Abstract
miRNAs are small noncoding RNAs with critical roles in a large variety of biological processes such as development and tumorigenesis. miRNA expression profiling has been reported to be a powerful tool to classify tissue samples, including cancers, based on their developmental lineage. In this study, we have profiled the expression of miRNAs in bladder carcinoma in situ (CIS) and distinct cell compartments of the normal bladder, namely umbrella and basal-intermediate urothelial cells, as well as the muscularis propria. We identified several miRNAs differentially expressed between umbrella and basal-intermediate cells (miR-133a, miR-139-3p, miR-142-3p, miR-199b-5p, and miR-221). In situ hybridization confirmed the expression of miR-133a and miR-139-3p in umbrella cells, and miR-142-3p in basal-intermediate cells. Strikingly, miRNA expression levels of CIS most closely resembled the miRNA profile of umbrella cells. Finally, we examined well-established umbrella and basal-intermediate cell immunohistochemical biomarkers in an independent series of CIS samples. Again, this analysis revealed the significant expression of umbrella-specific markers in CIS when compared to non-CIS lesions. Overall, our studies represent a comprehensive and accurate description of the different miRNAs expressed in CIS tumors and three distinct histological areas of the urinary bladder. Notably, this study provides evidence of the possible origin relationship between CIS and normal umbrella cells.
The urothelium is composed of specialized epithelial cells that coat the entire urinary tract, including the renal pelvis, ureter, bladder, and female proximal urethra. In humans, the urothelium of the bladder is a stratified epithelium, comprised of three different cell layers: i) basal, ii) intermediate, and iii) superficial umbrella cells. The basal membrane separates the urothelium from the stroma of the bladder, which includes the lamina propria and the muscularis propria.
Umbrella (UM) cells are distinct from basal-intermediate (B/i) cells in both morphology and expression of specific biomarkers. UM cells express high levels of uroplakin II (UPKII),1 low molecular weight cytokeratins (CKs; eg, CK18 and CK20),2 and Lewis X determinant,3 however, none of these markers are expressed in B/i cells. In contrast, B/i cells stain strongly for high molecular weight cytokeratins (eg, CK5, CK10, CK14), p63,4 and mature A and B blood group antigens, all of which are negative in umbrella (UM) cells. Although it has been proposed that UM cells mature from the underlying B/i cells in a manner similar to epithelial development of the skin, the vast number of differences between the two cell types suggests that UM cells may originate from a precursor distinct from B/i cells.5
The urothelium functions as a permeability barrier between blood and urine and is constantly exposed to potential carcinogens in urine. It is not surprising, therefore, that 90% of bladder tumors originate from the urothelium.6 Bladder tumors are classified into two groups based on phenotypic and molecular profiles. Low-grade tumors are always papillary (ie, protruding into the lumen), and usually superficial and nonmuscle invasive. In contrast, high-grade tumors can be papillary or nonpapillary, such as carcinoma in situ (CIS), and are often invasive. Nonmuscle invasive bladder tumors (low-grade tumors and carcinoma stages Ta, Tis, T1) comprise 75% to 85% of total bladder cancer cases at initial presentation.6 CIS presents as a flat lesion, found in association with 75% to 90% of high-grade papillary cases and in 45% to 65% of all invasive urothelial tumors.7,8 Presence of CIS greatly increases the risk of disease progression to invasive urothelial carcinoma; once the tumor is metastatic, the median survival rate is approximately 7 to 20 months.6,9 CIS is characterized by aneuploidy, and its histological abnormalities include enlarged nuclei and nucleoli and hyperchromasia.7 Given the heterogeneity of bladder tumors, it is possible that the two distinct populations of normal urothelium (ie, UM cells and B/i cells) may have different implications in bladder tumorigenesis. A better characterization of both normal urothelium and bladder cancer cases is therefore needed to delineate the role of these cells in tumorigenesis.
miRNAs have been implicated in development and tumorigenesis, as reviewed in Zhang et al,10 and He and Hannon,11 and Esquela-Kerscher and Slack.12 To date, more than 21,000 distinct mature miRNA sequences have been identified in 193 different species (miRBase 19) with more than 2000 miRNAs identified in humans.13,14 Transcribed from independent miRNA genes or as introns of protein-coding mRNAs, miRNAs repress specific mRNA expression through 3′-UTR base pairing.15 Furthermore, miRNA profiles can classify tumors based on their respective developmental lineage.16 To the best of our knowledge, miRNA profiles of UM and B/i cells have not been examined. Moreover, several groups have compared miRNA expression between normal mucosa and bladder tumors,17–25 but none has assessed miRNA expression in CIS samples relative to subpopulations of normal urothelium.
For the study described herein, we used laser capture microdissection to obtain UM and B/i cell populations from normal bladder urothelium, as well as pure CIS cells. Then we profiled the expression of both miRNAs and mRNAs in CIS tumors and three distinct histological areas of the urinary bladder: i) the UM (lumen-facing cells), ii) the B/i cells, and iii) the muscularis propria (MP; stromal). We identified several miRNAs differentially expressed between UM and B/i cells. Furthermore, and for the first time, through miRNA profiling and immunohistochemical analyses, we established a correlation between CIS and UM cells.
Materials and Methods
Cell Culture
The immortalized normal urothelial cell line, Human Urothelial Cell (HUC) from ScienCell Research Laboratories, was grown in Urothelial Cell Medium (ScienCell Research Laboratories, Carlsbad, CA) at 37°C in a humidified air atmosphere at 5% CO2. RNA was extracted from the cells using the miRVANA Kit (Ambion, Carlsbad, CA).
Laser Capture Microdissection of Normal Bladder Urothelium and CIS Biopsies
Human normal (n = 3) and tumor bladder (CIS, n = 4) tissue samples were obtained through approved protocols of the International Review Board. The three normal bladders were obtained from organ donors (males: ages 15, 25, and 28 years old) with no history of bladder cancer through the International Institute for the Advancement of Medicine. Fresh tissues were processed and frozen in optimal cutting temperature (OCT) molds, which were used for laser capture microdissection. CIS samples were obtained from patients who had undergone radical cystectomy for an invasive urothelial carcinoma and sections that did not include the invasive area were chosen for further processing. Frozen normal bladder urothelium was microdissected to obtain pure populations of UM cells, B/i cells, and MP. Frozen CIS samples were also microdissected from the bladder tissue samples to obtain pure populations of tumor cells. To rule out the possibility of normal urothelial cell contamination in CIS samples, only cases in which the CIS lesion permeated the whole thickness of the epithelium were chosen. Cases in which we could histologically identify UM or B/i cells surrounding the malignant cells by H&E stain, and cases with pagetoid spread or denuded CIS were not used. Briefly, 20-μm sections from OCT blocks were cut and mounted on MembraneSlide NF 1.0 PEN (Zeiss, Munich, Germany) and stored at −80°C until use. Slides were fixed in ethanol at −20°C for 2 minutes, washed in diethyl-pyrocarbonate-H2O, stained with hematoxylin for 1 minute, washed in diethyl-pyrocarbonate-H2O, and air-dried. PALM Microbeam IV Laser Capture Microscope (Zeiss) was used to perform laser microdissection. A total of 25 20-μm sections were prepared from each sample, in which the microdissected tissue was pooled per patient to extract RNA. Formalin-fixed paraffin-embedded (FFPE) CIS samples (n = 6) were macrodissected by scratching 40 10-μm sections in which dissected tissue was pooled per patient.
RNA Extraction and Microarray Profiling and Data Analysis
RNA was extracted from microdissected samples using RNAquesous-Micro Kit (Ambion). RNA was extracted using RecoverAll Total Nucleic Acid Isolation Kit for FFPE (Ambion) for macrodissected samples. The quality of all RNA samples was verified using a RNA Pico Kit (Agilent, Santa Clara, CA). Only sample qualities with RNA Integrity Number >6.0 and with clean 18S and 28S peaks were used, as previously described.26
Agilent Human miRNA Microarray (V3, based on Sanger miRbase release 12.0) were used for measuring miRNA expression in bladder samples. Samples used for the Agilent miRNA microarray were labeled as described by the manufacturer.
Affymetrix Human U133 Plus 2.0 (Santa Clara, CA) arrays were used for measuring gene expression in bladder samples. Samples used for Affymetrix microarray were amplified using the Ovation RNA Amplification System (NuGen, San Carlos, CA) and labeled using FL-Ovation cDNA Biotin Module V2 (NuGen) according to the manufacturer’s protocol. Raw intensity miRNA data were normalized and median transformed using GeneSpring GX 12 (Agilent). The raw mRNA data were log transformed and analyzed using Partek Genomics Suite 6.6 (Partek, Saint Louis, MO). Only detected probesets were used, and compromised or undetected probesets were filtered out. One-way analysis of variance and Tukey’s honestly significant difference post hoc test were performed across all samples to obtain miRNAs or mRNAs differentially expressed (P < 0.05). Unsupervised hierarchical cluster analysis was performed on the list of differentially expressed probesets with a fold change ≥2 (295 miRNAs, 1113 mRNAs).
Quantitative Real-Time RT-PCR miRNA Expression Profiling
TaqMan microRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) was used to convert miRNA to cDNA. Forty-four–nucleotide (nt) reverse transcription primers were designed so that the first 36 nt formed an internal stem loop and the last 8 nt were complementary to the mature miRNA sequence of interest. Qiagen Reverse Transcriptase System (Qiagen, Valencia, CA) was used to produce cDNA from mRNA. Qiagen QuantiTect PCR (Qiagen) was used to measure quantitative expression of miRNA and mRNA. PCR assays were performed as described by the manufacturer using a Stratagene MX3005P PCR system (Agilent). For normalization, we used RNU6B or ACTIN for miRNA and mRNA, respectively.
Immunohistochemical Detection of Biomarkers
Immunohistochemical analyses were conducted on FFPE tissue sections from 10 cases of normal human bladder and 19 CIS specimens according to the standard avidin-biotin protocol. These 10 cases of normal bladder included the three cases used for the laser microdissection (organ donors) and seven cases of histologically normal urothelium obtained from patients who had undergone a radical cystectomy for an invasive carcinoma. From the 19 CIS specimens, 10 corresponded to biopsies of patients with primary CIS and the other nine were obtained from cystectomy specimens of patients with a diagnosis of invasive bladder cancer. Briefly, 5-μm sections were deparaffinized and submitted to antigen retrieval by steamer treatment for 15 minutes in 10 mmol/L citrate buffer at pH 6.0. Subsequently, slides were incubated in 10% normal serum for 30 minutes, followed by primary antibody incubation overnight at 4°C. Primary antibodies used were anti-UPKII goat polyclonal (Santa Cruz Biotechnology, Dallas, TX), anti-CK5 rabbit polyclonal (AF138, Covance, Princeton, NJ), anti-CK20 mouse monoclonal (clone Ks20.8, Dako, Carpinteria, CA), and anti-high molecular weight cytokeratin mouse monoclonal (clone 34βE12, Dako). Then slides were incubated with biotinylated immunoglobulins at 1:500 dilution for 30 minutes (Vector Laboratories, Inc., Burlingame, CA) followed by avidin-biotin peroxidase complexes at a 1:25 dilution (Vector Laboratories, Inc.) for 30 minutes. Diaminobenzidine was used as chromogen and hematoxylin as the nuclear counterstain. Slides were analyzed with a Nikon Eclipse 50i microscope (Nikon Instruments, Melville, NY) equipped with a SPOT Insight2MP Mosaic camera and SPOT Advanced software version 5.0 (Diagnostic Instruments, Sterling Heights, MI).
In Situ Detection of miRNAs
In situ detection of miRNAs was performed on 10-μm frozen tissue sections from normal human bladders obtained from healthy donor patients (n = 3) and B6-wt mouse bladders (n = 15). Sections were fixed in 4% paraformaldehyde, acetylated, and prehybridized in hybridization solution (50% formamide, 5× saline-sodium citrate, 0.5 mg/mL yeast tRNA, 1X Denhardt’s solution) for 3 hours at 25°C below the predicted Tm of the probe. Five pmol probe (LNA-modified and FITC-labeled oligonucleotide; Exiqon, Woburn, MA) complementary to U6 (positive control), miR-133a, miR-139-3p, and miR-142-3p was hybridized to the sections overnight in a humidified chamber at prehybridization temperature. Posthybridization slides were washed with 5× and 0.2× saline-sodium citrate, blocked in 10% normal goat serum for 30 minutes, and incubated in anti-fluorescein isothiocyanate/alkaline-phosphatase antibody (Vector Laboratories, Inc.) at 1:400 at 4°C overnight. Five pmol sense-strand of the primary transcript of the miR-424/503 cluster (Table 1) was used as negative control. In situ hybridization signals were detected using BM purple (Roche, Indianapolis, IN). Slides were counterstained with Nuclear Fast Red (Sigma-Aldrich, St. Louis, MO), mounted using 80% glycerol, and analyzed with a Nikon Eclipse 50i microscope (Nikon Instruments) equipped with a SPOT Insight2MP Mosaic camera and SPOT Advanced software version 5.0 (Diagnostic Instruments, Sterling Heights, MI).
Table 1.
Sequence of the Sense-Strand of the Primary Transcript of miR-424/503 Cluster
| Probe sequence | Hybridization temperature |
|---|---|
| 5′-GGGCAGTCAACGACATTTTTCTCCATTAATCCCAAAGTCAGTGTAAAAGTTGTTTGTAAATATCATTGCC-AAAACAAGATGGAATGTAGAGATACTTGATTGTCGCAATTTAGGGAATGGGGTTATTGTTACTAATGACTT-TTTTTTTTTACCATGACATAGTGTATTTGTTACCACTGGATCTCAGTCTGGATGTTATAAATTCGATACTT-AATCTTATTAAATACTCTGCACATTTTATTAACTAATAAATAGGACACTTAACATTCATTGTACTGGTTTT-GAAAAACCGACTTAATTTGTAAGTGTAACAAGATACTTACAGTTGGATAATTT-3′ | 65°C |
This probe was 3′ digoxigenin-labeled and used as a negative control in miRNA ISH stainings.
Results
Isolation of Pure Populations of UM, B/i, and MP by Laser Capture Microdissection
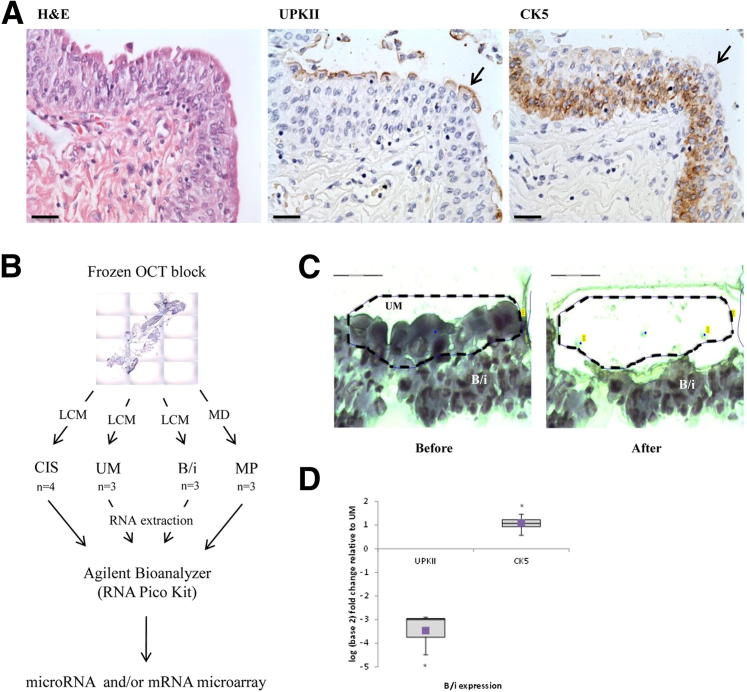
As previously described,5,27 UPKII and CK5 were shown to be differentially expressed in UM versus B/i cells in normal bladder urothelium (Figure 1A). Membranous expression of UPKII was observed exclusively in the UM layer, whereas cytoplasmic CK5 expression was specific for the cells in the B/i layers. Three cases of normal bladder samples and four CIS lesions were used to perform the study (Figure 1B). The urothelium was microdissected to separate UM from B/i cells as explained in Material and Methods (Figure 1C). The MP population was obtained by removing the urothelium and lamina propria through macrodissection. Quantitative real-time RT-PCR (RT-qPCR) showed on average an eightfold decrease in UPKII levels in B/i versus UM, and a twofold increase in CK5 in B/i versus UM (Figure 1D). These results demonstrated that the distinct urothelial subpopulations could be efficiently separated by microdissection.
Figure 1.
Experimental design and isolation of UM and B/i cells from normal bladder urothelium. A: Representative immunohistochemical analyses of UPKII and CK5 in normal urothelium; prominent UM cells (arrows). Scale bar = 50 μm. B: Microphotograph of frozen tissue section was taken with PALM Microbeam IV Laser Capture Microscope (LCM; Zeiss). UM, B/i, and CIS samples were microdissected and analyzed with both miRNA and mRNA microarrays. MP samples were macrodissected (MD) and analyzed only in the miRNA microarray. C: Delineation of UM cells, before and after laser capture. Scale bar = 75 μm. D: RT-qPCR on UM and B/i cells for UPKII and CK5. Graph shows B/i log2 fold expression; B/i expression normalized to UM. Horizontal line indicates the sample median. Square indicates the sample mean. *P < 0.05 by Student’s t-test.
miRNA Microarray Profiling Associates CIS with Umbrella Cells
Once we had isolated pure populations of urothelial subtypes and stroma, we examined the miRNA expression in these cells, along with four CIS cases and an HUC line. Of the 984 human miRNAs evaluated, there were 295 miRNAs differentially expressed (fold change ≥2) across the normal bladder, HUC line, and CIS. Interestingly, hierarchical cluster analysis of these 295 miRNAs separated samples were based on cell type of origin instead of normal versus cancer. Endoderm-derived cells, including the urothelium, CIS, and HUC, clustered into one arm, whereas mesoderm-derived MP branched into another arm (Supplemental Figure S1). Expression of individual samples was averaged into groups, with the exception of the HUC line. Notably, the level of expression of miRNAs in CIS most closely resembled the level observed in normal UM cells, whereas the HUC line shared greater similarity in miRNA expression with B/i cells. So far, an analysis of a broad sampling of miRNAs indicates a correlation between UM and CIS cells.
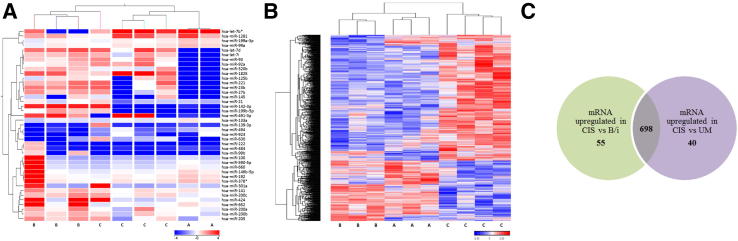
Then we focused our analyses on the individual miRNA and mRNA profiles of UM, B/I, and CIS, because MP is not epithelial in origin and HUC is a cell line. With the removal of MP and HUC, there were 41 miRNAs and 1113 mRNAs differentially expressed across UM, B/i, and CIS samples. A hierarchical cluster analysis of individual cases revealed distinct miRNA expression profiles between UM cells and B/i cells; interestingly, the majority of CIS samples clustered with UM cells (Figure 2A). From these 41 miRNAs, four (ie, let-7b*, miR-1281, miR-133a, and miR-139-3p) exhibited higher levels of expression in both CIS and UM cells when compared to B/i cells. Conversely, nine miRNAs (ie, miR-142-3p, miR-199b-5p, miR-200a, miR-200b, miR-205, miR-21, miR-27b, miR-424, and miR-491-3p) displayed higher levels of expression in B/i cells than CIS and UM cells. Importantly, only five of 41 miRNAs were significantly altered between UM and B/i cells (Table 2).
Figure 2.
Hierarchical clustering of normal urothelium and CIS. A: Samples are in columns and miRNAs are in rows. Individual samples of normal urothelium and CIS were clustered according to the expression signature of 41 differentially expressed miRNAs (P ≤ 0.05). B: Samples are in columns, mRNAs are in rows. Normal urothelium and CIS samples were clustered according to the expression signature of 1113 differentially expressed mRNAs (P ≤ 0.05). C: Venn diagram illustrating 698 mRNAs upregulated in common between CIS compared to normal UM and B/i cells. There were 753 mRNAs upregulated in CIS compared to B/i cells, and 738 mRNAs upregulated in CIS compared to UM cells. Arbitrary assignment of cluster groups is as follows: A, UM cells; B, B/i cells; C, CIS.
Table 2.
Statistical Results and Location of a Set of miRNAs Significantly Differentially Expressed Between UM and B/i Cells in the Normal Urothelium
| miRNA | Chromosome | Fold change (UM vs B/i) | P value | RT-qPCR verification | ISH verification |
|---|---|---|---|---|---|
| hsa-miR-139-3p | 11q13.4 | 2.30 | 0.006 | Yes | Yes |
| hsa-miR-199b-5p | 9q34.11 | −4.03 | 0.024 | Yes | ND |
| hsa-miR-142-3p | 2q35 | −3.12 | 0.030 | Yes | Yes |
| hsa-miR-133a | 18q11.2 | 2.26 | 0.037 | Yes | Yes |
| hsa-miR-221 | Xp11.3 | −5.04 | 0.048 | Yes | ND |
ISH, in situ hybridization; ND, not detectable.
In contrast, based on mRNA expression levels, normal urothelium (UM and B/i) and CIS formed two separate arms (Figure 2B). From the 1113 mRNAs differently expressed, 23 were commonly down-regulated in UM cells and CIS in comparison to B/i cells, whereas another 23 followed the opposite trend (Supplemental Table S1). Notably, TP63 (specific to B/i cells, but absent in UM cells) was down-regulated in UM cells and CIS compared to B/i cells. Interestingly, among the genes that were upregulated in UM cells and CIS compared to B/i cells, we found CD24, a proposed marker of tumor-initiating cells (reviewed by Gires28), and genes involved in metabolism, such as GFOD2, NDUFB2, and NQO1.
In fact, more than half of the differentially expressed mRNAs (698) were upregulated in CIS compared to UM and B/i samples (Figure 2C). Gene ontology enrichment was performed on the 698 mRNAs to investigate biologically driven effects. This analysis revealed 74 biological processes with enrichment scores >3 (P < 0.05), and as expected, the top seven (P < 0.005) are involved in genomic alterations, transcription regulation, and cell cycle (Supplemental Table S2). Furthermore, we examined differentially expressed pathways in the group of 698 mRNAs upregulated in CIS using Pathway analysis of variance (Partek Genomics Suite 6.6). Statistically significant pathways with more than 10 genes involved include metabolic pathways and pathways in cancer (Supplemental Table S3). Not surprisingly, in this list (Supplemental Table S4), the bladder cancer pathway was enriched (enrichment score, 3.2). As supported by gene ontology analysis, hierarchical clustering by global gene expression separated samples based on normal (UM and B/i) versus cancer (CIS).
Validation of miRNA Arrays in UM and B/i Cells
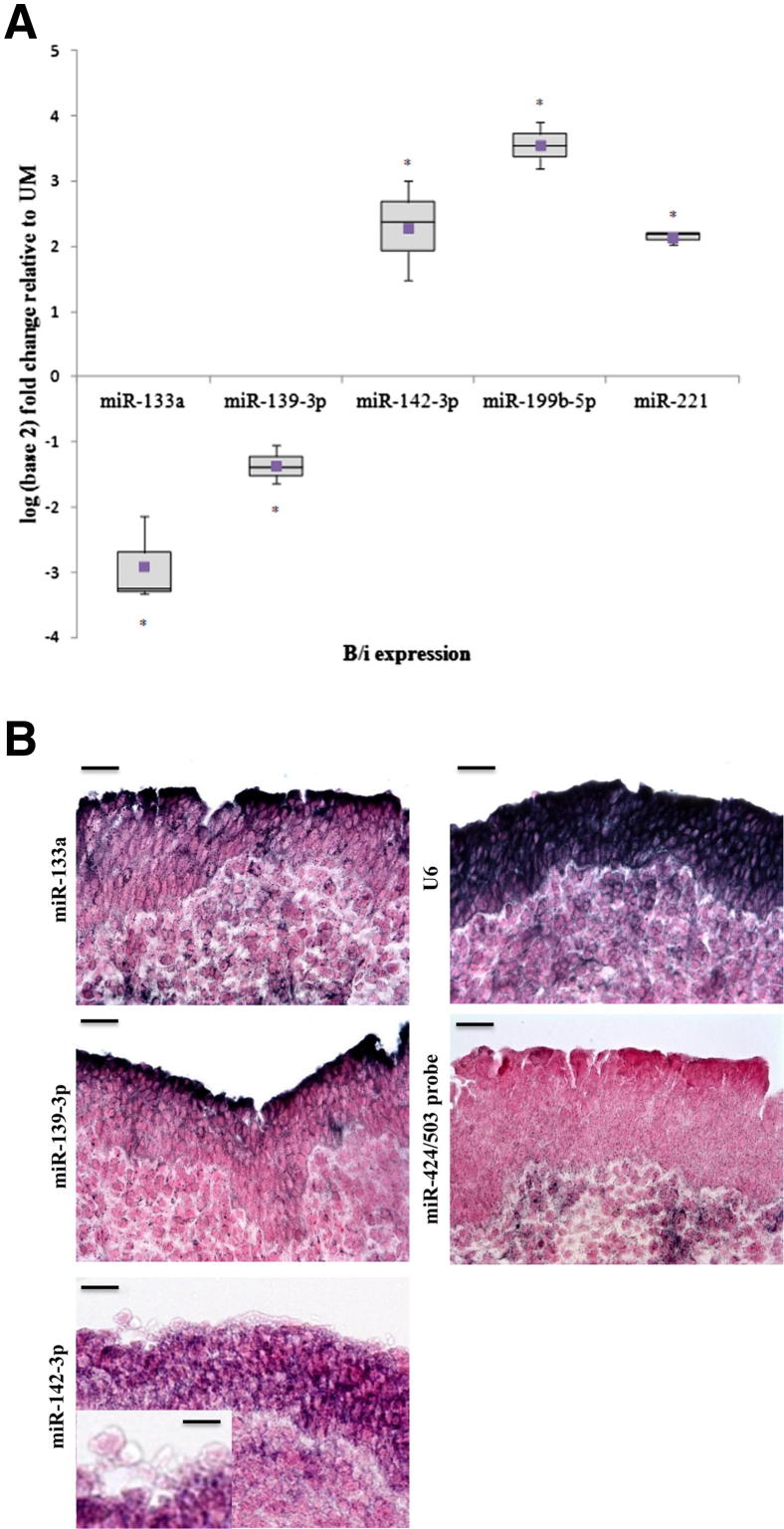
Having examined miRNA and mRNA profiles globally in both normal urothelium and in CIS cases, we decided to further explore the five miRNAs that were statistically differentially expressed between UM cells and B/i cells (Table 2), according to the microarray data. We performed RT-qPCR to determine the expression of the mature form of these five miRNAs (Figure 3A). RT-qPCR results showed an average Pearson correlation to the microarray data of 0.95. The miR-133a and miR-139-3p were upregulated in UM cells compared to B/i cells, whereas miR-142-3p, miR-199b-5p, and miR-221 were down-regulated in UM cells compared to B/i cells. Taken together, the RT-qPCR results confirmed that these five miRNAs were differentially expressed between UM and B/i cell populations.
Figure 3.
Validation of differentially expressed miRNAs from microarray data. A: RT-qPCR on UM and B/i cells for various miRNAs. Graph shows B/i log2 fold expression; B/i expression is normalized to UM. Horizontal line indicates the sample median. Square indicates the sample mean. *P < 0.05 by Student’s t-test. B: Localization of miR-133a, miR-139-3p, and miR-142-3p in human bladder samples by in situ hybridization. Scale bar = 100 μm. Inset: miR-142-3p shows UM cells. Scale bar = 25 μm. No staining was observed in a negative control in situ hybridization using the sense strand of miR-424/503, whereas a strong signal was observed in a positive control in situ hybridization using U6 in representative cases of normal bladder. Scale bar = 100 μm.
Then we verified the localization of our miRNAs of interest via in situ hybridization. We used LNA oligonucleotide probes for detection of the miRNAs of interest in human and mouse bladders. A positive control using U6 gave strong signals in human urothelium, whereas no signal was detected using a nonspecific probe for the miR-424/503 cluster. Robust in situ hybridization conditions were established for 3 of 5 miRNAs. Expression of miR-133a and miR-139-3p was observed in UM cells, whereas miR-142-3p expression was localized to B/i cells with low expression in UM cells (Figure 3B). To further validate the phenotypes described for human urothelial tissues, we extended the study to the analysis of mouse urothelium. We observed that the miRNA phenotypes of both human and murine bladder samples were consistent (Supplemental Figure S2).
CIS Samples Express UM But Not B/i Urothelium-Specific Protein Markers
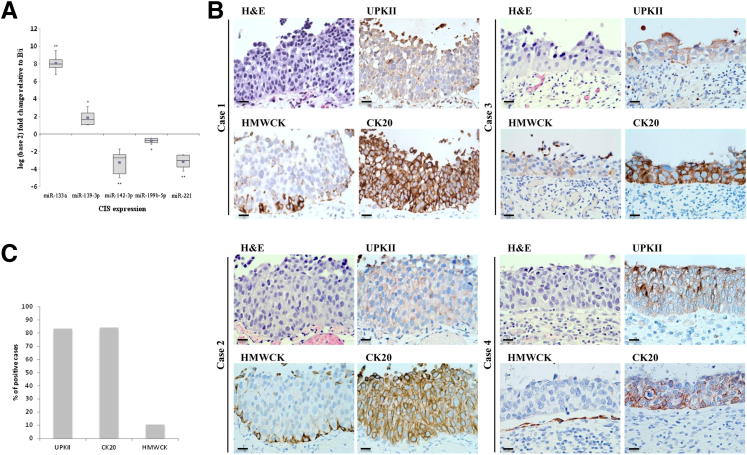
To further explore whether CIS samples were more closely related to UM cells than B/i cells, as suggested by the miRNA microarray results, we examined the expression of miRNAs specific to UM cells in CIS. Compared to B/i cells, expression of the five miRNAs (Table 2) by RT-qPCR in six independent CIS biopsies were similar to that of UM cells. miR-133a and miR-139-3p were upregulated in CIS compared to B/i, whereas miR-142-3p, miR-199b-5p, and miR-221 were down-regulated in CIS compared to B/i (Figure 4A).
Figure 4.
Phenotypic association of CIS with UM cells by RT-qPCR and immunohistochemistry. A: RT-qPCR results on B/i and CIS samples for various miRNAs. Graph shows CIS log2 fold expression; CIS expression is normalized to B/i. Horizontal line indicates the sample median. Square indicates the sample mean. *P < 0.05, **P < 0.005 by Student’s t-test. B: Representative immunohistochemical analyses of UPKII, CK20, and high molecular weight cytokeratins (HMWCK) in four different CIS tissue samples. Scale bar = 100 μm. C: Quantification of the percentage of cases with expression of the respective biomarkers.
Because miRNA expression revealed a correlation between CIS and UM cells, we decided to assess the expression of several well-established differentiation biomarkers of the urothelium in a cohort of CIS lesions. We have previously reported B/i cells to be positive for high molecular weight cytokeratins, whereas UM cells are characterized by low molecular weight cytokeratins and UPKII.5,27 Indeed, a high percentage of CIS lesions revealed a positive UPKII (83.3%) and CK20 (84.2%) phenotype similar to that of UM cells, as well as a negative high molecular weight cytokeratins (10.5%) phenotype (Figure 4, B and C). Table 3 summarizes the individual immuno-anatomical profiles of the 19 CIS tissue samples.
Table 3.
Summary of Immunohistochemical Results in Individual CIS Tissue Samples
| Case No. | UPKII | CK20 | HMWCK |
|---|---|---|---|
| 1 | − | + | − |
| 2 | + | + | − |
| 3 | + | + | − |
| 4 | + | + | + |
| 5 | + | + | − |
| 6 | + | + | − |
| 7 | + | + | − |
| 8 | + | − | − |
| 9 | − | − | − |
| 10 | + | + | − |
| 11 | + | + | − |
| 12 | + | + | − |
| 13 | − | + | − |
| 14 | NA | + | − |
| 15 | + | + | − |
| 16 | + | + | + |
| 17 | + | + | − |
| 18 | + | + | − |
| 19 | + | − | − |
| % Positive | 83.3 | 84.2 | 10.5 |
NA, core not available.
Discussion
To date, there have not been any studies examining miRNA expression within normal bladder urothelium and bladder CIS. Despite a limited number of samples, our study reveals that the phenotype of CIS most closely resembles that of UM cells, rather than B/i cells.
A novel approach undertaken in this study is the micro-anatomical isolation of the normal bladder into three compartments: i) UM cells, ii) B/i cells, and iii) MP tissue. Thus far, there is no consensus and there are several inconsistencies between different microarray studies for the majority of miRNAs reported as misexpressed. For instance, Lin et al24 reported miR-125b and miR-199b to be significantly down-regulated in tumors compared to normal tissue, whereas Veerla et al29 reported high expression of the same two miRNAs in invasive (T2/T3) tumors. One explanation for this discrepancy is that different populations of normal bladder tissue were used. It is extremely difficult to obtain pure urothelium by macrodissection because it often leads to contamination of the urothelial sample with the lamina propria. Furthermore, although some groups use normal bladder mucosa,20,21,23 others use normal tissue adjacent to the tumor,24,25 which can include nonurothelium components, such as the muscularis propria. Microdissection is necessary, therefore, to ensure clean urothelial samples, and it is the only method to dissect single-layer UM cells.
We profiled the expression of 984 human miRNA and 22,277 human mRNA genes in the subpopulations of normal bladder, CIS tumors, and a normal urothelial cell line (HUC). Surprisingly, miRNA expression profile revealed CIS tumors to share a molecular profile displayed by UM cells, although mRNA expression profile clustered B/i and UM cells together, but separate from CIS. These data suggest that gene expression analysis is able to separate normal from tumor tissues. In fact, more than half of the differentially expressed genes that were upregulated in CIS, when compared to normal UM and B/i cells, were associated with proliferative phenotypes (eg, cell cycle, metabolic pathways, and pathways in cancer). In contrast, miRNA expression has been shown to classify tissue samples based on histogenesis,16 and our data show that CIS samples share more similarity with UM cells than B/i cells in the normal urothelium.
There were five miRNAs that were significantly altered between B/i and UM cells. RT-qPCR and in situ hybridization confirmed miR-133a and miR-139-3p to be specific to UM cells, whereas miR-142-3p was expressed only in B/i cells. Interestingly, the two UM cell specific miRNAs, miR-133a and miR-139-3p, are located on chromosome 18q and 11q, respectively. It is well known that deletions of chromosomes 18 and 11 are associated with invasive bladder carcinomas,30 which mainly arise from high-grade lesions such as CIS. Conversely, B/i specific miR-199b-5p is located on chromosome 9q, the deletion of which is associated almost exclusively with papillary, nonmuscle invasive tumors.6,31–33 It is plausible that deletion of these miRNAs may affect downstream targets involved in bladder tumorigenesis.
It is well accepted that there are two pathways of bladder tumor progression.6,34–36 The development of papillary, nonmuscle invasive tumors is largely independent from that of CIS lesions, as demonstrated through transgenic mouse models.37–39 Although bladder-specific expression of mutant H-RAS led to the growth of papillary tumors,38 simultaneous deletion of tumor suppressors p53 and Pten gave rise to CIS with progression to muscle-invasive tumors and metastasis.37 Furthermore, the double inactivation of p53 and Rb through UPKII-driven expression of SV40T also led to the development of high-grade CIS and muscle-invasive tumors.39 A clonal origin of bladder cancer has been suggested,40 although the study did not include CIS tumors in the analyses. It remains unclear, therefore, whether oncogenic changes that give rise to two phenotypic variants occur within one cell, or whether they arise in different progenitors (UM versus B/i). Our current study identifies a strong association between CIS and UM cells in miRNA expression and the expression of a few well-established differentiation markers of the UM cell, and provides evidence of the possible relationship between the two.
Acknowledgments
We thank Dr. Antonio Lopez-Beltran for providing the CIS FFPE tissue samples, Veronica Castro and Dr. David Llobet-Navas for their technical assistance, the members of the Dr. Cordon-Cardo Laboratory for their support, and the staff at Mount Sinai Hospital and the Herbert Irving Cancer Center, Columbia University (New York, NY).
Footnotes
Supported in part by NIH grant P01 CA087497 (C.C.C.).
Contributor Information
Mireia Castillo-Martin, Email: mireia.castillo-martin@mssm.edu.
Carlos Cordon-Cardo, Email: carlos.cordon-cardo@mssm.edu.
Supplemental Data
Hierarchical clustering of normal urothelium and CIS. Samples are in columns and miRNAs are in rows. Expression of individual samples were averaged into groups, with the exception of HUC, and clustered according to the expression signature of 295 differentially expressed miRNAs (P ≤ 0.05). One-way analysis of variance, Tukey honestly significant difference was performed as post hoc (false positive rate ≤ 0.05).
Validation of differentially expressed miRNAs in mouse bladder samples. Localization of miR-133a, miR-139-3p, and miR-142-3p in mouse bladder samples by in situ hybridization. No staining was observed in a negative control (sense strand of miR-424/503), whereas a strong signal was observed in a positive control (U6). Scale bars: 100 μm.
References
- 1.Wu X.R., Lin J.H., Walz T., Haner M., Yu J., Aebi U., Sun T.T. Mammalian uroplakins. A group of highly conserved urothelial differentiation-related membrane proteins. J Biol Chem. 1994;269:13716–13724. [PubMed] [Google Scholar]
- 2.Harnden P., Eardley I., Joyce A.D., Southgate J. Cytokeratin 20 as an objective marker of urothelial dysplasia. Br J Urol. 1996;78:870–875. doi: 10.1046/j.1464-410x.1996.23511.x. [DOI] [PubMed] [Google Scholar]
- 3.Sheinfeld J., Reuter V.E., Fair W.R., Cordon-Cardo C. Expression of blood group antigens in bladder cancer: current concepts. Semin Surg Oncol. 1992;8:308–815. doi: 10.1002/ssu.2980080510. [DOI] [PubMed] [Google Scholar]
- 4.Di Como C.J., Urist M.J., Babayan I., Drobnjak M., Hedvat C.V., Teruya-Feldstein J., Pohar K., Hoos A., Cordon-Cardo C. p63 expression profiles in human normal and tumor tissues. Clin Cancer Res. 2002;8:494–501. [PubMed] [Google Scholar]
- 5.Castillo-Martin M., Domingo-Domenech J., Karni-Schmidt O., Matos T., Cordon-Cardo C. Molecular pathways of urothelial development and bladder tumorigenesis. Urol Oncol. 2010;28:401–408. doi: 10.1016/j.urolonc.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 6.Cordon-Cardo C. Molecular alterations associated with bladder cancer initiation and progression. Scand J Urol Nephrol Suppl. 2008:154–165. doi: 10.1080/03008880802291915. [DOI] [PubMed] [Google Scholar]
- 7.Sesterhenn I.A. Urothelial carcinoma in situ. In: Eble J.N., Sauter G., Epstein J.I., Sesterhenn I.A., editors. Wold Health Organization Classification of Tumors: Pathology and Genetics of Tumors of the Urinary System and Male Genital Organs. IARC Press; Lyon: 2004. pp. 119–120. [Google Scholar]
- 8.McKenney J.K., Gomez J.A., Desai S., Lee M.W., Amin M.B. Morphologic expressions of urothelial carcinoma in situ: a detailed evaluation of its histologic patterns with emphasis on carcinoma in situ with microinvasion. Am J Surg Pathol. 2001;25:356–362. doi: 10.1097/00000478-200103000-00010. [DOI] [PubMed] [Google Scholar]
- 9.Wolf H., Melsen F., Pedersen S.E., Nielsen K.T. Natural history of carcinoma in situ of the urinary bladder. Scand J Urol Nephrol Suppl. 1994;157:147–151. [PubMed] [Google Scholar]
- 10.Zhang W., Dahlberg J.E., Tam W. MicroRNAs in tumorigenesis: a primer. Am J Pathol. 2007;171:728–738. doi: 10.2353/ajpath.2007.070070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He L., Hannon G.J. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 12.Esquela-Kerscher A., Slack F.J. Oncomirs — microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 13.Griffiths-Jones S., Saini H.K., van Dongen S., Enright A.J. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozomara A., Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwarz D.S., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 16.Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., Downing J.R., Jacks T., Horvitz H.R., Golub T.R. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 17.Puerta-Gil P., Garcia-Baquero R., Jia A.Y., Ocana S., Alvarez-Mugica M., Alvarez-Ossorio J.L., Cordon-Cardo C., Cava F., Sanchez-Carbayo M. miR-143, miR-222, and miR-452 are useful as tumor stratification and noninvasive diagnostic biomarkers for bladder cancer. Am J Pathol. 2012;180:1808–1815. doi: 10.1016/j.ajpath.2012.01.034. [DOI] [PubMed] [Google Scholar]
- 18.Wang M., Chu H., Li P., Yuan L., Fu G., Ma L., Shi D., Zhong D., Tong N., Qin C., Yin C., Zhang Z. Genetic variants in microRNAs predict bladder cancer risk and recurrence. Cancer Res. 2012;72:6173. doi: 10.1158/0008-5472.CAN-12-0688. [DOI] [PubMed] [Google Scholar]
- 19.Catto J.W., Miah S., Owen H.C., Bryant H., Myers K., Dudziec E., Larre S., Milo M., Rehman I., Rosario D.J., Di Martino E., Knowles M.A., Meuth M., Harris A.L., Hamdy F.C. Distinct microRNA alterations characterize high- and low-grade bladder cancer. Cancer Res. 2009;69:8472–8481. doi: 10.1158/0008-5472.CAN-09-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dyrskjot L., Ostenfeld M.S., Bramsen J.B., Silahtaroglu A.N., Lamy P., Ramanathan R., Fristrup N., Jensen J.L., Andersen C.L., Zieger K., Kauppinen S., Ulhoi B.P., Kjems J., Borre M., Orntoft T.F. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009;69:4851–4860. doi: 10.1158/0008-5472.CAN-08-4043. [DOI] [PubMed] [Google Scholar]
- 21.Gottardo F., Liu C.G., Ferracin M., Calin G.A., Fassan M., Bassi P., Sevignani C., Byrne D., Negrini M., Pagano F., Gomella L.G., Croce C.M., Baffa R. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007;25:387–392. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 22.Hanke M., Hoefig K., Merz H., Feller A.C., Kausch I., Jocham D., Warnecke J.M., Sczakiel G. A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol Oncol. 2010;28:655–661. doi: 10.1016/j.urolonc.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 23.Ichimi T., Enokida H., Okuno Y., Kunimoto R., Chiyomaru T., Kawamoto K., Kawahara K., Toki K., Kawakami K., Nishiyama K., Tsujimoto G., Nakagawa M., Seki N. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009;125:345–352. doi: 10.1002/ijc.24390. [DOI] [PubMed] [Google Scholar]
- 24.Lin T., Dong W., Huang J., Pan Q., Fan X., Zhang C., Huang L. MicroRNA-143 as a tumor suppressor for bladder cancer. J Urol. 2009;181:1372–1380. doi: 10.1016/j.juro.2008.10.149. [DOI] [PubMed] [Google Scholar]
- 25.Wang G., Zhang H., He H., Tong W., Wang B., Liao G., Chen Z., Du C. Up-regulation of microRNA in bladder tumor tissue is not common. Int Urol Nephrol. 2010;42:95–102. doi: 10.1007/s11255-009-9584-3. [DOI] [PubMed] [Google Scholar]
- 26.Fleige S., Pfaffl M.W. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med. 2006;27:126–139. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Karni-Schmidt O., Castillo-Martin M., Shen T.H., Gladoun N., Domingo-Domenech J., Sanchez-Carbayo M., Li Y., Lowe S., Prives C., Cordon-Cardo C. Distinct expression profiles of p63 variants during urothelial development and bladder cancer progression. Am J Pathol. 2011;178:1350–1360. doi: 10.1016/j.ajpath.2010.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gires O. Lessons from common markers of tumor-initiating cells in solid cancers. Cell Mol Life Sci. 2011;68:4009–4022. doi: 10.1007/s00018-011-0772-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Veerla S., Lindgren D., Kvist A., Frigyesi A., Staaf J., Persson H., Liedberg F., Chebil G., Gudjonsson S., Borg A., Mansson W., Rovira C., Hoglund M. MiRNA expression in urothelial carcinomas: important roles of miR-10a, miR-222, miR-125b, miR-7 and miR-452 for tumor stage and metastasis, and frequent homozygous losses of miR-31. Int J Cancer. 2009;124:2236–2242. doi: 10.1002/ijc.24183. [DOI] [PubMed] [Google Scholar]
- 30.Presti J.C., Jr., Reuter V.E., Galan T., Fair W.R., Cordon-Cardo C. Molecular genetic alterations in superficial and locally advanced human bladder cancer. Cancer Res. 1991;51:5405–5409. [PubMed] [Google Scholar]
- 31.Cairns P., Mao L., Merlo A., Lee D.J., Schwab D., Eby Y., Tokino K., van der Riet P., Blaugrund J.E., Sidransky D. Rates of p16 (MTS1) mutations in primary tumors with 9p loss. Science. 1994;265:415–417. doi: 10.1126/science.8023167. [DOI] [PubMed] [Google Scholar]
- 32.Dalbagni G., Presti J., Reuter V., Fair W.R., Cordon-Cardo C. Genetic alterations in bladder cancer. Lancet. 1993;342:469–471. doi: 10.1016/0140-6736(93)91595-d. [DOI] [PubMed] [Google Scholar]
- 33.Orlow I., Lacombe L., Hannon G.J., Serrano M., Pellicer I., Dalbagni G., Reuter V.E., Zhang Z.F., Beach D., Cordon-Cardo C. Deletion of the p16 and p15 genes in human bladder tumors. J Natl Cancer Inst. 1995;87:1524–1529. doi: 10.1093/jnci/87.20.1524. [DOI] [PubMed] [Google Scholar]
- 34.McConkey D.J., Lee S., Choi W., Tran M., Majewski T., Siefker-Radtke A., Dinney C., Czerniak B. Molecular genetics of bladder cancer: emerging mechanisms of tumor initiation and progression. Urol Oncol. 2010;28:429–440. doi: 10.1016/j.urolonc.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dinney C.P., McConkey D.J., Millikan R.E., Wu X., Bar-Eli M., Adam L., Kamat A.M., Siefker-Radtke A.O., Tuziak T., Sabichi A.L., Grossman H.B., Benedict W.F., Czerniak B. Focus on bladder cancer. Cancer Cell. 2004;6:111–116. doi: 10.1016/j.ccr.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 36.Wu X.R. Urothelial tumorigenesis: a tale of divergent pathways. Nat Rev Cancer. 2005;5:713–725. doi: 10.1038/nrc1697. [DOI] [PubMed] [Google Scholar]
- 37.Puzio-Kuter A.M., Castillo-Martin M., Kinkade C.W., Wang X., Shen T.H., Matos T., Shen M.M., Cordon-Cardo C., Abate-Shen C. Inactivation of p53 and Pten promotes invasive bladder cancer. Genes Dev. 2009;23:675–680. doi: 10.1101/gad.1772909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z.T., Pak J., Huang H.Y., Shapiro E., Sun T.T., Pellicer A., Wu X.R. Role of Ha-ras activation in superficial papillary pathway of urothelial tumor formation. Oncogene. 2001;20:1973–1980. doi: 10.1038/sj.onc.1204315. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Z.T., Pak J., Shapiro E., Sun T.T., Wu X.R. Urothelium-specific expression of an oncogene in transgenic mice induced the formation of carcinoma in situ and invasive transitional cell carcinoma. Cancer Res. 1999;59:3512–3517. [PubMed] [Google Scholar]
- 40.Sidransky D., Frost P., Von Eschenbach A., Oyasu R., Preisinger A.C., Vogelstein B. Clonal origin bladder cancer. N Engl J Med. 1992;326:737–740. doi: 10.1056/NEJM199203123261104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Hierarchical clustering of normal urothelium and CIS. Samples are in columns and miRNAs are in rows. Expression of individual samples were averaged into groups, with the exception of HUC, and clustered according to the expression signature of 295 differentially expressed miRNAs (P ≤ 0.05). One-way analysis of variance, Tukey honestly significant difference was performed as post hoc (false positive rate ≤ 0.05).
Validation of differentially expressed miRNAs in mouse bladder samples. Localization of miR-133a, miR-139-3p, and miR-142-3p in mouse bladder samples by in situ hybridization. No staining was observed in a negative control (sense strand of miR-424/503), whereas a strong signal was observed in a positive control (U6). Scale bars: 100 μm.