Significance
For proteins, function is generally associated with order. Some proteins, however, are at least partially disordered. Because proteins tend to evolve into disorder in the absence of selection, it has been difficult to establish any significance of disorder for protein function. Here, we isolate a constitutively active variant of the normally acid-activated, conditionally disordered chaperone HdeA. We find this mutant to be largely destabilized, partially unstructured, and monomeric at a concentration at which it prevents the aggregation of a client protein. Our data therefore provide experimental evidence for the significance of partial disorder in protein function.
Keywords: acid activation, conditional disorder, intrinsically disordered protein, constant pH molecular dynamics
Abstract
Conditionally disordered proteins can alternate between highly ordered and less ordered configurations under physiological conditions. Whereas protein function is often associated with the ordered conformation, for some of these conditionally unstructured proteins, the opposite applies: Their activation is associated with their unfolding. An example is the small periplasmic chaperone HdeA, which is critical for the ability of enteric bacterial pathogens like Escherichia coli to survive passage through extremely acidic environments, such as the human stomach. At neutral pH, HdeA is a chaperone-inactive dimer. On a shift to low pH, however, HdeA monomerizes, partially unfolds, and becomes rapidly active in preventing the aggregation of substrate proteins. By mutating two aspartic acid residues predicted to be responsible for the pH-dependent monomerization of HdeA, we have succeeded in isolating an HdeA mutant that is active at neutral pH. We find this HdeA mutant to be substantially destabilized, partially unfolded, and mainly monomeric at near-neutral pH at a concentration at which it prevents aggregation of a substrate protein. These results provide convincing evidence for direct activation of a protein by partial unfolding.
The highly acidic nature of the mammalian stomach serves as an effective barrier against food-borne microbial pathogens (1). Certain gastrointestinal pathogens like Escherichia coli, however, have developed protection systems that enable them to survive in this hostile environment, which is a prerequisite for their colonization of the host’s intestine (2, 3). Such bacterial acid resistance systems are based, in part, on the removal of intracellular protons by amino acid decarboxylases (4, 5) and the ability of many enteric bacteria to reverse their cytoplasmic membrane potential (6), slowing down proton influx into the cell. These measures allow enterobacteria to maintain a more tolerable pH of 4.5 in the cytoplasm (6, 7). In contrast to the cytoplasm, the bacterial periplasm quickly equilibrates to pH values around 2 when these microorganisms are exposed to gastric acid. This is because porins in the outer cell envelope permit unhindered influx of molecules smaller than 600 Da into the periplasmic space, including protons (8). Such extremely acidic conditions can lead to massive unfolding and aggregation of periplasmic proteins (9–11), and are therefore fatal for many microorganisms. Enteric bacterial pathogens, however, elegantly use this quick drop in periplasmic pH to activate rapidly a crucial acid resistance system: the periplasmic chaperone HdeA.
At neutral pH, HdeA is an abundant, well-folded, but chaperone-inactive dimer (10, 12). On a shift to low pH, within 1 s, HdeA partially unfolds, monomerizes, and becomes active (13). In its active state, HdeA apparently uses disorder to bind to multiple interaction partners through structural flexibility (13–15). Adopting different conformations seems to allow HdeA to bind tightly to a variety of different substrate proteins at low pH, preventing their aggregation (13). On pH neutralization, which occurs on passage into the small intestine, HdeA slowly releases its client proteins over the course of minutes (16). This slow release is postulated to keep the concentration of aggregation-prone substrate species low. Because aggregation reactions are very concentration-dependent, slow release should disfavor aggregation while favoring refolding at the same time.
HdeA is an example of a pH sensor: a protein whose activity is regulated by changes in environmental pH. Diffusion-limited reactions are generally fast, but diffusion of protons is particularly fast, being partially facilitated through quantum tunneling in water, a process called Grotthuss diffusion (17). Notwithstanding their extraordinarily small size, protons can promote substantial yet reversible changes in protein conformation, stability, enzymatic activity, and ability to interact with other proteins simply by modulating electrostatic interactions (17). Posttranslational regulation through protons therefore allows for an exceptionally fast response to acid stress and is not subject to the significant delays associated with transcription, translation, or protein translocation that apply to many other stress responses (18).
Despite the myriad of cellular processes that are affected by changes in the extra- or intracellular pH and the number of diseases associated with a dysregulation in pH, the molecular basis of pH-dependent regulation of protein activity is not very well understood (17). To elucidate the mechanism of acid-triggered HdeA activation, we sought to identify which of HdeA’s acid-titratable residues are the key players in sensing environmental pH changes. Although we, as well as others, had previously observed that HdeA activation coincides with monomerization and partial unfolding, it was still unclear which structural changes are necessary to allow for HdeA activation and which changes are just byproducts of the massive drop in pH (10, 13, 19).
More generally, HdeA is a member of the family of conditionally disordered proteins. These proteins form a major subset of the larger family of intrinsically disordered proteins (14). Although even for intrinsically disordered proteins, structure is often associated with protein function, for at least some proteins, including HdeA, the disordered form of the protein seems to be the functional one. Even though a large amount of circumstantial evidence has accumulated that links disorder and function, establishing a direct connection between these two seemingly disparate properties has proven experimentally difficult (14). Here, we report on the construction of constitutively active variants of HdeA that show chaperone activity at pH values significantly higher than those required for the activity of WT HdeA, including one mutant that shows substantial chaperone activity even at neutral pH. We found this mutant to be substantially destabilized, partially unfolded, and monomeric at near-neutral pH at a concentration at which it prevents aggregation of a substrate protein. Our results thus suggest a direct link between the acquisition of partial disorder and HdeA activation.
Results
HdeA as a Model to Study the Relationship Between Disorder and Protein Function.
For most proteins, a high degree of order is thought to be necessary for protein function. However, an entire class of proteins, termed intrinsically disordered proteins, appears to exhibit a high degree of disorder, at least in vitro (20–22). It is easy to rationalize that such marked flexibility may be relevant for the various tasks these proteins carry out, for instance, binding to multiple target molecules. However, hard evidence for the functional importance of disorder is more difficult to obtain (14). Fortunately, conditionally disordered proteins whose flexibility can be modulated by changes in external conditions represent an excellent opportunity to dissect the relationship between flexibility and function because their activity can be assayed under both order- and disorder-promoting conditions. We have previously shown that on exposure to low pH, HdeA becomes activated, a process that is accompanied by monomerization and a significant loss in secondary structure (13). Although the correlation between activation and acquisition of disorder in HdeA is some of the best available evidence for a functional role of disorder in proteins (14), it remains just a correlation. It is currently not entirely clear if these structural changes that occur at low pH are actually required for chaperone activity or if HdeA is active as a chaperone despite these unfolding and monomerization events.
To determine which structural changes in HdeA are necessary for HdeA to function as a chaperone, we aimed to construct a constitutively active variant of HdeA. The use of constitutively active mutants to determine activation mechanisms has a long-standing history of success in many genetic systems (23). In the case of HdeA, constitutively active mutants are those that prevent aggregation of substrate proteins at neutral or near-neutral pH, where WT HdeA is highly structured and completely chaperone-inactive (24). We hypothesized that if constitutively active mutants in HdeA were also disordered at these elevated pH values, this would provide good evidence that disorder is necessary for HdeA activation. Importantly, these mutant proteins are likely to reflect the minimal increase in disorder that is necessary for activation because they are not subject to the global structural changes that occur in HdeA on a shift to pH 2. To generate such a constitutively active mutant, we first needed to identify which of the residues in HdeA are responsible for the activation process. In its inactive form at neutral pH, HdeA forms a dimer that is stabilized not only by hydrophobic interactions between residues in the dimer interface but by a network of attractive electrostatic interactions between the two monomers (12, 24). It has been suggested that the exposure of the dimer interface as a result of the dissociation of the monomers on a shift to low pH is crucial for substrate binding, and therefore for activity of the chaperone (10). Given that HdeA activation is mediated by a change in pH, we suspected that one or several of the pH-titratable residues in HdeA could serve as potential pH switches: residues that would activate the protein by becoming protonated, and hence disrupting electrostatic interactions crucial for the structural stability of the dimer (12, 24).
Identification of pH Switch Residues in HdeA.
Each HdeA monomer contains 28 pH-titratable residues: 12 lysine residues and 16 aspartic and glutamic acid residues (HdeA does not contain any histidine or arginine residues). Isolated lysine side chains have a pKa value of 10.5 (25), suggesting that these residues are permanently protonated at pH values lower than 8. Therefore, changes in the protonation state of lysine residues on the shift from neutral to acidic pH are very unlikely to be responsible for the acid-induced activation of HdeA. The pKas of the carboxyl group of isolated aspartic acid and glutamic acid, on the other hand, are 4.0 and 4.4, respectively (25). On the shift to pH values lower than 3, most of the glutamic and aspartic acid side chains are thus expected to undergo a transition from a negatively charged species to a neutral species. As a result of this protonation, these residues should lose their attraction to the positively charged lysines in the opposite monomer (26). This would result in a decrease in the strength of the monomer–monomer interaction, facilitating the separation of monomers and enabling exposure of the substrate binding site (10).
Because functional residues are often conserved among protein homologs, we examined the degree of conservation for all acidic residues in HdeA homologs. Residues E19, D20, D25, E26, E37, E46, D47, and D51 are conserved as acidic residues among all species, with D20 being absolutely conserved as an acidic residue in all HdeA sequences aligned (Fig. 1 and Fig. S1). Of these conserved acidic residues, E46, D47, and D51 are in close proximity to conserved basic clusters in the opposite monomer in the HdeA crystal structure (E46 is in proximity to K42, and D47 and D51 are in proximity to K10), suggesting that electrostatic interactions involving these residues could be conserved as well and are potentially functionally relevant. Residue D20 is also adjacent to a conserved patch of basic residues located in the same monomer in an adjacent helix (K10 and K11). However, this does not eliminate the possibility that D20 is involved in the pH-dependent monomerization. Protonation of D20, for example, might cause the two involved helices to exhibit greater flexibility, facilitating a loss in HdeA structure that then leads to activation.
Fig. 1.
WebLogo (75) representation of a ClustalW (65) sequence alignment of HdeA sequences. Only the part of the alignment corresponding to the mature E. coli HdeA protein is shown. Numbers indicate residue numbers for mature E. coli HdeA. The residue that is present in the alignment but missing in E. coli HdeA is indicated with “−”. Acidic residues are indicated in red, and basic residues are indicated in blue.
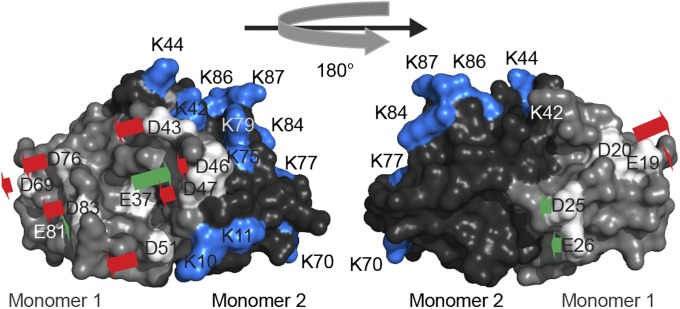
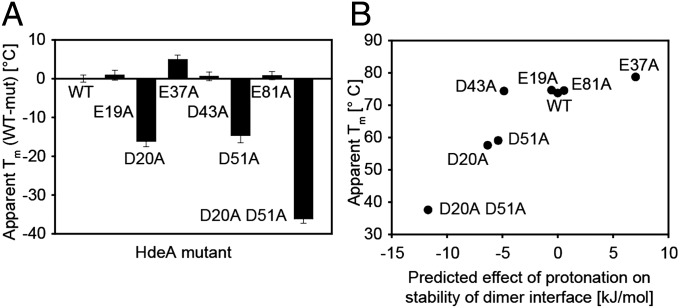
Although amino acid conservation often indicates functional relevance, in the case of HdeA, such conservation might not be related to pH-dependent activation. Conserved residues could alternatively be involved in substrate binding, substrate release, or some other important property of the chaperone. To determine which of HdeA’s acidic residues are crucial for the activation of HdeA’s chaperone activity, we performed constant pH molecular dynamics (CpHmd) calculations based on the published crystal structure of HdeA (12). The CpHmd method is a technique to simulate biomolecules in specific pH environments. It is a powerful tool to calculate the pKa values of titratable residues, as well as a simulation protocol with the ability to capture the coupling between the protonation of titratable groups and the conformational changes of the biomolecules (25, 27). For HdeA in particular, we calculated the relative contribution of each residue’s protonation to the destabilization of the dimer interface (Fig. 2 and Table 1). Because of a high degree of disorder, HdeA’s termini are not resolved in the crystal structure, and thus were excluded in the CpHmd calculations (12). Based on the CpHmd results, we predict that the largest destabilizing effect on the dimer interface is caused by protonation of residues D20, D43, and D51. Our calculations therefore suggest that these residues might be directly involved in triggering HdeA monomerization on a shift to low pH (Fig. 2 and Table 1). On the opposite end of the spectrum, the neutralization of several of the titratable groups we examined is actually predicted to cause an increase in the stability of the dimer interface, with protonation of E37 having the largest predicted stabilizing effect (Table 1). Given their high degree of conservation as acidic residues among HdeA homologs (100% for D20 and 75% for D51) and their large predicted destabilizing effect on the dimer interface on protonation, we selected residues D20 and D51 as candidates for pH switches. We were particularly interested in whether the elimination of any charge change of these residues by mutation that made them permanently neutral would lead to changes in the stability of the protein as predicted by our CpHmd calculations. We were also interested if these changes would alter the pH-induced transition from the folded state to the unfolded state or facilitate chaperone activity at elevated pH values.
Fig. 2.
Predicted effect of protonation of a particular acidic residue on the thermodynamic stability of the dimer interface. Acidic residues in monomer 1 are shown in white, and lysine residues in monomer 2 are shown in blue. Red arrows indicate a destabilization of the dimer interface on protonation of the indicated residue, and green arrows indicate a stabilization of the dimer interface. The length of the arrow indicates the extent of stabilization/destabilization.
Table 1.
Predicted destabilization of the HdeA dimer by protonation of acidic residues and experimentally determined apparent melting temperatures of HdeA and its mutants
| Residue | Average pKa value of dimer | Average pKa value of monomer | ΔΔG stability interface, kJ/mol | ΔΔG stability interface, % | HdeA mutant | Apparent melting temperature (Tm), °C | ΔΔTm (WT-mutant), °C |
| WT | 73.8 ± 0.9 | — | |||||
| E19 | 3.75 | 3.85 | −0.56 | −1.9 | E19A | 74.7 ± 0.4 | 0.9 ± 1.3 |
| D20 | 2.6 | 3.8 | −6.33 | −20.9 | D20A | 57.6 ± 0.6 | −16.1 ± 1.4 |
| D25 | 3.1 | 2.7 | 2.02 | 17.9 | |||
| E26 | 4.35 | 4.05 | 1.70 | 15.0 | |||
| E37 | 6.7 | 5.3 | 7.03 | 62.1 | E37A | 78.7 ± 0.2 | 5.0 ± 1.1 |
| D43 | 2.85 | 3.75 | −4.85 | −16.0 | D43A | 74.4 ± 0.2 | 0.6 ± 1.1 |
| E46 | 3.45 | 3.75 | −1.67 | −5.5 | |||
| D47 | 3.85 | 4.25 | −2.26 | −7.5 | |||
| D51 | 2.8 | 3.8 | −5.38 | −17.8 | D51A | 59.1 ± 1.0 | −14.7 ± 1.9 |
| D69 | 3.5 | 3.8 | −1.67 | −5.5 | |||
| D76 | 2.75 | 3.55 | −4.23 | −14.0 | |||
| E81 | 4.1 | 4 | 0.57 | 5.0 | E81A | 74.5 ± 0.2 | 0.8 ± 1.1 |
| D83 | 3.1 | 3.7 | −3.28 | −10.9 | |||
| D20/D51 | −11.71 | −38.7 | D20A D51A | 37.6 ± 0.3 | 36.2 ± 1.2 |
CpHmd calculations performed on the HdeA crystal structure (12) were used to predict the pKa values of acidic residues in the monomer and the dimer (26), as well as the extent of destabilization of the dimer interface on protonation of these residues. Because HdeA is slightly asymmetrical, pKa values differ slightly for a given residue in the two monomers, and the average value is given here. The acidic HdeA residues D2, D8, and D88 are not resolved in the crystal structure, and were therefore not considered in the CpHmd calculations (12).
Generation of a Constitutively Active HdeA Variant.
Consistent with our calculations, introduction of nonionizable residues at position 20 or position 51 did indeed lead to a large destabilization of the protein (Fig. 3 and Table 1): HdeA D20A and D51A exhibit melting temperatures that are decreased by 16.1 °C and 14.7 °C, respectively, compared with the WT protein. To exclude the possibility that alanine substitutions of acidic residues in HdeA tend to have a destabilizing effect, we measured the stability of several mutants in addition to D20 and D51, namely: (i) E37A, which is conserved but whose protonation, as mentioned, is predicted to stabilize the dimer interface substantially; (ii) E19, which is conserved among HdeA homologs and in close structural proximity to D20 but whose protonation is predicted to destabilize the dimer interface only marginally; and (iii) E81, which is not conserved and is predicted to have only a small stabilizing effect on the dimer interface on protonation (Table 1). Consistent with our CpHmd calculations, HdeA variants E19A, D43A, and E81A had melting temperatures that were essentially unaltered compared with WT HdeA and E37A was actually stabilized. Apart from mutation of residues D20 and D51, we thus found HdeA to be rather tolerant to alanine substitutions independent of the degree of conservation of these residues among HdeA homologs. Our results suggest that mutating an acidic residue to an alanine, by itself, does not lead to destabilization of HdeA; instead, the specific location of the residue in the protein is critical. Overall, our prediction of dimer destabilization by protonation of particular acidic residues correlates very well with the experimentally determined apparent melting temperatures of our HdeA alanine mutants (Fig. 3B). Surprisingly, however, substitution of D43, although not strongly conserved but predicted to destabilize the dimer interface substantially on protonation, only had a small effect on HdeA stability.
Fig. 3.
Difference in apparent melting temperature (Tm) for different HdeA mutants (mut) compared with WT HdeA. Apparent Tm values were determined by monitoring the CD signal of 20 μM HdeA protein at 222 nm. (A) Difference in apparent Tm compared with WT. Data are presented as mean ± SD from three independent experiments. (B) Apparent Tm of different HdeA alanine mutants plotted against the predicted effect on the stability of the dimer interface on protonation (Table 1).
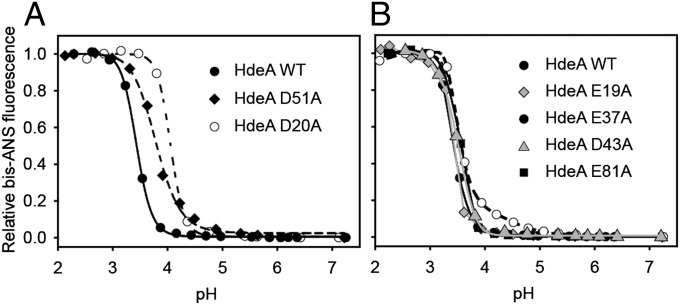
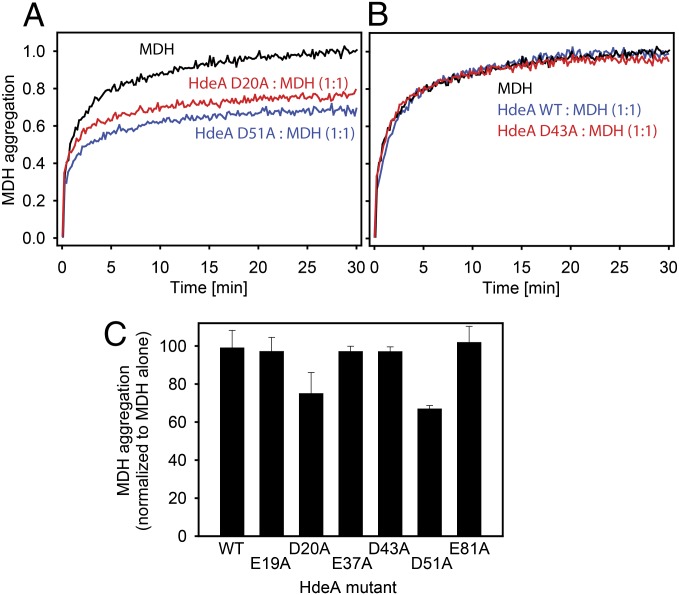
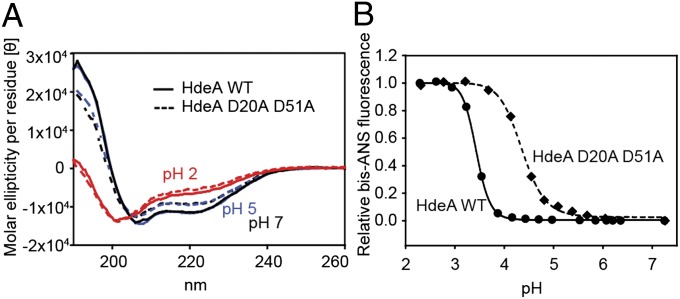
To examine if HdeA’s pH-induced unfolding transition was altered in the HdeA mutants, we performed pH titrations in the presence of 4,4′-bis(1-anilinonaphthalene 8-sulfonate) (bis-ANS). The dye bis-ANS is essentially nonfluorescent in solution and becomes fluorescent on binding to hydrophobic surfaces (including hydrophobic patches in proteins); it can therefore be used to assess the degree of unfolding in polypeptides (28). Both of the destabilized variants D20A and D51A showed pH midpoints of transition from the folded to the unfolded state at higher pH values compared with WT HdeA (Fig. 4A). These results imply that these variants may have a higher pH of activation. Most satisfyingly, both also showed chaperone activity at pH 4, a pH at which WT HdeA is completely inactive in preventing aggregation of the substrate malate dehydrogenase (MDH) (Fig. 5). None of the other HdeA variants with alanine substitutions (E19A, E37A, D43A, and E81A) showed midpoints of pH-induced unfolding that were significantly different from the value obtained for WT HdeA (Fig. 4), nor did any of these mutants have chaperone activity at pH 4 (Fig. 5). These data suggest that HdeA variants D20A and D51A are at least partially constitutively active as chaperones. Interestingly, both HdeA D51A and HdeA E37A showed decreased slopes in their pH-dependent transitions from the folded state to the unfolded state compared with HdeA WT, suggesting that the cooperativity of the overall unfolding and dimer dissociation transition is weakened for these mutants (Fig. 4). At this point, however, a much more detailed investigation of the structural changes that occur in HdeA on pH denaturation will be required to obtain a more thorough understanding of the observed changes in cooperativity for the different mutants.
Fig. 4.
pH-dependent unfolding of HdeA as measured by bis-ANS fluorescence. Binding of 15 μM bis-ANS to 3 μM HdeA was measured in buffer, which was titrated from about pH 7 to pH 2. The midpoints for the pH-induced transition from the state at neutral pH to the state at low pH for the different HdeA mutants tested are 3.4 (WT), 3.8 (D51A), 4.0 (D20A), 3.4 (E19A), 3.5 (E37A), 3.4 (D43A), and 3.5 (E81A).
Fig. 5.
Chaperone activity of various HdeA variants at pH 4. (A–C) Guanidine denatured MDH was diluted into aggregation buffer to a final concentration of 4 μM. MDH aggregation was measured by monitoring light scattering at 350 nm in the presence or absence of various HdeA variants. (C) Extent of MDH aggregation in the presence of various HdeA variants at pH 4 after 30 min, normalized to the aggregation of MDH in the absence of HdeA. Data are presented as mean ± SD from at least three independent experiments.
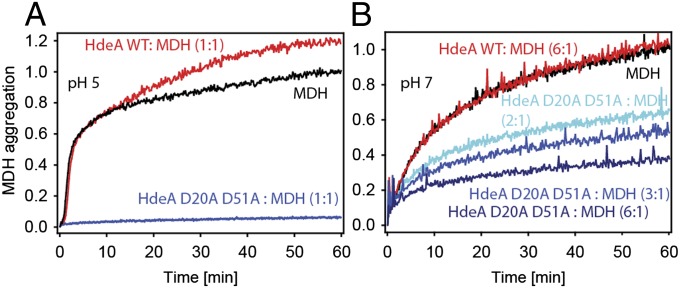
To obtain an HdeA variant that is active at near-neutral pH, we combined alanine mutations of the two pH switch residues (D20 and D51) in a single HdeA variant. Strikingly, the resulting mutant HdeA D20A D51A prevented aggregation of MDH completely at pH 5, even when the chaperone-to-substrate ratio was only 1:1 (Fig. 6A). HdeA D20A D51A was also partially active at pH 7, with increasing concentrations of chaperone leading to an increased suppression of MDH aggregation (Fig. 6B). Thus, by using alanine substitutions to neutralize only two acidic residues permanently, which we specifically predicted would reduce the attractive forces that stabilize the dimeric protein at neutral pH, we succeeded in turning the acid-specific chaperone HdeA into a constitutively active chaperone that is also active at neutral pH. This constitutively active variant, in turn, allowed us to assess which structural changes in HdeA are actually required for its function.
Fig. 6.
Chaperone activity of HdeA D20A D51A at pH 5 and pH 7. Guanidine denatured MDH was diluted into aggregation buffer at pH 5 (A) or pH 7 (B) to a final concentration of 4 μM. MDH aggregation was measured by monitoring light scattering at 350 nm in the presence or absence of various HdeA variants. Aggregation is normalized to the extent of aggregation of MDH in the absence of HdeA after 60 min.
Constitutively Active Variant HdeA D20A D51A Is Partially Unfolded, Largely Destabilized, and Monomeric in Its Active State.
Far-UV CD spectra suggest that HdeA D20A D51A exhibits a partial loss in secondary structure compared with WT HdeA both at pH 7 and pH 5, pH values at which HdeA D20A D51A shows chaperone activity but WT HdeA does not (Figs. 6 and 7A). Predictions of secondary structural fractions in HdeA using the software CDSSTR (available at http://dichroweb.cryst.bbk.ac.uk/html/home.shtml) (29) suggest a loss in α-helical content for HdeA D20A D51A in the range of 20–25% compared with WT HdeA at both pH 7 and pH 5. Note that this loss in secondary structure for the variant vs. WT at pH 7 and pH 5 is smaller than the loss observed for both WT HdeA and D20A D51A on the shift to pH 2 (Fig. 7A), where both proteins show similar chaperone activity (Fig. S2). The predicted fraction of α-helical content is very similar for both proteins at pH 2 and about 60% smaller than the α-helical content of WT HdeA at pH 7. This additional loss in structure on moving from pH 5 or pH 7 to pH 2 may reflect structural changes that accompany the activation of the protein at low pH but are not actually crucial for chaperone function. Consistent with the idea that partial unfolding activates HdeA, HdeA D20A D51A is significantly destabilized compared with WT at neutral pH (Fig. 3 and Table 1). The midpoint of thermal denaturation of HdeA D20A D51A is reduced by 37 °C (from about 74 °C to about 37 °C). Further, HdeA D20A D51A has an elevated midpoint of pH-induced unfolding (Fig. 7B), consistent with our prediction that D20 and D51 act to stabilize the dimeric configuration of the protein at neutral pH. Like HdeA D51A, HdeA D20A D51A exhibits a decreased slope in its pH-dependent unfolding transitions compared with HdeA WT, indicating a decrease in cooperativity of the overall unfolding and dimer dissociation transition for this mutant.
Fig. 7.
Structural characterization of HdeA D20A D51A. (A) Far-UV CD spectra of 20 μM WT HdeA and HdeA D20A D51A at pH 2, pH 5, and pH 7. (B) Binding of 15 μM bis-ANS to 3 μM HdeA was measured in buffer, which was titrated from about pH 7 to pH 2. The midpoints for the pH-induced transition from the state at neutral pH to the state at low pH for the different HdeA mutants tested are 3.4 for WT HdeA and 4.4 for HdeA D20A D51A.
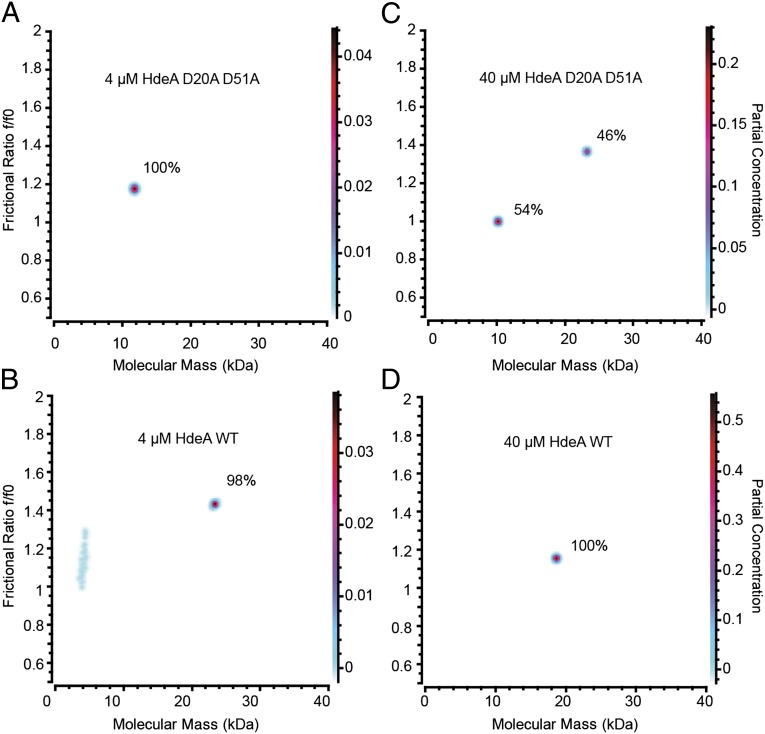
To characterize the structural oligomeric properties of our constitutively active mutant further, we performed analytical ultracentrifugation experiments. Our data suggest that at pH 5, HdeA D20A D51A has a Kd of dimerization in the range of ∼30 μM and populates the monomeric and dimeric states at concentrations between 24 and 70 μM (Fig. 8C and Fig. S3 E, G, and I). WT HdeA, on the other hand, almost exclusively populates the dimeric state at all tested concentrations between 4 and 70 μM at pH 5 (Fig. 8 B and D and Fig. S3 B, D, F, H, J). Taken together with the published Kd values of dimerization for WT HdeA at pH 4 (about 1 μM) and at pH 7 (about 0.25 μM) (24), these results imply that the binding affinity between the two monomers at pH 5 is significantly weaker for HdeA D20A D51A compared with HdeA WT. Remarkably, the Kd for HdeA D20A D51A at pH 5 (where this mutant is chaperone-active) is comparable to the Kd of dimerization for WT HdeA (about 45 μM) at pH 2, its natural pH of activation (21). At concentrations of 4 and 8 μM at pH 5, HdeA D20A D51A mainly populates a monomeric state (Fig. 8A and Fig. S3C). Note that 4 μM is the same concentration at which HdeA D20A D51A completely prevents aggregation of the substrate protein MDH (Fig. 6), providing additional evidence that the active species of HdeA is monomeric. Interestingly, the observed frictional ratio for the dimeric species is consistently larger for HdeA D20A D51A than for HdeA WT at all protein concentrations at which a dimeric species could be observed for both protein variants (Fig. 8 C and D, Fig. S3, and Table S1). This larger frictional ratio for the HdeA D20A D51A dimer indicates a larger degree of unfolding, and therefore hydration, for this species, resulting in a higher partial specific volume. Small errors in the partial specific volume estimation for either WT or mutant will translate to systematic errors in the molecular mass transformation. A derivation from the assumed partial specific volume for the HdeA D20A D51A dimer could explain why the molecular mass for the HdeA D20A D51A dimer is predicted to be slightly higher than the molecular mass of the HdeA WT dimer at all protein concentrations at which a dimeric species could be observed for both protein variants (Fig. 8 C and D, Fig. S3, and Table S1). Overall, the increase in frictional ratio for the dimeric species of HdeA D20A D51A is consistent with our overall observation that the introduction of alanines at positions D20 and D51 of HdeA leads to a destabilization of the dimer and partial unfolding of the protein. Although there appears to be a significant difference in the apparent molecular mass of the HdeA WT dimer compared with the HdeA D20A D51A dimer, within any one protein, measurements of the apparent molecular masses fall within a fairly narrow range that is well within the accuracy of the method.
Fig. 8.
Analytical ultracentrifugation SV analysis of WT HdeA (B and D) and HdeA D20A D51A (A and C) at pH 5. The molecular mass of WT HdeA is 9.7 kDa (monomer) and 19.5 kDa (dimer). Fractions of the monomeric and dimeric species are indicated. The Kd of dimerization for HdeA D20A D51A at pH 5 is in the range of 30 μM. (A and B) Due to low signal extensity, the fractional ratio and molecular mass are approximate for these very low protein concentrations.
Taken together, these data provide further support that monomerization and partial unfolding of HdeA are required for its activation. However, based on our far-UV CD data, the degree of unfolding necessary for activity is smaller than the degree of unfolding that HdeA is subjected to on exposure to low pH. This suggests that some of the pH-induced structural changes that accompany HdeA activation are byproducts of acidification rather than an actual requirement for activity.
Discussion
Changes in the intra- and extracellular pH influence and regulate a magnitude of cellular processes in both eukaryotes and bacteria, ranging from metabolic reactions, gene expression, cellular proliferation (16, 30–33), cell cycle progression (34, 35), cell motility (36), and apoptosis (37) to ameboid (38) and bacterial chemotaxis (39, 40) and activation of toxins. On a molecular level, changes in the environmental pH can significantly affect the strength of electrostatic interactions by directly influencing the ionization/protonation state of solvent-accessible functional groups like amino acid side chains. Such a pH-dependent modulation of charges on a protein’s surface can, in turn, greatly affect the strength of the protein’s interactions with other proteins or ligands, as well as the structure of the protein itself or its catalytical activity. Not surprisingly, very acidic conditions are detrimental to the structure and function of most proteins. Host organisms, including humans, thus frequently use these conditions to protect themselves against pathogens (1). Intriguingly, however, many viral and bacterial pathogens have adapted to use the low pH present in endosomes, phagosomes, and the mammalian stomach to activate pH-sensing proteins crucial for their defense or for infection of the host. Examples are as follows: (i) the integral membrane protein HA, which undergoes acid-induced conformational changes necessary for the uptake of the influenza A virus (41); (ii) pore-forming bacterial toxins like the diphtheria toxin, which undertakes an acid-triggered transformation from a monomeric and soluble protein to a multimeric transmembrane protein (42); and (iii) the acid-sensing chemoreceptor TlpB, which helps the gastric pathogen Helicobacter pylori to colonize its preferred habitat within the human stomach, the epithelial lining, which is more neutral in pH than the acidic lumen (40).
Despite the importance of pH homeostasis for a large number of cellular processes and a magnitude of known pH-sensing proteins, the molecular basis of such pH sensing is not very well understood (17). In the work reported here, we investigated the pH-induced chaperone activation of HdeA, a periplasmic chaperone vital for the survival of the gastric pathogen E. coli during its passage through the human stomach. E. coli is the most common cause of the up to 4 billion episodes of infectious diarrhea that occur each year, with infectious diarrhea being the second leading cause of death in children under 5 y of age (43). Understanding the structural basis for the relationship between protonation, conformation, and activation of HdeA is therefore not only critical in understanding the virulence of this bacterium but could aid in the design of specific drugs targeting this vital protein in a mission to attenuate E. coli’s virulence as well as that of other pathogenic enteric bacteria. In addition, a better understanding of pH-induced protein regulation in general will be instrumental in designing predictable pH switches for therapeutic or scientific purposes. A recent example of an engineered pH switch that shows therapeutic potential is a histidine switch that was engineered into troponin in an effort to prevent myocardial ischemia (44).
In this work, we show that a combined approach of evolutionary conservation assessments and CpHmd simulations is effective in identifying key residues crucial for the acid-induced activation of HdeA. By specifically and permanently neutralizing only two residues (D20 and D51) by mutation, we were able to generate an HdeA variant that is active as a chaperone at neutral pH. The notion that only a very small number of titratable groups within a protein are mainly responsible for triggering the protein’s pH-induced activation is consistent with previous studies (45, 46). In the crystal structure of HdeA, residue 51 is in close proximity to a fairly well-conserved lysine cluster composed of residues K10 and K11 in the other monomer (12, 24). Interestingly, D20 is also located in close proximity to the lysine cluster K10/K11 and forms electrostatic interactions with K10, but within the same monomer. It is therefore possible that neutralization of both D20 and D51 through either mutation or protonation would cause this lysine cluster to lose electrostatic interactions simultaneously to both an adjacent helix and a residue in the other monomer. This could lead to significant conformational flexibility in this part of the protein, consistent with the loss in secondary structure and large destabilization we observe for HdeA D20A D51A. It is worth noting that even in the dimer, residues K10 and K11 are located directly adjacent to disordered regions: Both residues exhibit elevated B factors in the HdeA crystal structures (12, 24), and the adjacent residues 1–8 are not resolved in either crystal structure due to disorder. The close proximity to the already disordered N terminus could therefore enhance any additional structural flexibility resulting from the loss of electrostatic interactions between the lysine cluster K10/K11 and D20/D51.
Many proteins have been postulated to be active in a partially unfolded configuration (47, 48). These so-called “intrinsically disordered” proteins are surprisingly common: About 40% of human proteins are predicted to contain at least one region of disorder (49). However, the mere presence of disordered regions, by itself, does not indicate that disorder plays a functional role. This is because only a tiny percentage of all possible amino acid sequences are predicted to fold into stable structures (50). Further, the vast majority of random mutations are destabilizing (51). As a result, amino acid sequences, in the absence of any selection, are expected to devolve rapidly into disorder. Although there are many good reasons to think that disorder can play an important role in proteins (20, 52, 53), firmly establishing a functional role for disorder has been experimentally difficult. As a conditionally disordered protein, HdeA opens up a unique approach to dissecting the contribution of disorder to protein function. With its small size (active conformation <10 kDa), its independence from any cochaperones or ATP, and our demonstration that it is active as a monomer, HdeA is an example of the minimal protein unit necessary for chaperone function. The fact that HdeA’s activity is regulated by pH makes it particularly attractive, because recent developments have made it possible to predict the effects of pH on protein structure accurately on a residue-by-residue basis (54, 55). Such calculations, coupled with site-specific mutagenesis, have allowed us to design and isolate constitutively active mutants of HdeA. The alanine substitution mutants we generated for HdeA show a clear correlation between the increase in unfolding, loss in secondary structure, decrease in thermostability, and increase in monomerization on the one hand and chaperone activity at elevated pH values on the other hand. These results indicate that partial disorder is indeed necessary for HdeA’s chaperone function.
A role of partial unfolding of HdeA for its activity is consistent with our previous observation that HdeA adopts different conformations when bound to different substrates (13). HdeA is the main acid protection mechanism in the periplasm of enterobacteria; thus, this structural promiscuity might be necessary for HdeA to be able to protect a variety of substrate proteins from acid-induced aggregation and inactivation (56, 57). There is also evidence that disorder allows chaperones to provide a solubilizing effect to chaperone–substrate complexes (58–60). This might be particularly true for HdeA, where the N and C termini are disordered and highly charged at pH values between 7 and 2 (10). WT HdeA is activated by shifts to low pH, a process that is accompanied by a large increase in disorder. Interestingly, the extent of unfolding required for HdeA’s activation (as seen for HdeA D20A D51A) is smaller than the extent of unfolding observed on a drop to low pH. This implies that HdeA does not need to be fully disordered to be fully active. This is consistent with our prior observation that WT HdeA still has some residual secondary structure at pH 2 (13). Our results suggest that some of HdeA’s unfolding on exposure to low pH is a byproduct of acidification rather than a requirement for chaperone activity.
Permanent activation of HdeA as a chaperone coincides not only with unfolding but with monomerization of the protein. In the crystal structure, the HdeA dimer buries a hydrophobic interface of 2,370 Å2, which becomes exposed on monomerization. It is assumed that HdeA engages with unfolded substrates through its exposed hydrophobic residues, similar to how chaperones are thought to interact with their clients to prevent aggregation (13, 14, 56, 61). Consistent with that assumption, we previously observed that the introduction of a single charged residue in the dimer interface significantly reduces chaperone activity toward the two model substrates MDH and rhodanese (13). Although we cannot say definitely if the partial unfolding of HdeA in this permanently activated state is the result or the cause of the monomerization, it is likely that both processes occur simultaneously and that both are important for HdeA’s chaperone activity. Our data therefore support a theory in which both monomerization (exposing hydrophobic residues for substrate binding) and partial unfolding of HdeA (uncoupling specificity from binding strength to bind to a variety of substrate proteins) are necessary for chaperone function. We presume that HdeA has not evolved to be constitutively active because the unregulated tight binding of chaperones to unfolded proteins as soon as they emerge from the translocon could act to inhibit rather than facilitate their proper folding. Overall, our work suggests a rather nuanced view of the role of disorder in protein function. Clearly, complete disorder is not required for HdeA function; rather, it appears that only partial disorder is required. In nature, this partial disorder is conditionally imposed on the protein by pH shifts, but partial disorder can also be imposed on the protein by mutations that cause monomer dissociation at neutral or near-neutral pH.
Materials and Methods
Sequence Alignments.
HdeA homologs were identified by performing eight rounds of position-specific iterative basic local alignment search tool (PSI-BLAST) using E. coli HdeA (24) as a reference sequence. This yielded a set of 164 sequences, which were aligned with MUSCLE (62), adjusted by eye, and used to construct a phylogenetic tree using MEGA5 (63). Of the 164 sequences, 50 were assigned to the HdeA clade. The remaining sequences were either more similar to HdeB [which shares 13% sequence identity with HdeA in E. coli (64)] or clustered into additional clades that appeared to be about equally distant from HdeA and HdeB. From the 50 sequences identified as HdeA homologs, a single sequence was selected per strain per genus and aligned with ClustalW (65).
Constant pH Molecular Dynamics Calculations and pKa Value Predictions.
The effect of protonation of specific acidic residues on the stability of the HdeA dimer interface was based on previously published differences in pKa values between the HdeA dimer and a model of the monomeric state (26). These pKa differences were calculated using the methods of CpHmd (25, 27, 66) for the native WT dimer and a model of the monomer, constructed from the dimer structure and relaxed at different pH values. The pKa differences were then used in the Wyman–Tanford linkage equation to predict the effect of protonation of specific acidic residues on the stability of the HdeA dimer interface (67, 68):
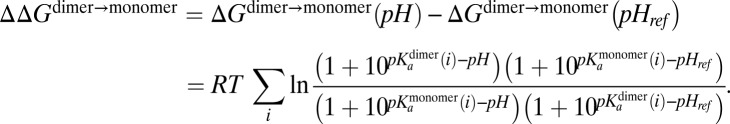 |
In the above expression, pHref = 7 and the change in the pH-dependent electrostatic stability of the dimer interface is calculated at pH = 2.5.
Protein Purification and HdeA Activity Assays.
HdeA and its variants were expressed as described previously (13). HdeA activity was assessed by measuring HdeA’s ability to prevent aggregation of a model protein, MDH (Roche). For aggregation assays at pH 4, pH 5, or pH 7, guanidine denaturated MDH was diluted into buffer containing 400 mM KHPO4, 150 mM NaCl, and 150 mM (NH4)2SO4 (aggregation buffer) to a final concentration of 4 μM in the presence or absence of 4 μM HdeA. Aggregation was monitored by observing light scattering at 350 nm at 25 °C using a Cary Eclipse fluorescence spectrophotometer (Varian).
To determine HdeA activity at low pH, 4 μM MDH was incubated at 25 °C in either the presence or absence of 4 μM HdeA in 150 mM KHPO4, 150 mM NaCl, and 150 mM (NH4)2SO4 (pH 2.2) buffer for 30 min. The solution was then neutralized by adding 0.34 V of 2 M K2HPO4, and light scattering was monitored as described above.
Bis-ANS Binding.
Binding of 15 μM bis-ANS to 3 μM HdeA was measured in buffer containing 10 mM citrate and 150 mM NaCl, which was titrated from about pH 7 to pH 2 by stepwise addition of 1 M HCl. Bis-ANS fluorescence was monitored at 510 nm (emission at 385 nm) using a Cary Eclipse fluorescence spectrophotometer. Bis-ANS fluorescence was then normalized to the signals observed at neutral pH (relative bis-ANS fluorescence of 0) and low pH (relative bis-ANS fluorescence of 1).
CD.
CD measurements were performed with a J-180 spectropolarimeter (Jasco) using quartz cuvettes with a path length of 0.1 cm. Spectra were recorded for 20 μM HdeA in 20 mM KHPO4 buffer (pH 7). For protein melting curves, the CD signal at 222 nm was monitored while heating up the protein sample at a rate of 1 °C/min.
Analytical Ultracentrifugation.
Sedimentation velocity (SV) experiments were carried out in a ProteomeLab XL-I (Beckman Coulter) centrifuge in 50 mM KHPO4, 90 mM NaCl buffer at pH 5. Protein concentrations were 4–70 µM. All data were collected in intensity mode at 280 nm and analyzed with Ultrascan III software, accessed at www.ultrascan.uthscsa.edu (69). SV experiments were designed as described by Demeler (70). They were analyzed first by using the 2D spectrum analysis (71) to remove time- and radially invariant noise and then by genetic algorithm analysis (72), followed by a combined genetic algorithm--Monte Carlo analysis (73) to determine the fitting statistics. All finite element modeling was performed with Ultrascan III (69, 74). Partial specific volumes of HdeA WT and HdeA D20A D51A were estimated based on amino acid composition with UltraScan III and found to be 0.729 mL/g and 0.732 mL/g, respectively. Buffer density and viscosity were estimated based on composition with UltraScan III.
Supplementary Material
Acknowledgments
We thank Dr. Thomas Duda for help with the sequence alignments. We also thank Borries Demeler, Philipp Koldewey, and Titus Franzmann for help with the analytical ultracentrifugation analysis. The Howard Hughes Medical Institute (J.C.A.B.) and National Institutes of Health [Grant GM057513 (to C.L.B.)] funded this work. Analytical ultracentrifugation calculations were performed on the UltraScan Laboratory Information Management System (LIMS) cluster at the Bioinformatics Core Facility at the University of Texas Health Science Center at San Antonio. Extreme Sciences and Engineering Discovery Environment (XSEDE) resources were supported by National Science Foundation XSEDE Grant MCB070038 (to Borries Demeler), and the Gateway is made possible by the use of XSEDE resources and the Extended Collaborative Support Service Program funded by the National Science Foundation through Award OCI-1053575.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 5279.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222458110/-/DCSupplemental.
References
- 1.Smith JL. The role of gastric acid in preventing foodborne disease and how bacteria overcome acid conditions. J Food Prot. 2003;66(7):1292–1303. doi: 10.4315/0362-028x-66.7.1292. [DOI] [PubMed] [Google Scholar]
- 2.Gorden J, Small PL. Acid resistance in enteric bacteria. Infect Immun. 1993;61(1):364–367. doi: 10.1128/iai.61.1.364-367.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Small P, Blankenhorn D, Welty D, Zinser E, Slonczewski JL. Acid and base resistance in Escherichia coli and Shigella flexneri: Role of rpoS and growth pH. J Bacteriol. 1994;176(6):1729–1737. doi: 10.1128/jb.176.6.1729-1737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hong W, Wu YE, Fu X, Chang Z. Chaperone-dependent mechanisms for acid resistance in enteric bacteria. Trends Microbiol. 2012;20(7):328–335. doi: 10.1016/j.tim.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Zhao B, Houry WA. Acid stress response in enteropathogenic gammaproteobacteria: An aptitude for survival. Biochem Cell Biol. 2010;88(2):301–314. doi: 10.1139/o09-182. [DOI] [PubMed] [Google Scholar]
- 6.Richard H, Foster JW. Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J Bacteriol. 2004;186(18):6032–6041. doi: 10.1128/JB.186.18.6032-6041.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foster JW. Escherichia coli acid resistance: Tales of an amateur acidophile. Nat Rev Microbiol. 2004;2(11):898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- 8.Koebnik R, Locher KP, Van Gelder P. Structure and function of bacterial outer membrane proteins: Barrels in a nutshell. Mol Microbiol. 2000;37(2):239–253. doi: 10.1046/j.1365-2958.2000.01983.x. [DOI] [PubMed] [Google Scholar]
- 9.Malki A, et al. Solubilization of protein aggregates by the acid stress chaperones HdeA and HdeB. J Biol Chem. 2008;283(20):13679–13687. doi: 10.1074/jbc.M800869200. [DOI] [PubMed] [Google Scholar]
- 10.Hong W, et al. Periplasmic protein HdeA exhibits chaperone-like activity exclusively within stomach pH range by transforming into disordered conformation. J Biol Chem. 2005;280(29):27029–27034. doi: 10.1074/jbc.M503934200. [DOI] [PubMed] [Google Scholar]
- 11.Fink AL, Calciano LJ, Goto Y, Kurotsu T, Palleros DR. Classification of acid denaturation of proteins: Intermediates and unfolded states. Biochemistry. 1994;33(41):12504–12511. doi: 10.1021/bi00207a018. [DOI] [PubMed] [Google Scholar]
- 12.Yang F, Gustafson KR, Boyd MR, Wlodawer A. Crystal structure of Escherichia coli HdeA. Nat Struct Biol. 1998;5(9):763–764. doi: 10.1038/1796. [DOI] [PubMed] [Google Scholar]
- 13.Tapley TL, et al. Structural plasticity of an acid-activated chaperone allows promiscuous substrate binding. Proc Natl Acad Sci USA. 2009;106(14):5557–5562. doi: 10.1073/pnas.0811811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bardwell JC, Jakob U. Conditional disorder in chaperone action. Trends Biochem Sci. 2012;37(12):517–525. doi: 10.1016/j.tibs.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reichmann D, et al. Order out of disorder: Working cycle of an intrinsically unfolded chaperone. Cell. 2012;148(5):947–957. doi: 10.1016/j.cell.2012.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tapley TL, Franzmann TM, Chakraborty S, Jakob U, Bardwell JC. Protein refolding by pH-triggered chaperone binding and release. Proc Natl Acad Sci USA. 2010;107(3):1071–1076. doi: 10.1073/pnas.0911610107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srivastava J, Barber DL, Jacobson MP. Intracellular pH sensors: Design principles and functional significance. Physiology (Bethesda) 2007;22:30–39. doi: 10.1152/physiol.00035.2006. [DOI] [PubMed] [Google Scholar]
- 18.Chuang SE, Blattner FR. Characterization of twenty-six new heat shock genes of Escherichia coli. J Bacteriol. 1993;175(16):5242–5252. doi: 10.1128/jb.175.16.5242-5252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu YE, Hong W, Liu C, Zhang L, Chang Z. Conserved amphiphilic feature is essential for periplasmic chaperone HdeA to support acid resistance in enteric bacteria. Biochem J. 2008;412(2):389–397. doi: 10.1042/BJ20071682. [DOI] [PubMed] [Google Scholar]
- 20.Tompa P, Fuxreiter M. Fuzzy complexes: Polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci. 2008;33(1):2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Cortese MS, Uversky VN, Dunker AK. Intrinsic disorder in scaffold proteins: Getting more from less. Prog Biophys Mol Biol. 2008;98(1):85–106. doi: 10.1016/j.pbiomolbio.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uversky VN. Intrinsically disordered proteins from A to Z. Int J Biochem Cell Biol. 2011;43(8):1090–1103. doi: 10.1016/j.biocel.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 23.Beckwith J, Rossow P. Analysis of genetic regulatory mechanisms. Annu Rev Genet. 1974;8:1–13. doi: 10.1146/annurev.ge.08.120174.000245. [DOI] [PubMed] [Google Scholar]
- 24.Gajiwala KS, Burley SK. HDEA, a periplasmic protein that supports acid resistance in pathogenic enteric bacteria. J Mol Biol. 2000;295(3):605–612. doi: 10.1006/jmbi.1999.3347. [DOI] [PubMed] [Google Scholar]
- 25.Lee MS, Salsbury FR, Jr, Brooks CL., III Constant-pH molecular dynamics using continuous titration coordinates. Proteins. 2004;56(4):738–752. doi: 10.1002/prot.20128. [DOI] [PubMed] [Google Scholar]
- 26.Zhang BW, Brunetti L, Brooks CL., III Probing pH-dependent dissociation of HdeA dimers. J Am Chem Soc. 2011;133(48):19393–19398. doi: 10.1021/ja2060066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khandogin J, Brooks CL., III Constant pH molecular dynamics with proton tautomerism. Biophys J. 2005;89(1):141–157. doi: 10.1529/biophysj.105.061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hawe A, Sutter M, Jiskoot W. Extrinsic fluorescent dyes as tools for protein characterization. Pharm Res. 2008;25(7):1487–1499. doi: 10.1007/s11095-007-9516-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitmore LL, Wallace BAB. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 30.Denker SP, Huang DC, Orlowski J, Furthmayr H, Barber DL. Direct binding of the Na–H exchanger NHE1 to ERM proteins regulates the cortical cytoskeleton and cell shape independently of H(+) translocation. Mol Cell. 2000;6(6):1425–1436. doi: 10.1016/s1097-2765(00)00139-8. [DOI] [PubMed] [Google Scholar]
- 31.Kapus A, Grinstein S, Wasan S, Kandasamy R, Orlowski J. Functional characterization of three isoforms of the Na+/H+ exchanger stably expressed in Chinese hamster ovary cells. ATP dependence, osmotic sensitivity, and role in cell proliferation. J Biol Chem. 1994;269(38):23544–23552. [PubMed] [Google Scholar]
- 32.Pouysségur J, Chambard JC, Franchi A, Paris S, Van Obberghen-Schilling E. Growth factor activation of an amiloride-sensitive Na+/H+ exchange system in quiescent fibroblasts: Coupling to ribosomal protein S6 phosphorylation. Proc Natl Acad Sci USA. 1982;79(13):3935–3939. doi: 10.1073/pnas.79.13.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pouysségur J, Franchi A, L’Allemain G, Paris S. Cytoplasmic pH, a key determinant of growth factor-induced DNA synthesis in quiescent fibroblasts. FEBS Lett. 1985;190(1):115–119. doi: 10.1016/0014-5793(85)80439-7. [DOI] [PubMed] [Google Scholar]
- 34.Putney LK, Barber DL. Na-H exchange-dependent increase in intracellular pH times G2/M entry and transition. J Biol Chem. 2003;278(45):44645–44649. doi: 10.1074/jbc.M308099200. [DOI] [PubMed] [Google Scholar]
- 35.Turchi L, Loubat A, Rochet N, Rossi B, Ponzio G. Evidence for a direct correlation between c-Jun NH2 terminal kinase 1 activation, cyclin D2 expression, and G(1)/S phase transition in the murine hybridoma 7TD1 cells. Exp Cell Res. 2000;261(1):220–228. doi: 10.1006/excr.2000.5060. [DOI] [PubMed] [Google Scholar]
- 36.Denker SP, Barber DL. Cell migration requires both ion translocation and cytoskeletal anchoring by the Na-H exchanger NHE1. J Cell Biol. 2002;159(6):1087–1096. doi: 10.1083/jcb.200208050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schelling JR, Abu Jawdeh BG. Regulation of cell survival by Na+/H+ exchanger-1. Am J Physiol Renal Physiol. 2008;295(3):F625–F632. doi: 10.1152/ajprenal.90212.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patel H, Barber DL. A developmentally regulated Na-H exchanger in Dictyostelium discoideum is necessary for cell polarity during chemotaxis. J Cell Biol. 2005;169(2):321–329. doi: 10.1083/jcb.200412145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kihara M, Macnab RM. Cytoplasmic pH mediates pH taxis and weak-acid repellent taxis of bacteria. J Bacteriol. 1981;145(3):1209–1221. doi: 10.1128/jb.145.3.1209-1221.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goers Sweeney E, et al. Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB. Structure. 2012;20(7):1177–1188. doi: 10.1016/j.str.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371(6492):37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 42.Chenal A, et al. Membrane protein insertion regulated by bringing electrostatic and hydrophobic interactions into play. A case study with the translocation domain of diphtheria toxin. J Biol Chem. 2002;277(45):43425–43432. doi: 10.1074/jbc.M204148200. [DOI] [PubMed] [Google Scholar]
- 43.Ochoa TJ, Contreras CA. Enteropathogenic escherichia coli infection in children. Curr Opin Infect Dis. 2011;24(5):478–483. doi: 10.1097/QCO.0b013e32834a8b8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Day SM, et al. Histidine button engineered into cardiac troponin I protects the ischemic and failing heart. Nat Med. 2006;12(2):181–189. doi: 10.1038/nm1346. [DOI] [PubMed] [Google Scholar]
- 45.Tang Y, Grey MJ, McKnight J, Palmer AG, 3rd, Raleigh DP. Multistate folding of the villin headpiece domain. J Mol Biol. 2006;355(5):1066–1077. doi: 10.1016/j.jmb.2005.10.066. [DOI] [PubMed] [Google Scholar]
- 46.Törnroth-Horsefield S, et al. Structural mechanism of plant aquaporin gating. Nature. 2006;439(7077):688–694. doi: 10.1038/nature04316. [DOI] [PubMed] [Google Scholar]
- 47.Minezaki Y, Homma K, Kinjo AR, Nishikawa K. Human transcription factors contain a high fraction of intrinsically disordered regions essential for transcriptional regulation. J Mol Biol. 2006;359(4):1137–1149. doi: 10.1016/j.jmb.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 48.Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: Introducing the D2 concept. Annu Rev Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- 49.Uversky VN, Dunker AK. Understanding protein non-folding. Biochim Biophys Acta. 2010;1804(6):1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lau KF, Dill KA. Theory for protein mutability and biogenesis. Proc Natl Acad Sci USA. 1990;87(2):638–642. doi: 10.1073/pnas.87.2.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo HH, Choe J, Loeb LA. Protein tolerance to random amino acid change. Proc Natl Acad Sci USA. 2004;101(25):9205–9210. doi: 10.1073/pnas.0403255101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen J. Towards the physical basis of how intrinsic disorder mediates protein function. Arch Biochem Biophys. 2012;524(2):123–131. doi: 10.1016/j.abb.2012.04.024. [DOI] [PubMed] [Google Scholar]
- 53.De Simone A, et al. Intrinsic disorder modulates protein self-assembly and aggregation. Proc Natl Acad Sci USA. 2012;109(18):6951–6956. doi: 10.1073/pnas.1118048109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alexov E, et al. Progress in the prediction of pKa values in proteins. Proteins. 2011;79(12):3260–3275. doi: 10.1002/prot.23189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wallace JA, Shen JK. Predicting pKa values with continuous constant pH molecular dynamics. Methods Enzymol. 2009;466:455–475. doi: 10.1016/S0076-6879(09)66019-5. [DOI] [PubMed] [Google Scholar]
- 56.Lin S, et al. Site-specific incorporation of photo-cross-linker and bioorthogonal amino acids into enteric bacterial pathogens. J Am Chem Soc. 2011;133(50):20581–20587. doi: 10.1021/ja209008w. [DOI] [PubMed] [Google Scholar]
- 57.Zhang M, et al. A genetically incorporated crosslinker reveals chaperone cooperation in acid resistance. Nat Chem Biol. 2011;7(10):671–677. doi: 10.1038/nchembio.644. [DOI] [PubMed] [Google Scholar]
- 58.Kim TD, Paik SR, Yang CH. Structural and functional implications of C-terminal regions of alpha-synuclein. Biochemistry. 2002;41(46):13782–13790. doi: 10.1021/bi026284c. [DOI] [PubMed] [Google Scholar]
- 59.Pasta SY, Raman B, Ramakrishna T, Rao ChM. Role of the C-terminal extensions of alpha-crystallins. Swapping the C-terminal extension of alpha-crystallin to alphaB-crystallin results in enhanced chaperone activity. J Biol Chem. 2002;277(48):45821–45828. doi: 10.1074/jbc.M206499200. [DOI] [PubMed] [Google Scholar]
- 60.Tompa P, Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18(11):1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- 61.Lin Z, Rye HS. GroEL-mediated protein folding: Making the impossible, possible. Crit Rev Biochem Mol Biol. 2006;41(4):211–239. doi: 10.1080/10409230600760382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang W, et al. Salt bridges regulate both dimer formation and monomeric flexibility in HdeB and may have a role in periplasmic chaperone function. J Mol Biol. 2012;415(3):538–546. doi: 10.1016/j.jmb.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Khandogin J, Brooks CL., III Toward the accurate first-principles prediction of ionization equilibria in proteins. Biochemistry. 2006;45(31):9363–9373. doi: 10.1021/bi060706r. [DOI] [PubMed] [Google Scholar]
- 67.Wyman J., Jr Linked functions and reciprocal effects in hemoglobin: A second look. Adv Protein Chem. 1964;19:223–286. doi: 10.1016/s0065-3233(08)60190-4. [DOI] [PubMed] [Google Scholar]
- 68.Tanford C. Protein denaturation. C. Theoretical models for the mechanism of denaturation. Adv Protein Chem. 1970;24:1–95. [PubMed] [Google Scholar]
- 69.Demeler B. UltraScan—A comprehensive data analysis software package for analytical ultracentrifugation experiments. In: Scott DJ, Harding SE, Rowe AJ, editors. Modern Analytical Ultracentrifugation: Techniques and Methods. Cambridge, UK: Royal Society of Chemistry; 2005. pp. 210–229. [Google Scholar]
- 70. Demeler B (2010) Methods for the design and analysis of sedimentation velocity and sedimentation equilibrium experiments with proteins. Curr Protoc Protein Sci, 10.1002/0471140864.ps0713s60. [DOI] [PMC free article] [PubMed]
- 71.Brookes E, Cao W, Demeler B. A two-dimensional spectrum analysis for sedimentation velocity experiments of mixtures with heterogeneity in molecular weight and shape. Eur Biophys J. 2010;39(3):405–414. doi: 10.1007/s00249-009-0413-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brookes EH, Demeler B. Parsimonious regularization using genetic algorithms applied to the analysis of analytical ultracentrifugation experiments. Gecco 2007: Genetic and Evolutionary Computation Conference (ACM, New York) 2007;Vol 1 and 2:361–368. [Google Scholar]
- 73.Demeler B, Brookes E. Monte Carlo analysis of sedimentation experiments. Colloid Polym Sci. 2008;286(2):129–137. [Google Scholar]
- 74.Demeler B, et al. UltraScan-III (version 1482). A Comprehensive Analysis Software for Analytical Ultracentrifugation experiments. San Antonio, TX: University of Texas Health Science Center; 2011. [Google Scholar]
- 75.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.