Abstract
Chamomile (Matricaria chamomilla L.), tarragon (Artemisia dracunculus L.) and Mexican arnica (Heterotheca inuoides) are common compositae spices and herbs found in the US market. They contain flavonoids and hydroxycinnamates that are potentially beneficial to human health. A standardized LC-PDA-ESI/MS profiling method was used to identify 51 flavonoids and 17 hydroxycinnamates. Many of the identifications were confirmed with authentic standards or through references in the literature or the laboratory’s database. More than half of the phenol compounds for each spice had not been previously reported. The phenolic profile can be used for plant authentication and to correlate with biological activities.
Keywords: Chamomile, Tarragon, Mexican arnica, Flavonoids, Caffeoylquinic acids, LC-PDA-ESI/MS
Chamomile (Matricaria chamomilla L.) flowers and tarragon (Artemisia dracunculus L.) leaves are among the common compositae spices and herbs that are used worldwide. They are used as home spices, health foods, and herb teas, and their extracts are used in some pharmaceutical preparations [1–4]. As a traditional medicine, Mexican arnica (Heterotheca inuloides Cass) has been taken internally for the treatment of nervous disorders, stomach ailments, and fever [5,6], and the flowers have been used as a spice in Mexico and the USA. Chemically, they contain volatile oils, flavonoids, and hydroxycinnamic acid derivatives [1,6–16]. The flavonoids and hydroxycinnamates are potentially beneficial to human health [17]. These plants have been previously studied using liquid chromatography-mass spectrometry [18–23], but their phenolic components have not been systematically studied.
The systematic identification and quantification of the phenolic compounds in food is necessary in order to determine their impact on human health. Liquid chromatography-photodiode-array-mass spectrometry (LC-PDA-MS) has been shown to be a powerful tool for on-line identification of plant phenolic compounds [24,25]. The only drawback is the inability to identify isomers, e.g. specific sites of attachment of the saccharides. As part of our project of systematic identification of the phenolic compounds in plant derived foods, including spices and herbs, over 200 standards and 400 food samples have been screened using a standardized LC-PDA-ESI/MS method. More than 1000 food phenolic compounds have been identified and stored in our food phenolic database. They are used as references to provide reliable identification of the compounds in subsequent analyzed samples [25–27]. In this study, as many as 37 phenolic compounds were identified in chamomile, tarragon and Mexican arnica. More than half are new for these spices.
Identification of flavonoids and caffeoylquinic acids
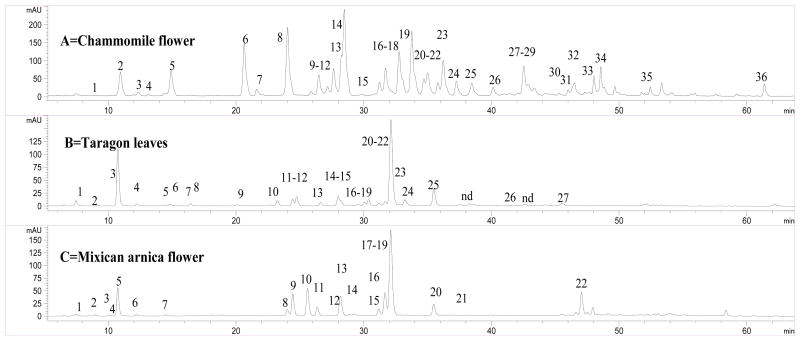
Chromatograms (350 nm) of the extracts of chamomile, tarragon and Mexican arnica are shown in Figure 1. The retention times (tR), wavelength of maximum absorbance (λmax), molecular ions ([M+H]+/[M−H]−), and major fragment ions (PI/NI) are listed in Table 1.
Figure 1.
The LC chromatograms of Chammomile flowers (A), tarragon leaves (B) and Mexican arnica flowers (C).
Table 1a.
The identification of the phenolic compounds in chamomile flower (C), tarragon leaf (T) and Mexican arnica flower (MA).
| Compound | Peak Number
|
tR (min) | [M+H]+ and [M−H]− m/z | Major fragments m/z | UV λmax (mn) | ||
|---|---|---|---|---|---|---|---|
| C | T | MA | |||||
|
| |||||||
| Hydroxycinnamates | |||||||
| 1-Caffeyolquinic acida | nd | 1 | 1 | 6.4 | ---/353 | 191, 179, 167, 135 | 240sh, 300sh, 330 |
| 3-Caffeoylquinic acida | 1c | 2 | 2 | 7.3 | ---/353 | 191, 179, 167, 135 | 240sh, 300sh, 330 |
| Chlorogenic acida | 2c | 3 | 5c | 11.5 | ---/355 | 191, 179, 167, 135 | 240sh, 300sh, 330 |
| 4-Caffeoylquinic acida | 3c | 4 | 6 | 12.3 | ---/356 | 191, 179, 167, 135 | 240sh, 300sh, 330 |
| Caffeic acida | nd | 5 | 7c | 14.7 | ---/179 | 163 | 240, 298sh, 326 |
| Caffeoyltartaric acida | nd | 7 | nd | 18.4 | ---/311 | 179 | nd |
| 5-Feruloylquinic acida | nd | 8 | nd | 18.6 | ---/367 | 193, 191 | nd |
| Ferulic acid glucosea | 5c | nd | nd | 14.4 | ---/355 | 193 | nd |
| Ferulic acid glucosea | 8c | nd | nd | 23.2 | ---/355 | 193 | 240, 298sh, 326 |
| 1,4-Dicaffeoylquinic acida | nd | 17 | nd | 30 | ---/515 | 353,191, 179, 167, 135 | nd |
| 3,4-Dicaffeoylquinic acida | 16 | 20 | 16 | 31.2 | ---/515 | 353,191, 179, 167, 135 | 240, 300sh, 328 |
| 1,5-Dicaffeoylquinic acida | 17c,d | 21 | 17 | 31.7 | ---/517 | 353,191, 179, 167, 135 | 240, 300sh, 328 |
| 3,5-Dicaffeoylquinic acida | nd | 22 | 18 | 32.1 | ---/517 | 353,191, 179, 167, 135 | 240, 298sh, 326 |
| 4,5-Dicaffeoylquinic acida | 24 | 25 | 20 | 35.2 | ---/519 | 353,191, 179, 167, 135 | 240, 300sh, 328 |
| 3-Caffeoyl-5-feruloyquinic acida | nd | 26 | nd | 38.3 | ---/529 | 367, 353, 191,179,135 | 240, 326 |
| 3,4,5-Tricaffeoylquinic acida | nd | 27 | nd | 48.8 | ---/667 | 515,353,515,191,179 | nd |
| Flavone 7-O-glycosides | |||||||
| Hexahydroxyflavone 3-O-hexosideb | 6 | nd | nd | 20 | 481/479 | 319/317 | 260, 354 |
| Pentahydroxyflavone 7-O-hexosideb | 7 | nd | nd | 20.9 | 465/463 | 303/301 | 282, 342 |
| Leteolin 7-O-rutinosidea | 9 | nd | nd | 24.9 | 595/593 | 287/285 | 256, 350? |
| Pentahydroxyflavone 7-O-hexosideb | 11 | nd | nd | 26.8 | 465/463 | 303/301 | 268, 338 |
| Luteolin 7-O-glucosidea | 12c | nd | nd | 27.5 | 449/447 | 287/285 | 256, 266, 348 |
| Hexahydroxyflavone 3-O-dihexosideb | 15 | nd | nd | 30.3 | 643/641 | 319/317 | nd |
| Apigenin 7-O-glucosidea | 19c | nd | nd | 33.6 | 433/431 | 287/285 | 256, 266, 348 |
| Apigenin 7-O-acetylglucosidea | 29c | nd | nd | 42.3 | 475/473 | 271/269 | 268, 338 |
| Apigenin 7-O-acetylglucosidea | 32c | nd | nd | 47.2 | 475/473 | 271/269 | 268, 338 |
| Apigenin 7-O-malonylglucosidea | 28 | nd | nd | 42.6 | 519/517 | 271/269 | 268, 338 |
| Apigenin 7-O-caffeoyllucosideb | 30 | nd | nd | 45 | 595/593 | 271/269 | nd |
| Apigenin 7-O-molonylglucosidea | 31 | nd | nd | 45.6 | 519/517 | 271/269 | 268, 338 |
| Apigenin 7-O-malonylacetylglucosideb | 33 | nd | nd | 47.9 | 561/559 | 271/269 | 268, 338 |
| Apigenin 7-O-malonylacetylglucosideb | 34 | nd | nd | 48.2 | 561/559 | 271/269 | 268, 338 |
| Apigenin 7-O-malonylacetylglucosideb | 35 | nd | nd | 49.1 | 561/559 | 271/269 | 268, 338 |
the identification was confirmed by comparison to standards or positively identified compounds in reference plant samples;
identified tentatively from the LC-MS data;
reported in the plant previously;
previous identification was not corrected; nd: not determined.
The LC-PDA-ESI/MS instrument offered the UV spectra, retention time, and mass data for each of the phenols in a plant extract in a single run. The molecular ions and their fragments, including the aglycone ions of a flavonoids and the acyls of the cinnamates, were obtained with positive and negative ionization at low (100 V or less) and high (250 V or higher) fragmentation energies. The positive and negative mass data were always used to confirm the mass of each compound in each chromatographic peak. Tentative identification was made based on the UV and MS spectra and retention times. Positive identification was achieved by comparison to data for either authentic standards or positively identified compounds in the reference plant samples. In Table 1, positively and tentatively identified compounds are indicated with superscript “a” and “b”, respectively.
All 17 of the hydroxycinnamates and 27 of the 46 glycosylated flavonoids were positively identified based on standards or reference compounds from previously tested Compositae plants [25–27]. The 19 remaining flavonoids were tentatively identified with reasonable confidence. The positive identification of the aglycones (chromatograms not shown) resulting from hydrolysis of the extracts confirmed the flavonoid glycoside identifications. Some of the compounds in Table 1 have been reported previously in Compositae plants (superscript “c” next to peak number) and were identified by comparison of the LC-MS spectra [1,6–16,18–23].
The main phenolic components of Chamomile flowers were the glycosides of flavones, while hydroxycinnamates were the main phenolic components of tarragon leaves. Mexican arnica flowers contained hydroxycinnamates and the glycosides of flavones and flavonols. All 3 plants can be distinguished easily.
A systematic LC-DAD-ESI/MS plant phenolic component analysis requires a gram or less of material and can be completed in several hours. Use of a standardized approach to compile retention times and UV and MS spectra greatly facilitates compound identification [24–27]. Characterization of the herb chemical component profile is valuable not only for identification and quality control, but will also enhance understanding of their biological activity and their benefit to human health.
Experimental
Plant materials and extraction
Dried chamomile flower, tarragon leaves, and Mexican arnica flowers were purchased from local food stores in Maryland. All were finely powdered and passed through a20-mesh sieve prior to extraction. Dried ground material (100 mg) was extracted with methanol-water (5.0 mL, 60:40, v/v) using a sonicator (Fisher Scientific, Pittsburg, PA, USA) at 40 KHz and 100 W for 60 min. at room temperature. The extract was filtered through a 0.45 μm nylon acrodisk 13 filter (Gelman, Ann Arbor, MI, USA), and a 10 μL of the extract was injected onto the analytical column for analysis [25].
LC-PDA-ESI/MS analysis
The LC-PDA-ESI/MS instrument and operating parameters have been previously described [25]. Briefly, the LC-PDA-ESI/MS consisted of an 1100 HPLC (with a diode array detector) coupled to a mass spectrometer (MSD, SL mode), both from Agilent (Palo Alto, CA). A 250 × 4.6 mm i.d., 5 μm Symmetry C18 column (C18, 5 μm,) (Waters Corp., Milford, MA) and a 20 × 3.9 mm i.d., 5 μm sentry guard column (Symmetry, 3.9 × 20 mm) (Waters Corp., Milford, MA) were used with a flow rate of 1.0 mL/min. Symmetry Shield column (250 × 4.6 mm i.d., 5 μm) was also used further to separate some overlapped peaks. The column oven temperature was set at 25°C. The mobile phase consisted of a combination of A (0.1% formic acid in water) and B (0.1% formic acid in acetonitrile). The gradient was varied linearly from 10% to 26% B (v/v) in 40 min, to 65% B at 70 min, to 100% B at 71 min, and held at 100% B to 75 min. The PDA was set at 350, 310 and 270 nm to provide real-time records of the peak intensity and UV spectra were recorded from 190–650 nm for plant component identification. Mass spectra were simultaneously acquired using electrospray ionization in the positive and negative ionization (PI and NI) modes at low and high fragmentation voltages (100 V and 250 V) over the range of m/z 100–2000. A drying gas flow of 13 L/min, a drying gas temperature of 350°C, a nebulizer pressure of 50 psi, and capillary voltages of 4000 V for PI and 3500 V for NI were used. The LC system was directly coupled to the MSD without stream splitting.
Acid hydrolyzed extracts
The filtered extract (0.50 mL) was mixed with concentrated HCl (37%, 0.10 mL) and heated in a covered tube at 85°C for 2 h. Then, 0.40 mL of methanol was added to the mixture and the solution was sonicated for 10 min. The solution was re-filtered prior to HPLC injection [25].
Table 1b.
The identification of the phenolic compounds in chamomile flower (C), tarragon leaf (T) and Mexican arnica flower (MA).
| Compound | Peak Number | tR | [M+H]+ and [M−H]− m/z | Major fragments m/z | UV λmax (mn) | ||
|---|---|---|---|---|---|---|---|
|
| |||||||
| C | T | MA | (min) | ||||
|
| |||||||
| Flavone C-glucoside | |||||||
| Apigenin 6,8-di-C-glucosidea | nd | 6 | nd | 15.3 | 595/593 | 577, 475, 455 | 270, 338 |
| Flavone aglycones | |||||||
| Luteolina | nd | nd | 22 | 47.9 | 287/285 | 256, 266, 348 | |
| Apigenina | 36c | nd | nd | 51.9 | 271/269 | 268, 338 | |
| Dihydroxy-tetramethoxyflavoneb | 37 | nd | nd | 60.2 | 375/373 | 256, 272, 352 | |
| Flavonol O-glycosides | |||||||
| Quercetin 3-O-glucuronide-7-O-galactosideb | nd | nd | 3 | 9 | 641/639 | /303 | 256, 266, 354 |
| Quercetin 3-O-glucuronide-7-O-glucosideb | nd | nd | 4 | 10.1 | 641/639 | /303 | 256, 266, 354 |
| Hexahydroxyflavone 3-O-dihexosidea | 4 | nd | nd | 13.7 | 641/639 | 479, 317/315 | 256, 266, 356 |
| Quercetin 3-O-rhamnosylgalactosidea | nd | nd | 8c | 23.1 | 611/609 | 303/301 | 256, 266, 354 |
| Quercetin 3- O-rutinosidea | nd | nd | 9c | 24.2 | 611/609 | 303/301 | 256, 266, 354 |
| Quercetin 7-O-glucosidea | nd | 10 | nd | 24.7 | 465/463 | 303/301 | 254, 266, 370 |
| Petuletin 3-O-robinobiosideb | nd | 13c | nd | 24.7 | 641/639 | 495, 333/331 | 258, 266sh, 350 |
| Pentahydroxyflavone 7-O-hexosidea | 10 | nd | nd | 25.7 | 465/463 | 303/301 | 256, 370 |
| Quercetin 3-O-galactosidea | nd | nd | 10c | 26.3 | 463/461 | 303/301 | 256, 266sh,352 |
| Petuletin 3-O-glucosidea | nd | 14c | nd | 26.6 | 495/493 | 333/331 | 258, 266sh, 350 |
| Quercetin 3-O-glucosidea | nd | nd | 11c | 27.1 | 463/461 | 303/301 | 256, 266sh,352 |
| Pentahydroxymethoxyflavone 7-O-glucosideb | 13 | nd | nd | 27.3 | 495/493 | 333/331 | 256, 370 |
| Pentahydroxymethoxyflavone glucosideb | 14 | nd | nd | 27.6 | 495/493 | 333/331 | 260, 360 |
| Petuletin 3-O- manolylrobinobiosidea | nd | 15 | nd | 28 | 727/725 | 495, 333/331 | 258, 266sh, 350 |
| Quercetin 3-O-glucuronidea | nd | nd | 12 | 28.2 | 479/447 | 303/301 | 256, 266sh,352 |
| Quercetin 3-arabinosidea | nd | nd | 14 | 29 | 435/433 | 303/301 | 256, 266, 354 |
| Kaempferol-3-O-rutinosidea | nd | nd | 13c | 29 | 595/593 | 287/285 | 266, 348 |
| Isorhamonetin-3-O-rhamnosylgalactosidea | 21 | 16 | nd | 29.5 | 625/623 | 479, 317/315 | 258, 266sh, 350 |
| Isorhamonetin-3-O-rutinosidea | nd | 18 | nd | 30.1 | 625/623 | 479, 317/315 | 258, 266sh, 350 |
| Syringenin 3-O-rhamnosylhexosideb | nd | 19 | nd | 30.4 | 655/653 | 509, 347/347 | 258, 266sh, 350 |
| Kaempferol 3-O-glucosidea | nd | nd | 15c | 32.1 | 449/447 | 287/285 | 266, 348 |
| Kaempferol-3-O-glucuronidea | nd | nd | 19 | 32.7 | 465/463 | 287/285 | nd |
| Isorhamnetin 3-O-glucosidea | 21 | 23 | nd | 32.9 | 479/477 | 317/315 | 256, 266, 350 |
| Petuletin 3-O-manolylrhamnosylhexosideb | nd | 24 | nd | 33.2 | 741/739 | 333/331 | 256, 266, 350 |
| Isorhamnetin 7-O-glucosidea | 20 | nd | nd | 33.6 | 479/477 | 317/315 | 254, 372 |
| Petuletin 7-O-glucosideb | 22c | nd | nd | 34.3 | 495/493? | 333/331 | 258, 368 |
| Tetrahydroxy-dimethooxyflavone 7-O-glucosideb | 23 | nd | nd | 34.6 | 509/507 | 347/345 | 258, 368 |
| Pentahydroxymethoxyflavone caffeoylglucosideb | 25 | nd | nd | 36.1 | 657/655 | 333/331 | 278, 338 |
| Pentahydroxymethoxyflavone caffeoylglucosideb | 26 | nd | nd | 37.4 | 657/655 | 333/331 | 278, 334 |
| Quercetin 3-O-caffeoylglucosidea | nd | nd | 21 | 37.3 | 627/625 | 303/301 | 256, 266sh, 336 |
the identification was confirmed by comparison to standards or positively identified compounds in reference plant samples;
identified tentatively from the LC-MS data;
reported in the plant previously;
previous identification was corrected; nd: not determined.
Acknowledgments
This research was supported by the Agricultural Research Service of the U.S. Department of Agriculture and the Office of Dietary Supplements at the National Institutes of Health under an Interagency Agreement.
References
- 1.Leung AY, Foster S. Chamomile, Tarragon. In: Leung AY, Foster S, editors. Encyclopedia of common natural ingredients used in food, drugs and cosmetics. 2. 145–147. John Wiley & Sons, Inc; New York: 1996. pp. 487–489. [Google Scholar]
- 2.Srivastava JK, Shankar E, Gupta S. Chamomile: a herbal medicine of the past with bright future. Molecular Medicine Report. 2010;3:895–901. doi: 10.3892/mmr.2010.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kato A, Minoshima Y, Yamamoto J, Adachi I, Watson AA, Nash RJ. Protective effects of dietary chamomile tea on diabetic complications. Journal of Agricultural & Food Chemistry. 2008;56:8206–8211. doi: 10.1021/jf8014365. [DOI] [PubMed] [Google Scholar]
- 4.Srivastava JK, Gupta S. Antiproliferative and apoptotic effects of chamomile extract in various human cancer cells. Journal of Agricultural & Food Chemistry. 2007;55:9470–9478. doi: 10.1021/jf071953k. [DOI] [PubMed] [Google Scholar]
- 5.Coballase-Urrutia E, Pedraza-Chaverri J, Camacho-Carranza R, Cárdenas-Rodríguez N, Huerta-Gertrudis B, Medina-Campos ON, Mendoza-Cruz M, Delgado-Lamas G, Espinosa-Aguirre JJ. Antioxidant activity of Heterotheca inuloides extracts and of some of its metabolites. Toxicology. 2010;276:41–48. doi: 10.1016/j.tox.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 6.Delgado G, del Socorro Olivares M, Chávez MI, Ramírez-Apan T, Linares E, Bye R, Espinosa-García FJ. Antiinflammatory constituents from Heterotheca inuloides. Journal of Natural Products. 2001;64:861–864. doi: 10.1021/np0005107. [DOI] [PubMed] [Google Scholar]
- 7.Harborne JB, Baxter H. Artemisia spp., Matricaria chamolilla, Heterotheca spp. In: Harborne JB, Baxter H, editors. The handbook of natural flavonoids. Vol. 1. John Wiley & Sons, LTD; New York: 1999. pp. 826–827.pp. 862pp. 854 [Google Scholar]
- 8.Kubo I, Kinst-Hori I, Chaudhuri SK, Kubo Y, Sánchez Y, Ogura T. Flavonols from Heterotheca inuloides: tyrosinase inhibitory activity and structural criteria. Bioorganic & Medicinal Chemistry. 2000;8:1749–1755. doi: 10.1016/s0968-0896(00)00102-4. [DOI] [PubMed] [Google Scholar]
- 9.Haraguchi H, Ishikawa H, Sanchez Y, Ogura T, Kubo Y, Kubo I. Antioxidative constituents in Heterotheca inuloides. Bioorganic & Medicinal Chemistry. 1997;5:865–871. doi: 10.1016/s0968-0896(97)00029-1. [DOI] [PubMed] [Google Scholar]
- 10.Jerga C, Merfort I, Willuhn G. Flavonoid glycosides and other hydrophilic compounds from flowers of Heterotheca inuloides. Planta Medica. 1990;56:413–415. doi: 10.1055/s-2006-960997. [DOI] [PubMed] [Google Scholar]
- 11.Ceska O, Chaudhary SK, Warrington PJ, Ashwood-Smith MJ. Coumarins of chamomile, Chamomilla recutita. Fitoterapia. 1992;63:387–394. [Google Scholar]
- 12.Logendra S, Ribnicky DM, Yang H, Poulev A, Ma J, Kennelly EJ, Raskin I. Bioassay-guided isolation of aldose reductase inhibitors from Artemisia dracunculus. Phytochemistry. 2006;67:1539–1546. doi: 10.1016/j.phytochem.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 13.Vienne M, Braemer R, Paris M, Couderc H. Chemotaxonomic study of two cultivars of Artemisia dracunculus L.: (“French” and “Russian” Tarragon) Biochemical Systematics and Ecology. 1989;17:373–374. [Google Scholar]
- 14.Carnat A, Carnat AP, Fraisse D, Ricoux L, Lamaison JL. The aromatic and polyphenolic composition of Roman chamomile tea. Fitoterapia. 2004;75:32–38. doi: 10.1016/j.fitote.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Švehlíková V, Richard N, Bennett RN, Mellon FA, Needs PW, Piacente S, Kroon PA, Bao Y. Isolation, identification and stability of acylated derivatives of apigenin 7-O-glucoside from chamomile (Chamomilla recutita [L.] Rauschert) Phytochemistry. 2004;65:2323–2332. doi: 10.1016/j.phytochem.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 16.Fonseca FN, Tavares MFM. Validation of a capillary electrophoresis method for the quantitative determination of free and total apigenin in extracts of Chamomilla recutita. Phytochemical Analysis. 2004;15:65–70. doi: 10.1002/pca.744. [DOI] [PubMed] [Google Scholar]
- 17.Scalbert A, Manach C, Morand C, Rémésy C, Jiménez L. Dietary polyphenols and the prevention of diseases. Critical Reviews in Food Science and Nutrition. 2005;45:287–306. doi: 10.1080/1040869059096. [DOI] [PubMed] [Google Scholar]
- 18.Justesen U. Negative atmospheric pressure chemical ionisation low-energy collision activation mass spectrometry for the characterisation of flavonoids in extracts of fresh herbs. Journal of Chromatography A. 2000;902:369–379. doi: 10.1016/s0021-9673(00)00861-x. [DOI] [PubMed] [Google Scholar]
- 19.Nováková L, Vildová A, Mateus JP, Gonçalves T, Solich P. Development and application of UHPLC-MS/MS method for the determination of phenolic compounds in chamomile flowers and chamomile tea extracts. Talanta. 2010;82:1271–1280. doi: 10.1016/j.talanta.2010.06.057. [DOI] [PubMed] [Google Scholar]
- 20.Srivastava JK, Gupta S. Extraction, characterization, stability and biological activity of flavonoids isolated from chamomile flowers. Molecular and Cellular Pharmacology. 2009;1:138. doi: 10.4255/mcpharmacol.09.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eisenman SW, Poulev A, Struwe L, Raskin I, Ribnicky DM. Qualitative variation of anti-diabetic compounds in different tarragon (Artemisia dracunculus L.) cytotypes. Fitoterapia. 2011;82:1062–1074. doi: 10.1016/j.fitote.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mulinacci N, Romani A, Pinelli P, Vincieri FF, Prucher D. Characterization of Matricaria recutita L. flower extracts by HPLC-MS and HPLC-DAD analysis. Chromatographia. 2000;51:301–307. [Google Scholar]
- 23.Fonseca FN, Tavares MFM, Horvath C. Capillary electrochromatography of selected phenolic compounds of Chamomilla recutita. Journal of Chromatography A. 2007;1154:390–399. doi: 10.1016/j.chroma.2007.03.106. [DOI] [PubMed] [Google Scholar]
- 24.Lin L-Z, Sun J, Chen P, Harnly JM. UPLC-PDA-EIS/HRMS/MSn analysis of anthocyanins, flavonol glycosides and hydroxycinnamic acid derivatives in red mustard green (Brassica juncea Coss variety) Journal of Agricultural & Food Chemistry. 2011;59:12059–12072. doi: 10.1021/jf202556p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin L-Z, Harnly JM. A screening method for the identification of glycosylated flavonoids and other phenolic compounds using a standard analytical approach for all plant materials. Journal of Agricultural & Food Chemistry. 2007;55:1084–1096. doi: 10.1021/jf062431s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin L-Z, Harnly JM. Identification of hydroxycinnamoylquinic acids of arnica flower and burdock roots using a standardized LC-DAD-ESI/MS method. Journal of Agricultural & Food Chemistry. 2008;56:10105–10114. doi: 10.1021/jf802412m. [DOI] [PubMed] [Google Scholar]
- 27.Lin L-Z, Harnly JM. Identification of the phenolic components of chrysanthemum flower (Chrysanthemum morifolium Ramat) Food Chemistry. 2010;120:319–326. [Google Scholar]