distribution plot of the high and low control nuclei count wells. (C) Histogram distribution of individual shRNA hairpin effect on nuclei count for the entire library. (D) Frequency distribution of number of active hairpins per essential gene identified using the Bhinder–Djaballah analysis (BDA) method. (E) Frequency distribution of number of active hairpins per rescuer gene identified using the BDA method.
The BDA Methodology Nominates 1,252 Essential and 6 Rescuer Candidates
To score for active genes, we used the BDA method for hit nomination encompassing a stringent and systematic approach.12 In the first step of active duplex identification, we selected 8,726 individual active shRNA hairpins that scored below +2σ from the mean of the LC (Supplementary Table S1). A major portion equivalent to 67% of these active shRNA hairpins correspond to <2 active shRNA hairpins per gene, and perhaps might be associated with false positives in the screen, while the remaining 33% of these active shRNA hairpins corresponded to ≥3 active shRNA hairpins per gene (Fig. 4D). In the second step of active gene identification, we calculated H score values for each gene based on the corresponding active shRNA hairpins and nominated a gene as active with an H score value of ≥60. We found that 6 of the genes with an H score of ≥60 had only <2 targeting shRNA hairpins in total in the TRC1 library; and these genes were excluded from the active gene identification step to maintain a consistent stringency in the hit identification process. As described earlier, the TRC1 library contains 4% of genes with coverage of >5 shRNA hairpins; which are likely to skew the results giving false negatives. Accordingly, we identified 13 such genes, which had a total of >5 shRNA hairpins in the TRC1 library out of which >4 shRNA hairpins scored as actives. To minimize the false negatives, these 13 genes were subjected to an additional analysis parameter according to which a P value was calculated by applying a t-test for the difference in means of the NUCL between the active versus the nonactive duplexes for these genes and a P value threshold was set at <0.05. Finally, 1,255 genes were selected based on an H score of ≥60 and 13 genes were selected based on a P value of <0.05. In total, we have identified 1,268 essential gene candidates with an overall hit rate of 8%. The remaining 2,638 genes fell to meet our stringent criteria and, hence, filtered out as inactive genes from the subsequent analysis.
In the third step of OTE filtering, the seed sequences of the 5,533 active shRNA hairpins corresponding to the 1,268 active genes were selected and scored for their enrichments in three categories: (1) hits versus library, (2) 3′UTR sequences, and (3) miRNA sequences. The seed sequence was found based on two methods as previously described;12 in short, (1) the ideal location of the seed on an oligonucleotides sequence as determined by its reverse complementary match with the given target sequence, and (2) using the ESS method. Based on a stringent threshold of qualification in all of the above criteria, 91 shRNA hairpins were selected as HC_OTEs and removed from subsequent analysis (Supplementary Table S2). There were 1,091 shRNA hairpins that qualified in at least two criteria and were therefore deemed as LC_OTEs, but were kept in the list of active hairpins. The remaining 4,351 shRNA hairpins did not have an obvious OTE based on our filtering criteria and were retained in the list of active shRNA hairpins. In the fourth step of rescoring, the remaining active shRNA hairpins after filtering out for the HC_OTEs were subjected to re-calculation of the H score with a threshold set at ≥60, thus yielding a final list of 1,252 nominated essential gene candidates (Supplementary Table S3).
Similarly, for the nomination of the rescuer genes, we first identified 636 individual active shRNA hairpins that scored above +2σ from the mean of the HC values (Supplementary Table S4). Like essential genes, a significant portion of 97% of these active shRNA hairpins corresponded to <2 active shRNA hairpins per gene, while the remaining 3% of these active shRNA hairpins corresponded to >3 active shRNA hairpins per gene (Fig. 4E). This was followed by active gene identification based on an H score threshold of ≥60. In total, we have identified seven candidate rescuer genes giving an overall hit rate of 0.04%, while the remaining 564 genes were filtered out. In the next step, the 21 active shRNA hairpins corresponding to the 7 active rescuer genes identified were subjected to OTE filtering, wherein 1 shRNA hairpin was selected as HC_OTE from both methods of seed selection and removed from the list of active shRNA hairpins (Supplementary Table S5). In the remainder, 11 shRNA hairpins were LC_OTE, while 9 shRNA hairpins did not qualify our selected OTE thresholds. Rescoring of these active hairpins yielded a refined list of 6 nominated rescuer gene candidates (Supplementary Table S6). The overall workflow for hit nomination using the BDA method is depicted (Fig. 5).
Fig. 5.
BDA method workflow to nominate 1,252 essential and 6 rescuer gene candidates.
OTE Filtering Revealed Potential microRNA Interference
Among the HC_OTEs identified during the OTE filtering step of the BDA method, we have found two distinct sets of miRNAs: (1) 31 unique miRNAs with seed heptamer matches in the HC_OTEs pertaining to the class of essential genes, and (2) two unique miRNAs with seed heptamer matches in the HC_OTEs pertaining to the class of rescuer genes (Supplementary Table S7). We have found no sequence overlap in the miRNA seed heptamer among the miRNAs selected in the essential versus the rescuer gene classes. Within the essential gene class, we have observed five seed heptamers (GGAGGCA, AUACAAG, CUCAGGG, UGGCCAC, and UGGGAGG) which were identical in at least two miRNAs. Briefly, some of the miRNA seed heptamers were conserved within each class, but all miRNA seed heptamers were distinct between the two classes corresponding to the active HC_OTE shRNA hairpins for the essential and rescuer genes, respectively. Perhaps, the effect on cell survival and the observed directionality in performance of the corresponding shRNA hairpins in the screen is merely due to a miRNA seed mimic-like activity, and consequentially silencing of a miRNA target. To explore this further, we searched for the experimentally validated targets of the miRNAs identified in our analysis and were able to find validated targets for only five miRNAs (hsa-let-7f-2-3p, hsa-miR-650, hsa-miR-654-3p, hsa-miR-218-5p, and hsa-miR-145; Table 2). Of note, the target genes of these five miRNAs did not match with the genes intended to be silenced by the corresponding shRNA hairpins in our screen. Furthermore, the miRNA targets obtained participate in some of the essential cellular processes. For example, three subunits of the COP9 signalosome (CSN) complex, namely, CSN1, CSN6, and CSN8, were found as known targets of hsa-let-7f, a member of the let-7 family of miRNAs, which participate in regulation of the CSN biosynthesis.29 The CSN complex is involved in the ubiquitin–proteasome pathway and plays an important role in regulation of cell cycle and gene expression.30 Also, three targets of hsa-miR-145, namely, MYC, PPP3CA, RASA1, are involved in the MAPK signaling pathway, which has an established role in cancer and various cellular processes.31 In summary, it is likely that the observed phenotype is an outcome of the perturbation of a miRNA target instead of the intended transcript, therefore accounting for an OTE, also highlighting the concerns of miRNA-based OTEs in RNAi screening.
Table 2.
Five Identified miRNAs Associated with Their Experimentally Validated Targets
| miRBase ID | Mature miRNA | Validated miRNA targetsa | Gene silenced in screen | Sequence (5'→3') |
|---|---|---|---|---|
| MIMAT0004814 | hsa-miR-654-3p | 1 gene (CDKN1A) | 4 genes (CDC34, RAMP3, IL4, SEMA6D) | UAUGUCUGCUGACCAUCACCUU |
| MIMAT0003320 | hsa-miR-650 | 1 gene (ING4) | 2 genes (LYRM1, DSCAM) | AGGAGGCAGCGCUCUCAGGAC |
| MIMAT0004487 | hsa-let-7f-2-3p | 21 genes (ASS1, CCND1, CDKNIA, CSN1, CSN6, CSN8, EEFIA2, ESM1, FDPS, IL-13, KIF1A, KLK10, KLK6, MYC, NIS, NTS, PRDM1, TFF1, TG, TITF1, VIM) | 4 genes (LILRA2, IGSF10, I L1RAP, OR2T27) | CUAUACAGUCUACUGUCUUUCC |
| MIMAT0000275 | hsa-miR-218-5p | 63 genes (ACO26713.5, ACTN1, AGPAT5, AHI1, AKAP8, ATP6V0E, BIRC6, BLCAP, BTBD7, BTF3L4, C130RF7, C5ORF5, CAMTAI, CDKNIB, CENPO, CNTNAP2, COL1A1, COL4A1, DUSP18, EBP, EFNA1, EFNB2, ETS2, FOXN2, GLCE, GPM6A, HECTD2, IKBKB, KATNALI, KIAA0391, KIAA1462, KLF9, KLHL13, LAMBS, LASP1, MAFG, MBNL2, MRPS27, NACC1, NFE2L1, NUP93, OLIG2, PHC3, POGK, POZ, PTPN11, PUM2, RARA, RHBDD1, RITZ, RNF38, SERINC3, SFRS12, SLC11A2, SLC38A1, SP1, SSH1, STAM2, THOC2, TNIP2, TPD52, TTC33, VOPP1) | 5 genes (SOX30, IL12RB2, IL18BP, MY018A, CDK10) | UUGUGCUUGAUCUAACCAUGU |
| MIMAT0000437 | hsa-miR-145 | 88 genes (ACBD3, AKR1B10, ALDH3A1, ALPPL2, AP1G1, APHIA, ARL6IP5, BNIP3, C110RF58, C110RF65, C3ORF34, C6ORF115, CBFB, CCDC25, CCDC43, CCNA2, CDKNIA, CIQTNF5, CLINTI, DDR1, DFFA DTD1, ELK-1, ERLINI, F11R, FAM108C1, FAM3C, FAM45A, FAM79B, FBXO28, FLI1, FLJ21308, FSCN1, GMFB, GOLPH2, HLTF, HOXA9, IFNBI, IGFIR, IGF-IR, IRSI, IRS-I, JAM-A, KLF4, KLF5, KREMENI, KRT7, LOC203547, LOC340888, LOC342705, LOC400011, LYPLA2, MAP2K6, MEST, MIXL1, MMP1, MMP14, MTMR14, MUC1, MYC, MYO6, NDRG2, NDUFA4, NHEDCI, NIPSNAPI, OCT4, PARP8, PHF17, PIGF, PODXL, POU5F1, PPP3CA, PTP4A2, RASA1, ROBO2, RTKN, S0CS7, SERINC5, SOX2, STAT1, SWAP70, TIRAP, TMEM9BMMP12, TMOD3, TPM3, TSPAN6, USP46, YES1) | 5 genes (RABI3, KISS1, LCP1, ENC1, KDELR1) | GUCCAGUUUUCCCAGGAAUCCCU |
Seed heptamer sequences within the mature miRNA sequence are presented in boldface.
Information on experimentally validated targets was obtained from Tarbase 6.0.20
Nominated Essential Genes Modulate Fundamental Cellular Functions
The biological classification of the 1,252 candidates revealed their involvement in some fundamental cellular process. A network analysis to identify highly interconnected gene clusters revealed 11 gene clusters (Supplementary Fig. S3). The top-scoring cluster of genes, namely cluster 1, could be broken down into two major functional categories. The first category is associated with RNA processing and is comprised of 12 genes involved in mRNA splicing predominantly via the spliceosome, while three others are involved in transcription regulation. The second category is associated with protein translation and is comprised of 12 genes encoding ribosomal proteins and five genes encoding translation initiation factors. Cluster 2 is composed of 13 components of the proteasome, which are proteolysis sites for degradation of misfolded proteins and play an essential regulatory role in vital cellular functions, such as cell cycle regulation, cellular stress, and immune response. Cluster 3 is composed of genes with oxidoreductase activity and predominantly involved in a respiratory electron transport chain. The remaining clusters were associated with functions in protein transport, endocytosis, cell–cell signaling, growth factor activity, cell communication, cytoskeleton organization, mRNA processing, cell cycle, and apoptosis.
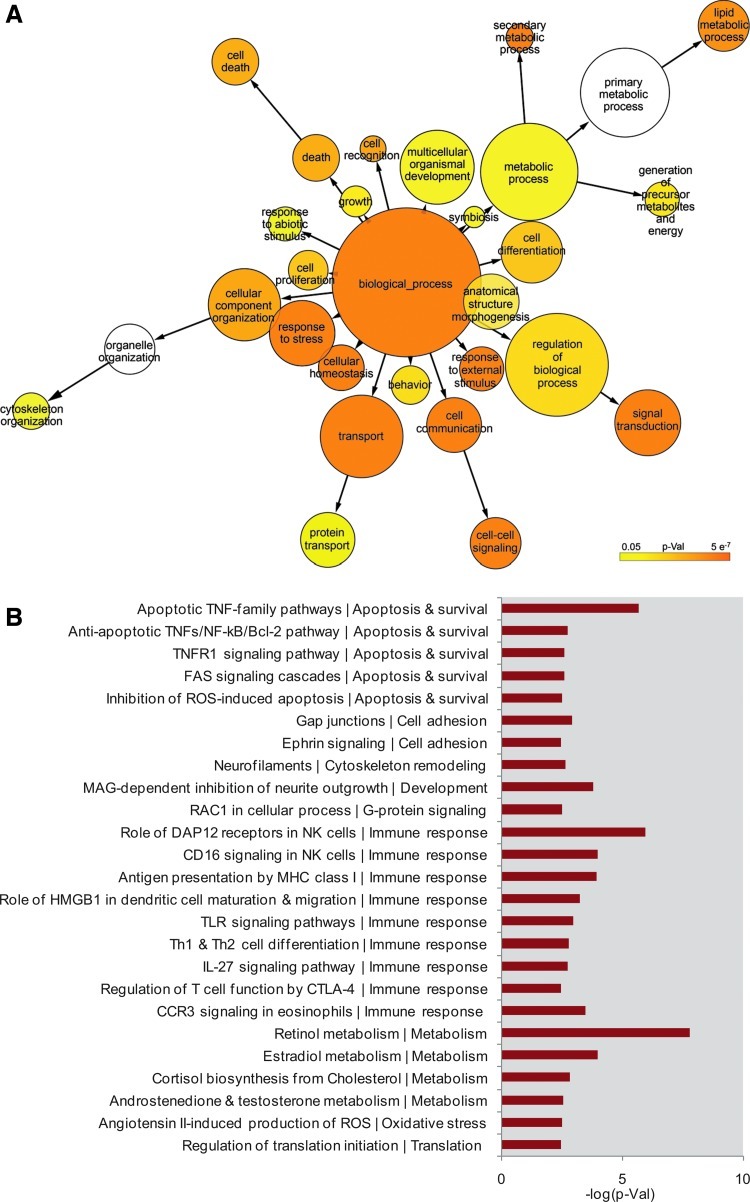
A functional heat map based on the enriched GO terms is illustrated (Fig. 6A) and an overview of the canonical pathways modulated is shown (Fig. 6B). The functional heat map highlighted enrichments in protein transport, cell signaling, cell homeostasis, lipid metabolism, cytoskeleton organization, and cell death. Furthermore, the PANTHER classifications of the essential genes revealed more specific functional categories in: cytochrome P450 genes (45 genes) having a role in lipid and xenobiotic metabolism, splicing factors (15 genes), vesicle docking, and fusion SNAP/SNARE proteins (13 genes), ABC transporters (9 genes), and AP complex members related to membrane trafficking and protein sorting in the Trans-Golgi Network (9 genes). Among the receptors identified, the 4 pronounced receptor families were interleukin receptor (14 genes), glutamate receptor (13 genes), GABA receptor (6 genes), and acetylcholine receptor (4 genes). Also, 18 genes were involved in 3 signaling pathways namely, fibroblast growth factor signaling (8 genes), MAPKKK cascade (6 genes), and JNK cascade (4 genes). The immune response-related genes (205 in total), had prominence in natural killer (NK) cell activation (11%), MHC complex-mediated immunity (11%), B cell-mediated immunity (9%), chemokines (4%), and macrophage activation (4%).
Fig. 6.
Biological classification of 1,252 nominated essential candidates in the shRNA screen. (A) Biological function classification of essential gene candidates as a heatmap based on gene ontology term enrichments. (B) Pathway mapping for essential gene candidates from Metacore. P-Val, P value.
The canonical pathway analysis for these genes revealed members of key apoptosis pathways, including apoptosis TNF-family pathways, FAS signaling cascades, caspase cascade, and curiously the anti-apoptotic TNFs/NF/Bcl2 pathway. Hits enriched in the cellular immune response included TLR signaling, IL-27 signaling, and DAP12 receptors in NK cells. Also of prominence are those genes that belong to components of the pathways associated with metabolism specifically retinol and estradiol. A small proportion of genes, six in number, were identified as rescuers; out of which only two genes (ASB5 and CROCC) have been characterized to have a cellular function. ASB5 is a member of ankyrin repeat and the SOCS box-containing (ASB) protein family with a potential role in suppression of cytokine signaling, while CROCC plays a role in mitosis.
Overlap Analysis with Essential Genes Identified in the Murine TRC1 Library Screen
Colombi and coworkers have performed the only other mammalian genome-scale arrayed shRNA hairpin screen against the murine TRC1 library in the 6.5 cell line; they reported 800 essential genes, the knockdown of which was lethal as measured by Alamar Blue viability assay over a course of ∼5 days.4 Thus, we have compared the screen parameters used in their study for essential genes in the 6.5 murine cells with ours (Supplementary Fig. S4). For the purpose of performing an overlap analysis between the human and murine essential gene data sets, we first performed a cross-library comparison between the two TRC libraries used in both screens. We found that only 722 genes out of the 800 total murine genes were present in the human TRC library. Similarly, 1,165 genes out of the 1,252 total nominated essential gene candidates were present in the murine TRC library screened by Colombi (Supplementary Fig. S4). We have used the mouse ortholog information as an effort to estimate the best degree of commonality with highest accuracy and have identified only 71 essential genes common in both data sets (P valueoverlap=0.003; Supplementary Table S8).
Discussion
To identify functionally important genes for cellular viability, we have conducted the first human whole-genome shRNA screen in an arrayed format of one hairpin per well. The technical ease and feasibility of using arrayed shRNA over pooled formats has only recently been reported by small-scale studies that screened less than 1,000 genes and measured hairpin activity for cellular viability using the Promega Cell Titer-Glo luminescent assay technology.2,3,5 Moreover, Colombi and coworkers reported the only other mammalian genome-scale arrayed shRNA-conditional lethal screen, performed against the murine Pten mutant mast cell line 6.5 and using Alamar Blue as cell viability readout for the screen.4 Interestingly, all of the published shRNA arrayed screens used low content readouts measuring cellular metabolic activities; in contrast, our screen used whole-well imaging automated microscopy of Hoechst-stained nuclei as the readout, thus measuring residual cellular fitness per well. This approach offers many advantages over low content methods, the main one being the ability to measure cell by cell data, as opposed to an average response for the well. Therefore, our automated microscopy-based strategy allows for better hairpin activity visualization and scoring (Fig. 3B), especially in view of the inherent heterogeneity problem associated with established cell lines used in screening (Fig. 4B).12,32,33 Furthermore, high content screening approaches provide the ultimate advantage of identifying those genes essential for cell viability and at the same time score for those rescuers, the knockdown of which enhances cellular growth, which would have been missed if relying on a low content approach.
The individual hairpin performance in the screen was deemed successful with a good control separation of puromycin-treated versus untreated wells, and exhibiting an S/B ratio of 5. The calculated Z′ value for the screen was 0.36—attributes of a marginal assay due mainly to the observed spread in the controls attributed, in part, to the inherent heterogeneity of cell populations; not surprising, since we and others have also reported on the Z′ factor as being a poor indicator of assay performance given that these high content image-based data sets do not follow normal distributions; and very often yielding low Z′ values for high content cell-based assays.17,34 Automated microscopy-based assays rely on segmentation algorithms to extract multiple features within an image to assess biological phenotypes. As such, they have a tendency to suffer from noise as a consequence of biological variation within the cell population.17
Frequency distribution of hairpin activity reveals two distinct populations of those severely hampering cell growths; therefore, essentials and the others enhancing cell proliferation, therefore rescuers (Fig. 4C). Of note, this observation highlights one of the many advantages of arrayed RNAi screening and the opportunity it provides to nominate hits based on individual hairpin performance rather than an average of sometimes opposite effects when using pooled shRNA libraries. Thus, to identify genes that were either essential or rescuer, we have implemented our recently developed BDA method to analyze the screening data systematically.12 To minimize false positives, we have set a stringent bar for a nominated hit gene as having a minimal H score value of at least 60 for its active shRNA hairpins against the same gene that have yielded the same growth suppression or growth advantage phenotype. An H score value of 60 translates into 3 active shRNA hairpins per gene rule based on an average of 5 shRNA hairpins per gene (Fig. 5). However, a subpopulation (4%) of the library contains more than 5 shRNA hairpins targeting one gene by virtue of which a number-based threshold determination is not comprehensive and, hence, the hairpin activity ratio in the form of H score renders optimal stringency. Nonetheless, we cannot undermine the plausible significance of scoring >4 active shRNA hairpins for a gene irrespective of the total number of shRNA hairpins for that gene in the library. Thus, we have subjected such genes to a t-test to determine if the performance average of the corresponding active shRNA hairpins is significantly different from the performance average of their inactive counterparts. Thirteen genes, C8G, CDK10, CSNK1E, FMO2, HDAC1, HRAS, IGF2, KRAS, LCK, MAPK15, PLK1, POLA1, and TRIB3, were nominated as essential gene candidates on the basis of this analysis method (Supplementary Table S9).
We also performed an OTE filtering analysis to further assess and attenuate false positives that originate from unintended target silencing by a hairpin. Notably, OTEs are becoming omnipresent and inherent to RNAi screens and thus we propose, as a part of the BDA method, to incorporate a structured OTE filtering step in a standardized hit nomination workflow to tag potential OTEs (Fig. 5). However, it must be noted that a hairpin is introduced into the cell through a lentiviral-enabled plasmid vector and undergoes intracellular processing to yield a functional duplex by a mechanism believed to be similar to the miRNA biogenesis pathway.35 Accordingly, the processing of an shRNA hairpin into an active duplex inside the cell depends on the specificity and efficiency of dicer cleavage, which brings into question the exact location of guide strand after a dicer-mediated intracellular splicing of an shRNA hairpin.36,37 To account for a possible shift in guide strand location and a consequent shift in seed heptamer location during the intracellular shRNA hairpin processing, we have incorporated the ESS method to determine the optimal seed heptamer location based on its correlation with the screen data output as previously described.12 We performed an OTE filtering on two sets of seed heptamers and merged the results to obtain a final list of HC_OTEs. Furthermore, we identified 33 miRNAs in total that are likely to play a regulatory role in cell fate. Our observations are supported by numerous other similar findings, where a miRNA-like activity of an shRNA hairpin leads to silencing of off-target transcripts.38,39 The systematic hit nomination workflow by the BDA method clearly highlights the benefit of arrayed RNAi screening; whereby analysis of an individual hairpin performance provides an additional layer of information that can be leveraged into the identification of high-confidence gene candidates.
Finally, The BDA method nominated a total of 1,252 essential and 6 rescuer gene candidates (Fig. 5). As illustrated in the frequency distribution plot for the controls versus the library (Fig. 4C), there is a wide range of growth variability in the cell population per well, therefore accounting for a small proportion of genes nominated as rescuers that confer a significant growth advantage to the HeLa cells. Among the 1,252 candidates are known oncogenes as well as genes with reported oncogenic properties, including v-Ki-ras2 Kirsten rat sarcoma viral oncogene (KRAS), Kinase suppressor of ras1 (KSR1), the baculoviral IAP repeat-containing 6 (BIRC6), E2F transcription factor 5 (E2F5), Ras-related C3 botulinum toxin substrate 1 (RAC1), and receptor tyrosine kinase-like orphan receptor 2 (ROR2). RAC1, a member of the Ras GTPase super family, is a well-characterized oncogene in several carcinomas including skin tumor, lung cancer, breast cancer, and cervical cancer.40–43 E2F5 has also been reported to be dysregulated in hepatocellular carcinoma and breast tumors.44,45 Also, E2F5 has been shown to be a direct activator of HPV18 E6/E7 transcription in HeLa cells.46 Based on the fact that we identify these genes implicated in specific cancer as essential in our screen, it could be argued that, perhaps, these genes are generally essential for cell survival and not specific for a cancer type.
However, a surprising result pertaining to the TRC1 library performance was the total absence of several known essential genes, such as KIF11 and WEE1 to name a few, from the nominated list of actives. Of note, we have marginally identified PLK1 as a hit in our screen; PLK1 has previously been reported a potential drug target for cancer therapy.47,48 Notably, PLK1 is also routinely used as a reliable positive control in multiple published RNAi screening studies49–51 and in several siRNA screens from our own group.* A highly unusual number of shRNA hairpins target the PLK1 gene in the TRC1 library with 23 different sequences; and in our screen only 4 of them were deemed active and consequently lethal (Supplementary Table S9). In addition, 20 of the 23 clones targeting PLK1 were deemed validated as they reduce its message by at least 50% and with clone # 121325 being the most active and achieving a 95% knockdown (Supplementary Table S9). Therefore, it is rather puzzling that from these 20 validated clones, only 4 of them exhibit a lethal phenotype considering that PLK1 is indeed an essential target in HeLa cells. Furthermore, none of the published shRNA screening studies using the TRC library have reported PLK1 as a hit across many different cell lines.2–5,11 Of note, the lack of phenotype associated with validated clones was also observed for other gene candidates (Supplementary Table S9). Obviously, this raises a fundamental concern as to the intracellular efficiency of processing shRNA hairpins into desired and active duplexes; and potentially, questions the merits of utilizing shRNA in RNAi screening.
Since the TRC library is hairpin based, we sought to compare it to the Elledge-Hannon library, which is a miRNA adapted and based on the miR30 backbone; and asked whether PLK1 was identified in any of their published screens. Lou and coworkers, although reported on identifying PLK1 as a high-value target as a result of a genome-scale pooled shRNA screen using an isogenic pair of DLD-1 cells.52 However, close examination of their screening data revealed that PLK1 was indeed inactive. Their pooled library contained three hairpins (#19708, #19709, and #19711) with one active hairpin (#19708) in the primary screen. Surprisingly, it did not meet the qualifying criteria in their secondary competition assay, but kept as a hit for some unknown reason.52 Additionally, Schlabach and coworkers screened a similar pooled library harboring the three PLK1 hairpins and reported on their differential activities across four cell lines screened with 3/3 actives in HCT116, 2/3 actives in both DLD-1 and HCC1954 cell lines, and 1/3 active in the HMEC cell line; PLK1 hairpin #19708 was active in all four cell lines.53 Furthermore, Silva and coworkers using the same pooled library reported on only one active hairpin (#19708) in both MCF-10A and MDA-MB-435 cell lines.54 It is interesting that only one hairpin (#19708) targeting PLK1 was active across the eight cell lines. Therefore, it seems that the concerns as to the efficiency of intracellular processing of either hairpin- or miR30 backbone-based shRNAs are universal.
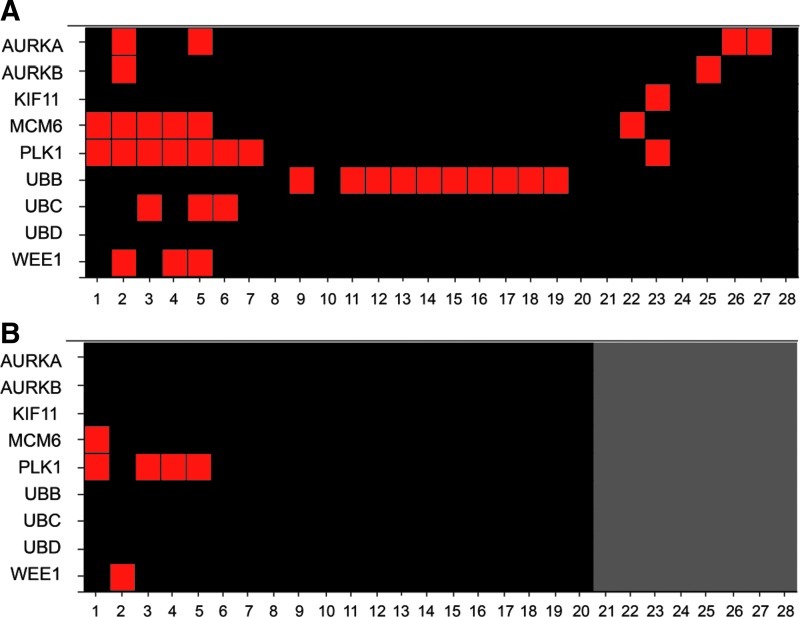
To further assess the overall performance of the TRC library, we selected nine representative genes deemed essential (AURKA, AURKB, KIF11, PLK1, MCM6, UBB, UBC, UBD, and WEE1)55–60 and checked their activities in seven published shRNA hairpin screens, four of which were performed against the TRC library2,3,11,61 and the remaining three were performed against the Elledge-Hannon library52–54 (Supplementary Table S10). We initially observed a rather poor overlap with none of the nine essential genes being consistently nominated across multiple cell lines, especially for PLK1 (Fig. 7A); upon applying a high-stringency filter of an H score of ≥60, and with the exception of very few, most of them became inactive leading to a negligible overlap (Fig. 7B). Such a lack in activity of known essential genes across the board is baffling and, perhaps, brings into question the widely overlooked aspects of differential intracellular processing leading to active or inactive duplexes; a nonissue it seems for the use of siRNA duplexes by passing this extra step.
Fig. 7.
Heat map plot of nine known essential genes from seven published shRNA hairpin screens. (A) Heat map plot showing the activity of the nine genes based on published reports. (B) Heat map plot analysis after applying a high-stringent activity criteria of an H score threshold set at ≥60. Red indicates genes scored as active; black indicates genes scored as inactive; gray indicates unavailability of active hairpin numbers per gene. Heatmap rows are numbered studies as defined in Supplementary Table S10.
Colombi and coworkers reported on a murine genome-scale arrayed shRNA hairpin screen identifying approximately 800 essential genes in the murine pten 6.5 cell line.4 Of note, KIF11, PLK1, and WEE1 were not reported as essential. The human orthologs to mouse essential genes would provide useful insights toward profiling human essential genes; mouse models are routinely used for human studies, and gene essentiality is believed to be conserved during evolution to some extent.62 Since this is the only other mammalian genome-scale arrayed shRNA screen published, we took this as an opportunity to assess the TRC library performance across species. We have identified 71 genes common to both species and reported as essential in the Colombi study; these include members of the ribosomal complex and proteasome. It is a rather low overlap between the two screens potentially due to inherent differences in intracellular hairpin processing between human and mouse cells.
In summary, we report on the successful execution of the first human arrayed genome-scale shRNA screen in HeLa cells and describe a high-stringency hit nomination methodology, with an attempt to standardize the process of hit nomination in RNAi screen data outputs. We have identified a set of gene candidates essential for cell survival and important processes associated with them that include in prominence components of the proteasome, ribosome, and spliceosome complexes. We also report on rescuer gene candidates that conferred growth advantage to HeLa cells; and a plausible role of endogenous miRNA interference in modulating the on-target hairpin performance. We also provide a list of 239 high-confidence essential gene candidates (Supplementary Table S11). As to the performance of the TRC library, several known essential genes were not identified as actives raising concerns as to the intracellular efficiency of processing both hairpin- and miR30 backbone-based shRNA plasmids, Finally, it is our hope that such an effort serves as the first key step toward standardizing RNAi screen hit nomination.
Supplementary Material
Abbreviations
- BDA
the Bhinder–Djaballah analysis
- ESS
empirical seed selection
- GFP
green fluorescent protein
- GO
gene ontology
- HC
high control
- H score
hit rate per gene
- INCA2000
IN Cell Analyzer 2000
- LC
low control
- MOI
multiplicity of infectivity
- NK
natural killer
- NUCL
residual Hoechst-stained nuclei count per well, a cytotoxicity index used for quantification of remaining cells
- OTE
off-target effect
- RNAi
RNA interference
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
- S/B ratio
signal-to-background ratio
- TRC
The RNAi Consortium.
Footnotes
Djaballah H, Bhinder B, Shum D, unpublished observations, High-Throughput Screening Core Facility, Memorial Sloan-Kettering Cancer Center, New York, NY, 2012.
Acknowledgments
The authors wish to thank members of the HTS Core Facility for their assistance during the course of this study. The HTS Core Facility is partially supported by Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research, the Experimental Therapeutics Center of the Memorial Sloan-Kettering Cancer Center, the William Randolph Hearst Fund in Experimental Therapeutics, the Lillian S. Wells Foundation, and by an NIH/NCI Cancer Center Support Grant 5 P30 CA008748-44.
Disclosure Statement
The authors declare no competing financial interests.
References
- 1.Mohr S. Bakal C. Perrimon N. Genomic screening with RNAi: results and challenges. Annu Rev Biochem. 2010;79:37–64. doi: 10.1146/annurev-biochem-060408-092949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scholl C. Fröhling S. Dunn IF, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137:821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 3.Barbie DA. Tamayo P. Boehm JS, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colombi M. Molle KD. Benjamin D, et al. Genome-wide shRNA screen reveals increased mitochondrial dependence upon mTORC2 addiction. Oncogene. 2011;30:1551–1565. doi: 10.1038/onc.2010.539. [DOI] [PubMed] [Google Scholar]
- 5.Delmore JE. Issa GC. Lemieux ME, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitehurst AW. Bodemann BO. Cardenas J, et al. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature. 2007;446:815–819. doi: 10.1038/nature05697. [DOI] [PubMed] [Google Scholar]
- 7.Cherry S. What have RNAi screens taught us about viral-host interactions? Curr Opin Microbiol. 2010;12:446–452. doi: 10.1016/j.mib.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaelin WG., Jr. Use and abuse of RNAi to study mammalian gene function. Science. 2012;337:421–422. doi: 10.1126/science.1225787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Babij C. Zhang Y. Kurzeja RJ, et al. STK33 kinase activity is nonessential in KRAS-dependent cancer cells. Cancer Res. 2011;71:5818–5826. doi: 10.1158/0008-5472.CAN-11-0778. [DOI] [PubMed] [Google Scholar]
- 10.Naik G. Scientists' Elusive Goal: Reproducing Study Results. The Wall Street Journal. 2011. online.wsj.com online.wsj.com
- 11.Cheung HW. Cowley GS. Weir BA, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci USA. 2011;108:12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhinder B. Djaballah H. A simple method for analyzing actives in random RNAi screens: introducing the “H score” for gene nomination & prioritization. Comb Chem High Throughput Screen. 2012;15:686–704. doi: 10.2174/138620712803519671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antczak C. Takagi T. Ramirez CN, et al. Live cell imaging of caspase activation for high content screening. J Biomol Screen. 2009;14:956–969. doi: 10.1177/1087057109343207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sigma-Aldrich. MISSION® shRNA Library. www.sigmaaldrich.com/life-science/functional-genomics-and-rnai/shrna/library-information.html. [May 8;2012 ]. www.sigmaaldrich.com/life-science/functional-genomics-and-rnai/shrna/library-information.html
- 15.Ramirez CN. Ozawa T. Takagi T, et al. Validation of a high-content screening assay using whole-well imaging of transformed phenotypes. Assay Drug Dev Technol. 2011;9:247–261. doi: 10.1089/adt.2010.0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antczak C. Bermingham A. Calder P, et al. Domain-based biosensor assay to screen for EGFR modulators in live cells. Assay Drug Dev Technol. 2012;10:24–36. doi: 10.1089/adt.2011.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shum D. Bhinder B. Radu C, et al. An image-based biosensor assay strategy to screen for modulators of the microRNA 21 biogenesis pathway. Comb Chem High Throughput Screen. 2012;15:529–541. doi: 10.2174/138620712801619131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karolchik D. Hinrichs AS. Furey TS, et al. The UCSC table browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozomara A. Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vergoulis TI. Vlachos P. Alexiou G, et al. Tarbase 6.0: capturing the exponential growth of miRNA targets with experimental support. Nucleic Acids Res. 2012;40:D222–D229. doi: 10.1093/nar/gkr1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao J. Ade AS. Tarcea VG, et al. Integrating and annotating the interactome using the MiMI plugin for cytoscape. Bioinformatics. 2009;25:137–138. doi: 10.1093/bioinformatics/btn501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bader GD. Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maere S. Heymans K. Kuiper M. BiNGO: a cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 24.Huang DW. Sherman BT. Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 25.Thomas PD. Kejariwal A. Campbell MJ, et al. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003;31:334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mouse Genome Informatics (MGI) Web, The Jackson Laboratory, Bar Harbor, Maine. www.informatics.jax.org. [May;2012 ]. www.informatics.jax.org
- 27.Blake JA. Eppig JT. Richardson JE, et al. The Mouse Genome Database (MGD): integration nexus for the laboratory mouse. Nucleic Acids Res. 2001;29:91–94. doi: 10.1093/nar/29.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fury W. Batliwalla F. Gregersen PK, et al. Overlapping probabilities of top ranking gene lists, hypergeometric distribution, and stringency of gene selection criterion. Conf Proc IEEE Eng Med Biol Soc. 2006;1:5531–5534. doi: 10.1109/IEMBS.2006.260828. [DOI] [PubMed] [Google Scholar]
- 29.Leppert U. Henke W. Huang X, et al. Post-transcriptional fine-tuning of COP9 signalosome subunit biosynthesis is regulated by the c-Myc/Lin28B/let-7 pathway. J Mol Biol. 2011;409:710–721. doi: 10.1016/j.jmb.2011.04.041. [DOI] [PubMed] [Google Scholar]
- 30.Wei N. Serino G. Deng XW. The COP9 signalosome: more than a protease. Trends Biochem Sci. 2008;33:592–600. doi: 10.1016/j.tibs.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 31.Zhang W. Liu HT. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002;12:9–18. doi: 10.1038/sj.cr.7290105. [DOI] [PubMed] [Google Scholar]
- 32.Ramirez C. Antczak C. Djaballah H. Cell viability assessment: toward content-rich platforms. Expert Opin Drug Discov. 2010;5:223–233. doi: 10.1517/17460441003596685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krausz E. Korn K. High-content siRNA screening for target identification and validation. Expert Opin Drug Discov. 2008;3:551–564. doi: 10.1517/17460441.3.5.551. [DOI] [PubMed] [Google Scholar]
- 34.Kummel A. Gubler H. Gehin P, et al. Integration of multiple readouts into the z' factor for assay quality assessment. J Biomol Screen. 2010;15:95–101. doi: 10.1177/1087057109351311. [DOI] [PubMed] [Google Scholar]
- 35.Rao DD. Vorhies JS. Senzer N, et al. siRNA vs. shRNA: similarities and differences. Adv Drug Deliv Rev. 2009;61:746–759. doi: 10.1016/j.addr.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 36.Park JE. Heo I. Tian Y, et al. Dicer recognizes the 5' end of RNA for efficient and accurate processing. Nature. 2011;475:201–205. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vermeulen A. Behlen L. Reynolds A, et al. The contributions of dsRNA structure to dicer specificity and efficiency. RNA. 2005;11:674–682. doi: 10.1261/rna.7272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Birmingham A. Anderson EM. Reynolds A, et al. 3' UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- 39.Sudbery I. Enright AJ. Fraser AG, et al. Systematic analysis of off-target effects in an RNAi screen reveals microRNAs affecting sensitivity to TRAIL-induced apoptosis. BMC Genomics. 2010;11:175. doi: 10.1186/1471-2164-11-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z. Pedersen E. Basse A, et al. Rac1 is crucial for Ras-dependent skin tumor formation by controlling Pak1-Mek-Erk hyperactivation and hyperproliferation in vivo. Oncogene. 2010;29:3362–3373. doi: 10.1038/onc.2010.95. [DOI] [PubMed] [Google Scholar]
- 41.Akunuru S. Palumbo J. Zhai QJ, et al. Rac1 targeting suppressing human non-small cell lung adenocarcinoma cancer stem cell activity. PLoS One. 2011;6:e16951. doi: 10.1371/journal.pone.0016951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan Y. Greer PM. Cao PT, et al. RAC1 GTPase plays an important role in gamma-irridiation induced G2/M checkpoint activation. Breast Cancer Res. 2012;14:R60. doi: 10.1186/bcr3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mendoza-Catalán MA. Cristóbal-Mondragón GR. Adame-Gómez J, et al. Nuclear expression of Rac1 in cervical premalignant lesions and cervical cancer cell lines. BMC Cancer. 2012;12:116. doi: 10.1186/1471-2407-12-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang Y. Yim SH. Xu HD, et al. A potential oncogenic role of the commonly observed E2F5 overexpression in hepatocellular carcinoma. World J Gastroenterol. 2011;17:470–477. doi: 10.3748/wjg.v17.i4.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Polanowska J. Le Cam L. Orsetti B, et al. Human E2F5 gene is oncogenic in primary rodent cells and is amplified in human breast tumors. Genes Chromosomes Cancer. 2000;28:126–130. [PubMed] [Google Scholar]
- 46.Teissier S. Pang CL. Thierry F. The E2F5 repressor is an activator of E6/E7 transcription and of the S-phase entry in HPV18-associated cells. Oncogene. 2010;29:5061–5070. doi: 10.1038/onc.2010.246. [DOI] [PubMed] [Google Scholar]
- 47.Ding Y. Huang D. Zhang Z, et al. Combined gene expression profiling and RNAi screening in clear cell renal cell carcinoma identify PLK1 and other therapeutic kinase targets. Cancer Res. 2011;71:5225–5234. doi: 10.1158/0008-5472.CAN-11-0076. [DOI] [PubMed] [Google Scholar]
- 48.Duan Z. Ji D. Weinstein EJ. Liu X, et al. Lentiviral shRNA screen of human kinases identifies PLK1 as a potential therapeutic target for osteosarcoma. Cancer Lett. 2010;293:220–229. doi: 10.1016/j.canlet.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 49.Sarthy AV. Morgan-Lappe SE. Zakula D, et al. Survivin depletion preferentially reduces the survival of activated K0-Ras-transformed cells. Mol Cancer Ther. 2007;6:269–276. doi: 10.1158/1535-7163.MCT-06-0560. [DOI] [PubMed] [Google Scholar]
- 50.Zheng M. Morgan-Lappe SE. Yang J, et al. Growth inhibition and radiosensitization of glioblastoma and lung cancer cells by small interfering RNA silencing of tumor necrosis factor receptor-associated factor 2. Cancer Res. 2008;68:7570–7578. doi: 10.1158/0008-5472.CAN-08-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cole KA. Huggins J. Laquaglia M, et al. RNAi screen of the protein kinome identifies checkpoint kinase 1 (CHK1) as a therapeutic target in neuroblastoma. Proc Natl Acad Sci USA. 2011;108:3336–3341. doi: 10.1073/pnas.1012351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luo JL. Emanuele MJ. Li D, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schlabach MR. Luo J. Solimini NL, et al. Cancer proliferation gene discovery through functional genomics. Science. 2008;319:620–624. doi: 10.1126/science.1149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Silva JM. Marran K. Parker JS, et al. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science. 2008;319:617–620. doi: 10.1126/science.1149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fu J. Bian M. Jiang Q, et al. Roles of Aurora kinases in mitosis and tumorigenesis. Mol Cancer Res. 2007;5:1–10. doi: 10.1158/1541-7786.MCR-06-0208. [DOI] [PubMed] [Google Scholar]
- 56.Harborth J. Elbashir SM. Bechert K, et al. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J Cell Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- 57.Yim H. Erikson RL. Polo-like kinase 1 depletion induces DNA damage in early S prior to caspase activation. Mol Cell Biol. 2009;29:2609–2621. doi: 10.1128/MCB.01277-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chuang CH. Yang D. Bai G, et al. Post-transcriptional homeostasis and regulation of MCM2-7 in mammalian cells. Nucleic Acids Res. 2012;40:4914–4924. doi: 10.1093/nar/gks176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu YX. Tiedemann R. Shi C, et al. RNAi screen of the druggable genome identifies modulators of proteasome inhibitor sensitivity in myeloma including CDK5. Blood. 2011;117:3847–3857. doi: 10.1182/blood-2010-08-304022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Domínguez-Kelly R. Martín Y. Koundrioukoff S, et al. Wee1 controls genomic stability during replication by regulating the Mus81-Eme1 endonuclease. J Cell Biol. 2011;194:567–579. doi: 10.1083/jcb.201101047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luo B. Cheung HW. Subramanian A, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci USA. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dickerson JE. Zhu A. Robertson DL, et al. Defining the role of essential genes in human disease. PLoS One. 2011;6:e27368. doi: 10.1371/journal.pone.0027368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.