Abstract
BRAF is the most mutated gene in melanoma, with approximately 50% of patients containing V600E mutant protein. V600EB-RAF can be targeted using pharmacological agents, but resistance develops in patients by activating other proteins in the signaling pathway. Identifying downstream members in this signaling cascade is important to design strategies to avoid the development of resistance. Unfortunately, downstream proteins remain to be identified and therapeutic potential requires validation. A kinase screen was undertaken to identify downstream targets in the V600EB-RAF signaling cascade. Involvement of aurora kinase B (AURKB) and Wee1-like protein kinase (WEE1) as downstream proteins in the V600EB-RAF pathway was validated in xenografted tumors, and mechanisms of action were characterized in size- and time-matched tumors. Levels of only AURKB and WEE1 decreased in melanoma cells, when V600EB-RAF, mitogen-activated protein kinase 1/2, or extracellular signal–regulated kinase 1/2 protein levels were reduced using siRNA compared with other identified kinases. AURKB and WEE1 were expressed in tumors of patients with melanoma at higher levels than observed in normal human melanocytes. Targeting these proteins reduced tumor development by approximately 70%, similar to that observed when inhibiting V600EB-RAF. Furthermore, protein or activity levels of AURKB and WEE1 decreased in melanoma cells when pharmacological agents targeting upstream V600EB-RAF or mitogen-activated protein kinase were used to inhibit the V600EB-RAF pathway. Thus, AURKB and WEE1 are targets and biomarkers of therapeutic efficacy, lying downstream of V600EB-RAF in melanomas.
Melanoma remains the most common cause of skin cancer–related deaths worldwide.1 The incidence of melanoma increases with age, with a 28% probability of disease for individuals <40 years and a ≥70% probability for those >60 years.2 Approaches to manage advanced melanoma include surgery, radiation, immunotherapy, chemotherapy, or combinations of these approaches. Patients in the advanced stages of this disease have few treatment options for long-term management of the disease, with average 5-year survival being 10%.3 Therefore, a better understanding of the genes and processes regulating melanoma that could be used for selection of therapeutic targets as biomarkers for particular drug efficacy or prognostic indicators to assist in therapeutic agent selection and for overcoming resistance to targeted agents is needed.
Kinases play a key role regulating cellular proliferation and drug resistance development.4 In the mitogen-activated protein (MAP) kinase pathway, 50% and 25% of sporadic melanomas harbor BRAF or NRAS mutations, respectively, which activate the MAP kinase pathway measured through the activation of extracellular signal–regulated kinase (ERK).5 These mutations rarely occur in the same cell, but both mutations activate pathways to regulate diverse cellular processes aiding cancer development, with the most prominent being regulation of cellular proliferation.6 The most frequent BRAF mutation is a valine to glutamic acid substitution at residue 600 (V600E), which increases basal kinase activity.7 The most common NRAS mutation is a glutamine to leucine substitution (Q61L), which impairs GTP hydrolysis and maintains a constitutively active protein.8
Pharmacological agents have been developed to inhibit the activity of various proteins in the deregulated MAP kinase signaling pathway.9–12 Recent FDA approval of Zelboraf (vemurafenib; formerly known as PLX4032), is a major breakthrough for individuals with mutant V600EB-RAF.13–16 Vemurafenib leads to a high response rate in patients, but in most cases, more invasive resistant disease eventually recurs by circumventing V600EB-RAF, leading to mortality.13,16,17 Therefore, a better understanding of downstream members of the V600EB-RAF pathways is needed so that these proteins could be targeted together with vemurafenib or inhibited after the development of resistance to more effectively manage this disease.
To identify novel kinases regulating the proliferative potential of melanoma cells and then pinpoint those lying downstream of V600EB-RAF in this signaling cascade, an siRNA-based screen of a library of 636 kinases was undertaken. AURKB, Wee1-like protein kinase (WEE1), glycogen synthase kinase-3α (GSK3A), thiamin pyrophosphokinase 1 (TPK1), and B-RAF were identified as potential modulators of melanoma cell survival. The aurora kinase family consists of aurora kinase A (AURKA), aurora kinase B (AURKB), and aurora kinase C (AURKC).18 Involvement of AURKA in melanoma development has been reported, but it is not known whether AURKB and AURKC play roles in melanoma pathogenesis or development of drug resistance.19 WEE1 is a dual-specificity protein kinase involved in regulating cell cycle progression by phosphorylating and deactivating cyclin-associated CDKs.20,21 WEE1 currently has no known role in melanoma development. Two isoforms of GSK-3, called GSK3A and GSK-3β (GSK3B), have been identified. Although GSK3B has been shown to play a role in melanoma development and drug resistance,22 GSK3A has not been identified as a melanoma therapeutic target. The TPK catalyzes phosphorylation of thiamin to thiamin pyrophosphate and also has no known role in melanoma development.23
This study shows that AURKB, WEE1, GSK3A, and TPK1 were all expressed in tumors of patients with melanoma at higher levels than observed in normal human melanocytes. However, only AURKB and WEE1 levels decreased when V600EB-Raf, mitogen-activated protein kinase (MEK) 1/2, or ERK1/2 were targeted using siRNA, demonstrating that these proteins were downstream of V600EB-RAF in the deregulated MAP kinase signaling pathway. Subsequent studies confirmed that targeting AURKB or WEE1 reduced melanoma tumor development and led to a phenotype similar to that observed when inhibiting V600EB-RAF in this deregulated signaling cascade. Furthermore, AURKB or WEE1 levels decreased when pharmacological agents inhibiting V600EB-Raf or MEK were used to target melanoma cells. Thus, AURKB and WEE1 can be used as downstream therapeutic targets and as biomarkers of efficacy of agents targeting the V600EB-RAF signaling cascade in melanomas.
Materials and Methods
Cell Lines and Culture Conditions
Normal human primary melanocytes FOM 103 (provided by Dr. Meenhard Herlyn, Wistar Institute, Philadelphia, PA) were cultured as previously described.24 Human fibroblast FF2441 cells (provided by the laboratory of Dr. Craig Myers, Penn State College of Medicine, Hershey, PA), metastatic melanoma cell lines UACC 903 (provided by Mark Nelson, University of Arizona, Tucson, AZ), A375M (CRL-1619; ATCC, Manassas, VA), and 1205 Lu (provided by Dr. Herlyn) were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies, Grand Island, NY), supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT) and 1% GlutaMAX from Gibco (Life Technologies). Radial (WM35 and WM3211) and vertical (WM115 and WM278) growth phase melanoma cell lines (provided by Dr. Herlyn) were maintained in Tu 2% medium, as previously described.25 Cell lines were maintained in a 37°C humidified 5% CO2 atmosphere incubator and periodically monitored for cell phenotype, genetic biomarkers, and growth potential in culture and xenografts in mice to confirm the identity of the individual cell lines.
siRNA Screening to Identify Kinases Regulating Melanoma Cell Proliferation
To identify kinases that regulate the proliferative potential of melanoma cells, an siRNA screen was undertaken using the human StealthRNAi collection from Invitrogen (Life Technologies), containing three independent validated siRNAs for each of 636 kinase targets. Each plate was supplied with appropriate positive, negative, and transfection controls, including one fluorescent siRNA control and scrambled siRNA controls for low, medium, and high guanine-cytosine (GC) content. A primary screen was performed by transfecting 100 pmol of pooled siRNA (three individual siRNAs targeting the same kinase) into 2 × 104 UACC 903 melanoma cells using an Amaxa Nucleofector 96-well shuttle system, program CM-130, and solution SF (Lonza, Walkersville, MD). After 24 to 48 hours of recovery in 10% FBS containing culturing media, transfected cells were grown in serum-free media for an additional 3 days and viable cells were measured using the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) assay (CellTiter 96 AQueous Cell Proliferation Assay; Promega, Madison, WI). A minimum 20% decrease in cell viability compared with control-transfected cells was considered as a positive hit in the primary screen. siRNA-mediated inhibition of V600EB-Raf served as a positive control for the screen. The second validation step involved evaluating individual siRNAs of the pool from the primary screen. A minimum of two siRNAs had to inhibit cell survival to proceed. The next step was to validate growth-inhibitory effects in two additional melanoma cell lines, 1205 Lu and A375M. A candidate kinase only underwent further study after two independent siRNAs showed similar growth-inhibitory effects in three independent melanoma cell lines.
Western Blot Analysis
Cell lysates were collected and processed for Western blot analysis, as previously described.9,10,24,26,27 A total of 1 to 1.5 × 106 melanoma cells were plated in 100-mm culture dishes, treated 48 hours later with 0.2 to 20 μmol/L V600EB-Raf inhibitor; vemurafenib (ChemieTek, Indianapolis, IN) or 2.5 to 50 μmol/L MEK1/2 inhibitor; and U0126 (Cell Signaling Technologies, Beverly, MA) for 6 to 48 hours. Protein lysates were collected for Western blot analysis. Blots were probed with antibodies, according to manufacturer’s recommendations. Antibodies used in this study were as follows: AURKB, cyclin D1, ERK2, B-RAF, and α-enolase (Santa Cruz Biotechnology, Santa Cruz, CA); phospho-AURKB (LifeSpan Biosciences, Seattle, WA) and phospho-H3 (Millipore, Billerica, MA); TPK1 (Proteintech, Chicago, IL); and WEE1, phospho-WEE1, GSK3A, total ERK1/2, total MEK, phospho-MEK1/2, and phospho-ERK1/2 (Cell Signaling Technology). Secondary antibodies conjugated with horseradish peroxidase were obtained from Santa Cruz Biotechnology. Immunoblots were developed using the enhanced chemiluminescence detection system (Amersham Pharmacia Biotech, Piscataway, NJ).
siRNA Protein Knockdown Studies
A total of 100 pmol of siRNA was introduced into 1 × 106 melanoma cells via nucleofection using an Amaxa Nucleofector with Solution R/program K-17 for UACC 903 and 1205 Lu or Solution R/program A-23 for A375M. Transfection efficiency after nucleofection was >90%, with 80% to 90% cell viability. After siRNA transfection, cells were left to recover for 2 days and replated in 96-well plates to assess viability and proliferation. For duration of siRNA-mediated protein knockdown studies in vitro, 1.0 × 106 UACC 903, 1205 Lu, or A375M cells were nucleofected with small-interfering (si) AURKB#1, siAURKB#3, siWEE1#2, siWEE1#3, siRNA against mutant B-RAF, siMEK1 + MEK2, siERK1 + ERK2, siCYCLIN D1, and scrambled siRNA and protein lysates harvested at day 4 or 2, 4, 6, or 8 days later for Western blot analysis for siRNA knockdown time course experiments. The following Duplexed Stealth siRNA (Invitrogen) sequences were used:AURKB#1, 5′-CCAACAUCCUGCGUCUCUACAACUA-3′; AURKB#2, 5′-GGAUCUGUGGUGCAUUGGAGUGCUU-3′; AURKB#3, 5′-ACUUGUCCUCAUGAG-CCGCUCCAAU-3′; WEE1#1, 5′-GGGCAUCCAACAAAGUUAUGUUUAA-3′; WEE1#2, 5′-CCAGAGUAAUAGAACAUCUCGACUU-3′; WEE1#3, 5′-CCUCUUCCGAG-AAAUGGAGAUCAAU-3′; GSK3A#1, 5′-CCAAGGCCAAGUUGACCAUCCCUAU-3′; GSK3A#2, 5′-GGAGUUCAAGUUCCCUCAGAUUAAA-3′; GSK3A#3, 5′-CCAGGG-AACUAGUCGCCAUCAAGAA-3′; TPK1#1, 5′-GCUACUAAGGGAUGUGAGCUCAUUU-3′; TPK1#2, 5′-CCACACUGACUUUACUAGUGCCUU-3′; and TPK1#3, 5′-UG-GAACAUUGGUCAGUACUUCCAAU-3′. The siRNA validated and published sequences for Scrambled, V600EB-RAF, MEK1, MEK2, ERK1, ERK2, and CYCLIN D1 were as previously reported.9
Cell Viability and Proliferation Studies
For experiments using siRNA, 1 × 106 UACC 903 or 1205 Lu cells were nucleofected with 100 pmol of AURKB#1, siAURKB#3, siWEE1#2, siWEE1#3, siV600EB-Raf (as a positive control), scrambled siRNA (as a negative control), or transfection buffer (as a transfection reagent control). Cells were allowed to recover for 48 hours in 60-mm culture dishes and then 5 to 10 × 103 cells were seeded into 96-well plates. At 3 and 5 days later, cell viability was measured by MTS assay (Promega), or cell proliferation using the 5-bromo-2′-deoxyuridine (BrdU) enzyme-linked immunosorbent assay kit (Roche Applied Sciences, Indianapolis) was measured.
For studies using aurora kinase inhibitor (VX-680; LC Laboratories, Woburn, MA), viability and inhibitory concentration of 50% (IC50) of UACC 903, 1205 Lu, or A375M melanoma cells were assessed by MTS assay. Briefly, 5 × 103 melanoma or human fibroblast (FF2441) cells per well in 100 μL DMEM containing 10% FBS were grown in a 96-well plate for 24 to 76 hours and treated with either control dimethyl sulfoxide (DMSO) vehicle or increasing concentrations (1 to 100 μmol/L) of VX-680. Cell viability compared with vehicle control–treated cells was measured using the MTS assay. IC50 values for each compound in respective cell lines were estimated from three independent experiments using GraphPad Prism software version 4.01 (GraphPad Software Inc., La Jolla, CA).
Cell Cycle Analysis
For cell cycle analysis using siRNA, 1 × 106 UACC 903, 1205 Lu, and A375M cells were nucleofected, as previously detailed.27 To determine the effects of aurora kinase inhibitor on the cell cycle, UACC 903 cells were treated with VX-680 for 48 hours at a concentration ranging between 2.5 and 7.5 μmol/L. Cells were trypsinized, centrifuged (500 × g, for 5 minutes), and stained with 1 mL of propidium iodide. Stained cells were analyzed using the FACScan analyzer (Becton Dickinson Biosciences, San Jose, CA), and data were processed using ModFit LT software version 3.3 (Verity Software House, Topsham, ME).
Analysis of Tumors from Patients with Melanoma
Melanoma tumor specimens from human patients (n = 39) were randomly selected according to the protocols approved by the Institutional Review Board at Pennsylvania State University (Hershey, PA), and the Cooperative Human Tissue Network (Columbus, OH, and Philadelphia, PA). Informed consent was provided according to the Declaration of Helsinki. Tissue samples were collected from patients at surgery, immediately snap frozen in liquid nitrogen, and stored at −80°C until protein lysate collection. To collect protein for Western blot analysis, tumors were pulverized using a mortar and pestle chilled in liquid nitrogen. Protein lysates were extracted from tumors, as previously reported,26 and analyzed by using Western blot analysis to assess levels of AURKB, WEE1, GSK3A, and TPK1. Protein levels in tumors were normalized to α-enolase loading control, and relative AURKB, WEE1, GSK3A, and TPK1 expression levels were quantified using ImageJ software version 1.46r (NIH, Bethesda, MD), compared with melanocyte control, and graphed with the beeswarm package in R package version 0.1.5 (http://CRAN.R-project.org/package=beeswarm, last accessed February 15, 2013).
Animal Tumorigenicity Assessments Using siRNA
Animal experimentation was performed according to protocols approved by the Institutional Animal Care and Use Committee at Pennsylvania State University. Tumor kinetics studies were undertaken on athymic-Foxn1nu nude mice purchased from Harlan Sprague Dawley (Harlan Laboratories, Indianapolis, IN). A total of 100 pmol of siRNA was nucleofected into 20 × 106 cells, and after 48 hours of recovery, 1 × 106 cells were fractionated in 0.2 mL of 10% FBS-DMEM and then injected s.c. above both the left and right rib cages of 4- to 6-week-old female mice (three to five mice per group). Dimensions of developing tumors were measured on alternate days up to day 17.5, using calipers (Thomas Scientific, Swedesboro, NJ) by LxWxD (mm3).
Analysis of AURKB and WEE1 Expression, Cell Proliferation, and Apoptosis Rates in Time- and Size-Matched Tumors
To generate tumors of the same size developing at parallel time points, 1 × 106 UACC 903 melanoma cells nucleofected with either control buffer or scrambled siRNA or 10 × 106 cells nucleofected with AURKB#1 and AURKB#3, WEE1#2, and WEE1#3 siRNAs were injected into nude mice. Tumors were harvested at day 11 to measure AURKB and WEE1 expression and activity using Western blot analysis. For analyzing the cell proliferation index and apoptosis in these tumors, mouse anti-human Ki-67 from BD Pharmingen (BD Biosciences, San Diego, CA) and a TUNEL TMR Red Apoptosis kit (Roche, Mannheim, Germany) were used, respectively, as previously described.26 The number of Ki-67– and TUNEL-stained cells were quantified as the percentage of total cells in tumors. To quantify AURKB and WEE1 expression by immunohistochemistry (IHC) in formalin-fixed, paraffin-embedded tumor sections, tissue sections were deparaffinized and rehydrated in PBS, after which antigen retrieval was undertaken by incubation in 0.01 mol/L citrate buffer, pH 6.0, for 20 minutes in a 95°C water bath. Slides were cooled for 20 minutes, rinsed in PBS, and incubated in 3% H2O2 for 10 minutes to quench endogenous peroxidase activity. Next, sections were blocked with 1% bovine serum albumin for 30 minutes and incubated with a 1:50 dilution of anti-AURKB (Abcam, Cambridge, MA) or WEE1 (Abgent, Inc., San Diego, CA) antibody overnight at 4°C. After rinsing in PBS, sections were incubated with biotinylated anti-rabbit IgG (BD Pharmingen) for 1 hour and treated with peroxidase-labeled streptavidin (BD Pharmingen) for 30 minutes. Visualization was accomplished using 3, 3′-diaminobenzidine for 5 to 10 minutes (BD Pharmingen), and nuclei were counterstained with hematoxylin (Dako, Carpentaria, CA). The percentage of cells that stained positive for AURKB and WEE1 was measured from a minimum of three to five different tumors (four to five fields per tumor). Sections were imaged using a Nikon Eclipse 600 camera (Nikon, Melville, NY), photographed at ×400 magnification, and quantified using Image Processing laboratory imaging software version 4.0.14 (Scanalytics, Fairfax, VA).
Tumor Studies Using VX-680, Vemurafenib, or U0126
A total of 1.5 × 106 UACC 903 cells in 0.2 mL of DMEM, supplemented with 10% FBS, were s.c. injected above both left and right rib cages of 3- to 4-week-old female athymic nude-Foxn1nu mice (Harlan Sprague Dawley). Six days later, when a fully vascularized tumor of 50 to 75 mm3 had formed, mice were randomly divided into DMSO vehicle control and experimental groups (three mice per group and two tumors per mouse) and treated i.p. with 50 or 75 mg/kg body weight VX-680 on alternate days for 3 to 4 weeks. Twenty-six days later, tumors were harvested and analyzed by IHC and Western blot analysis, as previously detailed. vemurafenib or U0126 was dissolved in 10 mg/kg body weight DMSO and injected i.p. every day for 6 days. Body weight (in grams) and dimensions of developing tumors (in mm3) were measured at drug administration. Tumors were harvested and analyzed for AURKB and WEE1 expression using IHC, as previously detailed. Tumors from animals treated with VX-680 were analyzed for pAURKB, AURKB, and pHistone-3 using Western blot analysis, as previously mentioned.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism Software version 4.0 and R version 2.15.1 (http://www.r-project.org). One- or two-way analysis of variance was used for groupwise comparisons, followed by the Tukey’s or Bonferroni’s post hoc tests. For comparison between two groups, the Student’s t-test (two tailed) was used. The two-sided, one-sample Wilcoxon signed-rank test was used to analyze tumor samples from patients with melanoma. Results represent at least two to three independent experiments and are shown as averages ± SEM. Results with a P < 0.05 (95% CI) were considered significant. Sample sizes and number of times experiments were repeated are indicated in the figure legends. The level of statistical significance is given in the figures.
Results
siRNA Screening Identifies Kinases Regulating Melanoma Cell Survival
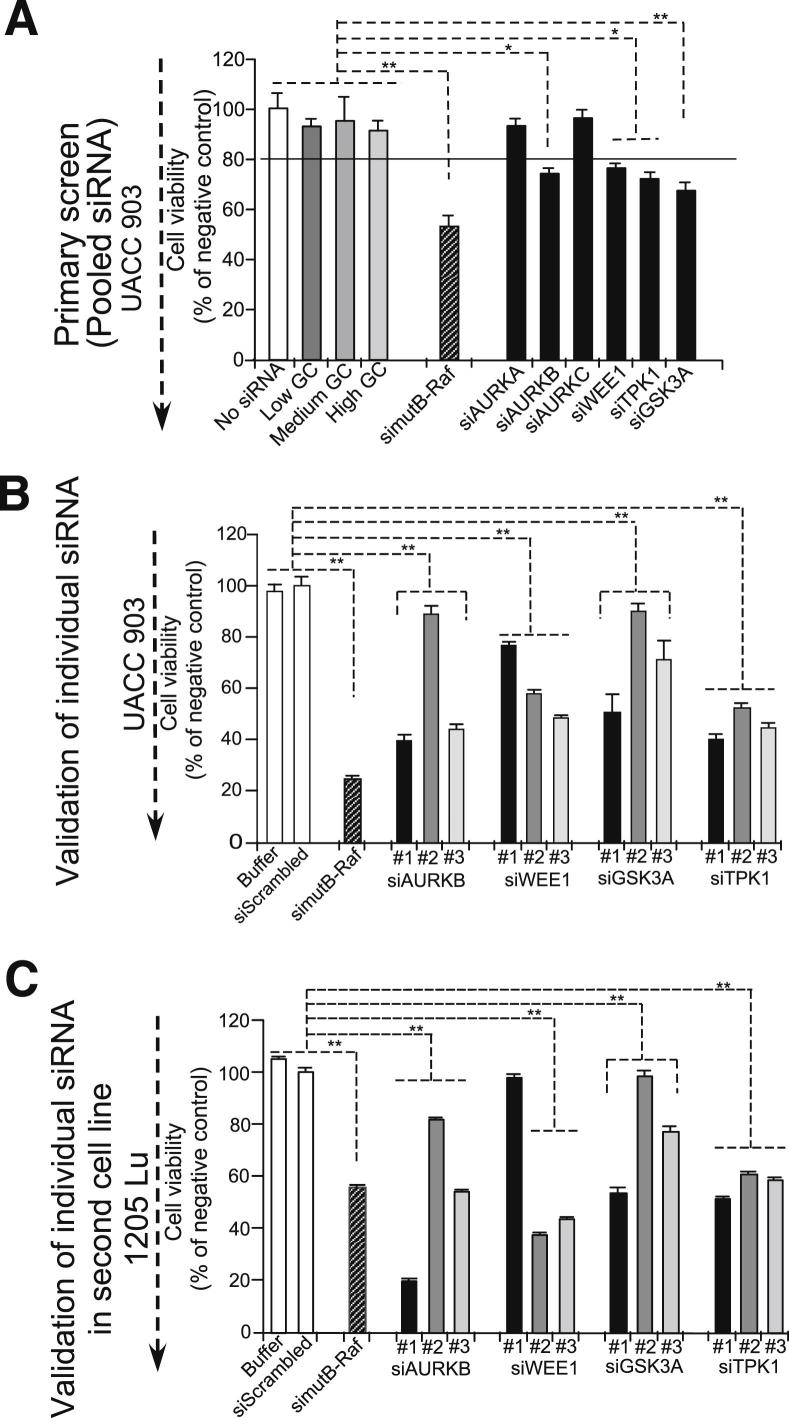
To identify kinases that regulate melanoma cell survival, an siRNA library screen was undertaken using the human Stealth RNAi collection (Life Technologies). The primary screen involved transfecting 100 pmol of siRNA pool (three individual siRNAs targeting the same kinase) into UACC 903 metastatic melanoma cells using the Amaxa Nucleofector 96-well shuttle system. The primary screen identified 33 of 636 kinases (hit rate, 5%). Of the 33 hits, AURKB, WEE1, GSK3A, TPK1, and B-RAF were identified among the possible targets in melanoma development. The identification of B-RAF as one of the targets validated the efficacy of the primary screen for identifying potentially important proteins involved in melanoma cell proliferation (Figure 1A). AURKA and AURKC were used as controls for related family members that did not decrease UACC 903 cell survival. The secondary validation step was to evaluate whether individual siRNAs to each target would have a similar inhibitory effect to the pooled siRNA in UACC 903 cells (Figure 1B). At least two of the three siRNAs targeting different regions of each respective mRNA decreased UACC 903 cell survival and protein expression (Supplemental Figure S1). Although all three siRNAs decreased the expression of target protein, only two siRNAs decreased the proliferative potential (Supplemental Figure S1). AURKB, WEE1, GSK3A, and TPK1 had at least two siRNAs that reduced the proliferative potential of melanoma cells (Figure 1B). The third validation step involved evaluating the inhibitory efficacy in two additional cell lines, 1205 Lu (Figure 1C) and A375M (data not shown), which showed similar results to those observed for UACC 903 cells. siRNAs targeting AURKB, WEE1, GSK3A, and TPK1 had similar growth-inhibitory effects in all three independently derived melanoma cell lines.
Figure 1.
Identification of kinases regulating melanoma cell survival. A: Screen showing identification of siAURKB, siWEE1, siGSK3A, siTPK1, and simutB-RAF as potential key targets in melanoma. A cellular viability test using MTS was used as the end point assay. B: A secondary validation step involved measuring inhibitory effects of three individual siRNAs on UACC 903 cell viability. C: Confirming effect of individual siRNA in a second melanoma cell line. siRNAs targeting siAURKB, siWEE1, siGSK3A, and siTPK1 reduced cell viability in multiple cell lines. siRNAs targeting V600EB-Raf were used as positive controls. Data are representative of a single experiment done in triplicate (A) and two to three independent experiments done in triplicate (B and C). Data are represented as means ± SEM percentage of cell viability. *P < 0.05, **P < 0.01.
Expression Levels of AURKB, WEE1, and GSK3A Are Elevated in Tumors of Patients with Melanoma
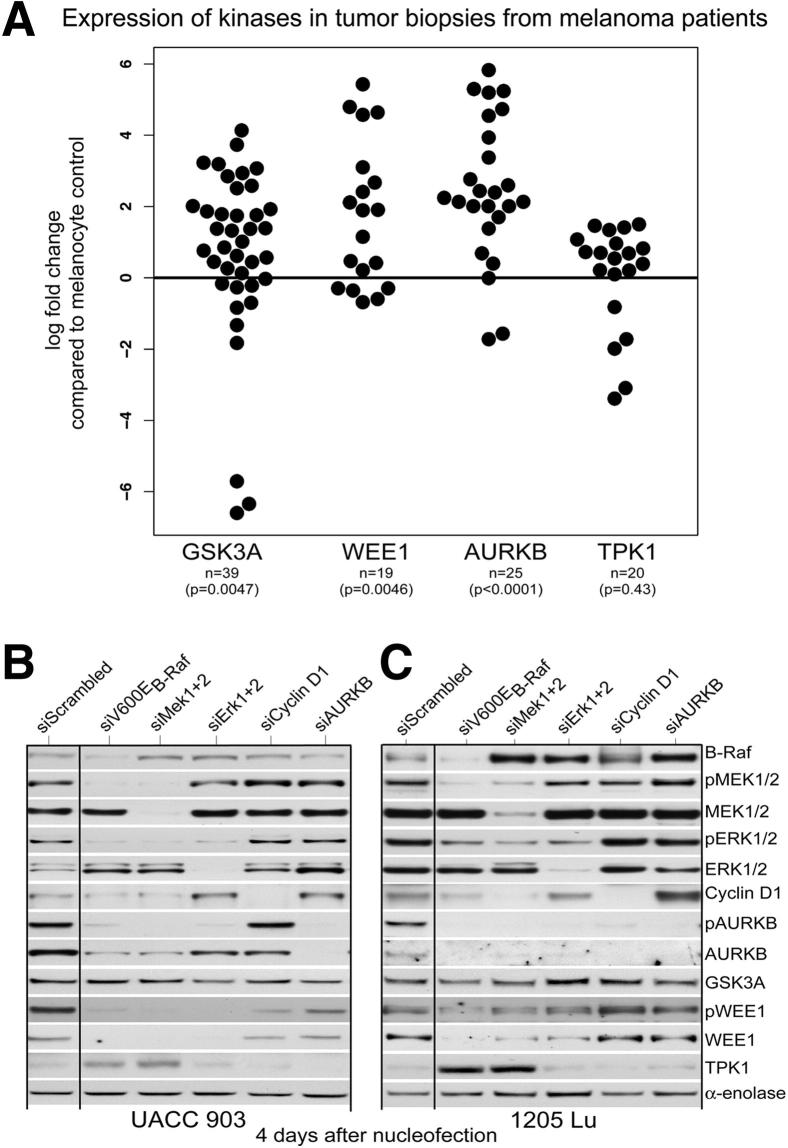
To validate involvement of AURKB, WEE1, GSK3A, and TPK1 in melanoma, protein from tumors of patients with melanoma (n = 39) was analyzed for AURKB, WEE1, GSK3A, and TPK1 expression by using Western blot analysis. Melanoma tumor specimens from human patients were randomly selected. All of the tumor specimens used were derived from patients with malignant or metastatic melanoma. Results were normalized to α-enolase loading control and compared with normal human melanocyte controls. The fold changes, relative to melanocytes, were analyzed and graphed on the log scale for improved visualization and increased robustness in the analysis. The two-sided, one-sample Wilcoxon signed-rank test was used to determine whether the distribution of log fold changes was statistically different from 0 (melanocytes were normalized to a fold change of one or a log fold change of zero). A graph shows significant up-regulation of AURKB, WEE1, and GSK3A compared with melanocytes. However, TPK1 showed no significant (P = 0.43) differences compared with melanocyte control (Figure 2A). Representative Western blot analyses showing expression and activity of WEE1 and AURKB, compared with melanocyte control, can be seen (Supplemental Figure S2). Advanced-stage melanoma cell line UACC 903 was used as a positive control. Increased expression of these kinases in melanomas suggested that they might play a potentially important role in melanoma development. The next goal was to determine which of the kinases lay downstream of V600EB-RAF in this important signaling cascade.
Figure 2.
Expression and demonstration that AURKB and WEE1, but not GSK3A or TPK1, lie downstream of V600EB-RAF in the MAP kinase–signaling pathway. A: Expression of AURKB, WEE1, GSK3A, and TPK1 in tumor biopsy specimens from patients with melanoma. Protein isolated from tumors of patients with melanoma (n = 39) was analyzed for AURKB, WEE1, GSK3A, and TPK1 expression by using Western blot analysis. Results were normalized to α-enolase for equal protein loading and compared with normal human melanocyte controls. Data represent fold changes, relative to melanocytes (melanocytes were normalized to a fold change of one or a log fold change of zero), and were analyzed and graphed on the log scale. Number of samples for AURKB (n = 25, P < 0.0001), WEE1 (n = 19, P = 0.0046), GSK3A (n = 39, P = 0.0047), and TPK1 (n = 20, P = 0.43), two-sided one-sample Wilcoxon signed-rank test. B and C: Decreasing levels of protein expression for each member of the MAP kinase pathway inhibit AURKB or WEE1 protein or activity levels. siRNA was introduced into UACC 903 or 1205 Lu cells to reduce protein levels of V600EB-Raf, MEK1/2, ERK1/2, cyclin D1, and AURKB. The effect on AURKB, pAURKB, WEE1, GSK3A, or TPK1 protein levels was measured. Compared with cells treated with scrambled siRNA, decreasing protein levels of each member of the MAP kinase pathway led to a decrease in AURKB and WEE1 levels. No changes were observed in GSK3A or TPK1 levels. α-Enolase served as a control for equal protein loading. Western blot was reproduced at least two to three times (B).
AURKB and WEE1 Lie Downstream of V600EB-Raf in the MAP Kinase Pathway
The MAP kinase pathway is constitutively active in 50% to 60% of melanomas because of a single-base mutation in Braf converting T to A at nucleotide 1799, which substitutes a valine for glutamic acid at codon 600 (V600E).7 It is unknown whether the V600EB-Raf signaling cascade mediates its proliferative effects through AURKB, WEE1, GSK3A, or TPK1 expression or activity. To determine whether these kinases were regulated by V600EB-Raf signaling, siRNA-targeting V600EB-Raf, MEK1/2, or ERK1/2 were nucleofected into UACC 903 or 1205 Lu melanoma cells, and the effect on expression or activity (ie, phosphorylated protein levels) of the kinases was examined. siRNA to cyclin D1 was used to rule out that the kinases are simply being regulated in a cell cycle–dependent manner. These siRNAs have been previously validated as targeting MAP kinase proteins in these cell lines.9
siRNA-mediated knockdown of V600EB-Raf, MEK1/2, or ERK1/2 genes decreased the expression and activity of AURKB and WEE1 in both UACC 903 and 1205 Lu cell lines (Figure 2, B and C). In contrast, only AURKB protein levels decreased with the knockdown of cyclin D1, which is an important downstream transcription factor of the B-Raf/MEK/ERK cascade.28 No change was observed in GSK3A levels, which is consistent with its role in regulating apoptosis through the phosphatidylinositol 3-kinase pathway.29 TPK1 protein levels were up-regulated on knockdown of V600EB-Raf and MEK1/2 proteins; however, knockdown of neither ERK1/2 nor cyclin D1 altered TPK1 levels, indicating that another cascade downstream of MEK1/2 protein might be regulating TPK1 protein levels (Figure 2, B and C). In a well-established cell line tumor progression model30 (data not shown), all melanoma cell lines had decreased TPK1 expression compared with the melanocyte control; however, no statistically significant difference was observed in patient tumors (Figure 2A). Therefore, the effect observed in cell culture (Figure 2, B and C) is likely an artifact. Decreased cyclin D1 levels had no effect on AURKB or WEE1 expression in UACC 903 cells and no effect on WEE1 levels in 1205 Lu cells (Figure 2, B and C). Based on these observations, subsequent studies focused on AURKB and WEE1 to determine whether these proteins could be used as downstream therapeutic targets of the V600EB-Raf signaling cascade or as biomarkers of therapeutic efficacy when using agents targeting this pathway.
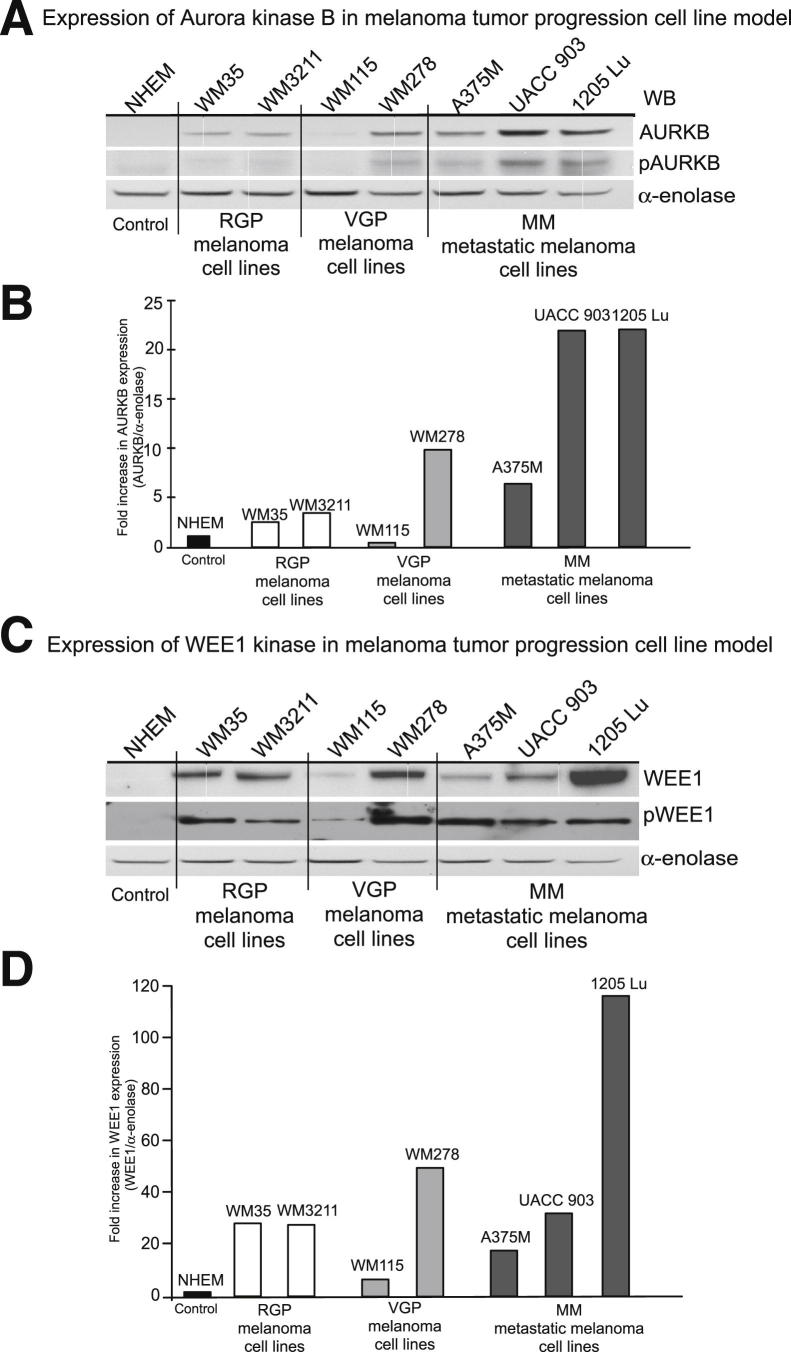
Increased Levels of AURKB and WEE1 Protein Are Observed in Melanoma Cell Lines
Protein activity and expression levels of AURKB and WEE1 in cell lines isolated from the different stages of melanoma tumor progression were compared with normal human melanocytes. Compared with the melanocyte control, higher AURKB levels were observed in all cell lines, except WM115 cells (Figure 3, A and B). Elevated levels of WEE1 were observed in all cell lines, with the highest occurring in 1205 Lu melanoma cells (Figure 3, C and D). Thus, levels of AURKB and WEE1 protein expression were increased in most cell lines compared with melanocytes.
Figure 3.
Expression of AURKB and WEE1 protein in advanced-stage melanoma cell lines. A and C: Protein lysates collected from normal human melanocytes, radial (WM35 and WM3211), vertical (WM115 and WM278), and metastatic (A375M, UACC 903, and 1205 Lu) stage cell lines were subjected to Western blot (WB) analysis and probed for AURKB and WEE1. B and D: Levels were normalized to α-enolase loading control and fold increase with respect to normal human melanocytes quantified. The bar graph shows fold increase in expression levels of AURKB and WEE1 in melanoma cell lines compared with normal human melanocytes. Western blot data were reproduced at least twice. NHEM, normal human melanocytes.
Targeting AURKB or WEE1 Protein Levels in Melanoma Cells Decreases the Tumorigenic Potential of Melanoma Cells
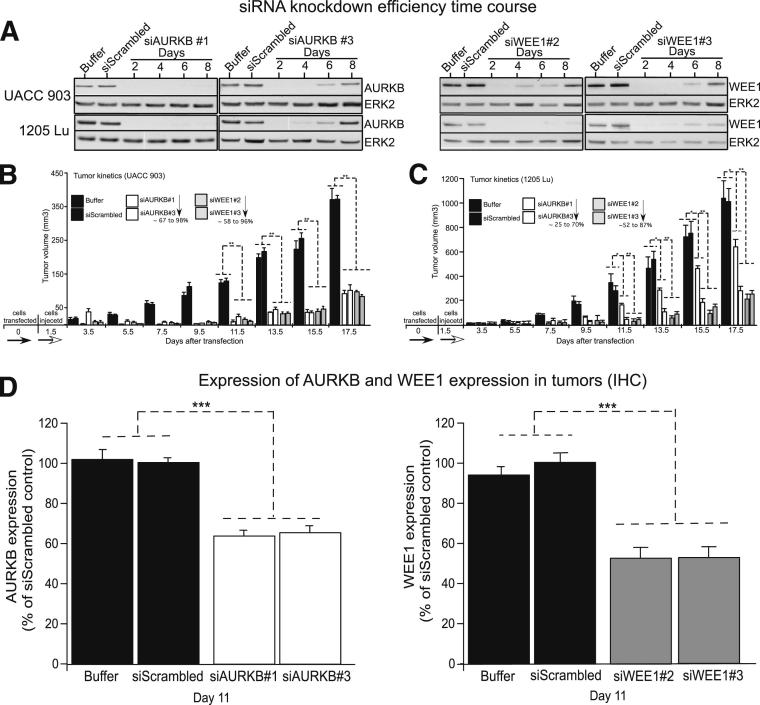
To determine the consequence of targeting AURKB or WEE1 in melanoma cells, siRNAs targeting these genes were introduced into UACC 903 and 1205 Lu cells. siRNA efficiency and duration of protein knockdown were validated by showing decreased protein levels for 6 to 8 days after transfection (Figure 4A). Examining duration of in vitro protein knockdown is essential for evaluation of the effect of siRNA-mediated targeting of genes for tumor development studies in animals.9,10,31 After siRNA transfection, cells were injected s.c. above both the left and right rib cages of 4- to 6-week-old female nude mice, and dimensions of developing tumors were measured on alternate days up to day 17.5. For both cell lines, tumor development was reduced by up to 70% compared with buffer or scrambled siRNA controls at day 17.5 (Figure 4, B and C). The IHC analysis showed an approximately 40% decrease in tumor cell AURKB or WEE1 protein expression compared with buffer or scrambled siRNA-treated cells 11 days after injection in mice9,10,26,27,31,32 (Figure 4D and Supplemental Figure S3). Thus, decreasing AURKB or WEE1 protein levels reduced the tumorigenic potential of melanoma cells. Next, the mechanism of action of targeting either of these proteins downstream of V600EB-RAF was investigated.
Figure 4.
siRNA-mediated targeting of AURKB or WEE1 decreased the tumorigenic potential of melanoma cells. A: A total of 100 pmol of siRNA to AURKB or WEE1 was introduced into melanoma cells via nucleofection and protein expression, assessed 2, 4, 6, or 8 days later, and compared with cells treated with control buffer or scrambled siRNA. Erk2 was used as a control for equal protein loading. Western blot analysis data were reproduced at least twice. B and C: UACC 903 or 1205 Lu melanoma cells were nucleofected with siRNA targeting AURKB or WEE1, and 48 hours later, viable cells were s.c. injected into left and right flanks of nude mice. Developing tumors were measured on alternate days for 17.5 days. The bar graph presents tumor volume (mm3). Black bars represent buffer and scrambled controls; white bars, AURKB siRNA#1 and AURKB siRNA#3; and gray bars, WEE1 siRNA#2 and WEE1 siRNA#3. Data were obtained from duplicate experiments with three mice per group, containing two tumors per mouse. Each point represents average data obtained from six nude mice. Data represent means ± SEM. *P < 0.05, **P < 0.01, two-way analysis of variance, followed by the post hoc test. D: IHC staining of xenografted tumors from animals injected with UACC 903 melanoma cells transfected with buffer or siScrambled controls, or siRNA to AURKB or WEE1. Day 11 tumors were harvested and stained, and the expression of AURKB or WEE1 was quantified and compared with control tumors. Data were obtained from three to four tumors, with four to five fields per tumor. Data represent means ± SEM. ***P < 0.001, t-test.
AURKB and WEE1 Inhibition Reduces Melanoma Tumor Development by Decreasing Cellular Proliferation
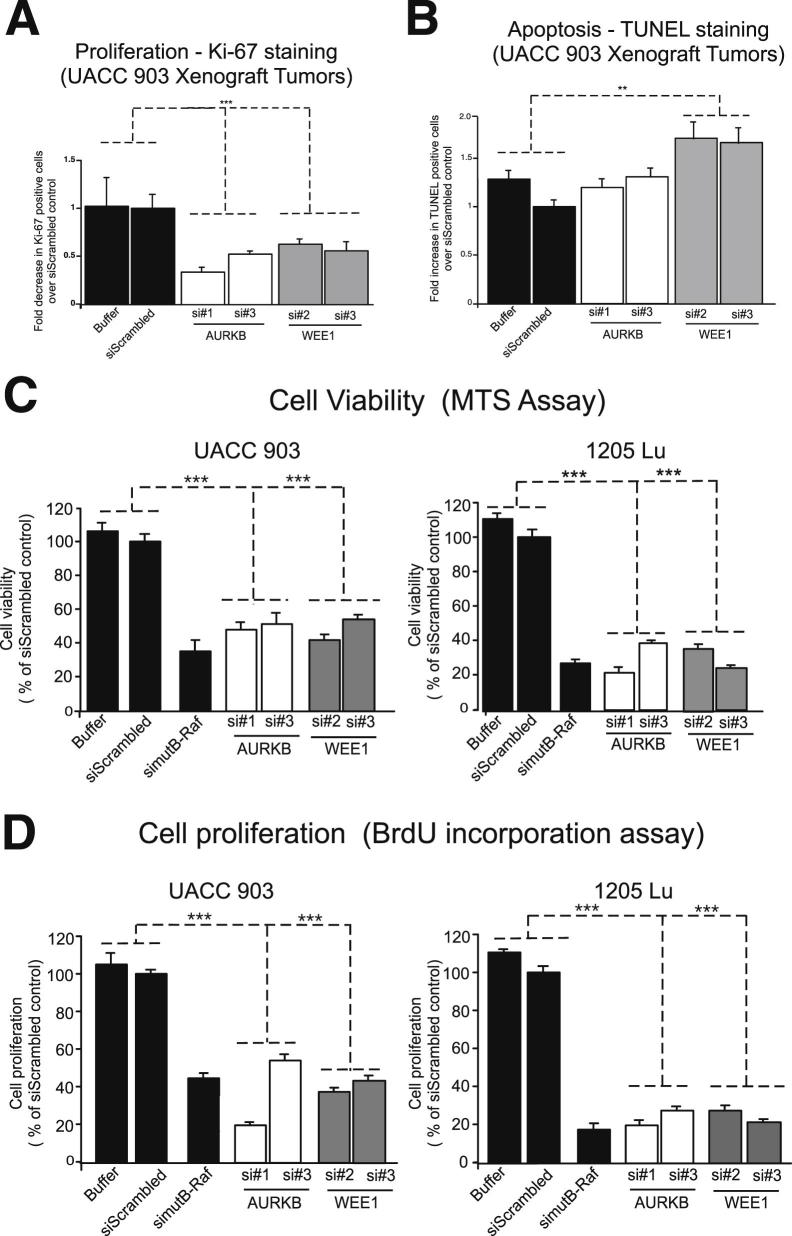
To identify the mechanistic basis leading to tumor inhibition after decreased AURKB or WEE1 protein levels, proliferation and apoptosis levels in tumors of the same size developing at day 11 were examined. Formalin-fixed, paraffin-embedded tumor sections were examined by Ki-67 staining to assess proliferation (Supplemental Figure S4A) and TUNEL analysis to estimate apoptosis rates (Supplemental Figure S4B). Reducing AURKB or WEE1 protein levels led to a statistically significant 47% to 66% decrease in Ki-67–positive tumor cell proliferation (Figure 5A). In contrast, apoptosis rates of tumor cells were not significantly different between control and xenografted tumors harvested from animals injected with cells nucleofected with AURKB siRNA (Figure 5B). A slight increase in apoptotic tumor cells was observed after knockdown of WEE1 protein levels as well (Figure 5B). Thus, decreasing AURKB or WEE1 protein expression levels in melanoma cells reduced tumor development by decreasing cellular proliferation, consistent with these proteins lying downstream of V600EB-Raf.
Figure 5.
Targeting AURKB or WEE1 reduced melanoma cell survival by decreasing the proliferative potential of melanoma cells. A and B: Size- and time-matched tumors from animals injected with UACC 903 melanoma cells transfected with buffer or siScrambled controls or siRNA to AURKB or WEE1. Tumor sections were immunostained for Ki-67 (A) or TUNEL (B) for proliferation and apoptosis, respectively. Images were quantified and plotted as fold decrease or increase in Ki-67– or TUNEL-positive cells compared with controls. Data were obtained from three to four tumors, with four to five fields averaged per tumor. C and D: siRNA was introduced in UACC 903 or 1205 Lu melanoma cells and an MTS or BrdU incorporation assay undertaken after 3 days to measure cell viability or proliferation, respectively. V600EB-RAF served as a control of a gene decreasing cell viability. Results represent pooled data from three independent experiments. Data represent means ± SEM. **P < 0.01, ***P < 0.01. ns, not statistically significant. An analysis of variance, followed by a post hoc test, was used.
Inhibition of AURKB or WEE1 Decreases the Viability of Cultured Melanoma Cells by Reducing Cellular Proliferative Potential
To demonstrate that AURKB and WEE1 inhibition reduced melanoma cell survival by decreasing the proliferative potential of melanoma cells, viability by MTS and proliferation using BrdU incorporation was measured after siRNA-mediated protein knockdown in cells. Decreasing either AURKB or WEE1 reduced melanoma cell growth in UACC 903 and 1205 Lu cells by 50% to 60% (Figure 5C). Decreased survival was mediated by reduced cellular proliferation because targeting AURKB or WEE1 led to a 30% to 80% decrease in BrdU incorporation in both the cell lines (Figure 5D). V600EB-Raf was used as the gene control for inhibiting this pathway. Thus, decreasing AURKB or WEE1 protein levels in cultured melanoma cells decreased cell survival, mediated by a reduction in proliferation.
Targeting AURKB or WEE1 Induces a G2/M Block, Leading to Increased Rates of Cellular Apoptosis
AURKB regulates a crucial spindle checkpoint during cell division, whose inhibition can cause a premature exit from mitosis, preventing proper chromosome segregation and cytokinesis, resulting in a G2/M block in the cell cycle.33,34 WEE1 regulates cell cycle progression by inhibiting entry into mitosis, and its absence leads to division at a premature stage and subnormal cell size.20,35 To evaluate the disruption of the cell cycle mediated by targeting these proteins, cell cycle analysis using the fluorescence-activated cell sorter was undertaken on cells after knockdown of AURKB or WEE1 protein levels. Control UACC 903, 1205 Lu, or A375M cells treated with buffer or scrambled siRNA had a G2/M cell population of approximately 7% to 15% compared with cells transfected with siAURKB having levels ranging from 25% to 60% (Table 1). Thus, decreasing levels of AURKB or WEE1 protein in melanoma cells causes an increase in the G2/M population.
Table 1.
Effect of Targeting AURKB or WEE1 on Cell Cycle Using FACS Analysis
| Variable | Buffer | siScrambled | siB-Raf | siAURKB#1 | siAURKB#3 | siWEE1#2 | siWEE1#3 |
|---|---|---|---|---|---|---|---|
| UACC 903 | |||||||
| G0/G1 | 65.7 ± 0.1 | 65.6 ± 0.2 | 85.7 ± 2 | 30.5 ± 0.2 | 57.5 ± 2.2 | 31.2 ± 3.0 | 37.8 ± 0.2 |
| S | 26.5 ± 2.9 | 27.1 ± 2.9 | 8.2 ± 1 | 9.6 ± 0.1 | 17.1 ± 2.4 | 45.7 ± 5.7 | 44.0 ± 0.2 |
| G2/M | 7.8 ± 3.0 | 7.3 ± 3.1 | 6.0 ± 0.9 | 59.9 ± 0.2 | 25.3 ± 4.3 | 23.0 ± 6.5 | 18.1 ± 1.0 |
| Sub-G0/G1 | 0.5 ± 1.1 | 0.4 ± 1.1 | 0.7 ± 0.1 | 15.2 ± 0.0 | 6.0 ± 1.0 | 7.8 ± 3.6 | 14.9 ± 0.1 |
| 1205 Lu | |||||||
| G0/G1 | 68.9 ± 1.1 | 66.6 ± 2.3 | 70.05 ± 1.0 | 37.7 ± 0.7 | 60.4 ± 2.2 | 35.5 ± 8.2 | 61.6 ± 7.2 |
| S | 20.0 ± 2.2 | 24.0 ± 4.5 | 28.78 ± 1.2 | 10.4 ± 1.6 | 12.3 ± 2.4 | 41.1 ± 2.5 | 13.3 ± 4.4 |
| G2/M | 10.2 ± 1.4 | 9.3 ± 2.6 | 1.29 ± 0.1 | 51.9 ± 0.9 | 27.3 ± 4.3 | 23.4 ± 5.6 | 25.0 ± 4.6 |
| Sub-G0/G1 | 0.5 ± 0.2 | 0.5 ± 0.2 | 21.1 ± 0.0 | 4.0 ± 0.4 | 3.4 ± 1.0 | 27.8 ± 4.7 | 45.1 ± 7.6 |
| A375M | |||||||
| G0/G1 | 66.28 ± 1.2 | 69.03 ± 1.0 | 71.91 ± 0.9 | 46.26 ± 0.2 | 50.68 ± 1.3 | 50.15 ± 0.8 | 43.28 ± 1.2 |
| S | 16.10 ± 0.1 | 15.44 ± 0.5 | 13.49 ± 0.5 | 30.29 ± 1.2 | 19.19 ± 0.6 | 22.91 ± 0.9 | 25.06 ± 3.0 |
| G2/M | 16.61 ± 0.3 | 14.53 ± 0.5 | 11.60 ± 0.6 | 24.94 ± 3.0 | 31.12 ± 0.6 | 25.44 ± 1.4 | 29.15 ± 1.8 |
| Sub-G0/G1 | 2.33 ± 0.3 | 2.85 ± 0.8 | 9.13 ± 0.1 | 23.00 ± 3.0 | 7.9 ± 0.4 | 9.14 ± 0.1 | 22.82 ± 3.8 |
Data are given as means ± SEM percentages, from three to four separate experiments. siRNAs targeting AURKB or WEE1 were introduced into UACC 903, 1205 Lu, or A375M cells, and populations of the cell cycle were analyzed by FACS analysis.
FACS, fluorescence-activated cell sorter.
AURKB and WEE1 Serve as Indicators of the Therapeutic Efficacy of Drugs Targeting the MAP Kinase Pathway
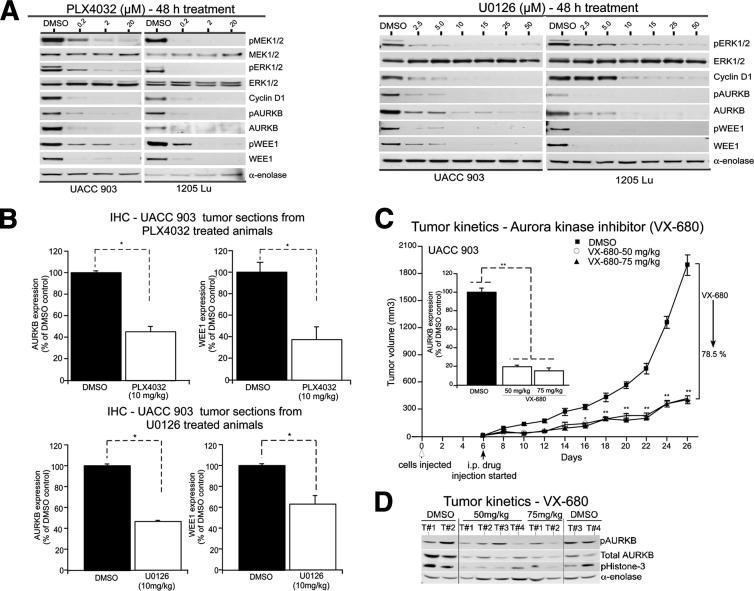
To establish whether AURKB or WEE1 could be used as biomarkers of the efficacy of pharmacological agents targeting the V600EB-RAF–signaling cascade, the pathway was targeted using vemurafenib or U0126, known inhibitors of V600EB-Raf and Mek1/2, respectively.2,9 Treatment of UACC 903 or 1205 Lu with vemurafenib decreased levels of phosphorylated Mek and Erk (Figure 6A). AURKB and WEE1 protein expression and/or activity levels decreased with reduction of the MAP kinase–signaling cascade after vemurafenib treatment (Figure 6A) in a manner similar to that of cyclin D1, which is an established biomarker of proliferation.36 Similarly, treatment with U0126 decreased pErk1/2 and cyclin D1 levels, which were mirrored by a reduction in AURKB and WEE1 protein and/or phosphorylation levels (Figure 6A). Tumors in animals treated with either vemurafenib or U0126 also exhibited decreased AURKB or WEE1 expression after IHC staining of tumors treated with the drugs compared with animals exposed to control DMSO (Figure 6B and Supplemental Figure S5). Thus, AURKB and WEE1 levels can be used as biomarkers to measure the therapeutic efficacy of MAP kinase pathway inhibitors.
Figure 6.
AURKB or WEE1 serve as biomarkers of the activity of pharmacological agents targeting the MAP kinase pathway. A: UACC 903 or 1205 Lu cells were treated with 0.2 to 20 μmol/L vemurafenib, a B-RafV600E inhibitor (left panel), or 2.5 to 50 μmol/L U0126, an MEK1/2 inhibitor (right panel), for 48 hours. Protein lysates were collected and analyzed for MEK1/2, ERK1/2, cyclin D1, and AURKB or WEE1 expression by using Western blot analysis. α-Enolase served as a control for equal protein loading. A representative blot from two to three independent experiments is shown. B: Nude mice were injected with 5 × 106 UACC 903 melanoma cells, and 7 days later, vemurafenib or UO126 treatment at a dose of 10 mg/kg body weight was initiated, once per day for 6 days. Tumors were removed after last treatment and divided into sections for IHC staining of AURKB or WEE1. The expression of AURKB (top left panel) or WEE1 (top right panel) was quantified in tumor sections from vemurafenib-treated animals. Similarly, the expression of AURKB (bottom left panel) or WEE1 (bottom right panel) was quantified in tumor sections from U0126-treated animals compared with DMSO-treated controls and plotted. Data were obtained from three to four tumors, with four to five fields per tumor analyzed. C: A total of 1 × 106 UACC 903 melanoma cells were injected s.c. into the right and left flanks of nude mice, and 6 days later, they were treated daily i.p. with a dose of 50 or 75 mg/kg body weight of VX-680. Developing tumors were measured on alternate days up to day 26 and compared with controls treated with DMSO. Each point represents data obtained from five nude mice (two tumors on each side). Tumor sections were immunostained for AURKB and compared with DMSO control–treated tumors (inset). Data represent three to four tumors, with four to five fields per tumor analyzed. D: Protein lysates were analyzed by using Western blot analysis for AURKB, pAURKB, and pHistone-3. α-Enolase served as a control for equal protein loading. Data represent means ± SEM. *P < 0.01 (t-test) (B); *P < 0.01, **P < 0.001, analysis of variance, followed by a post hoc test (C).
Demonstration That Pharmacological Inhibition of Therapeutic Targets Downstream of V600EB-RAF Can Effectively Inhibit Melanoma Development
To demonstrate the efficacy of a pharmacological agent targeting AURKB downstream in the V600EB-Raf–signaling cascade, the efficacy of VX-680 (a small-molecule pan-Aurora kinase inhibitor37), which inhibits cellular proliferation by disrupting the cell cycle without negatively affecting normal cell survival, was evaluated.38,39 IC50 values of UACC 903, A375M, and 1205 Lu melanoma cells treated with VX-680 were 8.3, 11.45, and 8.10 μmol/L, respectively (Supplemental Figure S6A). At 24 hours after drug treatment, melanoma cells were approximately 3.5- to 5-fold more sensitive than fibroblasts to the agent. The drug caused a G2/M block, with the highest accumulation occurring at 2.5 μmol/L VX-680 (Supplemental Figure S6B). Higher concentrations led to polyploidization due to continued cell cycle progression in the absence of cell division, leading to a G0/G1 block; hence, fewer cells were observed in G2/M than at lower concentrations, which would eventually lead to disappearance of the G2/M population.40 The i.p. administration of VX-680 at 50 and 75 mg/kg body weight reduced melanoma tumor development by 78% compared with DMSO control-treated mice and decreased AURKB expression in tumor cells measured by IHC (Figure 6C). In addition, decreased expression and activity of AURKB, as measured by pHistone-3 levels, were observed in VX-680–treated tumors harvested at day 26 (Figure 6D). Thus, pharmacological inhibition of AURKB decreased melanoma cell proliferation by inducing a G2/M block, which reduced melanoma tumor development.
Discussion
BRAF is the most mutated gene in melanoma constitutively activating the MAP kinase–signaling cascade.7 Vemurafenib preferentially binds to V600EB-Raf to inactivate the pathway.11,41 Although the drug is initially effective at reducing the tumor burden of patients, resistance quickly develops in the initial responders, leading to disease progression and death.17,42 Therefore, new and novel approaches are needed to overcome this drug-induced resistance. One approach could be to target proteins downstream in the V600EB-RAF signaling cascade.
This report identifies AURKB and WEE1 as two kinases lying downstream of V600EB-RAF in the MAP kinase–signaling cascade, which can be used as therapeutic targets or biomarkers of drug efficacy for agents inhibiting this pathway. A series of siRNA-based screens were undertaken using a library of 636 kinases, which identified AURKB, WEE1, GSK3A, TPK1, and B-RAF as potential modulators of melanoma cell survival. However, only AURKB and WEE1 protein levels decreased when V600EB-RAF, MEK1/2, or ERK1/2 were targeted using siRNA, demonstrating that these proteins were downstream in this signaling cascade.
AURKB and WEE1 protein levels were increased in tumors of patients with melanoma and in cell lines with highest amounts found in those derived from advanced disease, thereby further validating the potential importance of these proteins in melanoma development. In accordance with our results, Magnussen et al43 recently reported up-regulation of WEE1 in human malignant melanomas compared with benign nevi, and normal melanocyte–increased expression also occurs in breast cancer and glioblastoma.44,45 Studies in this report have demonstrated that siRNA-mediated reduction of AURKB or WEE1 expression in melanoma cells reduced tumor development by 80% to 90% compared with controls, which showed that these downstream MAP kinase–signaling proteins could be potentially important therapeutic targets.
Reducing AURKB or WEE1 protein levels led to a statistically significant 47% to 66% decrease in Ki-67–positive tumor cells, which is a phenotype similar to that observed when inhibiting V600EB-RAF.10 Fluorescence-activated cell sorter analysis of cells after knockdown of AURKB or WEE1 protein levels led to an increase in the G2/M population, which ultimately increased apoptotic cell death. AURKB is a chromosomal passenger protein regulating early mitotic stage transition of prophase to metaphase.46,47 Inhibition of AURKB has been reported to halt a crucial spindle checkpoint causing premature exit from mitosis-disrupting chromosome segregation and cytokinesis, which occurred in this study when the gene was targeted.47 WEE1 regulates cell cycle progression by phosphorylating and deactivating cyclin-associated CDK1 and CDK2 at Tyr15.20 Inhibition of tumor cell proliferation and induction of apoptosis have been reported by targeting WEE1 using siRNA or small-molecule inhibitors either alone or in combination with DNA damaging agents for several malignancies,20,48 and small molecule WEE1 inhibitors are being evaluated in phase I clinical trials.20
Pharmacological agents can inhibit these proteins to target melanoma development. Targeting AURKB using VX-680, which is a small molecule pan-Aurora kinase inhibitor, decreased melanoma tumor development by 78% compared to controls. The drug inhibited cell proliferation by disrupting the cell cycle causing a G2/M block and increasing apoptosis rates. Inhibition of WEE1 with PD0166285 or siRNA to reduce WEE1 protein levels and combined with irradiation decreased the G2/M cell population and triggered apoptosis.19,49
This is also the first study to show that AURKB and WEE1 can serve as biomarkers of the therapeutic efficacy of drugs targeting the MAP kinase pathway. Treatment of melanoma cells in culture or in animals with vemurafenib or U0126 decreased levels of phosphorylated Mek and Erk and downstream AURKB or WEE1 expression and/or activity levels. For these studies, cyclin D1 served as a control because it is commonly used as an indicator of cellular proliferation.50 Levels of AURKB and WEE1 were decreased in a manner similar to that observed for cyclin D1, indicating that these proteins could be used in a likewise manner. Thus, AURKB and WEE1 levels can be used as biomarkers to measure the therapeutic efficacy of MAP kinase pathway inhibitors.
Targeting AURKB or WEE1 using siRNA decreased cellular proliferation, inducing a G2/M block, and increased the apoptotic sub-G0/G1 cell population, which significantly decreased tumor development. Consistent with these observations, several reports in the literature document that WEE1 or AURKB inhibition using siRNA or pharmacological agents, combined with DNA-damaging therapy (either irradiation or cytostatic agents), can effectively decrease cellular proliferation and induce apoptosis by triggering mitotic catastrophe.51,52
In conclusion, WEE1 and AURKB are potentially important therapeutic targets downstream of V600EB-Raf in the MAP kinase signaling cascade. These proteins could be inhibited alone or in combination with B-RAF–targeting agents to more effectively treat patients having the V600E mutation or overcome resistance encountered when treating patients with inhibitors of this pathway. Furthermore, WEE1 or AURKB could be used as biomarkers to assess the efficacy of agents targeting the deregulated MAP kinase pathway in melanomas.
Acknowledgment
We thank Rebecca Watters for providing technical support for this article.
Footnotes
Supported by NIH grants R01 CA-136667-02 and RO1 CA-1138634-02 (G.P.R.) and RO3 CA-142060-2 (A.S.); and the Melanoma Research Foundation (S.V.M.).
Supplemental Data
Individual siRNAs targeting AURKB, WEE1, GSK3A, or TPK1 reduce protein levels in melanoma cells. siRNAs targeting AURKB, WEE1, GSK3A, or TPK1 were nucleofected into melanoma cell line UACC 903, and 3 days later, protein was isolated for Western blot analysis. Western blots are represented from a set of at least three to four different experiments.
AURKB or WEE1 expression in tumors from patients with melanoma. Representative Western blot analysis of protein isolated from tumors from patients with melanoma. Western blots were probed for AURKB, pAURKB, WEE1, and pWEE1. α-Enolase served as a loading control. Western blot analysis was undertaken independently two to three times. NHEM, normal human melanocytes.
Targeting AURKB or WEE1 decreased AURKB and WEE1 protein levels in xenograft melanoma tumors. A and B: Representative images of size- and time-matched tumors from UACC 903 xenografts, immunostained for AURKB (A) and WEE1 (B). Black arrows indicate strong positive staining; white arrows, weak or absent staining in tumor cells. A representative image is shown from three to four tumors, of which four to five fields per tumor were obtained. Original magnification, ×400.
Targeting AURKB or WEE1 decreased the proliferative potential of melanoma cells. A and B: Representative images of size- and time-matched tumors from UACC 903 xenografts, immunostained for Ki-67 (for proliferation) (A) and TUNEL (for apoptosis) (B). Black arrows indicate Ki-67–positive cells; white arrows, TUNEL-positive cells. A representative image is shown from three to four tumors, of which four to five fields per tumor were obtained. Original magnification, ×200.
Vemurafenib or U0126 treatment decreased AURKB or WEE1 expression in xenograft tumors. A and B: Representative images of tumors from animals injected with UACC 903 melanoma cells and treated with vemurafenib or U0126. Tumors were immunostained for AURKB and WEE1 protein. Black arrows indicate strong positive staining; white arrows, weak or absent staining of tumor cells. A representative image is shown from three to four tumors, of which four to five fields per tumor were obtained. Original magnification, ×400.
VX-680 decreased the survival of melanoma cells more effectively than normal cells and induced a G2/M arrest. A: Normal human fibroblasts, UACC 903, 1205 Lu, or A375M melanoma cell lines were plated into a 96-well plate and treated with VX-680 for 24 hours. An MTS assay was used to calculate IC50 using GraphPad Prism Software, version 4.01. Data represent the means ± SEM of IC50 values from three independent experiments. B: Histograms showing cell cycle analysis of UACC 903 melanoma cells treated with 2.5 to 7.5 μmol/L of Aurora kinase inhibitor VX-680 for 48 hours. Control was treated with DMSO alone.
References
- 1.Markovic S.N., Erickson L.A., Rao R.D., Weenig R.H., Pockaj B.A., Bardia A., Vachon C.M., Schild S.E., McWilliams R.R., Hand J.L., Laman S.D., Kottschade L.A., Maples W.J., Pittelkow M.R., Pulido J.S., Cameron J.D., Creagan E.T. Malignant melanoma in the 21st century, part 1: epidemiology, risk factors, screening, prevention, and diagnosis. Mayo Clin Proc. 2007;82:364–380. doi: 10.4065/82.3.364. [DOI] [PubMed] [Google Scholar]
- 2.Purdue M.P., Freeman L.E., Anderson W.F., Tucker M.A. Recent trends in incidence of cutaneous melanoma among US Caucasian young adults. J Invest Dermatol. 2008;128:2905–2908. doi: 10.1038/jid.2008.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chudnovsky Y., Khavari P.A., Adams A.E. Melanoma genetics and the development of rational therapeutics. J Clin Invest. 2005;115:813–824. doi: 10.1172/JCI24808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Libra M., Malaponte G., Navolanic P.M., Gangemi P., Bevelacqua V., Proietti L., Bruni B., Stivala F., Mazzarino M.C., Travali S., McCubrey J.A. Analysis of BRAF mutation in primary and metastatic melanoma. Cell Cycle. 2005;4:1382–1384. doi: 10.4161/cc.4.10.2026. [DOI] [PubMed] [Google Scholar]
- 6.Curtin J.A., Fridlyand J., Kageshita T., Patel H.N., Busam K.J., Kutzner H., Cho K.H., Aiba S., Brocker E.B., LeBoit P.E., Pinkel D., Bastian B.C. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 7.Davies H., Bignell G.R., Cox C., Stephens P., Edkins S., Clegg S. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 8.Dahl C., Guldberg P. The genome and epigenome of malignant melanoma. Apmis. 2007;115:1161–1176. doi: 10.1111/j.1600-0463.2007.apm_855.xml.x. [DOI] [PubMed] [Google Scholar]
- 9.Sharma A., Tran M.A., Liang S., Sharma A.K., Amin S., Smith C.D., Dong C., Robertson G.P. Targeting mitogen-activated protein kinase/extracellular signal-regulated kinase kinase in the mutant (V600E) B-Raf signaling cascade effectively inhibits melanoma lung metastases. Cancer Res. 2006;66:8200–8209. doi: 10.1158/0008-5472.CAN-06-0809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma A., Trivedi N.R., Zimmerman M.A., Tuveson D.A., Smith C.D., Robertson G.P. Mutant V599EB-Raf regulates growth and vascular development of malignant melanoma tumors. Cancer Res. 2005;65:2412–2421. doi: 10.1158/0008-5472.CAN-04-2423. [DOI] [PubMed] [Google Scholar]
- 11.Madhunapantula S.V., Robertson G.P. Is B-Raf a good therapeutic target for melanoma and other malignancies? Cancer Res. 2008;68:5–8. doi: 10.1158/0008-5472.CAN-07-2038. [DOI] [PubMed] [Google Scholar]
- 12.Smalley K.S. PLX-4032, a small-molecule B-Raf inhibitor for the potential treatment of malignant melanoma. Curr Opin Investig Drugs. 2010;11:699–706. [PubMed] [Google Scholar]
- 13.Flaherty K.T., Puzanov I., Kim K.B., Ribas A., McArthur G.A., Sosman J.A., O’Dwyer P.J., Lee R.J., Grippo J.F., Nolop K., Chapman P.B. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rubinstein J.C., Sznol M., Pavlick A.C., Ariyan S., Cheng E., Bacchiocchi A., Kluger H.M., Narayan D., Halaban R. Incidence of the V600K mutation among melanoma patients with BRAF mutations, and potential therapeutic response to the specific BRAF inhibitor PLX4032. J Transl Med. 2010;8:67. doi: 10.1186/1479-5876-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang H., Higgins B., Kolinsky K., Packman K., Go Z., Iyer R., Kolis S., Zhao S., Lee R., Grippo J.F., Schostack K., Simcox M.E., Heimbrook D., Bollag G., Su F. RG7204 (PLX4032), a selective BRAFV600E inhibitor, displays potent antitumor activity in preclinical melanoma models. Cancer Res. 2010;70:5518–5527. doi: 10.1158/0008-5472.CAN-10-0646. [DOI] [PubMed] [Google Scholar]
- 16.Bollag G., Hirth P., Tsai J., Zhang J., Ibrahim P.N., Cho H. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solit D., Sawyers C.L. Drug discovery: how melanomas bypass new therapy. Nature. 2010;468:902–903. doi: 10.1038/468902a. [DOI] [PubMed] [Google Scholar]
- 18.Lens S.M., Voest E.E., Medema R.H. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat Rev Cancer. 2010;10:825–841. doi: 10.1038/nrc2964. [DOI] [PubMed] [Google Scholar]
- 19.Wang X., Moschos S.J., Becker D. Functional analysis and molecular targeting of aurora kinases A and B in advanced melanoma. Genes Cancer. 2010;1:952–963. doi: 10.1177/1947601910388936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stathis A., Oza A. Targeting Wee1-like protein kinase to treat cancer. Drug News Perspect. 2010;23:425–429. doi: 10.1358/dnp.2010.23.7.1490760. [DOI] [PubMed] [Google Scholar]
- 21.Hashimoto O., Shinkawa M., Torimura T., Nakamura T., Selvendiran K., Sakamoto M., Koga H., Ueno T., Sata M. Cell cycle regulation by the Wee1 inhibitor PD0166285, pyrido [2,3-d] pyimidine, in the B16 mouse melanoma cell line. BMC Cancer. 2006;6:292. doi: 10.1186/1471-2407-6-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaidanovich-Beilin O., Woodgett J.R. GSK-3: functional insights from cell biology and animal models. Front Mol Neurosci. 2011;4:40. doi: 10.3389/fnmol.2011.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao R., Gao F., Goldman I.D. Molecular cloning of human thiamin pyrophosphokinase. Biochim Biophys Acta. 2001;1517:320–322. doi: 10.1016/s0167-4781(00)00264-5. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen N., Sharma A., Sharma A.K., Desai D., Huh S.J., Amin S., Meyers C., Robertson G.P. Melanoma chemoprevention in skin reconstructs and mouse xenografts using isoselenocyanate-4. Cancer Prev Res (Phila) 2011;4:248–258. doi: 10.1158/1940-6207.CAPR-10-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quong R.Y., Bickford S.T., Ing Y.L., Terman B., Herlyn M., Lassam N.J. Protein kinases in normal and transformed melanocytes. Melanoma Res. 1994;4:313–319. doi: 10.1097/00008390-199410000-00008. [DOI] [PubMed] [Google Scholar]
- 26.Madhunapantula S.V., Sharma A., Robertson G.P. PRAS40 deregulates apoptosis in malignant melanoma. Cancer Res. 2007;67:3626–3636. doi: 10.1158/0008-5472.CAN-06-4234. [DOI] [PubMed] [Google Scholar]
- 27.Sharma A., Sharma A.K., Madhunapantula S.V., Desai D., Huh S.J., Mosca P., Amin S., Robertson G.P. Targeting Akt3 signaling in malignant melanoma using isoselenocyanates. Clin Cancer Res. 2009;15:1674–1685. doi: 10.1158/1078-0432.CCR-08-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Musgrove E.A., Caldon C.E., Barraclough J., Stone A., Sutherland R.L. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11:558–572. doi: 10.1038/nrc3090. [DOI] [PubMed] [Google Scholar]
- 29.Cohen P., Frame S. The renaissance of GSK3. Nat Rev Mol Cell Biol. 2001;2:769–776. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- 30.Gowda R., Madhunapantula S.V., Desai D., Amin S., Robertson G.P. Simultaneous targeting of COX-2 and AKT using selenocoxib-1-GSH to inhibit melanoma. Mol Cancer Ther. 2013;12:3–15. doi: 10.1158/1535-7163.MCT-12-0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huh S.J., Chung C.Y., Sharma A., Robertson G.P. Macrophage inhibitory cytokine-1 regulates melanoma vascular development. Am J Pathol. 2010;176:2948–2957. doi: 10.2353/ajpath.2010.090963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stahl J.M., Sharma A., Cheung M., Zimmerman M., Cheng J.Q., Bosenberg M.W., Kester M., Sandirasegarane L., Robertson G.P. Deregulated Akt3 activity promotes development of malignant melanoma. Cancer Res. 2004;64:7002–7010. doi: 10.1158/0008-5472.CAN-04-1399. [DOI] [PubMed] [Google Scholar]
- 33.Carmena M., Earnshaw W.C. The cellular geography of aurora kinases. Nat Rev Mol Cell Biol. 2003;4:842–854. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- 34.Adams R.R., Carmena M., Earnshaw W.C. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 2001;11:49–54. doi: 10.1016/s0962-8924(00)01880-8. [DOI] [PubMed] [Google Scholar]
- 35.Kellogg D.R. Wee1-dependent mechanisms required for coordination of cell growth and cell division. J Cell Sci. 2003;116:4883–4890. doi: 10.1242/jcs.00908. [DOI] [PubMed] [Google Scholar]
- 36.Sherr C.J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 37.Keen N., Taylor S. Aurora-kinase inhibitors as anticancer agents. Nat Rev Cancer. 2004;4:927–936. doi: 10.1038/nrc1502. [DOI] [PubMed] [Google Scholar]
- 38.Matthews N., Visintin C., Hartzoulakis B., Jarvis A., Selwood D.L. Aurora A and B kinases as targets for cancer: will they be selective for tumors? Expert Rev Anticancer Ther. 2006;6:109–120. doi: 10.1586/14737140.6.1.109. [DOI] [PubMed] [Google Scholar]
- 39.Harrington E.A., Bebbington D., Moore J., Rasmussen R.K., Ajose-Adeogun A.O., Nakayama T., Graham J.A., Demur C., Hercend T., Diu-Hercend A., Su M., Golec J.M., Miller K.M. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat Med. 2004;10:262–267. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- 40.Keen N., Taylor S. Mitotic drivers: inhibitors of the Aurora B kinase. Cancer Metastasis Rev. 2009;28:185–195. doi: 10.1007/s10555-009-9184-9. [DOI] [PubMed] [Google Scholar]
- 41.Ribas A., Flaherty K.T. BRAF targeted therapy changes the treatment paradigm in melanoma. Nat Rev Clin Oncol. 2011;8:426–433. doi: 10.1038/nrclinonc.2011.69. [DOI] [PubMed] [Google Scholar]
- 42.Kaplan F.M., Shao Y., Mayberry M.M., Aplin A.E. Hyperactivation of MEK-ERK1/2 signaling and resistance to apoptosis induced by the oncogenic B-RAF inhibitor, PLX4720, in mutant N-RAS melanoma cells. Oncogene. 2011;30:366–371. doi: 10.1038/onc.2010.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Magnussen G.I., Holm R., Emilsen E., Rosnes A.K., Slipicevic A., Florenes V.A. High expression of Wee1 is associated with poor disease-free survival in malignant melanoma: potential for targeted therapy. PLoS One. 2012;7:e38254. doi: 10.1371/journal.pone.0038254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iorns E., Lord C.J., Grigoriadis A., McDonald S., Fenwick K., Mackay A., Mein C.A., Natrajan R., Savage K., Tamber N., Reis-Filho J.S., Turner N.C., Ashworth A. Integrated functional, gene expression and genomic analysis for the identification of cancer targets. PLoS One. 2009;4:e5120. doi: 10.1371/journal.pone.0005120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mir S.E., De Witt Hamer P.C., Krawczyk P.M., Balaj L., Claes A., Niers J.M., Van Tilborg A.A., Zwinderman A.H., Geerts D., Kaspers G.J., Peter Vandertop W., Cloos J., Tannous B.A., Wesseling P., Aten J.A., Noske D.P., Van Noorden C.J., Wurdinger T. In silico analysis of kinase expression identifies WEE1 as a gatekeeper against mitotic catastrophe in glioblastoma. Cancer Cell. 2010;18:244–257. doi: 10.1016/j.ccr.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dar A.A., Goff L.W., Majid S., Berlin J., El-Rifai W. Aurora kinase inhibitors: rising stars in cancer therapeutics? Mol Cancer Ther. 2010;9:268–278. doi: 10.1158/1535-7163.MCT-09-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carmena M., Ruchaud S., Earnshaw W.C. Making the Auroras glow: regulation of Aurora A and B kinase function by interacting proteins. Curr Opin Cell Biol. 2009;21:796–805. doi: 10.1016/j.ceb.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y., Li J., Booher R.N., Kraker A., Lawrence T., Leopold W.R., Sun Y. Radiosensitization of p53 mutant cells by PD0166285, a novel G(2) checkpoint abrogator. Cancer Res. 2001;61:8211–8217. [PubMed] [Google Scholar]
- 49.Davies K.D., Cable P.L., Garrus J.E., Sullivan F.X., von Carlowitz I., Huerou Y.L., Wallace E., Woessner R.D., Gross S. Chk1 inhibition and Wee1 inhibition combine synergistically to impede cellular proliferation. Cancer Biol Ther. 2011;12:788–796. doi: 10.4161/cbt.12.9.17673. [DOI] [PubMed] [Google Scholar]
- 50.Colozza M., Azambuja E., Cardoso F., Sotiriou C., Larsimont D., Piccart M.J. Proliferative markers as prognostic and predictive tools in early breast cancer: where are we now? Ann Oncol. 2005;16:1723–1739. doi: 10.1093/annonc/mdi352. [DOI] [PubMed] [Google Scholar]
- 51.Niermann K.J., Moretti L., Giacalone N.J., Sun Y., Schleicher S.M., Kopsombut P., Mitchell L.R., Kim K.W., Lu B. Enhanced radiosensitivity of androgen-resistant prostate cancer: AZD1152-mediated Aurora kinase B inhibition. Radiat Res. 2011;175:444–451. doi: 10.1667/RR2317.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Witt Hamer P.C., Mir S.E., Noske D., Van Noorden C.J., Wurdinger T. WEE1 kinase targeting combined with DNA-damaging cancer therapy catalyzes mitotic catastrophe. Clin Cancer Res. 2011;17:4200–4207. doi: 10.1158/1078-0432.CCR-10-2537. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Individual siRNAs targeting AURKB, WEE1, GSK3A, or TPK1 reduce protein levels in melanoma cells. siRNAs targeting AURKB, WEE1, GSK3A, or TPK1 were nucleofected into melanoma cell line UACC 903, and 3 days later, protein was isolated for Western blot analysis. Western blots are represented from a set of at least three to four different experiments.
AURKB or WEE1 expression in tumors from patients with melanoma. Representative Western blot analysis of protein isolated from tumors from patients with melanoma. Western blots were probed for AURKB, pAURKB, WEE1, and pWEE1. α-Enolase served as a loading control. Western blot analysis was undertaken independently two to three times. NHEM, normal human melanocytes.
Targeting AURKB or WEE1 decreased AURKB and WEE1 protein levels in xenograft melanoma tumors. A and B: Representative images of size- and time-matched tumors from UACC 903 xenografts, immunostained for AURKB (A) and WEE1 (B). Black arrows indicate strong positive staining; white arrows, weak or absent staining in tumor cells. A representative image is shown from three to four tumors, of which four to five fields per tumor were obtained. Original magnification, ×400.
Targeting AURKB or WEE1 decreased the proliferative potential of melanoma cells. A and B: Representative images of size- and time-matched tumors from UACC 903 xenografts, immunostained for Ki-67 (for proliferation) (A) and TUNEL (for apoptosis) (B). Black arrows indicate Ki-67–positive cells; white arrows, TUNEL-positive cells. A representative image is shown from three to four tumors, of which four to five fields per tumor were obtained. Original magnification, ×200.
Vemurafenib or U0126 treatment decreased AURKB or WEE1 expression in xenograft tumors. A and B: Representative images of tumors from animals injected with UACC 903 melanoma cells and treated with vemurafenib or U0126. Tumors were immunostained for AURKB and WEE1 protein. Black arrows indicate strong positive staining; white arrows, weak or absent staining of tumor cells. A representative image is shown from three to four tumors, of which four to five fields per tumor were obtained. Original magnification, ×400.
VX-680 decreased the survival of melanoma cells more effectively than normal cells and induced a G2/M arrest. A: Normal human fibroblasts, UACC 903, 1205 Lu, or A375M melanoma cell lines were plated into a 96-well plate and treated with VX-680 for 24 hours. An MTS assay was used to calculate IC50 using GraphPad Prism Software, version 4.01. Data represent the means ± SEM of IC50 values from three independent experiments. B: Histograms showing cell cycle analysis of UACC 903 melanoma cells treated with 2.5 to 7.5 μmol/L of Aurora kinase inhibitor VX-680 for 48 hours. Control was treated with DMSO alone.