Abstract
Sulfolobus turreted icosahedral virus (STIV) was isolated in acidic hot springs where it infects the archeon Sulfolobus solfataricus. We determined the STIV structure using near-atomic resolution electron microscopy and X-ray crystallography allowing tracing of structural polypeptide chains and visualization of transmembrane proteins embedded in the viral membrane. We propose that the vertex complexes orchestrate virion assembly by coordinating interactions of the membrane and various protein components involved. STIV shares the same coat subunit and penton base protein folds as some eukaryotic and bacterial viruses, suggesting that they derive from a common ancestor predating the divergence of the three kingdoms of life. One architectural motif (β-jelly roll fold) forms virtually the entire capsid (distributed in three different gene products), indicating that a single ancestral protein module may have been at the origin of its evolution.
Keywords: Archaea, electron microscopy, virus assembly, single-particle reconstruction, PRD1-Adeno viral lineage
The discovery of Archaea is a recent highlight of evolutionary biology that led to the definition of three (instead of two) domains of life (1, 2). Many Archaea are isolated from ecosystems characterized by extreme conditions of temperature, pH, or salinity, such as hot springs, geysers, or deep-sea hydrothermal vents (3). Despite the hostile nature of these ecological niches, viruses infecting extremophile organisms are ubiquitous, and their discovery supports the postulate that all forms of life are targeted by viral attacks. Archaeal viruses exhibit exceptional morphological (droplets, fusiforms, and bottle-shaped) and genetic features (sequences encoded in their genomes are generally unrelated to those of other known viruses) (3). These characteristics reflect their position in the Archaeal domain of life, and perhaps their adaptation to hostile ecosystems and the resulting isolation from atypical environments. Beyond these remarkable features, the association of extreme environments with the physico-chemical conditions of the “primordial soup” further fosters interest in Archaea and their viruses. Indeed, their study may shed light onto the emergence of life as well as its evolution (4).
Recent structural studies revealed that apparently unrelated viruses, infecting organisms belonging to different domains of life (Archaea, Bacteria, or Eukarya) share common architectural principles and topologically identical coat subunit folds. The latter is the two jelly-roll motif fused in a single polypeptide chain that assembles as quasi-hexagonal, trimeric capsomers. This observation strongly suggests a common evolutionary origin for a protein component belonging to viruses infecting organisms of the three domains of life. Based on these results, viruses harboring this double jelly-roll fold are proposed to be in the PRD1-Adenovirus lineage that comprises Adenovirus (5–7), Paramecium bursaria Chlorella virus-1 (PBCV-1) (8), vaccinia virus (9), Sputnik (10), PRD1 (11, 12), PM2 (13), virus—Salisaeta icosahedral phage 1 (SSIP-1) (4), P23-77 (14), Bam35 (15), SH1 (16), Haloarcula hispanica icosahedral virus 2 (HHIV-2) (17), and Sulfolobus turreted icosahedral virus (STIV) (18).
STIV infects the hyperthermo-acidophilic archeon Sulfolobus solfataricus, which grows optimally at pH 3 and 80 °C (19). This system is considered a model for hyperthermophilic Archaea and their viruses due to the relatively extensive genetic and biochemical data available in the literature compared with other archaeal virus/host systems (20, 21). STIV is a 75-MDa icosahedral virus that has a 17.6-kbp circular dsDNA genome encoding 36 predicted ORFs and nine structural proteins (19, 22). Electron microscopy and mass-spectrometry of virions, as well as crystallographic studies of its major capsid subunit, demonstrated that STIV is formed by a proteinaceous icosahedral capsid with turrets at its vertices. The capsid surrounds a host-derived internal membrane that protects the viral genome (19, 22, 23). Tight control of virus assembly is achieved by modulating the expression pattern of viral and host genes to produce quasi-crystalline lattices made of mature virions in the Sulfolobus cytoplasm (24, 25). At the end of the infection cycle, STIV particles induce host lysis and are released through pyramid-like structures perforating the S-layer at the cell surface (24, 26). This stunning virus exit mechanism is unique to the archaeal world and has only been observed for Sulfolobus islandicus rod-shaped virus (SIRV2) in addition to STIV (27).
Despite extensive efforts carried out to decipher the STIV life cycle, little is known about the detailed architecture of this large membrane-containing virion or that of any related archaeal viruses. Furthermore, the mechanisms governing its assembly are also poorly understood due to the limited resolution of the structural data available to date. We report here the atomic structure of the mature STIV virion obtained by a combination of high-resolution single-particle electron cryomicroscopy and X-ray crystallography. We present an atomic model for the virion capsid and analyze the interactions underlying the formation of this 75-MDa hyperthermo-acidophilic macromolecular machine. Integrating these results with data from the literature, we hypothesize that the vertex complexes coordinate the coassembly of the lipid membrane and the coat protein shell to form a provirion into which the genome is subsequently packaged. Finally, comparisons of the different STIV structural proteins lead us to posit that the single ancestral virus jelly-roll module may have given rise to the STIV virion via duplication and diversification events.
Results
Electron Cryomicroscopy Reconstruction of a 75-MDa Viral Molecular Machine.
We obtained the structure of the mature STIV virion using near-atomic resolution single-particle electron microscopy assisted by X-ray crystallography of isolated capsid gene products. The final electron potential map allowed identification of many structural proteins and the tracing of their polypeptide chains due to well-resolved features in the density. The Fourier shell correlation (FSC) for the entire virus reconstruction indicates a resolution of 4.5 Å at a cutoff of 0.143, as defined by Rosenthal and Henderson (28) (Fig. S1). The capsid shell is best defined, probably due to the fact that it dominates alignment of particle images during refinement whereas the resolution appears to gradually decrease while moving away from this region. We estimated the resolution in the coat subunit region to 3.9 Å according to the FSC and to the observed features in the map (Fig. S1). Fifteenfold quasi-equivalent averaging of the coat subunit density within an icosahedral asymmetric unit only marginally improved map quality and did not result in any significant resolution improvement. This result is explained by the 0.75 correlation coefficients between quasi-equivalent related densities before averaging.
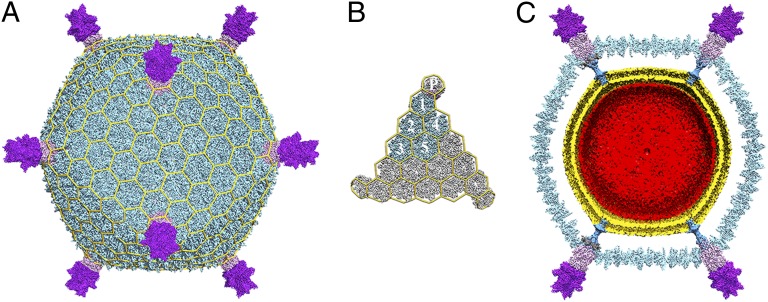
STIV virions have a total radius of 480 Å comprising an icosahedral capsid with a radius of 365 Å decorated with a turret structure at each fivefold vertex that extends 115 Å above the capsid (Fig. 1A). The virus architecture is based on a pseudo T = 31d capsid symmetry (Fig. 1B) with each icosahedral asymmetric unit encompassing 15 copies of the coat subunit (B345), one copy of the A223 penton base protein, one copy of the C381 turret protein, and one copy of the A55 membrane protein (A, B, and C denote different reading frames in the viral genome and the number corresponds to the number of amino acid residues in the ORF). A lipid membrane is present below the proteinaceous capsid shell and is composed of a specific subset of tetraether lipids present in the host cytoplasmic membrane (22). The 17.6-kbp circular dsDNA genome is enclosed within the viral membrane and fills the volume defined by it. A single ordered layer of dsDNA is adjacent to the lipid headgroups facing the virion interior (Fig. 1C).
Fig. 1.
Near-atomic resolution electron cryomicroscopy reconstruction of STIV. (A) The overall virus reconstruction is displayed with the different protein components individually colored (B345, light blue; A223, light pink; C381, purple) and with an icosahedral cage overlaid onto it. (B) Blow-up view of an icosahedral face with one capsid icosahedral asymmetric unit colored as in A and labeled (1–5 for the trimeric B345 capsomers and P for the A223 penton base). (C) Cross-section of the reconstruction revealing the presence of the viral membrane (gold) and the internal genome (red). The cement protein (dark gray) has been removed from the two vertex complexes located on the right-hand side to allow a better visibility of the β-pore.
Coat Subunit Shell.
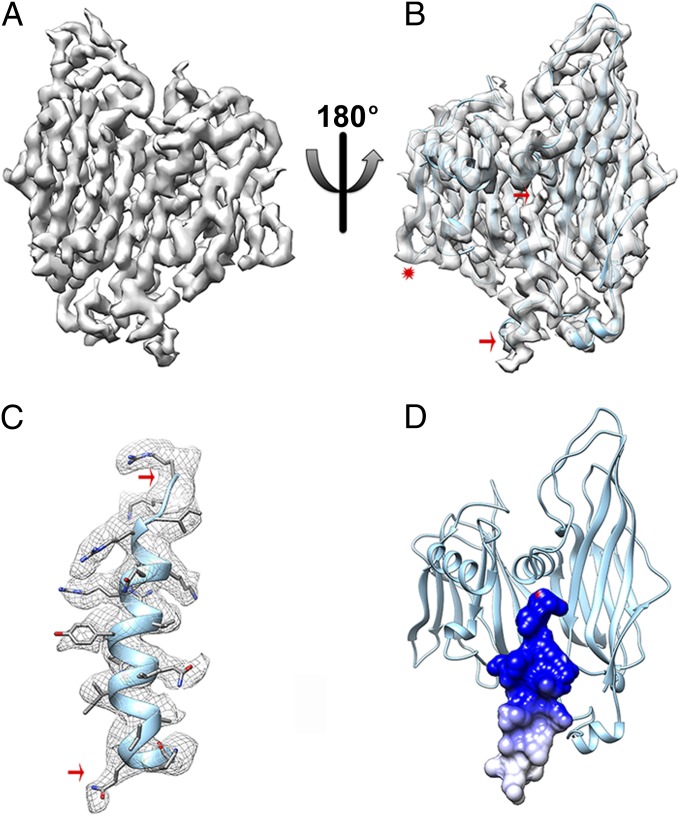
The B345 density is continuous from Gly2 to Arg345 (the C-terminal residue) with many side chains resolved, as expected for a 3.9-Å resolution structure. The monomeric B345 coat subunit crystal structure (18) fits well into the EM map (Fig. 2 A and B) with numerous side chains repositioned, at subunit interfaces due to the capsid shell formation. The conformation of the E-F loop in the smaller jelly-roll (residues 239–243) also exhibits various degrees of rearrangement in most B345 subunits to accommodate interactions with neighbors (Fig. 2B). This loop is projected either between the two jelly-rolls of a single B345 subunit or into the interface between two B345 subunits. The loop conformation is flexible and is dictated by its microenvironment. The overall B345 rigidity is emphasized by the exceptionally limited conformational changes between the crystal structure and assembled subunits. This rigidity is a typical characteristic of thermophilic proteins as they rely on increased thermodynamic stability to cope with the elevated temperature of the environment (29). As for all other viruses of the PRD1-Adenovirus lineage, the STIV B345 coat subunit capsomers are organized in trimers with quasi-sixfold appearance (Fig. 1B) due to the presence of two radially oriented jelly-rolls in each subunit (Fig. 2 B and D) (5–8, 13, 18, 30). B345 monomers bury an average surface of ∼3,700 Å2 per monomer, including all interfaces. The contacts have several conserved salt bridges and hydrogen bonds that ensure the specificity of assembly recognition and the stability of the shell (Table S1) (31).
Fig. 2.
The capsid shell. (A) CryoEM density of a single B345 coat subunit at 3.9 Å resolution. (B) Fit of the B345 crystal structure into the corresponding density. The crystal structure stops at residue 324 whereas the cryoEM density is continuous until the C-terminal residue and folds as an α-helix (delineated by the two red arrows). The red star indicates the position of the E-F loop belonging to the smaller jelly-roll of the B345 subunit. (C) The B345 C-terminal α-helix (residues 325–345) was modeled de novo in the cryoEM density. The red arrows are matching the region shown in B. (D) The electrostatic surface potential of the B345 C-terminal helix reveals that its most C-terminal moiety is strongly positively charged due to the presence of many basic residues. Electrostatics calculations were carried out at pH 3.0 and 80 °C, and the result is displayed colored from red (−5 kT/e) to blue (+5 kT/e).
The C-terminal 20 residues of B345 are fully resolved in the 15 monomers of the icosahedral asymmetric unit. We modeled, de novo, this protein segment (not visible in the crystal structure) and assigned the amino acid sequence relying on the presence of numerous bulky side chains (Fig. 2 B and C). These residues form an α-helix that projects vertically at the interface between the two jelly-roll domains of the protomer to which they belong (Fig. 2 B and D). The C-terminal half of this helix exhibits a largely positive electrostatic potential (Fig. 2D) that would be favorable for interactions with negatively charged lipid headgroups present in the adjacent viral membrane. A previously reported STIV reconstruction at 27 Å resolution supports such an interaction with the helices pointing down and contacting the membrane (19, 23). As mentioned in Discussion, the difference in the direction of the helices between the earlier reconstruction and the one presented here presents an unresolved conundrum that suggests the possibility of different states of maturation in the two virus samples. The B345 C terminus interaction with the membrane previously observed is reminiscent of a comparable interaction in phage PRD1 in which the coat subunit N terminus establishes such contacts (11, 12).
Penton Base.
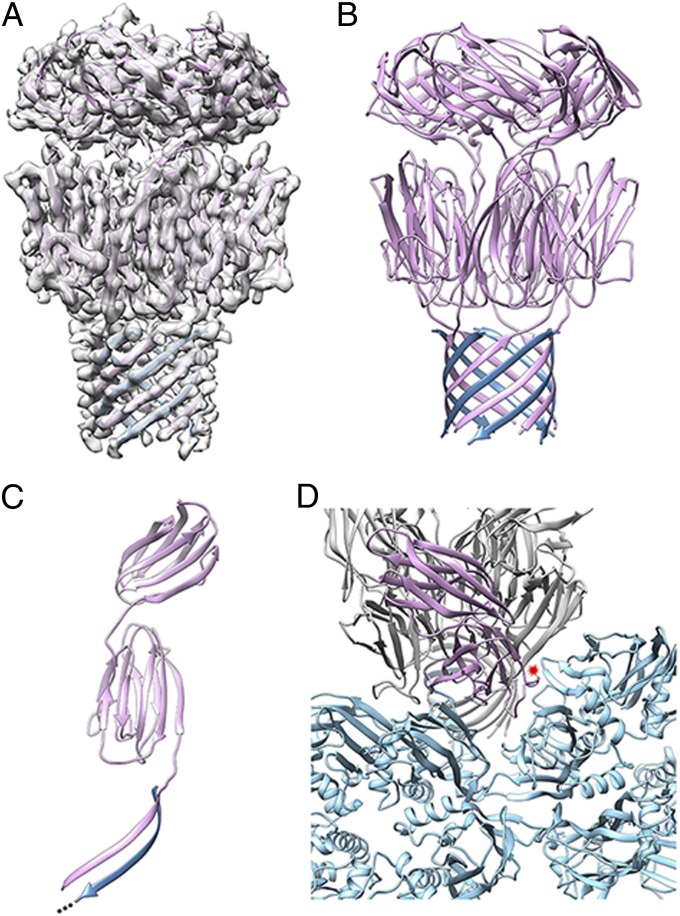
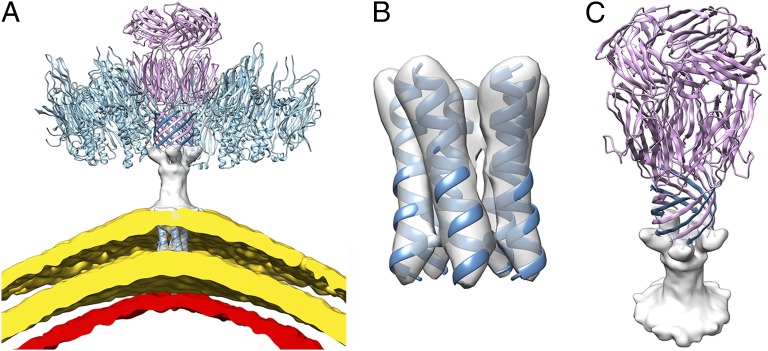
The quality of the reconstruction at the capsid vertices, occupied by A223, the penton base, is equivalent to that of the B345 subunits. We obtained a complete atomic model of A223 by combining de novo backbone modeling in the cryoEM density and high-resolution X-ray crystallography (Tables S2 and S3). Overall, the pentameric penton base is 94 Å high and 57 Å wide with three different layers (Fig. 3 A–C). The N termini are assembled as a 10-stranded β-pore with each protomer contributing one strand to a two-stranded antiparallel β-sheet formed with the A55 membrane protein. This pore constitutes the first internal layer and is reminiscent of the bacterial toxin α-hemolysin (32) although that pore is spanning a cytoplasmic membrane whereas in STIV it is exposed to the solvent just underneath the capsid. At the C terminus of the A223 strand contributing to the β-pore, each polypeptide folds into a canonical viral jelly-roll forming a 55-Å-wide pentameric ring that seals the vertices of the capsid shell (the penton base region) and forms the second layer. Each of these domains is supplemented by an additional C-terminal β-strand reaching the capsid outer surface where a second pentameric ring of jelly-rolls (forming the third layer) decorates the penton base structure and forms the first floor of the turret. These two pentameric rings have comparable widths but different heights as the penton base jelly-rolls forming the second layer are radially oriented relative to the capsid surface whereas the jelly-rolls forming the third layer are rotated by 78° and are thus nearly tangential relative to the capsid shell. The interactions between the B345 coat subunits and the A223 penton base rely on a complex network of contacts spanning neighboring asymmetric units. On one hand, each A223 jelly-roll interacts via its B-C face with the larger jelly-roll of B345 belonging to the same icosahedral asymmetric unit (Fig. 3D). On the other hand, the prominent E-F loop of each A223 fits in the concave surface defined by the two jelly-rolls of B345 from the next asymmetric unit (Fig. 3D). A few contacts occur directly between the proximal region of individual B345 subunits and the β-pore. Beyond these interactions, the cryoEM density reveals an additional fivefold symmetric cement protein forming an annulus around the external surface of the A223/A55 β-pore that bridges it to the B345 subunits (Fig. 1C and Fig. S2 A and B). This protein, whose identity remains to be defined, forms a short helical segment and contacts at two points the B345 subunit belonging to the same icosahedral asymmetric unit (the turn preceding the B345 coat subunit C-terminal helix and at the bottom of the smaller jelly-roll) as well as the bottom of the larger jelly-roll from a B345 subunit contributed by the adjacent asymmetric unit (Fig. S2C). Surprisingly, the density attributed to this protein is rather weak and poorly defined in comparison with the one corresponding to the protein components with which it interacts (B345 and A223). This apparent disorder might be either a result of partial occupancy or a consequence of the structural properties of the cementing protein that could serve as an adaptable protein to seal interactions at the vertex complexes.
Fig. 3.
The penton base structure. (A) The pentameric A223 structure was built by combining de novo modeling in the cryoEM density with X-ray crystallography of its C-terminal domain jelly-roll. (B) The A223 pentamer is formed of three layers from the N to the C terminus: (i) a 10-stranded β-pore formed with the A55 membrane protein (steel blue), (ii) the penton base jelly-roll ring, and (iii) the third layer, also forming a ring of jelly-rolls, which is connected to the rest of the structure via an additional strand added to the penton base jelly-roll domain. (C) A single A223 monomer. The A223 N-terminal β-strand is associated in an anti-parallel manner to the A55 N-terminal β-strand (steel blue). (D) Each A223 jelly-roll interacts via its B-C face with the larger jelly-roll of B345 belonging to the same icosahedral asymmetric unit whereas its prominent E-F loop fits in the concave surface defined by the two jelly-rolls of B345 from the next asymmetric unit (indicated by a red star). B345 subunits are shown in light blue. The β-strands B and C from the A223 penton base domain rendered in light pink are labeled for clarity.
Despite the absence of detectable sequence identity, the architecture of the second layer of the A223 penton-base is similar to the equivalent proteins of phages PRD1 (12) and PM2 (13) as well as to Adenovirus (33) and Sputnik virophage (10). Indeed, all these proteins exhibit a jelly-roll fold and they can be superimposed with rmsd values ranging between 2.5 and 3.5 Å (Fig. S3). Each penton base protein is, however, decorated in various ways with additional structural motifs appended on the outside and/or the inside of the capsid shell in some of these virions.
Architecture of the Turret.
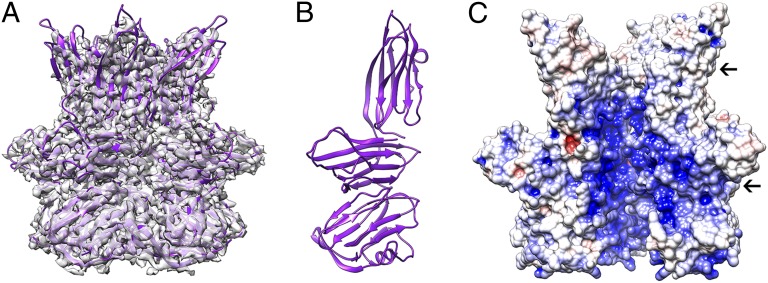
The cryoEM density of the turret subunits (C381) decreases gradually in quality with increasing distance above the capsid. To address this problem, we obtained an atomic model of the turret protein by solving its crystal structure in its biologically relevant pentameric state. We then used rigid-body fitting to place the coordinates into the STIV cryoEM map. The C381 pentamer is apposed on the penton base C-terminal jelly-roll ring and is 88 Å high and 114 Å wide (Fig. 4A). It is formed by three consecutive layers of jelly-rolls. Only the first domain is decorated at its N terminus by a short α-helix and three β-strands contributing to two different β-sheets formed with two neighboring C381 subunits. The three different jelly-rolls exhibit different orientations and therefore form rings of varying diameters: the N-terminal domain adopts an intermediate configuration whereas the central and C-terminal jelly-rolls are positioned tangentially and radially relative to the capsid surface, respectively (Fig. 4B). The interior of the C381 pentamer harbors two rather large cavities delineated by constrictions at the interface between the N-terminal and the central jelly-roll rings as well as at the turret distal extremity (Fig. 4C). These cavities exhibit an overall positive electrostatic potential, suggesting that the turrets are probably not involved in genome transit during packaging or infection (Fig. 4C) (34). Extensive interactions are observed at the interface between C381 turret proteins accounting for an average buried surface area of 2,420 Å2 per monomer (corresponding to 14% of the total protein surface). These contacts have an unusually high number of salt bridges (20), probably ensuring the thermodynamic stability of this structure (31).
Fig. 4.
Architecture of the turret. (A) The pentameric C381 turret protein X-ray structure is shown fit into the corresponding region of the cryoEM density. (B) Each C381 monomer is formed of three layers of jelly-rolls with markedly different orientations. (C) The interior of the C381 pentamer exhibits a positive electrostatic surface potential and several constrictions (indicated by black arrows). These characteristics are unfavorable to allow dsDNA genome transit during packaging or infection. Electrostatics calculations were carried out at pH 3.0 and 80 °C, and the result is displayed colored from red (−5 kT/e) to blue (+5 kT/e). One C381 monomer has been removed to allow visualization of the pentamer central cavities.
Analysis of the C381 turret protein structure using the DALI server (35) demonstrated that it shares similarity with many carbohydrate-binding modules as well as domains involved in protein–protein interactions. Based on these findings and on their exposed location, we hypothesize that the C381 turret proteins act as the STIV receptor-binding proteins carrying out recognition and attachment to the host receptors. This assumption is substantiated by comparison with the related phage PM2 that also uses the domains protruding from its capsid vertices to interact with the host (13).
Membrane Region.
STIV possesses an internal membrane vesicle located beneath the coat subunit shell and encapsulating the dsDNA genome. The lipid headgroups present on both sides of the membrane are observed in the electron potential map as two concentric layers of density of 455 Å and 515 Å diameter (along fivefold axes). The density corresponding to the lipid acyl chains has considerably lower intensity than the headgroups, in agreement with what has been observed by X-ray crystallography for bacteriophages PRD1 (11) and PM2 (13). The membrane is ∼30 Å thick (at the vertex complexes) and is composed of host-derived monolayer-forming tetraether lipids specifically found in thermophilic and hyperthermophilic archaea (22, 36). These types of lipids are crucial to protect the viral genome in the extreme acidic extracellular environment. The membrane architecture is markedly asymmetric as the lipid headgroups facing the virion interior have significantly stronger density than the outward-facing headgroups, and this probably reflects their different chemical nature as a result of selective orientation (Figs. 1B and 5A). Lipid segregation (in the case of bilayered membrane) or biased orientation (in the case of monolayered membrane) is a general trend observed in viruses of the PRD1-Adenovirus lineage and correlates with the requirement that the two sides of the membrane interact with different molecular entities, i.e., the genome or the proteinaceous capsid shell (11, 13). The membrane organization is perturbed by “sausages” spanning it at icosahedral fivefold axes and likely corresponding to transmembrane proteins (Fig. 5 A and B). We modeled an α-helical segment into each of these densities, resulting in the formation of a five-helix bundle at each vertex that interrupts the membrane continuity. This bundle extends in the outer direction and appears to merge into an electron-rich region contacting the membrane to form a 38-Å-high cylindrically shaped structure connecting to the proximal extremity of the β-pore (Fig. 5 A and C). Due to the moderate resolution of the reconstruction in the vicinity of the membrane, we did not attempt to model this latter region; however, we believe, based on size considerations, that it is likely to be a five-stranded β-structure (with each subunit contributing one strand) projecting knob-like extensions to anchor to the base of the β-pore. Sequence analysis using the TMHMM server (37) suggested that two out of the nine STIV structural proteins are likely to contain transmembrane helices: A55 and B130. As the former is predicted to have a single transmembrane span (in contrast to two predicted ones for B130), we postulate that A55 accounts for the unique transmembrane helix present within each icosahedral asymmetric unit as well as for the distal extension contacting the base of the β-pore and for the additional β-strand contributing to its formation (Figs. 3 A–C and 5 A–C). This assignment is supported by the estimation of the total number of residues required to assemble the observed structural motifs, by the possibility to identify into the cryoEM density the position of the numerous aromatic and bulky amino acid residues present into the A223 N-terminal region (distinguishing it from the A55 N terminus; Fig. S4 A and B) and the assignment of Phe17 as the first residue of the penton base jelly-roll domain based on the preliminary refinement results of X-ray data obtained from twinned crystals of full-length A223. Significantly, no contacts are observed between the capsid shell and the membrane except at each vertex complex where the A55 bundle and the pentameric A223 form a link bridging them.
Fig. 5.
Organization of the viral membrane. (A) The A55 protein (steel blue) accounts for the unique transmembrane helix present within each icosahedral asymmetric unit as well as for the 38-Å-high cylindrically shaped extension contacting the A223 penton base protein (light pink) and for the additional β-strand contributing to the formation of the β-pore structure. The lipid headgroups belonging to the monolayered viral membrane are visualized as two layers rendered in gold whereas the most external layer of the genome is displayed in red. Four out of five of the B345 capsomers surrounding A223 at the icosahedral fivefold axis are also shown (light blue). (B) The A55 pentameric bundle of transmembrane helices is shown within the corresponding cryoEM density. (C) Tilted view of the A223/A55 complex showing the N-terminal β-strand of each A55 monomer connecting to the density appended below the β-pore and reaching the transmembrane domain. Only the extramembranous region of the A55 membrane protein is represented.
Discussion
We described the atomic structure of mature STIV using a combination of cryoEM and X-ray crystallography. Analysis of the architecture of its structural proteins and their interactions and integration of data from the literature suggest a putative assembly pathway leading to the formation of this robust thermo-acidophilic archaeal virus. Previously, we demonstrated with electron cryotomography that STIV initially assembles as an empty procapsid that subsequently serves as a container for genome packaging (24). We also identified partially assembled particles in the cytoplasm of Sulfolobus formed by the capsid shell and the lipid membrane (24). These crescents, displaying the proper curvature as soon as they were recognizable, were proposed to represent an intermediate stage of particle formation. This model requires that the lipids, constituting the viral membrane, be recruited directly in the cytoplasm rather than by a budding mechanism from the host membrane. As the vertex complexes in the structure described mediate the only observed contacts between the capsid and the membrane, we envision that they are responsible for coordinating the interactions between these components. Association of the membrane protein with the penton base (and the cement protein) would allow nucleation promotion and subsequent growth of the capsid shell (by addition of monomeric B345 subunits) concomitantly with the formation of the internal lipid vesicle. Therefore, the vertex complexes would act as maestro orchestrating the interactions between the different viral building blocks to prevent unproductive formation of empty capsids or isolated lipid vesicles. Following procapsid formation in the cytoplasm, genome packaging would occur via yet uncharacterized mechanisms to yield mature STIV virions organized in quasi-crystalline arrays while awaiting release through the pyramids puncturing the host membrane and the S-layer (24, 26). As no change in the overall capsid structure and dimensions was detected between the procapsid and capsid states in the tomographic studies (24), we postulate that STIV undergoes a limited maturation compared with tailed phages, for instance (38). A more subtle maturation may involve the reorganization of the C-terminal helices in the B345 capsid proteins. As noted in Results, Coat Subunit Shell, they appeared to project into the viral membrane in the low-resolution structure previously reported (19), but in the current structure they project away from the membrane. Such a change in orientation might correspond to maturation transition. A second difference was the presence of the “petal” proteins that attached to the turrets in the previous structure. These petal proteins are not visible in the structure reported here.
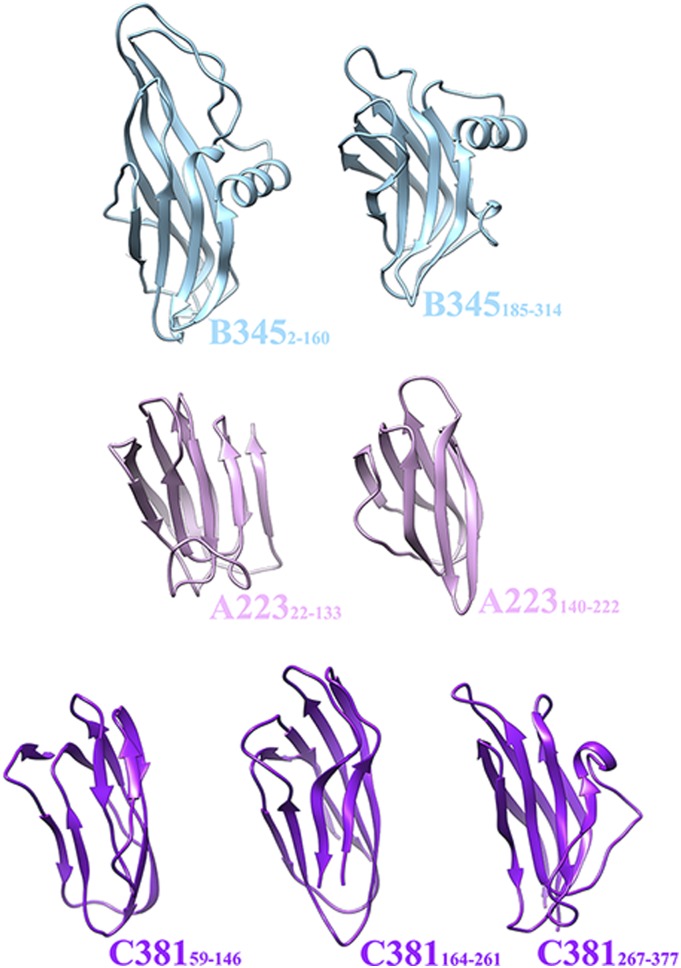
The conservation of the A223 penton base jelly-roll fold, as well as the B345 double jelly-roll fold, strongly suggests an evolutionary connection for these protein modules among phages, archaeal viruses, and eukaryotic viruses. This observation supports and reinforces the validity of the proposed PRD1-Adenovirus lineage encompassing viruses whose capsid shells are made of identical coat subunits and penton base proteins. Some aspects of the particle structure apply to only a subset of the viruses, such as the presence of an internal lipid-membrane in STIV, PRD1, and PM2 or the conserved structure of the Adenovirus and PRD1 spike shafts. The structural conservation of a large number of proteins playing key roles in the architecture and function of viruses belonging to this lineage constitutes a strong argument in favor of a common ancestor for these virions infecting organisms of the three domains of life. This observation suggests the possibility that STIV, PRD1-like phages, and Adenovirus all derive from a common viral ancestor present in the biosphere more than 3 billion years ago, before the divergence of Bacteria, Archaea, and Eukarya. The structural results presented here reveal that the STIV coat subunit, penton base, and turret proteins are made of a succession of fused jelly-roll motifs. Although not strictly identical, these domains exhibit similar folds with a common topology and can be superimposed with reasonable agreement (Fig. 6). Based on these observations, we speculate that a unique ancestral protein module gave rise to many of the STIV structural proteins by duplication and sequence diversification events that occurred during a lengthy evolutionary process. If this hypothesis is correct, a single primordial jelly-roll motif led to a 75-MDa viral molecular machine tuned to withstand the extreme conditions of pH and temperature to which STIV is exposed during its life cycle.
Fig. 6.
Assembly of the STIV virion from a single protein module. The B345 coat subunit, A223 penton base, and C381 turret protein are all formed of various numbers of jelly-roll domains fused on a single polypeptide chain. We therefore propose that a unique ancestral protein module gave rise to many of the STIV structural proteins by duplication and sequence diversification events.
Materials and Methods
Electron micrographs were acquired with a FEI Titan Krios electron microscope operating at 300 kV at liquid nitrogen temperature with a dose of 15–18 e−/Å2 using a FEI Falcon direct electron detector. Particles were automatically picked using FindEM (39) integrated into the Appion pipeline (40), and the parameters of the microscope contrast transfer function were estimated using CTFind3 (41) for each micrograph. The reconstruction was performed with Frealign (42) using the previously reported STIV reconstruction low-pass filtered at 50 Å (23). The resolution of the obtained map was determined by using the FSC, calculated between two reconstructions each containing half of the data, at a cutoff of 0.143 (28). A negative temperature factor of 400 Å2 was applied to the final reconstruction with the software Bfactor (http://emlab.rose2.brandeis.edu/bfactor). Model building was achieved by rigid-body fitting of individual crystal structures into the STIV reconstruction as well as de novo modeling using Chimera (43) and Coot (44).
Supplementary Material
Acknowledgments
We thank Prof. Paul Matsudaira (Department of Biological Sciences, National University of Singapore) for hosting J.E.J. as a visiting faculty member and for providing generous access to the Titan Krios electron microscope to collect the data used for the structure determination reported here. This work was supported by National Institutes of Health Grant R01GM54076 (to J.E.J.), Duke-National University of Singapore Block Grant R913-200-039-263 (to S.-M.L.), and National Science Foundation Grant MCB-0920312, as well as by a grant from the Murdock Foundation for support of the Macromolecular Diffraction Laboratory at Montana State University (to C.M.L.), National Institutes of Health Grant K99GM098393 (to C.-y.F.), and Seventh Framework Programme Marie Curie International Outgoing Fellowship 273427 (to D.V.). Part of this work was carried out at the Stanford Synchrotron Radiation Laboratory (SSRL), a national user facility operated by Stanford University on behalf of the US Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the US Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. Part of this research was also conducted at the National Resource for Automated Molecular Microscopy, which is supported by the National Institutes of Health though the National Center for Research Resources (2P41RR017573-11) and the National Institute of General Medical Sciences (9 P41 GM103310-11).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates and structure factors of the A223 C-terminal domain have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4IL7). The Sulfolobus turreted icosahedral virus cryoEM reconstruction has been deposited in the EMDataBank (accession no. EMD-5584). The modeled PDB has been deposited in the Protein Data Bank (PDB ID code 3J31).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300601110/-/DCSupplemental.
References
- 1.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc Natl Acad Sci USA. 1977;74(11):5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prangishvili D, Forterre P, Garrett RA. Viruses of the Archaea: A unifying view. Nat Rev Microbiol. 2006;4(11):837–848. doi: 10.1038/nrmicro1527. [DOI] [PubMed] [Google Scholar]
- 4.Aalto AP, et al. Snapshot of virus evolution in hypersaline environments from the characterization of a membrane-containing Salisaeta icosahedral phage 1. Proc Natl Acad Sci USA. 2012;109(18):7079–7084. doi: 10.1073/pnas.1120174109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reddy VS, Natchiar SK, Stewart PL, Nemerow GR. Crystal structure of human adenovirus at 3.5 A resolution. Science. 2010;329(5995):1071–1075. doi: 10.1126/science.1187292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu H, et al. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science. 2010;329(5995):1038–1043. doi: 10.1126/science.1187433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rux JJ, Kuser PR, Burnett RM. Structural and phylogenetic analysis of adenovirus hexons by use of high-resolution x-ray crystallographic, molecular modeling, and sequence-based methods. J Virol. 2003;77(17):9553–9566. doi: 10.1128/JVI.77.17.9553-9566.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nandhagopal N, et al. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc Natl Acad Sci USA. 2002;99(23):14758–14763. doi: 10.1073/pnas.232580699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bahar MW, Graham SC, Stuart DI, Grimes JM. Insights into the evolution of a complex virus from the crystal structure of vaccinia virus D13. Structure. 2011;19(7):1011–1020. doi: 10.1016/j.str.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X, et al. Structure of Sputnik, a virophage, at 3.5-Å resolution. Proc Natl Acad Sci USA. 2012;109(45):18431–18436. doi: 10.1073/pnas.1211702109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cockburn JJB, et al. Membrane structure and interactions with protein and DNA in bacteriophage PRD1. Nature. 2004;432(7013):122–125. doi: 10.1038/nature03053. [DOI] [PubMed] [Google Scholar]
- 12.Abrescia NGA, et al. Insights into assembly from structural analysis of bacteriophage PRD1. Nature. 2004;432(7013):68–74. doi: 10.1038/nature03056. [DOI] [PubMed] [Google Scholar]
- 13.Abrescia NGA, et al. Insights into virus evolution and membrane biogenesis from the structure of the marine lipid-containing bacteriophage PM2. Mol Cell. 2008;31(5):749–761. doi: 10.1016/j.molcel.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 14.Jaatinen ST, Happonen LJ, Laurinmäki P, Butcher SJ, Bamford DH. Biochemical and structural characterisation of membrane-containing icosahedral dsDNA bacteriophages infecting thermophilic Thermus thermophilus. Virology. 2008;379(1):10–19. doi: 10.1016/j.virol.2008.06.023. [DOI] [PubMed] [Google Scholar]
- 15.Laurinmäki PA, Huiskonen JT, Bamford DH, Butcher SJ. Membrane proteins modulate the bilayer curvature in the bacterial virus Bam35. Structure. 2005;13(12):1819–1828. doi: 10.1016/j.str.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 16.Jäälinoja HT, et al. Structure and host-cell interaction of SH1, a membrane-containing, halophilic euryarchaeal virus. Proc Natl Acad Sci USA. 2008;105(23):8008–8013. doi: 10.1073/pnas.0801758105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaakkola ST, et al. Closely related archaeal Haloarcula hispanica icosahedral viruses HHIV-2 and SH1 have nonhomologous genes encoding host recognition functions. J Virol. 2012;86(9):4734–4742. doi: 10.1128/JVI.06666-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khayat R, et al. Structure of an archaeal virus capsid protein reveals a common ancestry to eukaryotic and bacterial viruses. Proc Natl Acad Sci USA. 2005;102(52):18944–18949. doi: 10.1073/pnas.0506383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rice G, et al. The structure of a thermophilic archaeal virus shows a double-stranded DNA viral capsid type that spans all domains of life. Proc Natl Acad Sci USA. 2004;101(20):7716–7720. doi: 10.1073/pnas.0401773101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snyder JC, Young MJ. Advances in understanding archaea-virus interactions in controlled and natural environments. Curr Opin Microbiol. 2011;14(4):497–503. doi: 10.1016/j.mib.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 21.Lawrence CM, et al. Structural and functional studies of archaeal viruses. J Biol Chem. 2009;284(19):12599–12603. doi: 10.1074/jbc.R800078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maaty WSA, et al. Characterization of the archaeal thermophile Sulfolobus turreted icosahedral virus validates an evolutionary link among double-stranded DNA viruses from all domains of life. J Virol. 2006;80(15):7625–7635. doi: 10.1128/JVI.00522-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khayat R, Fu C-Y, Ortmann AC, Young MJ, Johnson JE. The architecture and chemical stability of the archaeal Sulfolobus turreted icosahedral virus. J Virol. 2010;84(18):9575–9583. doi: 10.1128/JVI.00708-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu C-Y, et al. In vivo assembly of an archaeal virus studied with whole-cell electron cryotomography. Structure. 2010;18(12):1579–1586. doi: 10.1016/j.str.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu C-Y, Johnson JE. Structure and cell biology of archaeal virus STIV. Curr Opin Virol. 2012;2(2):122–127. doi: 10.1016/j.coviro.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brumfield SK, et al. Particle assembly and ultrastructural features associated with replication of the lytic archaeal virus sulfolobus turreted icosahedral virus. J Virol. 2009;83(12):5964–5970. doi: 10.1128/JVI.02668-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quax TEF, Krupovic M, Lucas S, Forterre P, Prangishvili D. The Sulfolobus rod-shaped virus 2 encodes a prominent structural component of the unique virion release system in Archaea. Virology. 2010;404(1):1–4. doi: 10.1016/j.virol.2010.04.020. [DOI] [PubMed] [Google Scholar]
- 28.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333(4):721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 29.Xiao L, Honig B. Electrostatic contributions to the stability of hyperthermophilic proteins. J Mol Biol. 1999;289(5):1435–1444. doi: 10.1006/jmbi.1999.2810. [DOI] [PubMed] [Google Scholar]
- 30.Benson SD, Bamford JK, Bamford DH, Burnett RM. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell. 1999;98(6):825–833. doi: 10.1016/s0092-8674(00)81516-0. [DOI] [PubMed] [Google Scholar]
- 31.Ge M, Xia X-Y, Pan X-M. Salt bridges in the hyperthermophilic protein Ssh10b are resilient to temperature increases. J Biol Chem. 2008;283(46):31690–31696. doi: 10.1074/jbc.M805750200. [DOI] [PubMed] [Google Scholar]
- 32.Song L, et al. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science. 1996;274(5294):1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 33.Zubieta C, Schoehn G, Chroboczek J, Cusack S. The structure of the human adenovirus 2 penton. Mol Cell. 2005;17(1):121–135. doi: 10.1016/j.molcel.2004.11.041. [DOI] [PubMed] [Google Scholar]
- 34.Bebeacua C, et al. Visualizing a complete Siphoviridae member by single-particle electron microscopy: The structure of lactococcal phage TP901-1. J Virol. 2012;87(2):1061–1068. doi: 10.1128/JVI.02836-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konings WN, Albers S-V, Koning S, Driessen AJM. The cell membrane plays a crucial role in survival of bacteria and archaea in extreme environments. Antonie van Leeuwenhoek. 2002;81(1-4):61–72. doi: 10.1023/a:1020573408652. [DOI] [PubMed] [Google Scholar]
- 37.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 38.Veesler D, Johnson JE. Virus maturation. Annu Rev Biophys. 2012;41:473–496. doi: 10.1146/annurev-biophys-042910-155407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roseman AM. FindEM—a fast, efficient program for automatic selection of particles from electron micrographs. J Struct Biol. 2004;145(1-2):91–99. doi: 10.1016/j.jsb.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 40.Lander GC, et al. Appion: An integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166(1):95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142(3):334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 42.Grigorieff N. FREALIGN: High-resolution refinement of single particle structures. J Struct Biol. 2007;157(1):117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 43.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157(1):281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 44.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.