Abstract
Here we report the NMR solution structures of Mycobacterium tuberculosis (M. tuberculosis) thioredoxin C in both oxidized and reduced states, with discussion of structural changes that occur in going between redox states. The NMR solution structure of the oxidized TrxC corresponds closely to that of the crystal structure, except in the C-terminal region. It appears that crystal packing effects have caused an artifactual shift in the α4 helix in the previously reported crystal structure, compared to the solution structure. Based on these TrxC structures, chemical shift mapping, a previously reported crystal structure of the M. tuberculosis thioredoxin reductase (not bound to a Trx) and structures for intermediates in the E. coli thioredoxin catalytic cycle, we have modeled the complete M. tuberculosis thioredoxin system for the various steps in the catalytic cycle. These structures and models reveal pockets at the TrxR/TrxC interface in various steps in the catalytic cycle, which can be targeted in the design of uncompetitive inhibitors as potential anti-mycobacterial agents, or as chemical genetic probes of function.
Keywords: Thioredoxin, Thioredoxin reductase, Mycobacterium tuberculosis, NMR, solution structure
Introduction
Mycobacterium tuberculosis (M. tuberculosis) is the causative agent of tuberculosis, which has a mortality rate of 1.3 million people in 2008.1 It is estimated that a third of the world population is infected with a latent form of M. tuberculosis.2 The M. tuberculosis pathogen resides in the alveolar phagocytes of the host and can resist the oxidative killing of these immune cells, but the exact mechanism of this resistance is remains unclear.3 Disrupting this defense could be an effective means by which that pathogen could be killed, so the proteins involved in this defense are being pursued as potential targets for new anti-mycobacterial drugs. The method by which host phagocytes attempt to kill the invading M. tuberculosis pathogen is via production of reactive oxygen and nitrogen species, including superoxides and hydrogen peroxide, which can be toxic to the microbe.4 One method that M. tuberculosis uses to resist this oxidative attack is via the same process that eukaryotic cells use to combat oxidative stress, using the thiol reductants of the thioredoxin system (in addition to other enzymatic anti-oxidant systems). In other prokaryotic organisms, the thioredoxin system keeps cellular proteins in a reduced state, along with glutathione (mycothiol is used in M. tuberculosis).5–7
Thioredoxins are well-studied proteins, found in all organisms.8 They share common features such as a conserved catalytic motif of WCXXC, and have a low molecular weight of about 12 kDa.9 Thioredoxins catalyze thiol-disulfide exchange reactions using redox active cysteine thiols to reduce oxidized disulfide cysteines of proteins, including metabolically essential enzymes.10–12 The oxidized thioredoxins are then reduced by thioredoxin reductase (TrxR) in an NADPH-dependent reaction, via redox active cysteine or selenocysteine thiols, depending on the host organism.13–15 In plants, fungi and bacteria, the disulfide reductions in TrxR (~35 kDa) occur via a sequence of reactions that begins with hydride transfer from NADPH to a bound FAD cofactor, followed by electron transfer from FADH2 to the redox-active cysteines, to the thioredoxin, which forms a mixed disulfide with TrxR before the thioredoxin cysteines are fully oxidized. In higher organisms, such as mammals, the TrxR (~55 kDa) transfers the hydride from the NADPH to FAD, then from FADH2 to an amino-terminal TrxR active site, then to a carboxy-terminal active site of the TrxR/Trx dimer pair. This TrxR/Trx dimer contains a cysteine-selenocysteine redox-active pair, which then transfers electrons to the thioredoxin in a disulfide exchange reaction.14 Thus, the microbial and human thioredoxin mechanisms diverge significantly in the steps involving Trx binding and reduction. Disruption of this process, by selectively targeting the microbe over the host, could be used to kill the M. tuberculosis pathogen. Selective targeting should be possible since the Trx binding site is in a different region of the TrxR. Furthermore, the M. tuberculosis thioredoxins are only 35% identical to the human Trx, and the M. tuberculosis TrxR is only 28% identical to human TrxR. While these proteins are functionally related from microbe to man, the TrxR’s are structurally and mechanistically quite different.16,17
The catalytic cycle of the thioredoxin system from various prokaryotic sources has been extensively studied, with the best characterized being that from E. coli. The first E. coli Trx solution structures were reported over 20 years ago.18–22 Building on this rich literature, a comprehensive study from the Ludwig lab23 determined structures for the various steps in the E. coli thioredoxin system catalytic cycle, providing a structural complement to the mechanistic knowledgebase surrounding the microbial thioredoxin system. While the thioredoxin system from E. coli is well characterized, that of M. tuberculosis is not.3 Because the E. coli thioredoxin system is so similar to that of M. tuberculosis. (TrxR 45% identity, 63% homology, and Trx 50% identity and 64% homology), the structural studies presented herein build on what has been learned about the E. coli system, to provide a foundation for future structure-based design of inhibitors that disrupt the M. tuberculosis thioredoxin system, either as chemical probes of function, or as potential anti-mycobacterial agents. Analysis of structures and models for various steps in the M. tuberculosis thioredoxin catalytic cycle, as opposed to individual static snapshots of single proteins (ex. TrxR), may suggest novel strategies for inhibition.
The thioredoxin system in M. tuberculosis is comprised of three thioredoxins (TrxA, TrxB, and TrxC) and one thioredoxin reductase (TrxR).3, 24, 25 It has been shown that M. tuberculosis TrxB and TrxC behave as general disulfide reductases with near equal reduction potential (−262 mV vs. −269 mV), while TrxA is observed to have only a weak capacity to act as a disulfide reductase, and was found to not be a substrate for TrxR under the assay conditions tested.3 It has also been shown that the ability of M. tuberculosis to survive redox stress, while dependent on TrxR, is not dependent on the expression of just one thioredoxin. This suggests that having multiple thioredoxins in M. tuberculosis may provide a “redundant system” to ensure survival under oxidative conditions.3 The thioredoxin system of M. tuberculosis is increasingly viewed as a viable drug target for novel anti-mycobacterials, as long as a drug can be designed that knocks out the entire M. tuberculosis thioredoxin system (ex. inhibit TrxR catalytic turnover with all Trx’s).26 Of particular interest would be inhibitors that remain effective at high levels of competing substrate, either Trx or NADPH; this would be uncompetitive inhibition. Cells cannot achieve resistance to such uncompetitive inhibitors by increasing levels of substrates; but, design of such inhibitors requires structural information that enables more than just simple targeting the NADPH or Trx pockets on TrxR, as has been the focus to date.17 Design of uncompetitive inhibitors requires a knowledge of potential pockets that exist during the catalytic cycle and that could be occupied by inhibitors (in the presence of Trx substrate), and that stabilize reaction intermediates, thereby stopping catalytic turnover in a manner that cannot be overcome at high substrate. Studies presented herein will provide structures and models for various steps in the catalytic cycle of the M. tuberculosis thioredoxin system to facilitate design of such inhibitors.
Structural studies presented herein focus initially on M. tuberculosis TrxC (Fig. 1), and modeling of its interactions with TrxR throughout the catalytic cycle, based on NMR chemical shift perturbation studies (Fig. 2). While the crystal structure of TrxC in the oxidized state has been solved, there is no structure of TrxC in the reduced state, and no structural studies of interactions between TrxR and TrxC.25 Furthermore, it was previously suggested (but never proven) that the crystal structure of TrxC may have had distortions due to crystal packing forces that affect the C-terminal helix.25 Studies presented herein establish that this is in fact the case. Previous studies of the different redox states of prokaryotic thioredoxins have shown that there are subtle changes in structure in going from the oxidized to the reduced state.22, 27, 28 Most of the changes occur in the active site near the redox active cysteines, with some conformational flexibility observed in the active site loop.25, 29 Whether this is also true in M. tuberculosis TrxC is not known, since there is no structure of reduced TrxC. Studies presented herein will provide an analysis of differences between reduced and oxidized TrxC. Beyond this, studies will address how these forms of TrxC may interact with TrxR at different steps in the catalytic cycle (building on the E. coli work from the Ludwig lab), and whether these intermediates suggest unique opportunities for the design of uncompetitive inhibitors.
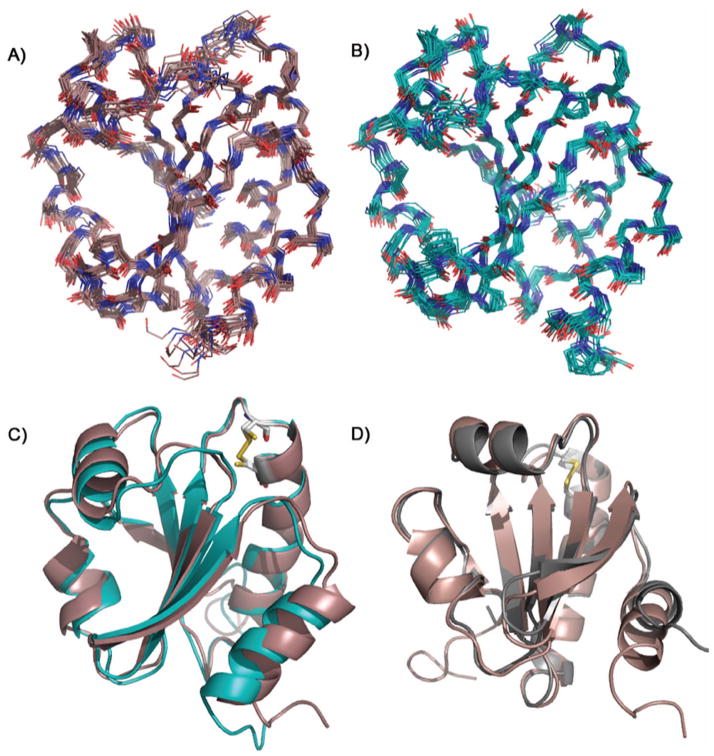
Figure 1.
Solution NMR structures of TrxC: (a) the ensemble for oxidized TrxC with a backbone RMSD of 0.54 Å, (b) the ensemble for the reduced state of TrxC with a backbone RMSD of 0.50 Å, and (c) overlay of the median structures for both redox states, colored as in panels (a) and (b). Note: disordered N- and C-terminal residues 1–6 and 113–116 have been omitted for clarity in panels (a) and (b). (d) Overlay of the median NMR solution structure for the oxidized TrxC (red) and the crystal structure of oxidized TrxC (gray), with the active site disulfides shown in yellow. The only significant deviation is in the C-terminal helix, α4, which in the crystal structure participated in an interaction with another TrxC in the asymmetric unit.
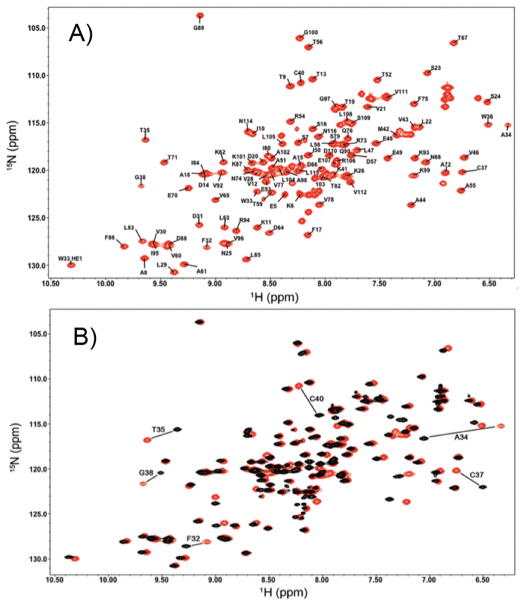
Figure 2.
2D 1H-15N HSQC spectra of (a) the oxidized state of TrxC (red) with all the backbone N-H chemical shifts assigned, and (b) the oxidized (red) and reduced (black) states of TrxC, with the large chemical shift changes associated with disulfide reduction labeled.
Materials and Methods
Protein Expression and Purification
The plasmids containing the Rv3914 gene (TrxC) in the pET22b vector and the Rv3913 gene (TrxR) in the pET23a vector were obtained from Tuberculosis Vaccine Testing and Research Materials at Colorado State University and from Dr. Mande from the Center for DNA Fingerprinting and Diagnostics in Hyderabad, India. Both expression constructs produced protein with a C-terminal poly-histidine tag (Supplementary Figure S4); plasmids were transformed into E. coli BL21-(DE3) Rosetta cells from Novagen and grown overnight at 37° C, then inoculated into 2 L of LB (Luria Bertani) media supplemented with a vitamin mix that included riboflavin (needed for TrxR flavin production). Cells containing the TrxR gene were grown until O.D. 600 nm reached 0.7, at which point cells were induced for 4 hours with 1 mM IPTG. For expression of TrxC protein cells were grown to 0.7 at O.D. 600 at which point the cells were spun down at 7000 rpm and washed with M9 minimal media salts (pH 7.0). Cell pellets were transferred to 0.5 L of M9 minimal media containing 0.5 grams 15NH4Cl and 2.0 grams of 13C-glucose as the only sources of nitrogen and carbon, to yield uniformly labeled protein.30 The cells were allowed to acclimate for 1 hour and then induced with 1 mM IPTG for an additional 4 hours at 37° C, then harvested. The bacterial pellet was resuspended in lysis buffer consisting of 50 mM Tris base, 300 mM NaCl, 5 mM imidazole, and 10% glycerol at pH of 7.8, and lysed using sonication. The lysate was clarified by centrifugation at 15,000 RPM for 30 minutes and the supernatant loaded onto 1 mL of nickel-Sepharose (Amersham Sciences) resin at 1 mL/minute. The bound protein (TrxC or TrxR) was then washed with lysate buffer, then with lysate buffer containing 25 mM imidazole, and then eluted using lysate buffer containing 300 mM imidazole. The eluted protein was than dialyzed with 40 mM potassium phosphate buffer at pH of 6.3 (for NMR structure studies) and pH of 7.0 (for TrxC/TrxR binding experiments) and concentrated to 1 mM using Amicon Ultra-4 centrifugal device (Millipore) with protein concentration determined by the absorbance at 280 nm using extinction coefficients of 11,000 and 14,440 L/mol-cm for TrxC and TrxR respectively, based on ProtParam calculations (http://expasy.org/tools/protparam.html). While estimation of protein concentration in this manner may slightly overestimate TrxR concentration (the FAD has some absorbance at 280 nm), exact stoichiometry is not required for titration of TrxC with TrxR, because we see complete disappearance of crosspeaks at < 1:1 stoichiometry due to exchange broadening. The purpose of the TrxR/TrxC titration is simply to identify interface crosspeaks, and the residues corresponding to those crosspeaks. Flavin incorporation into TrxR was verified using UV-Visible spectroscopy (Supplementary Fig. S5). For studies of reduced protein, 5 mM DTT was added.
NMR Spectroscopy and Structure Calculation
All NMR experiments were performed on a 600 MHz Varian NMR System at 599.515 MHz using a triple resonance cryoprobe with z-axis gradients at 25 °C. Backbone assignments were obtained using 2D 1H-15N HSQC, and 3D HNCO, HN(CA)CO, HNCA, HN(CO)CA, CBCA(CO)NH, and HNCACB spectra. Side chains were assigned using CC(CO)NH, HBHA(CO)NH, HCC(CO)NH, and HCCH-TOCSY spectra. Distance restraints were obtained from 3-D 15N-NOESY, 13C aliphatic NOESY, and 13C aromatic NOESY experiments, which were obtained using 150 ms mixing times. Structures were calculated using these distance restraints with the program CYANA,31–33 in conjunction with backbone phi and psi angle restraints as predicted from TALOS34 based on chemical shifts of Cα, Cβ, Hα, Hβ, HN, and CO atoms. Initial structures were obtained using the noeassign macro of CYANA,33 followed by manual refinement using the calibration of peaklists by the CYANA function caliba, with the disulfide bond added as a restraint for the oxidized structure. A final ensemble of 500 structures was calculated, and the 100 conformers with the lowest target function were then selected for water refinement using AMBER.35 Using implicit solvent, a 500 step energy minimization was first performed on the 100 structures (no restraints) followed by two cycles of 30 ps simulated annealing from 1000K to 0K, with distance and dihedral restraints applied. After the MD simulated annealing, a 2000 step minimization with restraints was applied in implicit solvent and analyzed for distance and dihedral penalties and rank ordered by lowest AMBER energy. The 20 lowest energy structures were then selected for the final ensemble (Fig. 1), and were analyzed using the Protein Structure Validation Software suite.36 NMR spectra were processed using NMRPipe37 and analyzed using NMRView38 and Sparky.39 Processing was performed with a cosine bell window function in the direct and indirect dimensions.
Structural Modeling of Intermediates in the M. tuberculosis Thioredoxin Reductase Catalytic Cycle
The M. tuberculosis thioredoxin reductase crystal structure (pdb code 2A87)17 had previously been solved in the oxidized state (in what is referred to as the FO conformation). To create a model of TrxR in the reduced state, where the disulfide is converted to surface free thiols (available to react with and reduce thioredoxins), the coordinates of the E. coli homolog of TrxR bound to a thioredoxin (pdb code 1F6M)(40) were used as template. Specifically, the M. tuberculosis TrxR (oxidized) structure was superimposed on the E. coli TrxR (reduced) structure. But, since the reduced E. coli TrxR is known to have a 66° domain rotation relative to the oxidized TrxR,17, 40 this same rotation was created in the M. tuberculosis structure. To do this, the pdb file of M. tuberculosis TrxR was split at amino acids 117 and 241 so that the FAD and NADPH domains could be separated. The separated NADPH and FAD domain coordinates were then superimposed onto the coordinates of the E. coli NADPH and FAD domain using Pymol41, then rejoined and then minimized using AMBER.35 This yielded what is referred to as the FR conformation. To model the various oxidation states of the TrxR/TrxC complexes, including the mixed disulfide between TrxR and TrxC, structures were minimized with a restrained disulfide bond between the cysteines (oxidized) or left with no restraint (reduced). All AMBER minimizations were performed with 2000 steps in implicit solvent.
Thioredoxin System NMR Binding Experiments
In order to establish which redox states of TrxC, TrxR and NADPH/NADP+ interact, and where interacts occur, chemical shift perturbations were monitored using 15N-labeled TrxC (reduced or oxidized) (Fig. 2). If binding occurs, based upon observation of chemical shift changes in HSQC spectra, chemical shifts are mapped onto the surface of TrxC to guide and confirm modeling of the TrxC/TrxR complexes, described in the previous section. These data were used, in part, to construct the models in Fig. 3, as described in Fig. 4 and Supplementary Fig. S2. TrxC and TrxR solutions were exchanged into 50 mM potassium phosphate buffer at pH 7.0, then concentrated to 1 mM each. TrxC was diluted to 320 μM in the absence of DTT, so that it was in the oxidized state. TrxR was then added to the TrxC NMR sample, so that both proteins were at equal concentrations of 250 μM. NADPH was then added to the sample at 1 mM so that TrxR would reduce the disulfide of TrxC. As a control to test for binding in the presence of the different redox states of the cofactor, NADP+ was added to the sample at 1 mM. 1H-15N HSQC experiments were acquired after every addition, and pH was measured to ensure that it remained at 7.0. As a control for the reduced state, 1H-15N HSQC spectra were acquired for TrxC in the oxidized state, and then reduced by DTT at pH 7.0.
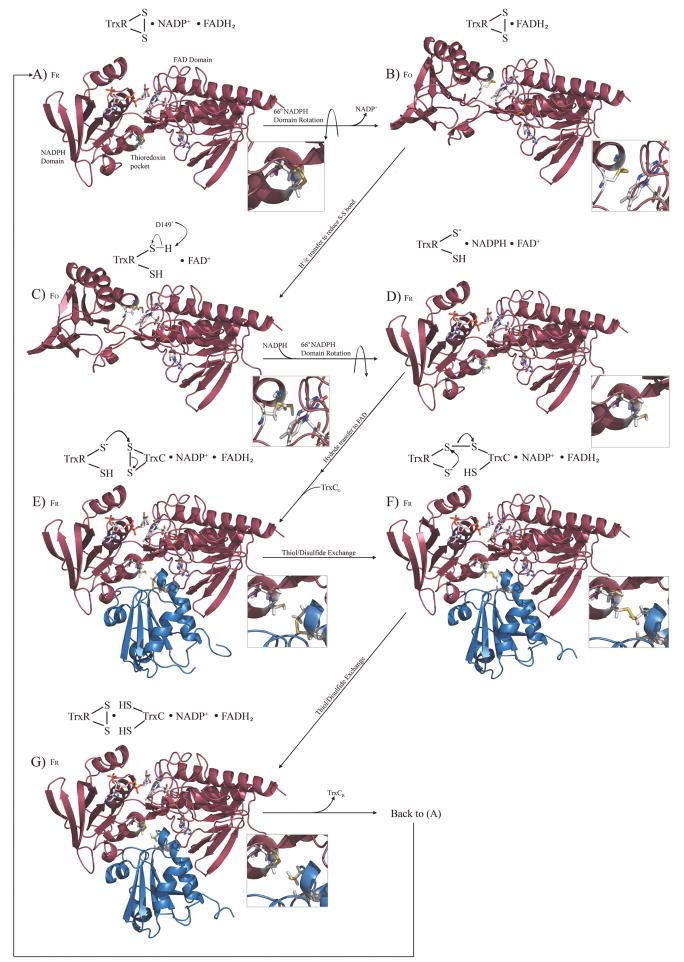
Figure 3.
The complete redox and catalytic cycle for the M. tuberculosis thioredoxin system, modeled after the E. coli thioredoxin catalytic cycle. The cycle begins with free TrxR (colored red) in the oxidized state, with bound NADP+, with the first step being release of NADP+ and a simple domain rotation to produce the FO conformation. FR and FO refer to the reduced or oxidized conformations (not the redox state) of TrxR, which are related by a 66° rotation of the NADPH and flavin domains. TrxC (ox and red) structures are those reported herein (colored blue), TrxR (FO) is the previously reported crystal structure, while models for the various complexes were constructed based on structural and functional data for the E. coli thioredoxin system, and chemical shift perturbation studies. Expansions show the redox state of relevant thiols or disulfides. Each modeled complex was minimized with AMBER. This cycle can also be visualized via a molecular animation (Supplementary materials), and structure models are available to be used in docking or structure-based drug design efforts.
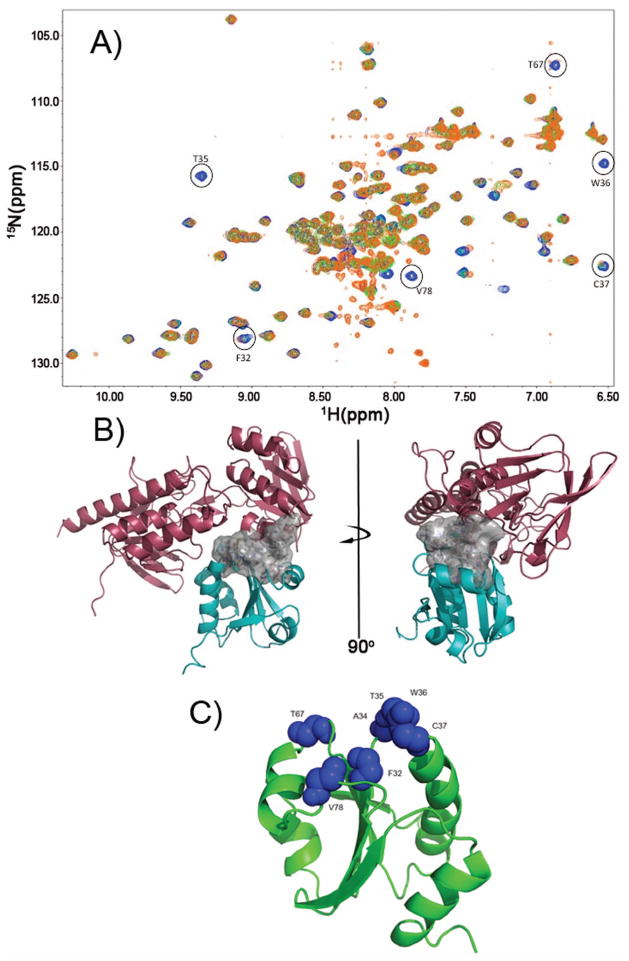
Figure 4.
Definition of the TrxR/TrxC interface using chemical shift perturbation studies. (a) Several HSQC spectra were overlaid, with blue crosspeaks corresponding to free reduced TrxC (in presence of 1 mM DTT). Green crosspeaks correspond to a ~1:1 complex between TrxC/TrxR at 250 μM (TrxC in slight excess over TrxR), with a fourfold excess of NADPH added. In cases where there was exchange broadening due to TrxR binding to TrxC, green peaks are no longer visible; so, the presence of blue identifies residues that are likely to be at the TrxC/TrxR interface. Orange crosspeaks correspond also to the complex between TrxC/TrxR at 250 μM, but now with a four-fold excess of NADP+ added in addition to 1 mM DTT. The exact match between green and orange crosspeaks suggests these two ternary complexes are similar, if not identical, and would correspond somewhat to intermediate F (but possibly with the mixed disulfide reduced) in Fig. 3. All spectra were acquired at pH 7.0. (b) Interface residues, which are largely hydrophobic, are shown as a surface rendering (white). This is the interface that forms when TrxR binds to and reduces an oxidized TrxC, and includes F142 and F143 on TrxR. (c) Residues on TrxC that show chemical shift perturbations or line broadening (from panel (a)) upon binding to TrxR have been mapped on the TrxC solution structure as blue Van der Waals spheres. These residues define the binding interface with TrxR (in panel (b)).
Thioredoxin System Interface Inhibition Docking and Titration Experiments
An in-house collection of 10,000 drug-like chemicals was docked into open spaces at the interface of the TrxC-TrxR complex using Autodock 4.2.42, 43 The grid box was centered at the interface and was 24.75 Å × 25.5 Å × 29.25 Å. Autodock 4.2 docked each ligand 50 times, scored the pose based on the program’s force field, and clustered each pose based on a root-mean-squared difference from the other poses. NMR screening of individual chemicals was prioritized by the cluster’s mean binding energy and population. Compounds that scored favorably were then dissolved in DMSO to a concnetration of 5 mM. Binding studies were performed by combining 200 μM TrxC, 250 μM TrxR, and 0.80 mM NADPH in the NMR buffer described above. Compounds were added at 200 μM (DMSO ~4% by volume) and HSQC spectra acquired before and after compound addition, and analyzed for any chemical shift perturbations.
Results and Discussion
NMR Solution Structures of Oxidized and Reduced Thioredoxin C
The solution structures of M. tuberculosis thioredoxin C were solved by NMR (Fig. 1), with 2036 NOE derived restraints and 168 angle restraints in the oxidized state and 2114 NOE restraints in the reduced state with 169 angle restraints. Statistics describing the restraints used in calculation, precision, and quality of the structures are summarized in Table I. The ensemble of structures for oxidized TrxC (PDB entry 2L59 and BMRB entry 17268) has a backbone RMSD of 0.54 Å, and 0.50 Å for the reduced state (PDB entry 2L4Q and BMRB entry 17242), shown in Figs. 1A and 1B. To ensure that the cysteines were reduced, 5 mM DTT was added to the protein sample and chemical shifts were monitored before and after each experiment, using a 1H-15N HSQC (Fig. 2A–B) spectrum. Reduction of the active site cysteines results in a decrease in the 13Cβ chemical shifts from 44.6 to 26.9 ppm and 35.1 to 27.6 ppm for Cys37 and Cys40 respectively, consistent with the additional shielding that is present with the electron rich thiol, compare to the disulfide. After addition of DTT to oxidized TrxC, those residues that are affected the most are located along the active site loop, including F32, A34, T35, C37, G38, and C40.
Table I.
NMR and water refinement statistics and analysis for solution structures of both redox states of Mycobacterium tuberculosis
| NMR Distance and Dihedral Constraints | ||
|---|---|---|
| TrxCox | TrxCred | |
| distance constraints | ||
| total NOEs | 2036 | 2114 |
| intra-residue | 207 | 257 |
| inter-residue | ||
| sequential (|i−j|=1) | 509 | 540 |
| medium-range (|i−j|<4) | 510 | 555 |
| long-range (|i−j|>5) | 810 | 762 |
| dihedral angle constraints | ||
| ϕ | 84 | 85 |
| ψ | 84 | 84 |
| Water Refinement and Structure Statistics | ||
| Mean AMBER energy (kcal/mol) | −3943 | −3940 |
| distance constraints (>0.3 Å) | 0 | 0 |
| dihedral angle constraints (>5.0°) | 1 | 1 |
| Ramachandran statistics | ||
| residues in most favored regions (%) | 92.2 | 90.5 |
| residues in additionally allowed regions (%) | 7.8 | 9.2 |
| residues in generously allowed regions (%) | 0 | 0.2 |
| residues in disallowed regions (%) | 0 | 0.1 |
| average pairwise RMSD (Å) 7–110 | ||
| backbone | 0.541 | 0.501 |
| heavy | 0.942 | 0.875 |
Analogous to the situation with the E. coli thioredoxin solution structures,18–22 the oxidized and reduced forms of TrxC are very similar to each other, showing only small differences between oxidized and reduced states, with a pairwise RMSD of only 0.53 Å between the ensembles (Fig. 1C). Consistent with other thioredoxins, TrxC in both states has the characteristic folding pattern of 4 α-helices surrounding a 5 stranded beta sheet core, consisting of three parallel strands and two antiparallel strands. The redox active cysteine residues C37 and C40 are located in the conserved active site loop (C37-G-P-C40) that leads into the α2 helix, which in both redox states is a bent helix. The active site of TrxC includes a predominately hydrophobic core including two Ala (34 and 44), two Pro (39 and 81), an Ile (80), and a Val (43) residue. Both Trp (33 and 36) indole rings are within 6 Å of the two Cys’s, with W36 also forming the interface surface that interacts with TrxR (Fig. 4). The disulfide in the oxidized state has the typical 90° χss angle for the C-S-S-C linkage. Interestingly, roughly the same geometry with the same χss angle is present in the reduced state. The most dramatic change in going from oxidized to reduced state is the distance between the Cys residue sidechains, with the S-S distance in the oxidized state of the median structure in the ensemble having a distance of 2.0 Å, compared with 3.5 Å in the reduced state (Fig. 1C). That is, overall geometry is roughly the same before and after S-S bond formation; but, the thiols move 1.5 Å closer together as the bond forms.
The N-terminal (α1) helix is somewhat irregular, but is nearly identical between the two redox states, and is the same as in the crystal structure of M. tuberculosis TrxCox25 as well as both E. coli Trx solution structures. The α4 helix spans residues 101–111, but begins to show fraying at L108. There are some differences between the two redox states in the turn region linking the α1 helix to the β3 sheet (residues 22–26), and the α3 helix connecting to the β4 sheet (residues75–81). The latter region closely contacts the active site region. These regions are probably dynamic in nature, and show similar effects in the E. coli solution structures of the different redox states (Fig. S1).
Comparison to the M. tuberculosis TrxC Crystal Structure
The M. tuberculosis TrxC crystal structure (pdb code 2i1u)25 was solved in the oxidized state, and is nearly identical in secondary structure to our solution structure, with a pairwise RMSD of 0.83 Å between the crystal structure and the median structure from the ensemble (Fig. 1D). But, there is one significant difference – and that is with regard to the orientation of the C-terminal α4 helix. It was noted in the crystal structure study that the protein crystals packed such that the C-terminus (residues 110–114) in the α4 helix rested in the active site groove of an adjacent protein in the crystal lattice, which the authors noted could cause a structural perturbation. Based on our solution structure of oxidized TrxC, this crystal packing effect does in fact seem to have caused the α4 helix to be pushed up 2.7 Å towards the β5 strand, and rotated in towards the β4 strand (Fig. 1D). Residues 108–115 in the crystal structure are disordered, perhaps so that those residues can occupy the active site of a neighboring TrxC protein. However, residues 108–111 still retain helical shape (as mentioned above) before becoming disordered at residues 112–115 in our solution structure. Although it is possible that some difference in the α4 helix may stem from the C-terminal histidine tag, we believe this is unlikely since the solution structure maintains the characteristic thioredoxin folding pattern, and is more compact than the crystal structure (Figure S1). Even though the distortion of the α4 helix and its ability to participate in a protein-protein interaction may be an artifact of crystal packing, its ability to participate in this interaction may also hint of a biologically relevant role. That is, since α4 is flexible, it is possible that it plays a role in protein-protein interactions with TrxR, which may require motion of this helix. In this regard, we have also performed calculations of generalized order parameter of TrxC, based on the “Random Coil Index”44, and found that several active site loop regions are predicted to have low order parameters (S2 < 0.8), including the loop that leads into the α4 helix (Supplementary Fig. S3). Flexibility in these loops may indicate they can function as hinges that enable motion of the N- and C-terminal helices α1 and α4, to optimally fit into the binding site presented by TrxR.
Structures and Models of M. tuberculosis Thioredoxin C (oxidized and reduced) and Thioredoxin Reductase throughout the Thioredoxin Catalytic Cycle
After having reduced other proteins in the cell, the oxidized TrxC must be regenerated to the reduced state by the enzyme thioredoxin reductase (TrxR). This process had been thoroughly studied in E. coli, culminating in a structural characterization of the various steps in the TrxR catalytic cycle.40 Fig. 3 presents the corresponding structures and structural models for the M. tuberculosis TrxR/TrxC system in different redox states, depicting the complete redox cycle between TrxR and TrxC, using the M. tuberculosis TrxR crystal structure17, our TrxC structures (oxidized and reduced), and the various E. coli structures40 as templates, with protein complexes created or confirmed based on chemical shift (and line-width) perturbation studies. Chemical shift mapping studies used to create these complexes are summarized in Fig. 4 and Supplementary Fig. S2. M. tuberculosis. TrxR is known to undergo a large domain rearrangement, to position the reactive cysteines so that they are able to react with the disulfide on the thioredoxin substrate. The crystal structure of the M. tuberculosis TrxR is in the oxidized conformation (FO), with its disulfide buried in the protein interior, near the isoalloxazine ring of FADH2; FADH2 provides the electrons to reduce this disulfide (step B to C in Fig. 3). The reduced state of M. tuberculosis TrxR (FR), with its cysteines now at the surface, and the NADPH close to the FAD (D in Fig. 3), was modeled using the E. coli thioredoxin reductase structure as a template (pdb code 1f6m).40 This structure was used to guide initial positioning of the M. tuberculosis TrxR FAD and NADPH domains. In this manner, structural models have been created for all the steps in the thioredoxin reaction sequence (Fig. 3), using chemical shift perturbation data to define the TrxR and TrxC interface (see below).
NMR Data Defining the Binding Interface between M. tuberculosis TrxC and TrxR
It has previously been proposed that the E. coli TrxR exists in a dynamic equilibrium between two conformations (FO and FR),23 irrespective of the state of oxidation for the flavin or cysteines. It is hypothesized that changing redox state or binding to the thioredoxin (Trx) substrate merely shifts the FO/FR equilibrium. To determine what oxidation state the proteins (TrxC and TrxR) must be in for binding to occur, a series of 1H-15N HSQC titration experiments were performed so that changes to the state of the proteins in different oxidative states could be monitored (Fig. 4; Supplementary Fig. S2). Chemical shift changes could be due to binding or to conformational/structural changes induced by redox changes; the latter effects are known from DTT titrations of isolated TrxC (Fig. 1B). When oxidized TrxR, in the absence of NADPH, was added to TrxC, there were no chemical shift changes to TrxC (Fig. S2), indicating that oxidized TrxR (that has no NADP(H) cofactor bound) cannot bind to TrxC. Interestingly, a small amount of TrxC that was present in a reduced state was converted to a fully oxidized state, indicating that even though the oxidized TrxC cannot bind, the reduced TrxC can bind to oxidized TrxR (lacking NADP(H)) and react in what would be the reverse of the biologically relevant direction (high NADPH concentrations ensure the reductive reaction is favored in vivo). When the reducing agent (NADPH) for the thioredoxin system was added in excess (4-fold), all of the oxidized TrxC was converted to the reduced state in the presence of TrxR, via the TrxR-catalyzed reduction (Supplementary Fig. S2); this is consistent with the reduced TrxR/NADPH being able to bind TrxC. While there were no chemical shift differences in comparing the TrxR/NADPH-reduced TrxC (TrxR present) with the DTT-reduced TrxC (TrxR absent), the presence of TrxR did cause disappearance of some TrxC crosspeaks due to exchange broadening (see below). This exchange broadening identifies residues at the TrxR/TrxC interface, thereby facilitating construction of properly docked TrxR/TrxC models (Fig. 3).
When the spectra for the TrxC/TrxR mixture, which has been reduced using NADPH or DTT (+ NADP+), were overlaid (Fig. 4), there were again no distinguishable chemical shift changes. When compared to the spectrum for unbound TrxC (reduced), there are a number of residues that disappear due to exchange broadening, due to binding interactions between TrxC and TrxR. The residues that showed exchange broadening were mapped onto the reduced state solution structure of TrxC in Fig. 4. The majority of these residues are in the active site region of the protein (F32, A34, T35, W36, C37, T67, V78), and these residues create a hydrophobic surface that interacts with TrxR, and that was used to define the protein-protein interface region for constructing the structural models in Fig. 3. Based on these results, although TrxR may be sampling both FO and FR states, TrxC will only bind to the reduced (free thiol) form of TrxR with cofactor (NADPH) bound, not to the FO state that is lacking NADPH cofactor.
Analysis of the M. tuberculosis Thioredoxin Cycle Models
As illustrated in Fig. 3, and by analogy to the E. coli system, the catalytic cycle for the M. tuberculosis thioredoxin system is shown starting in the conformation described as “FR” (intermediate A in Fig. 3), so that the active site cysteines have just finished reducing TrxC (which was released in the previous step), and there are bound NADP+ and FADH2 cofactors. It is called the FR state because of the relative orientation of the NADPH and FAD domains such that the NADPH binding site is close to the FAD (and the thiol/disulfide is distal). Assuming M. tuberculosis TrxR behaves as the E. coli reductase, it will then undergo a 66° rotation around the NADPH binding domain to bring the disulfide bonded cysteines (C145 and C148) proximal to the isoalloxazine ring of FADH2, while the NADPH binding pocket moves away from the FADH2, thereby allowing NADP+ to be released (this creates intermediate B). This also positions the isoalloxazine ring of FADH2 to within 3.7 Å of the S atom of Cys148 (now TrxR is in the “FO” state), which permits electron transfer from flavin to the disulfide (transfer will most likely be to the more proximal cysteine, Cys148). After the FADH2 reduces the disulfide, the sulfur inter-atomic distance increases from 2.0 Å to 3.7 Å, giving a slightly smaller χ angle across what was the disulfide. After reduction, a general base (likely to be D149, which is 5.3 Å from the thiol of C148) abstracts a proton from C148 to produce a thiolate that is ready to attack the disulfide of the thioredoxin (intermediate C converted to D).45 NADPH then binds to TrxR, and the NADPH domain undergoes another 66° rotation to bring the reduced active site cysteines away from the FAD isoalloxazine ring, and back to the surface (back to the FR conformation) of the protein, core that defines the TrxC binding pocket (the concave pocket on the bottom of intermediate D in Fig. 3). Elsewhere in the protein in intermediate D, the NADPH nicotinamide ring comes to within 3.4 Å of the isoalloxazine ring of FAD so that it can reduce the N5 position of FAD, again producing FADH2. The FADH2 can once again reduce the disulfide of TrxR, later in the catalytic cycle.
The TrxR is now in a conformation that can bind and then reduce TrxC (intermediate E). Inspection of this TrxC/TrxR structural model reveals that helix a4 is in contact with the mobile NADP(H) domain of TrxR (the α4 helix is on the far left side of the blue TrxC, as shown in Fig. 3, intermediates E–G). This same α4 helix was distorted in the crystal structure of TrxC, and is attached to the core TrxC structure via a flexible linker (predicted S2 < 0.8; Supplementary Fig. S3) based on our solution NMR studies. It appears that this mobile α4 helix permits binding of TrxC to the TrxR FR state (the FO state can’t bind Trx, because it has this TrxR interface rotated 66° away). Also of interest is that the α1 helix, which is likewise attached to the TrxC core via a flexible linker, makes contact on the opposite side of the TrxR pocket; and, the distortion (kink) in helixα1 creates a somewhat convex surface that may be essential for matching the concave shape of the TrxR pocket (the far right side of the blue TrxC structure in intermediate E of Fig. 3). Thus, a majority of the TrxR/TrxC binding interface is comprised of the α1 and α2 helices, which form a convex recognition surface on TrxC.
In addition to TrxR interactions with the TrxC α1 and α4 helices, the complex appears to be stabilized by a hydrophobic interface produced in the FR state by TrxR residues F142 and F143, which interacts with TrxC residues A34, P39, V43, A44, A72, V78, I80, and P81. Fig. 4 shows a view of the binding interface between TrxR and TrxC, identified using chemical shift mapping, with surface rendering showing the hydrophobic pocket. Of particular interest is that W36 of TrxC, which is conserved in all thioredoxins. W36 has its indole ring situated in this hydrophobic pocket, with the ring NH hydrogen bonding to the carboxyl group of Asp66, at a distance of 1.9 Å. This interaction is likely to be important for proper positioning of the TrxC C37, for reaction with the TrxR thiol/disulfide (C145, C148). The thiolate of C148 in TrxR is 4.9 Å from the disulfide-linked cysteine thiol (C37) of TrxC (intermediate E). Attack of C37 on C148 (disulfide) would produce the mixed disulfide between the two proteins (intermediate F), which is analogous to the mixed disulfide structure of E. coli TrxR/Trx that was structurally characterized by the Ludwig group.40 The free thiolate in TrxR (Cys145) then attacks Cys148 of the mixed disulfide, thereby producing fully reduced TrxC in the thiol reduced state (intermediate G). The newly reduced TrxC is released from the now oxidized TrxR, which is in the same state as Fig. 4A, thereby completing the thioredoxin catalytic cycle. The structural changes associated with this catalytic cycle can also be viewed as a molecular animation that was created using PyMol (supplementary materials). This series of thiol/disulfide exchanges is distinct from, and occurs in a different active site region, in the mammalian thioredoxin system. Accordingly, targeting this active site region in drug design efforts could provide compounds that are specific for microbial over mammalian enzymes.
Conclusion
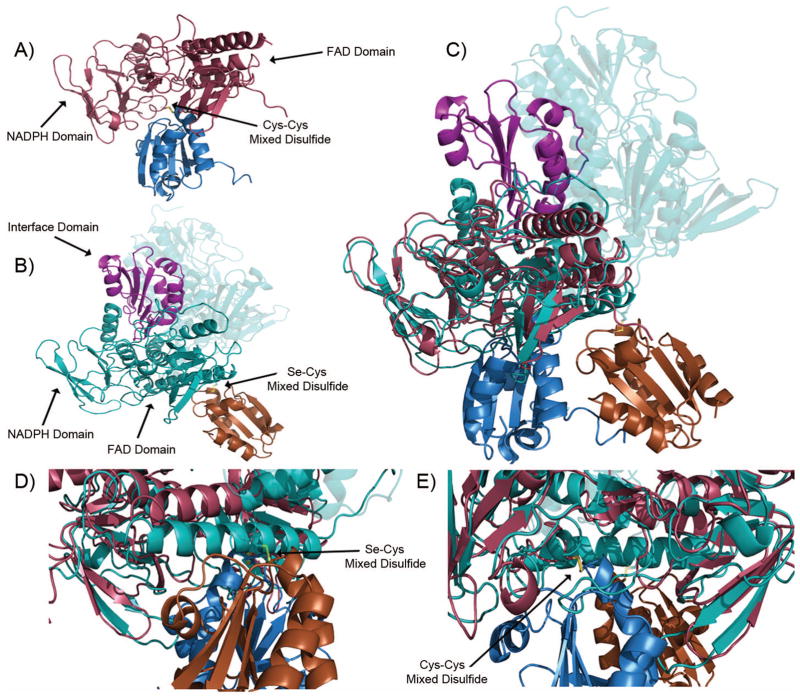
Developing an inhibitor that targets the M. tuberculosis. thioredoxin system rather than the human thioredoxin system would provide a novel route to anti-mycobacterial drugs. The function of the thioredoxin system, from microbe to man, is to maintain redox homeostasis by keeping cellular proteins reduced; and, in doing this, Trx becomes oxidized, so must be reduced by TrxR. The M. tuberculosis. TrxR undergoes a domain rotation in to move the NADPH and FAD(H2) in proximity to each other, and to move the reduced active site cysteines proximal to bound Trx.17 The human thioredoxin reductase instead uses a cysteine-selenocysteine active site on a flexible carboxy-terminal tail that is absent in the microbial enzyme, to shuttle FAD(H2) electrons to its bound thioredoxin (Figure 5B).14 In comparing the models from M. tuberculosis., described herein, and the recently published human thioredoxin system16, it becomes clear that there are significance structural differences (Fig. 5), along with the mechanistic differences, that can be exploited for selective inhibition of the microbial system. The human thioredoxin reductase has an additional domain that interacts with Trx (Fig. 6, interface domain, Fig. 5B and 5C), that is absent in the microbial system. Furthermore, the active site cysteines (that receive electrons from FADH2) are not directly exposed to the bound Trx, as they are in the microbial system. Of particular importance is the absence of the binding pocket between the Trx and TrxR that was observed in M. tuberculosis complex (compare Figs. 3B and 5B), because the human system uses the C-terminal domain for the final electron transfer step to bound Trx.
Figure 5.
Comparison of the M. tuberculosis. and human Trx-TrxR complex. (a) M. tuberculosis. Trx (blue)-TrxR (red) mixed disulfide. (b) Human Trx (orange)-TrxR (teal) mixed disulfide. Note the carboxy-terminus of the dimer pair is highlighted in violet and forms the mixed disulfide. (c) Overlay of the M. tuberculosis. and human thioredoxin systems. (d) Close up of M. tuberculosis thioredoxin complex disulfide from Figure 5(c). (e) Close up of the human thioredoxin complex disulfide from Figure 5(c).
Figure 6.
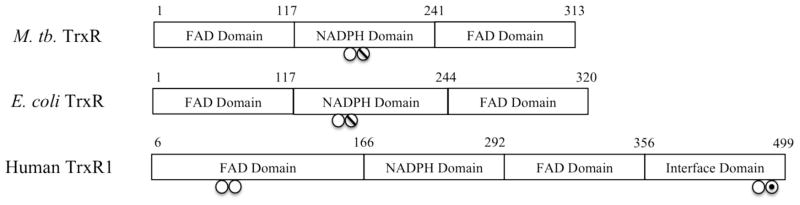
Comparison of the three thioredoxin reductases discussed in this study. The M. tuberculosis. TrxR shows 45% identity and 63% homology to the E. coli TrxR and 14% identity and 22% homology to human TrxR1. ○ denotes active site cysteines. ∅ denotes cysteines that form mixed disulfides between the thioredoxin and thioredoxin reductase. ⊙ denotes the selenocysteine from human thioredoxin reductase.
Besides providing insights into mechanism, and providing structural hypotheses that can be tested with future mechanistic studies (ex. using site-directed mutagenesis), the models presented herein (Fig. 3) can be used to facilitate structure-based design of inhibitors that selectively block the M. tuberculosis thioredoxin catalytic cycle, by targeting the Trx/TrxR interface. While inhibitors could be designed to block either the NADP(H) or TrxC substrate pockets on TrxR, the former class would yield inhibitors that might not be selective for the microbial system and which would be competitive versus cofactor. The latter approach is attractive, but suffers from the usual challenge of finding inhibitors that bind to broad protein-protein recognition surfaces; and, competitive inhibition can be overcome by the pathogen increasing expression levels of TrxC. A third approach might be to design inhibitors that occupy pockets that appear in the catalytic cycle at the Trx/TrxR interface, and are present even when Trx substrate is bound, so that binding of an inhibitor might stabilize that intermediate in the presence of substrate and thereby block catalytic turnover. This is the general concept behind uncompetitive inhibition. To illustrate the feasibility of the strategy of targeting interface pockets, a library of 10,000 compounds was docked into a pocket that is present in intermediates E, F and G (Fig. 3), at the interface of TrxR and TrxC. Compounds with reasonable drug-like appearance and properties (ex. adherence to Lipinski Rule of 5) are found that fit into this pocket (Fig. 7). Initial characterization of one of these compounds, by titrating onto the TrxR/TrxC complex, has shown some cross-peak perturbations, including the NH of interface residue W33. This result suggests feasibility of identifying compounds that bind at the TrxR/TrxC interface. More in-depth studies are needed to fully characterize the potential druggability of this binding site, by identifying and characterizing multiple inhibitors and establishing structure-activity relationships for binding at the TrxR/TrxC interface. These studies should include verification of binding by multiple techniques (direct binding and enzyme activity measurements), as well as MIC measurements, first in a model organism (ex. M. smegmatis) and ultimately in M. tuberculosis. Such structure-based drug design efforts are enabled by the results we have presented here, and are currently underway.
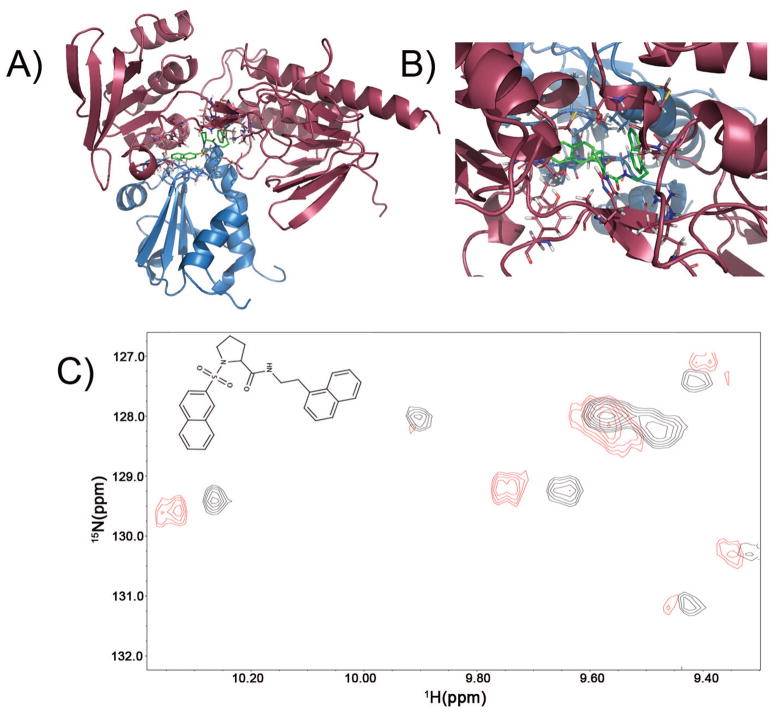
Figure 7.
(a) Result of docking into one of the intermediates in the thioredoxin catalytic cycle, to demonstrate the presence of potential binding pockets at the Trx/TrxR interface of sufficient size to accommodate potential inhibitors or drug leads. After docking 10,000 compounds, the best scoring (lowest energy) docked structure (shown in green, compound number CSDDD_6702) occupied a site at the interface of TrxR (red) and TrxC (blue) in Panels A and B, with energy = −10 kcal/mol using AutoDock4. (b) Expansion that shows the docking pose for CSDDD_6702. (c) Expansion showing the crosspeak perturbations in a section of the 15N-1H HSQC upon addition of 200 μM of compound CSDDD_6702 (blue) to a sample containing 200 μM TrxC, 250 μM TrxR, and 0.80 mM NADPH (black). Interface residue W33(HE1) is represented by the crosspeak at 10.2 ppm, which has been shifted upon binding CSDDD-6702.
In summary, we have determined high-resolution solution structures for both redox states of thioredoxin C (TrxC) from M. tuberculosis using NMR (backbone RMSD of 0.54 and 0.50 Å for TrxCox and TrxCred, respectively). The secondary structure and folding of the oxidized and reduced states of TrxC are nearly identical, with only subtle changes around the active site cysteines, and with some changes in loop regions. In the presence of the TrxR from M. tuberculosis, it appears that TrxC binds to TrxR only when TrxR has a reduced disulfide and cofactor (NADP(H)) bound. This was demonstrated by the disappearance of cross peaks for residues in the active site of TrxC, due to exchange broadening. The lack of other chemical shift changes (or peak shape changes) suggests that there are no significant structural changes to TrxC upon binding to TrxR. Using our structures and chemical shift perturbation data, along with the previously reported TrxR structure and structures for the thioredoxin system from E. coli,40 we constructed structural models for all the steps of the catalytic cycle of the M. tuberculosis thioredoxin system (Fig. 3). These structures, significantly different from the human thioredoxin system, should facilitate future mechanistic studies of the M. tuberculosis thioredoxin system, and aid in the design of inhibitors as potential drug leads. Indeed, initial analysis of these structures have identified at least one potential binding pocket, which is distinct from both substrate pockets, that could be pursued in structure-based drug design efforts. Inhibitors of the M. tuberculosis thioredoxin system may serve as chemical probes to study this pathogen’s ability to survive intracellular redox stress, in addition to providing a new class of anti-mycobacterial agents.
Supplementary Material
Acknowledgments
We thank Dr. Fancis Peterson at Medical College of Wisconsin for helpful discussions and advice on structure calculations, Elise Pellmann for assistance with UV-Vis spectral characterization of TrxR, as well as Chris Bohl and Kelsey Kalous for assistance in the production of TrxR. Some of this work was performed at Marquette University’s NMR facility with research supported in part by NIH research grants GM085739 and AI101975, NSF CREST grant CCLI #1022793, and shared instrumentation grant S10 RR019012.
Abbreviations
- DMSO
dimethylsulfoxide
- DTT
dithiothreitol
- HSQC
heronuclear single quantum coherence spectroscopy
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- LB
Luria Bertani
- MD
molecular dynamics
- NOESY
nuclear Overhauser effect spectroscopy
- RMSD
root mean square deviation
- Trx
Thioredoxin
- TrxR
Thioredoxin reductase
- TOCSY
total correlation spectroscopy
- Tris
tris(hydroxymethyl)aminomethane, NAD
- NAD
nicotinamide adenine dinucleotide
- FAD
flavin adenine dinucleotide.
References
- 1.World Health Organization. Global tuberculosis control: A short update to the 2009 report. 2009. [Google Scholar]
- 2.Jasmer RM, Nahid P, Hopewell PC. Latent Tuberculosis Infection. N Engl J Med. 2002;347:1860–1866. doi: 10.1056/NEJMcp021045. [DOI] [PubMed] [Google Scholar]
- 3.Akif M, Khare G, Tyagi AK, Mande SC, Sardesai AA. Functional studies of multiple thioredoxins from Mycobacterium tuberculosis. J Bacteriol. 2008;190:7087–7095. doi: 10.1128/JB.00159-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shinnick TM, King CH, Quinn FD. Molecular Biology, Virulence, and Pathogenicity of Mycobacteria. Am J Med Sci. 1995;309:92–98. doi: 10.1097/00000441-199502000-00007. [DOI] [PubMed] [Google Scholar]
- 5.Holmgren A. Antioxidant function of thioredoxin and glutaredoxin systems. Antioxid Redox Signal. 2000;2:811–820. doi: 10.1089/ars.2000.2.4-811. [DOI] [PubMed] [Google Scholar]
- 6.Arnér ESJ, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 7.Attarian R, Bennie C, Bach H, Av-Gay YG. Glutathione disulfide and S-nitrosoglutathione detoxification by Mycobacterium tuberculosis thioredoxin system. FEBS Letters. 2009;19:3215–3220. doi: 10.1016/j.febslet.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Holmgren A. Thioredoxin. Annu Rev Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- 9.Martin JL. Thioredoxin-a fold for all reasons. Structure. 1995;3:245–250. doi: 10.1016/s0969-2126(01)00154-x. [DOI] [PubMed] [Google Scholar]
- 10.Gleason FK, Holmgren A. Thioredoxin and related proteins in procaryotes. FEMS Microbiol Lett. 1998;54:271–297. doi: 10.1111/j.1574-6968.1988.tb02747.x. [DOI] [PubMed] [Google Scholar]
- 11.Ortenberg R, Gon S, Porat A, Beckwith J. Interactions of glutaredoxins, ribonucleotide reductase, and components of the DNA replication system of Escherichia coli. Proc Natl Acad Sci U S A. 2004;101:7439–7444. doi: 10.1073/pnas.0401965101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Powis G, Briehl M, Oblong J. Redox signalling and the control of cell growth and death. Pharmacol Ther. 1995;68:149–173. doi: 10.1016/0163-7258(95)02004-7. [DOI] [PubMed] [Google Scholar]
- 13.Williams CH, Arscott LD, Müller S, Lennon BW, Ludwig ML, Wang P, Veine DM, Becker K, Schirmer RH. Thioredoxin reductase. Eur J Biochem. 2000;267:6110–6117. doi: 10.1046/j.1432-1327.2000.01702.x. [DOI] [PubMed] [Google Scholar]
- 14.Zhong L, Arner ESJ, Holmgren A. Structure and mechanism of mammalian thoredoxin reductase: The active site is a redox-active selenolthiol/selenenylsulfide formed from the conserved cysteine-selenocysteine sequence. Proc Natl Acad Sci U S A. 2000;97:5854–5859. doi: 10.1073/pnas.100114897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gromer S, Urig S, Becker K. The Thioredoxin System--From Science to Clinic. Med Res Rev. 2004;1:40–89. doi: 10.1002/med.10051. [DOI] [PubMed] [Google Scholar]
- 16.Fritz-Wolf K, Kehr S, Stumpf M, Rahlfs S, Becker K. Crystal structure of the human thioredoxin reductase-thioredxin complex. Nat Commun. 2011;2:1–8. doi: 10.1038/ncomms1382. [DOI] [PubMed] [Google Scholar]
- 17.Akif M, Suhre K, Verma C, Mande SC. Conformational flexibility of Mycobacterium tuberculosis thioredoxin reductase: crystal structure and normal-mode analysis. Acta Crystallogr D Biol Crystallogr. 2005;61:1603–1611. doi: 10.1107/S0907444905030519. [DOI] [PubMed] [Google Scholar]
- 18.Dyson HJ, Holmgren A, Wright PE. Structural differences between oxidized and reduced thioredoxin monitored by two-dimensional 1H NMR spectroscopy. FEBS Lett. 1988;228:254–258. doi: 10.1016/0014-5793(88)80010-3. [DOI] [PubMed] [Google Scholar]
- 19.Dyson HJ, Holmgren A, Wright PE. Assignment of the proton NMR spectrum of reduced and oxidized thioredoxin: sequence-specific assignments, secondary structure, and global fold. Biochemistry. 1989;28:7074–7087. doi: 10.1021/bi00443a044. [DOI] [PubMed] [Google Scholar]
- 20.Dyson HJ, Gippert GP, Case DA, Holmgren A, Wright PE. Three-dimensional solution structure of the reduced form of Escherichia coli thioredoxin determined by nuclear magnetic resonance spectroscopy. Biochemistry. 1990;29:4129–4136. doi: 10.1021/bi00469a016. [DOI] [PubMed] [Google Scholar]
- 21.Forman-Kay J, Clore GM, Wingfield PT, Gronenborn AM. High-resolution three-dimensional structure of reduced recombinant human thioredoxin in solution. Biochemistry. 1991;30:2685–2698. doi: 10.1021/bi00224a017. [DOI] [PubMed] [Google Scholar]
- 22.Jeng M, Campbell AP, Begley T, Holmgren A, Case DA, Wright PE, Dyson HJ. High-resolution solution structures of oxidized and reduced Escherichia coli thioredoxin. Structure. 1994;2:853–868. doi: 10.1016/s0969-2126(94)00086-7. [DOI] [PubMed] [Google Scholar]
- 23.Lennon BW, Williams CH. Reductive Half-Reaction of Thioredoxin Reductase from Escherichia coli. Biochemistry. 1997;36:9464–9477. doi: 10.1021/bi970307j. [DOI] [PubMed] [Google Scholar]
- 24.Akif M, Chauhan R, Mande SC. Expression, purification, crystallization and preliminary X-ray crystallographic studies of Mycobacterium tuberculosis thioredoxin reductase. Acta Crystallogr D Biol Crystallogr. 2004;60:777–779. doi: 10.1107/S0907444904004366. [DOI] [PubMed] [Google Scholar]
- 25.Hall G, Shah M, McEwan PA, Laughton C, Stevens M, Westwell A, Emsley J. Structure of Mycobacterium tuberculosis thioredoxin C. Acta Crystallogr D Biol Crystallogr. 2006;62:1453–1457. doi: 10.1107/S0907444906038212. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z, Hillas PJ, Ortiz de Montellano PR. Reduction of Peroxides and Dinitrobenzenes by Mycobacterium tuberculosis Thioredoxin and Thioredoxin Reductase. Arch Biochem Biophys. 1999;363:19–26. doi: 10.1006/abbi.1998.1056. [DOI] [PubMed] [Google Scholar]
- 27.Weichsel A, Gasdaska JR, Powis G, Montfort WR. Crystal structures of reduced, oxidized, and mutated human thioredoxins: evidence for a regulatory homodimer. Structure. 1996;4:735–751. doi: 10.1016/s0969-2126(96)00079-2. [DOI] [PubMed] [Google Scholar]
- 28.Qin J, Clore GM, Gronenborn AM. The high-resolution three-dimensional solution structures of the oxidized and reduced states of human thioredoxin. Structure. 1994;2:503–522. doi: 10.1016/s0969-2126(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 29.Peterson FC, Lytle BL, Sampath S, Vinarov D, Tyler E, Shahan M, Markley JL, Volkman BF. Solution structure of thioredoxin h1 from Arabidopsis thaliana. Protein Science. 2005;14:2195–2200. doi: 10.1110/ps.051477905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marley J, Lu M, Bracken C. A method for efficient isotopic labeling of recombinant proteins. J Biomol NMR. 2001;20:71–75. doi: 10.1023/a:1011254402785. [DOI] [PubMed] [Google Scholar]
- 31.Güntert P, Braun W, Wüthrich K. Efficient computation of three-dimensional protein structures in solution from nuclear magnetic resonance data using the program DIANA and the supporting programs CALIBA, HABAS and GLOMSA. J Mol Biol. 1991;217:517–530. doi: 10.1016/0022-2836(91)90754-t. [DOI] [PubMed] [Google Scholar]
- 32.Güntert P, Mumenthaler C, Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program D. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 33.Herrmann T, Güntert P, Wüthrich K. Protein NMR Structure Determination with Automated NOE Assignment Using the New Software CANDID and the Torsion Angle Dynamics Algorithm DYANA. J Mol Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 34.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;4:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Case DA, Darden TA, Cheatham ITE, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Hayik S, Roitberg A, Seabra G, Swails J, Goetz AW, Kolossvai I, Wong KF, Paesani F, Vanicek J, Wolf RM, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh M, Cui G, Roe DR, Mathews DH, Seetin MG, Salomon-Ferrer R, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. Amber 11. University of California; San Francisco: 2010. [Google Scholar]
- 36.Bhattacharya A, Tejero R, Montelione GT. Evaluating Protein Structures Determined by Structural Genomics Consortia. Proteins: Struct Funct Bioinf. 2007;66:778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- 37.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 38.Johnson BA, Blevins RA. NMRView: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 39.Goddard TD, Kneller DG. SPARKY 3. University of California; San Francisco: [Google Scholar]
- 40.Lennon BW, Williams CH, Ludwig ML. Twists in Catalysis: Alternating Conformations of Escherichia coli Thioredoxin Reductase. Science. 2000;289:1190–1194. doi: 10.1126/science.289.5482.1190. [DOI] [PubMed] [Google Scholar]
- 41.The PyMOL Molecular Graphics System, Version 1.2.r3pre. Schrödinger, INC; [Google Scholar]
- 42.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19:1639–1662. [Google Scholar]
- 43.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berjanskii MV, Wishart DS. A simple method to predict protein flexibility using secondary chemical shifts. J Am Chem Soc. 2005;127:14970–14971. doi: 10.1021/ja054842f. [DOI] [PubMed] [Google Scholar]
- 45.Veine DM, Mulrooney SB, Wang P, Williams CH. Formation and properties of mixed disulfides between thioredoxin reductase from Escherichia coli and thioredoxin: Evidence that cysteine-138 functions to initiate dithiol-disulfide interchange and to accept the reducing equivalent from reduced flavin. Protein Science. 1998;7:1441–1450. doi: 10.1002/pro.5560070621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.