Summary
microRNAs (miRNAs) are emerging as key controllers of T-cell differentiation and function. Their expression is dynamically regulated by extracellular signals such as costimulation and cytokine signals. miRNAs set thresholds for gene expression and optimize protein concentrations of genetic networks. Absence of individual miRNAs can lead to severe immune dysfunction. Here we review emerging principles and provide examples of important functions exerted by miRNAs. Although our understanding of miRNA function in T-cell differentiation is still rudimentary, the available evidence leaves no doubt that these small posttranscriptional regulators are indispensable for proper functioning of the immune system.
Keywords: T-cell differentiation, microRNA, immune regulation, miR-155, miR-146a, miR-17-92
Introduction
T-cell development and differentiation is a highly complex process that occurs at multiple specialized microenvironments and involves multiple cellular intermediate stages (1). A large number of genes and developmental pathways are required for the controlled differentiation of hematopoietic stem cells into multiple highly differentiated effector T-cell subsets. Many extracellular signals delivered following cell-cell contact and soluble factors, like cytokines, drive intracellular signaling cascades. Genetic networks that integrate environmental signals have been deciphered in great detail (2). One of the key precepts in T-cell immunology has been the requirement that the system be exquisitely regulated. This is achieved through the balancing of various biochemical and cellular pathways that lead to immune homeostasis. Whether it is the differential signaling through the T-cell receptor (TCR), costimulatory, and negative signaling pathways or the balance of regulatory T cells (Tregs) versus memory T cells, for example, the immune system is controlled as a fine rheostat that allows for potent responses against foreign invaders while maintaining tolerance to self-antigens.
It has been revealed over the past decade that regulatory RNAs play a critical role in controlling gene expression (3). In fact, recent results of the ENCODE project and similar genome analyses suggest that RNA is not only an intermediate carrier of information between DNA and proteins but rather RNA elements, that do not encode proteins function as an important class of regulatory molecules (3–5). One particularly well-studied class of non-coding RNAs are microRNAs (miRNA), which are short posttranscriptional inhibitors of gene expression (6). About half of all protein-coding genes are regulated by miRNAs (7), and with a few rare exceptions (8, 9), almost all mammalian cells seem to engage miRNAs for gene regulation. Thus, it is not a surprise that the identification of miRNAs as fine regulators of gene expression play a critical role in maintaining this fine balance. In this review, we focus on our current understanding of the role of miRNAs in T-cell differentiation and function. Although much is to be learned and uncovered about the function of these endogenous short RNA sequences in biology overall, early studies suggest that these non-coding RNAs regulate the development and differentiation of T cells in general, in often subtle ways, providing a link to the fine specificity and delicate balance needed to control this robust system in the context of genomic regulatory elements.
Overall function of miRNAs in T cells
Several hundred miRNAs exist in the mammalian genome, and many of these are evolutionarily highly conserved (10). The growing list of miRNAs is curated in miRBase (11). Most miRNAs are transcribed as long transcripts that are cropped in a multistep process into the mature 21–23 nucleotide (nt) single stranded miRNAs (12). Nucleic DGCR8/DROSHA proteins are part of the important microprocessor complex that cleaves the primary transcripts into a stem loop shaped ~70nt precursor miRNA (12). After export to the cytoplasm DICER further crops the precursor miRNAs into their final length (13). The multi-step posttranscriptional miRNA processing allows for intricate regulation beyond transcription (14). In addition to the canonical miRNA biogenesis, alternative pathways involving the mRNA splicing machinery give rise to mirtrons that bypass the microprocessor and one miRNA, miR-451, can be cleaved by Argonaute (Ago) (15–20). Thus, multiple defined pathways contribute to the generation of the pool of miRNAs. Therefore, initial studies of the importance of miRNAs in the murine immune system utilized broad disruption of all miRNAs in specific cell types by deleting key proteins required for miRNA biogenesis.
Lck-cre-mediated ablation of Dicer during thymocyte development leads to severely reduced thymocyte numbers in part due to increased cell death, but surprisingly, the relative numbers of cells at the developmental stages remained intact (21). T-cell-specific Dicer deletion at a later time point (using CD4-cre) results in a mild lymphopenia (22) and a relatively mild multi-organ autoimmune disease (23). T-cell-specific miRNA deficiency led to an impaired balance of effector/suppressor T cells due to a propensity of CD4+ effector T cells committing to T-helper 1 (Th1) responses (22) and reduced Treg development (23), but T-effector cell function was not completely abrogated. Given that Dicer ablation is a sledgehammer approach, which removes the entire miRnome, it is surprising that miRNA-depleted T cells can survive and function. When using conditional gene ablation, selective outgrowth of Dicer-sufficient (i.e. escaped non-deleted) T cells suggests a competitive disadvantage of Dicer-deficient T cells (22, 24). However, there is evidence that Dicer is not essential for survival, and therefore the entire pathogenesis observed in CD4cre.Dicerlox/lox mice is not likely due to escaped Dicer-sufficient T cells. For instance, Dicer-deficient cells can proliferate and form tumors in a sarcoma transplant model, although tumor formation was slower than in Dicer heterozygous cells (25). Thus, DICER is dispensable for many tumor features and basic cellular processes including survival and proliferation of cells, although it is required for optimal cellular function. In addition to impaired proliferation and reduced survival, Dicer-deficient CD4+ T cells are strongly prone towards an interferon-γ (IFNγ)-producing Th1 phenotype, suggesting that miRNAs selectively repress the Th1 developmental program including expression of T-bet and IFN-γ (22). Similar results were observed in Drosha- and Dgcr8-deficient T cells (26, 27). Thus, in addition to controlling proliferation and survival, there are clear indications that T-cell differentiation is controlled by miRNAs. Specifically, DICER products are immunosuppressive by repressing Th1 responses. miRNAs can also control differentiation into other T-cell subsets. We employed an adoptive transfer system based on the use of cell tracker violet labeled cells to study T-cell differentiation of miRNA deficient T cells independent of any proliferation/survival defects. In this setting, we observed that Dgcr8-deficient naive CD4+ T cells were severely impaired in their ability to differentiate into T-follicular helper (Tfh) cells (Baumjohann, Ansel and Jeker, manuscript submitted). Thus, miRNAs promote Tfh but restrain Th1 differentiation.
In CD8+ T cells, DICER is required for accumulation after infection with Listeria monocytogenes (24). Surprisingly though, Dicer-deficient CD8+ T cells responded more rapidly than control cells in vitro, although their numbers were reduced. These results suggest that miRNAs inhibit the initial CD8+ T-cell activation. Of note, the proportion of cells expressing the early activation marker CD69 was increased in Dicer-deficient CD8+ T cells compared to control cells, and the miRNA-deficient cells showed substantially delayed CD69 downregulation during proliferation. Failure to properly downregulate CD69 led to altered migration.
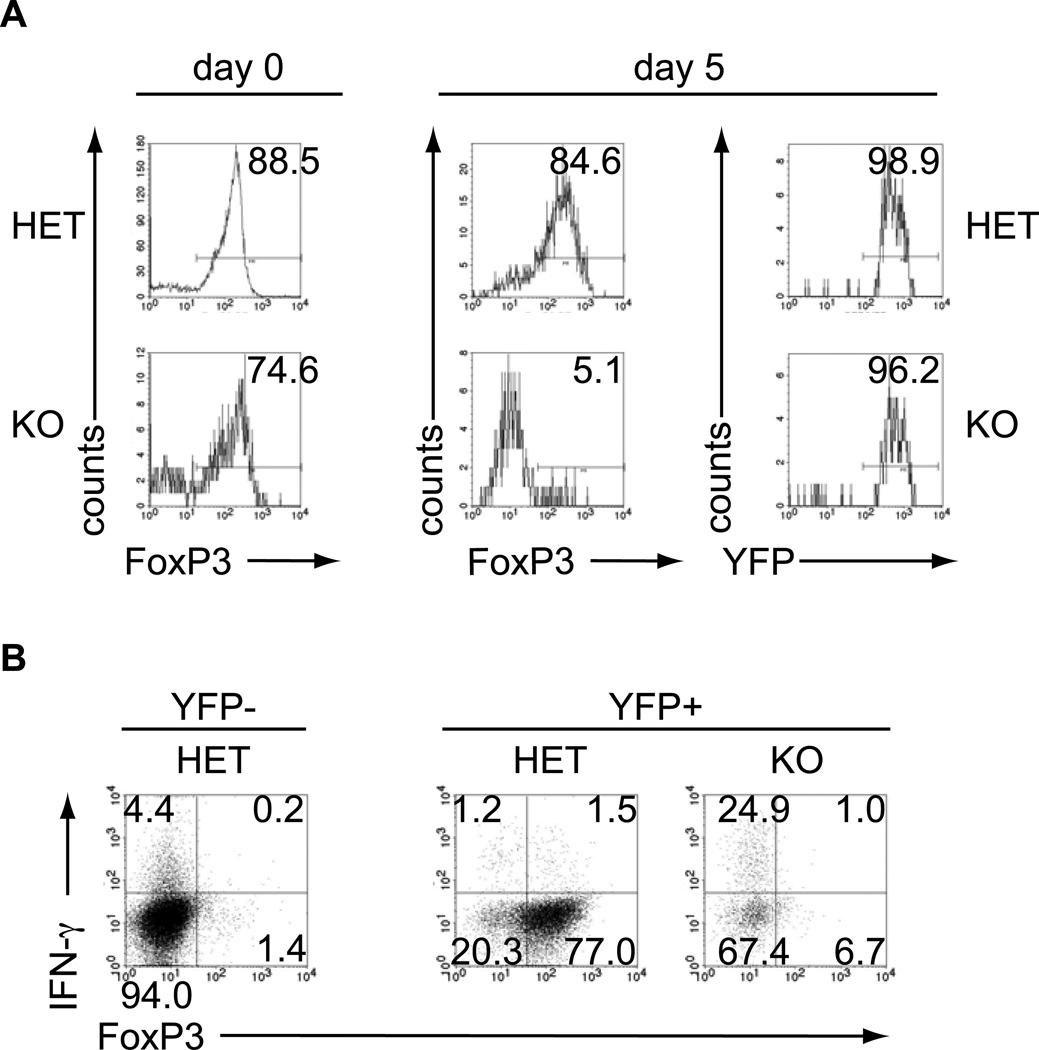
Analogous to findings in T-conventional cells (Tconv), CD4+forkhead box protein 3 (FoxP3)+ Tregs depend on miRNA regulation as Treg-specific Dicer- and Drosha-deficiency result in a scurfy-like disease (27–29). Importantly, Dicer-deficient Tregs lose expression of the hallmark Treg transcription FoxP3 (29). Like their Tconv counterparts, some Dicer-deficient Tregs produced IFN-γ despite the presence of FoxP3, arguing that FoxP3-mediated repression of IFN-γ on its own is incomplete and miRNAs add an extra layer of effector cytokine repression in Tregs (29). Treg-specific ablation of Dgcr8 results in a similar scurfy-like disease, as the ablation of Dicer or Drosha underscoring that canonical miRNAs are essential for Treg function (Jeker and Bluestone, manuscript submitted). To further investigate the fate of miRNA-deficient Tregs, we crossed the FoxP3-GFP-hCre mice to mice carrying a conditional ‘floxed’ Dgcr8 allele in combination with an additional YFP reporter allele that is only expressed after CRE-mediated excision of a stop cassette (R26-YFP) (30). In these mice, cells exposed to CRE driven by FoxP3 are permanently marked by YFP, which allows lineage-tracing studies (31). This approach revealed that loss of FoxP3 expression (GFP−YFP+) in vivo is much more pronounced in Dgcr8-deficient (KO) Tregs (GFP+YFP+) than Dgcr8 heterozygous (het) Tregs, suggesting that DGCR8-dependent miRNAs are required to maintain Treg identity (data not shown). Since we have previously demonstrated that cells that lose FoxP3 (termed exFoxP3 cells) are potentially pathogenic (31), we studied the kinetics of FoxP3 loss in the absence of miRNAs and tested the pathogenicity of miRNA-deficient exFoxP3 cells. FACS-sorted CD4+CD8−YFP+ KO cells from these mice had a higher frequency of exFoxP3 cells than het mice (Fig. 1A, left panel). After 5 days of culture, almost all Dgcr8-deficient cells had lost FoxP3 expression, while the majority of Dgcr8 heterozygous cells remained FoxP3+ (Fig. 1A, right panel). Importantly, all cells remained YFP+, indicating that the CRE recombinase had been active (Fig. 1A, right panel). Cells were resorted to ≥97% purity on day 8 and restimulated in vitro. A high fraction of miRNA-deficient exFoxP3 cells produced IFN-γ, in contrast to similar cells derived from heterozygous mice (Fig. 1B). To test the pathogenicity of miRNA-deficient exFoxP3 cells in vivo, we adoptively transferred the repurified lymphocytes into lymphopenic hosts. Two months after transfer, 2/2 recipients of miRNA-deficient exFoxP3 cells presented with cachexia, bloody diarrhea, and rectal prolapse, while 5/5 hosts receiving Dgcr8 heterozygous cells remained healthy. The Dgcr8 KO recipients had very severe kidney damage with destruction of tubuli and glomeruli and mild liver infiltration (data not shown). Thus, miRNAs are required to maintain Treg lineage identity by stabilizing FoxP3, which represses effector cell differentiation. In addition, miRNAs repress effector cytokine production in FoxP3-expressing Tregs. The result also further supports that miRNA-deficient effector T cells are functional. These data raise the possibility that the scurfy-like disease observed in mice with a Treg-specific lack of miRNAs may not only be passive through loss of Treg function but may have an active component through pathogenic exFoxP3 cells. Further studies are needed to test this hypothesis.
Fig. 1. DGCR8-deficient Tregs lose FoxP3 and turn into IFN-γ-producing miRNA-deficient exFoxP3 cells.
Tregs from FoxP3-GFP-hCre.R26YFP.DGCR8wt/lox (HET) and FoxP3-GFP-hCre.R26YFP.DGCR8lox/lox (KO) mice were used to investigate the contribution of miRNAs to Treg lineage identity. Flow cytometry purified CD4+CD8−YFP+ lymphocytes were cultured with anti-CD3 and anti-CD28 beads and 2000U IL-2/ml (Treg expansion conditions). (A) Intracellular FoxP3 staining of purified YFP+ cells at d0 and d5 and YFP purity 5 days after culture. (B) On day 8, CD4+CD8−YFP+ lymphocytes were resorted and then restimulated for 2h with PMA/ionomycin in the presence of monensin. Representative FACS plots of intracellular FoxP3 and IFN-γ staining. CD4+YFP− Tconv cells are shown as a comparison for CD4+YFP+ cells. Representative data from at least 2 experiments.
The half-life of mature miRNAs is not only regulated through control of transcription and posttranscriptional miRNA maturation but also by coordinated degradation through multiple mechanisms (32–34). Importantly, the exonuclease, Eri-1, regulates miRNA degradation in lymphocytes (35). Thus, another approach to modulating large families of miRNAs is to alter this degradation pathway. In this regard, elimination of Eri-1 gene expression resulted in accumulation of all miRNAs leading to the inhibition of natural killer (NK) cell development under homeostatic conditions. In addition, mixed bone marrow chimeras revealed that the development of Eri-1-deficient T cells were also at a competitive disadvantage as compared to wildtype T cells.
These examples demonstrate that global ablation of miRNAs is an effective approach to the study of T-cell development and differentiation. The results obtained with globally miRNA-deficient or -overexpressing T cells demonstrated that miRNAs are key regulators of T-cell proliferation, survival, migration, and differentiation but are surprisingly dispensable for other T-cell functions including cytokine secretion. Importantly, the observation that miRNA deficiency promotes differentiation into certain T-cell subsets (e.g. Th1) but inhibits differentiation into other subsets (Tfh) suggests that global modulation of the miRNA pool could help to regulate T-cell lineage choice. However, the interpretation of the resulting phenotypes is complex given the broad nature of the defects in miRNA-disrupted cells, the overarching effects on cell death and proliferation, and the unleashing of repressed programs in multiple T-cell subsets that can have both direct and indirect effects. In the future, the availability of embryonic stem (ES) cell lines with constitutively targeted miRNA loci (36) and conditional miRNA-gene targeted ES cells and mice (37) will undoubtedly enhance our understanding of miRNA-mediated control of T-cell differentiation and function.
Highly dynamic control of miRNA expression and miRNA-mediated gene regulation in T cells
Given the global effects of miRNA disruption on T-cell development and function, the next step would be to determine the role of individual miRNA loci in these settings. Thus, multiple laboratories, including our own, set out to determine the profile of miRNA expression in multiple T-cell types. Importantly, these analyses revealed that lymphocytes express a set of similar miRNAs (38–40), but unlike the liver (miR-122) and the heart (miR-1) which express characteristic specific individual miRNAs, no individual miRNAs were exclusively expressed in lymphocytes (39, 41, 42). For example, we and others (38, 43, 44) have found that miR-10a is enriched in Tregs, but it is also expressed in B cells and non-lymphoid tissues. miR-10a was expressed at low levels, but it remains possible that there is higher expression in a subset of Tregs that is masked by the population-based quantitative polymerase chain reaction (qPCR) analysis. In addition, as noted in multiple instances below, the expression levels may change during active immunity. There are other examples of individual miRNAs that are differentially expressed in T-cell subsets but the lack of defining miRNAs for individual T-cell subsets under homeostatic conditions, suggesting that major changes might exist during activation and differentiation of individual T-cell subsets.
miRNA expression did indeed change dramatically after T-cell activation (40, 45–48). An early study showed that Bic, which encodes miR-155, was induced by T-cell activation (49). Similar upregulation is observed for the miR-17-92 miRNA family (46, 47). miR-17-92 induction depends on maximal T-cell activation including CD28 costimulation in both mouse and human Tconv and Treg cells (de Kouchkovsky, Bluestone and Jeker, manuscript submitted). But as it turns out, these miRNAs are the exception. In fact, most miRNAs expressed in resting T cells are downregulated after T-cell activation, suggesting that miRNAs may play a general role in stabilizing gene expression and lineage determination including maintenance of a naive state (40, 47, 48). Detailed genome-wide miRNA profiling confirmed activation-induced rapid miRNA decline of most miRNAs, while miR-155 and miR-17-92 that belong to the few exceptions are induced (50). The group of miRNAs that was most rapidly and efficiently downregulated (e.g. miR-29a and miR-150) ceased transcription of their pri-miRNAs. In contrast, miR-19b expression, as a representative miRNA of the miR-17-92 cluster, was maintained or mildly induced through a disproportionally increased transcription compensating for the global miRNA depletion (50). Thus, transcriptional regulation contributes to the regulation of mature miRNA levels during T-cell activation. However, additional posttranscriptional mechanisms are involved. miRNAs function by physically interacting with their target mRNA in the miRNA-induced silencing complex (miRISC) serving as a guide for Argonaute (AGO) proteins (13, 51, 52). AGO2 is the key AGO protein in hematopoietic cells and is important to maintain physiologic miRNA levels (53). In fact, proteasomal degradation of AGO proteins is actively induced during T-cell activation, accounting for at least a partial explanation how T-cell activation induces the loss of miRNAs (50). Importantly, similar to Dicer- and Dgcr8-deficient T cells, which are prone to taking on a Th1 phenotype (22, 26), Ago2 heterozygosity or complete absence of Ago2 resulted in increased propensity to produce Th1 cytokines. In contrast to Dicer- and Dgcr8-deficient T cells, however, Ago2-deficient T cells proliferated normally despite a global reduction of miRNAs (50). Thus, this model uncoupled miRNA-mediated effects on proliferation from their function as suppressors of cytokine production. Cytokine suppression was likely in part indirect, because the Ago2-deficient cells expressed elevated Tbet. Furthermore, many Ago2-deficient T cells expressed increased IFN-γ, interleukin-4 (IL-4), or both. This indicates that miRNAs are required to restrain T cells from differentiation into Th1 and Th2 effector T cells. The emergence of significant numbers of cells co-producing IFN-γ and IL-4 suggests that some miRNAs are required to separate T-helper subsets during T-cell differentiation.
We note that miRNA-deficient T cells can still function. Thus, a major role of miRNAs may be to reinforce a given differentiation state by repressing multiple transcription factors and biochemical signaling pathways of alternative differentiation fates. As mentioned above, examples of miRNA-deficient T cells losing their identity and gaining features of other T-cell subsets support this notion (22, 29). In addition, during early T-cell differentiation, the activation-induced purging of the miRNA pool could ‘reset’ the miRNome and release repression of its targets facilitating acquisition of novel genetic programs and thus differentiation (40, 50). Importantly, downregulation of most miRNAs in conjunction with induction of select miRNAs leads to relative enrichment of those few miRNAs.
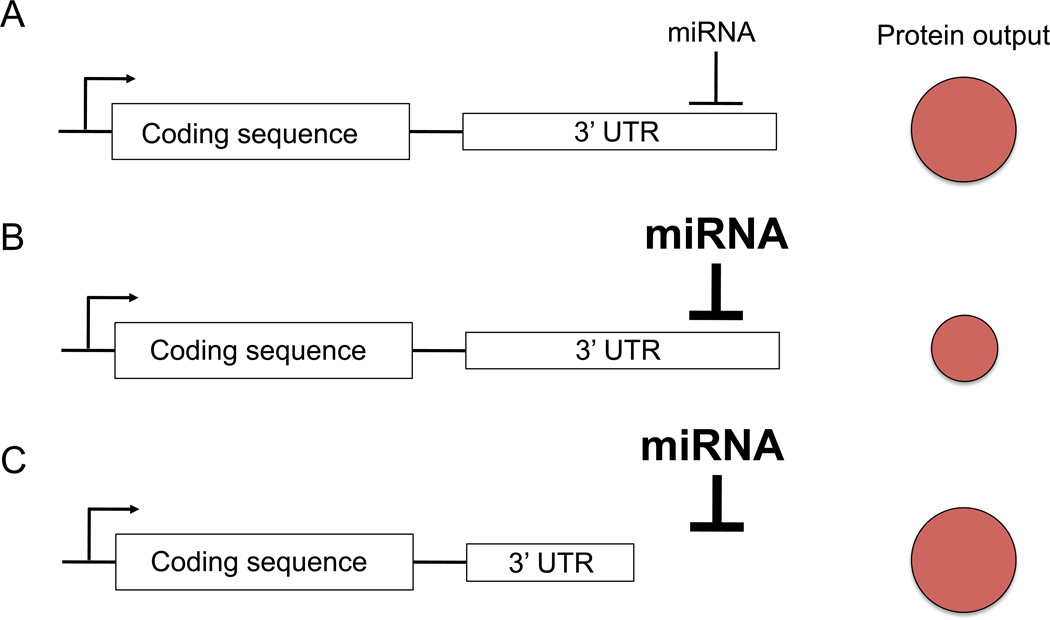
miRNA-mediated gene regulation occurs mainly through miRNA interaction with the 3′ untranslated region (3′UTR) of its target gene. In mammals, mRNA destabilization is the prevailing mechanism of miRNA regulatory function (54, 55), although there may be some contributions of translational inhibition (56). Alternative polyadenylation leads to mRNAs with varying length of their 3′UTR (57). Thus, alternative usage of different 3′UTR lengths is a way to allow or avoid miRNA-mediated repression (Fig. 2). Evolutionary pressure controls miRNA-mediated gene regulation by shaping the 3′UTR length and by inference miRNA regulation of genes, such that broadly expressed genes have short 3′UTRs while tissue-specific genes have longer 3′UTRs (58). Since lymphocyte activation is associated with 3′UTR shortening (46), the same gene that is subject to miRNA-mediated regulation when it has a long 3′UTR is less likely to be targeted by miRNAs once it shortens its 3′UTR. More than half of the miRNA-binding sites are downstream from the first polyadenylation site, suggesting that a large part of miRNA regulation could be lost during T-cell proliferation (57). Indeed, genes whose transcript expression increased during T-cell activation more frequently contain miRNA-binding sites exclusively in their extended 3′UTR than genes whose expression decreased, suggesting that they were repressed by miRNAs before activation. Importantly, predicted miR-155 and miR-17-92 target genes use the proximal polyadenylation site significantly more frequently than the distal one compared to non-targets in stimulated T cells (46). Thus, the model suggests that genes whose expression is needed but that are targeted by the activation-induced miRNAs miR-155 and miR-17-92 may use the short form of their 3′UTR to avoid repression during activation. The interplay of differential 3′UTR usage and target sequence conservation likely shapes the effectively targeted gene network among the predicted targets during T-cell activation, allowing one miRNA to exert differential functions in resting and activated T cells (Fig. 3). Thus, it is possible that differential regulation of short versus long 3′UTR usage by predicted miR-155 and miR-17-92 target genes poises a subset of genes for repression (maintaining long 3′UTR), while genes that require neutral or induced expression will switch to using the short 3′UTR (Fig. 3). A more comprehensive understanding of miRNA-mediated gene control will require deeper insight into alternative polyadenylation during T-cell activation and differentiation. Of note, knockdown of a single protein of the multi-protein 3′ end processing apparatus (57) in HEK293 cells is sufficient to switch a cell to the use of proximal polyadenylation sites (59).
Fig. 2. Alternative polyadenylation regulates miRNA-mediated gene repression.
Schematic representation of a model gene with constant transcription (arrow) of a coding sequence and its 3′ untranslated region (3′UTR). (A) During steady-state condition the interaction of a given miRNA with the 3′UTR of the target gene results in a defined protein output. (B) Induction of the miRNA leads to stronger repression of the gene leading to decreased protein output. (C) Shortening of the 3′UTR through alternative polyadenylation leads to avoidance of miRNA regulation despite the continued presence of the miRNA. Protein output increases as a result.
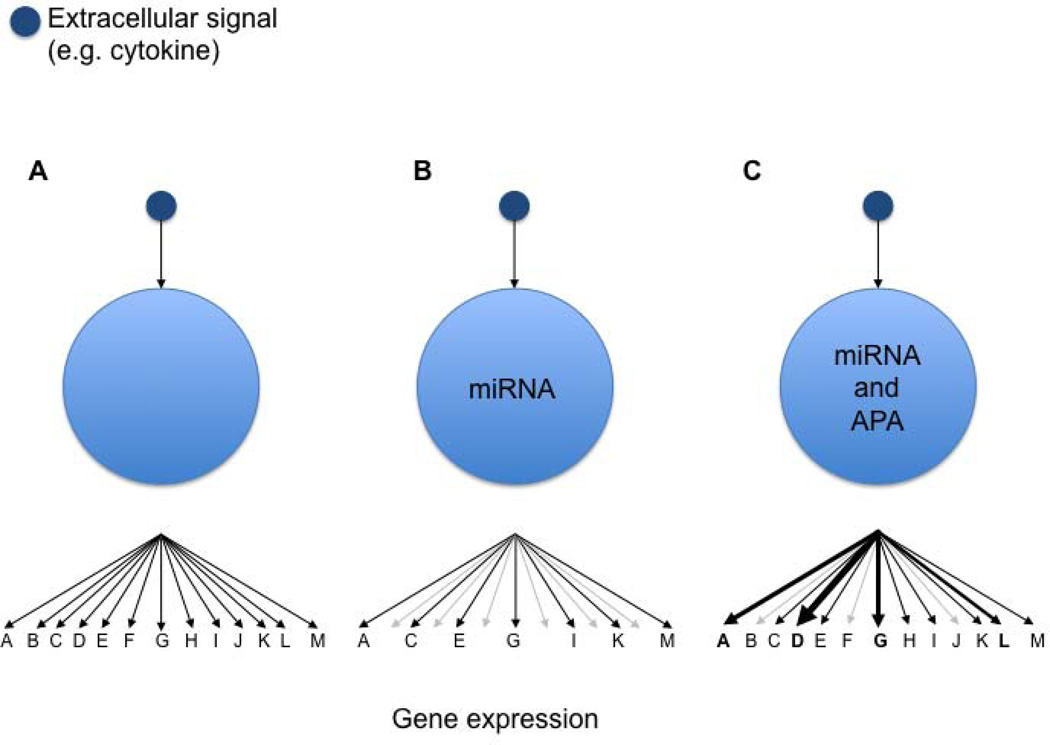
Fig. 3. Interplay of miRNA expression and 3′UTR usage shapes genetic networks.
Schematic representation of an extracellular cue (e.g. cytokine) on a T cell and its consequence on the genetic network. (A) A cytokine induces a network of genes A–M. (B) miRNA expression allows to shape the gene expression program induced by the cytokine. (C) The interplay of miRNA expression and alternative polyadenylation (APA) provides a cell additional flexibility to respond to the cytokine.
Multiple miRNA families, genomic miRNA clusters, and a multitude of mRNA targets
miRNA loci can be transcribed as transcripts coding for individual miRNAs or as polycistronic transcripts coding for multiple miRNAs. In addition, gene duplications led to multiple copies of similar miRNAs throughout the genome. The most important region in a miRNA to determine its target genes are nucleotide positions 2–7, termed the ‘seed’ sequence (6). Therefore, all miRNAs with an identical seed sequence likely target overlapping genes and are categorized as a family that can have multiple members. A prototypical and well-studied miRNA cluster is the miR-17-92 cluster which codes for 6 individual miRNAs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b, and miR-92a) (60). Interestingly, posttranscriptional processing of miRNA clusters can lead to differential expression of individual miRNAs within the same cluster (14). This can lead to synergistic or potentially even intra-cluster antagonistic functions (61). As an example, miR-19 is key for the well-documented oncogenic properties of miR-17-92 in B-cell lymphomas (62, 63), but miR-17 and miR-92a preferentially rescued proliferation of Dgcr8-deficient T cells (26). Thus, individual miRNAs among a cluster can be dominant for a particular function.
There is strong evidence that the miRNA target networks are complex, include many mRNAs and are under selective pressure (64). Analysis of miR-223-deficient neutrophils demonstrated that miR-223 represses dozens of genes in neutrophils (65). Furthermore, miR-29 and miR-17-92 miRNAs target dozens of genes in primary T cells (26, 47). Most gene expression changes were subtle but when cumulative gene expression changes of bioinformatically predicted direct targets are superimposed to all genes their preferential miRNA repression can be visualized using cumulative distribution plots (26, 54, 65). In addition, miRNA overexpression generally results in stronger repression than de-repression achieved by genetically or pharmacologically inhibiting miRNAs. Ansel and colleagues (66) transfected miR-29b into DGCR8-deficient (miRNA-depleted) CD4+ T cells, which led to much stronger repression of genes than inhibition of endogenous miR-29b, a strategy previously used in ES cells. Thus, an elegant approach to identify a set of genes regulated by miRNAs despite the modest regulation on a per gene basis is to combine overexpression and loss of function (26, 67). In addition, novel cross-linking based approaches allow genome-wide visualization of miRNA:mRNA binding sites and revealed that non-classical miR-155 targeting in T cells is widespread (68). Thus, it is now clear that individual miRNAs repress dozens of direct miRNA targets through different mechanisms and influence dozens of indirect targets in primary T cells.
Dynamic miRNA expression changes in response to environmental cues
Besides T-cell activation, a number of signals trigger substantial miRNA expression changes in T cells. We and others (43, 44) have shown that all-trans retinoic acid (RA) induces miR-10a expression in T cells likely through direct interaction with RA response elements in the miR-10a locus. This observation could mean that miR-10a is functionally relevant in the gut, where RA is important for induction of Tregs (69). Although controversial, one study suggested that miR-10a is also induced by transforming growth factor-β (TGFβ) (44). In this regard, there is clear evidence that TGFβ induces miR-21 in smooth muscle cells by promoting miR-21 maturation, not transcription of the primary transcript (70). SMAD proteins, the signaling molecules downstream of TGFβ and bone morphogenetic proteins (BMPs), recognize a specific consensus sequence in the primary miRNA transcript and interact with the DROSHA/DGCR8 microprocessor complex to specifically regulate processing of a number of miRNA precursors including the miR-21 precursor (70, 71). This regulation of miR-21 maturation in response to TGFβ is likely to have an important role in T cells, because TGFβ is a major immunosuppressive cytokine and miR-21 is highly expressed in T cells. Interestingly, TGFβ-mediated in vitro FoxP3 induction is inhibited by miR-17-92 (47), but miR-17-92 is required for Treg function in vivo (De Kouchkovsky, Bluestone, and Jeker, manuscript submitted). Since miR-17-92 is induced by CD28 costimulation (De Kouchkovsky, Bluestone, and Jeker, manuscript submitted), this might partially explain why strong T-cell activation inhibits Treg induction (69). In addition, miR-17-92 represses TGFβ responsiveness by targeting the TGFβRII (47). Finally, IL-2 induces miR-182 during T-cell clonal expansion (72) and IFN-γ induces miR-29ab-1 (73). Thus, expression of distinct miRNAs in T cells is highly dynamic in response to TCR/CD28 stimulation and different cytokines.
Individual miRNAs regulating T-cell differentiation and function
Among the best-studied miRNAs in T cells are miR-29ab, miR-146a, miR-155, and miR-17-92. miR-155, the first miRNA to be studied in miRNA-deficient mice, was found to have pleiotropic effects on dendritic, T, and B cells (74, 75). miR-155-deficient mice are immunodeficient, displaying a severely disturbed germinal center (GC) response and dysregulated transcription factor, cytokine, and chemokine expression. Subsequent studies demonstrated its function in Treg and effector T cells (76–78). Thus, from an immune homeostasis perspective, miR-155 has multiple opposing functions in various cell types by promoting Treg as well as T-effector function. In contrast, miR-146a is immunosuppressive in all cell types analyzed to date. miR-146a acts as a negative regulator of myeloid and T cells and promotes Treg cell function (79–82). Thus, individual miRNAs can have profound effects on immune regulation but often their functions are only revealed after specific challenges (83).
Role of miRNAs in CD8+ T cells
As in CD4+ T cells, miRNA expression in CD8+ T cells rapidly changes after activation (39, 48). miR-155 is induced during CD8+ T-cell activation but then rapidly declines and regulates CD8+ memory T-cell differentiation (84). Furthermore, miR-155 is required for normal CD8+ T-cell responses to lymphocytic choriomeningitis virus and Listeria monocytogenes infections (85). Similarly, miRNAs of the miR-17-92 cluster were induced but then declined after viral infection (86). During the initial expansion phase, miR-17-92 promoted proliferation, as miR-17-92-deficient T cells displayed mildly reduced proliferation, while miR-17-92 overexpression promoted expansion. Continued miR-17-92 overexpression inhibited memory CD8+ T-cell development, suggesting that miR-17-92 downregulation is required for normal memory formation. However, miR-17-92-deficiency only mildly affected the effector to memory ratio, possibly due to redundancy of similar miRNA clusters. Molecular analysis showed that miR-17-92 promoted short-lived terminal effector T cells at the expense of memory CD8+ T-cell formation. Analysis of miR-146a-deficient mice demonstrated that miR-146a was required to limit T-cell numbers (80). Adoptive transfer of TCR transgenic CD8+ miR-146a-deficient T cells and subsequent immunization with cognate antigen resulted in a strongly overshooting response and impaired contraction phase, particularly after a boosting immunization (81). Like in myeloid cells (82), miR-146a expression was regulated by nuclear factor-κB (NF-κB) and limited NF-κB activation in a negative feedback loop. Of note, naive CD8+ T cells were comparable between wildtype and miR-146a-deficient T cells. In contrast, after anti-CD3/anti-CD28 activation the miR-146a-deficient CD8+ T cells proliferated more, apoptosis was decreased, surface activation markers were increased, and the cells produced elevated levels of effector cytokines. Eliminating p50, the key component of most NF-κB complexes, did not alter the hyperactivated phenotype, indicating that the negative regulation of T-cell activation is mainly mediated through repressing NF-κB. Finally, since miR-155 and miR-146a have opposing effects on CD4+ and CD8+ effector T cells, O’Connell and colleagues (87) recently analyzed the effect of miR-155−/−miR-146a−/− double deficiency (DKO) on T-cell function. Interestingly, the effect of DKO followed the phenotypes of miR-155-deficient but not miR-146a-deficient T cells in several assays, indicating that miR-155 plays a dominant role over miR-146a. However, the interaction of genetic networks regulated by miR-155 and miR-146a, respectively, is likely complex and will require detailed analysis in various cell types and different conditions.
Role of miRNAs in CD4+ T cells: Th1, Th2, and Th17 cells
As described above, analysis of globally miRNA-deficient CD4+ T cells demonstrated clearly that miRNAs influence differentiation into and/or maintenance of T-cell subset identity (22). To identify individual miRNAs repressing the Th1-bias observed in miRNA-deficient T cells, Ansel and colleagues (66) adapted a screening approach used previously to identify miRNAs affecting ES cell proliferation. The principle is that DGCR8-deficient T cells are miRNA-deficient but have functional DICER. Therefore, reconstitution with individual miRNAs leads to loading into the intact miRISC complex and functional repression of target genes. Screening 110 miRNA candidates for their ability to repress IFN-γ production, the top two hits were miR-29a and miR-29b (26). Genome-wide target analysis of genes repressed after miR-29 transfection and derepressed after miR-29 inhibition revealed targeting of Tbx21 and Eomes, two well-known genes promoting Th1 differentiation. Furthermore, siRNA-mediated knockdown of TBET mildly phenocopied IFN-γ reduction but was less efficient than miR-29 transfection. Knockdown of EOMES alone did not affect IFN-γ production, while silencing TBET and EOMES simultaneously was more efficient than TBET knockdown alone. Thus, miR-29 is a potent repressor of IFN-γ production, likely acting indirectly by repressing TBET and EOMES. Interestingly, IFN-γ production and miR-29 expression inversely correlate in NK and T cells during infection with Listeria monocytogenes or bacillus Calmette-Guérin (BCG) (88). Among T-helper subsets, Th1 cells expressed the lowest miR-29 levels. T-cell activation led to downregulation of miR-29 through binding of NF-κB members to the miR-29 promoter. In this study, miR-29 was reported to directly target IFN-γ but neither TBET nor EOMES. Mice overexpressing a transgene containing multiple miR-29 binding sites to compete with endogenous miR-29 (‘sponges’) were used to demonstrate that inhibiting miR-29 led to increased IFN-γ production and increased resistance to Listeria infection. As expected, miR-29ab-1-deficient T cells express elevated TBET levels and increased IFN-γ production (73). This study reported direct miR-29 repression of TBET and IFN-γ. Importantly, the miR-29b2c locus did not regulate Th1 responses. Furthermore, miR-29 was induced by IFN-γ, suggesting a negative feedback loop. Unexpectedly, however, miR-29ab-1-deficient mice are resistant to experimental autoimmune encephalitis (EAE) due to a complete absence of central nervous system (CNS)-infiltrating lymphocytes. The reasons for this lack of infiltration will need to be carefully analyzed, because miR-29ab-1 is important in other cell types, such as thymic epithelial cells where it prevents thymic involution (89). Importantly, however, memory CD4+ T cells from patients with multiple sclerosis (MS) express increased miR-29b and T-bet mRNA (73). Thus, miR-29ab-1 is an important repressor of Th1 responses, but the underlying molecular mechanism remains controversial. miR-29 has likely other functions in the immune system. Given that miR-29ab represses IFN-γ in a negative feedback loop in Tconv cells, it might be particularly important to prevent Treg from producing IFN-γ in an inflammatory setting. It will therefore be interesting to test miR-29ab expression in human Tregs expressing IFN-γ (90).
Similar to EAE development in the absence of miR-29ab, miR-155-deficient mice are resistant to EAE and display strongly reduced lymphoid CNS infiltration (76). IFN-γ production was normal under some but reduced under different conditions. Importantly, cell-intrinsic expression of miR-155 was required for IL-17A production. In addition to miR-155, miR-17-92 also promotes Th1 responses (47). The miR-17-92 cluster not only promotes T-cell proliferation (26, 47, 91) but also is required for optimal IFN-γ production (47). Reconstitution experiments of miR-17-92-deficient T cells with individual miRNAs revealed that miR-19b was the only miRNA from all six miRNAs represented by this cluster that promoted IFN-γ production. Interestingly, while miR-19b was able to rescue defective proliferation, miR-19a, which differs by one nucleotide, failed to rescue. Intriguingly, miR-18a inhibited proliferation as well. Both miR-17 and miR-19b were able to rescue the susceptibility of miR-17-92-deficient cells for activation-induced cell death (AICD), while again miR-18a enhanced apoptosis. Overall, the miR-17-92 cluster supported a Th1 response, because mice with miR-17-92-deficiency in all T cells showed a reduced delayed-type hypersensitivity response and showed reduced tumor rejection. Absence of miR-17-92 impaired production of many cytokines including IFN-γ, IL-2, IL-4, IL-5, and TNFα, suggesting that multiple Th subsets (and possibly CD8+ T cells) were affected. As mentioned previously, miR-146a is a negative regulator of T-cell activation and function (81). The findings discussed for CD8+ T cells were essentially also true for CD4+ T cells. Similarly, miR-155 plays a dominant role over miR-146a in both CD4+ and CD8+ T cells (87). In summary, miR-29, miR-146a, miR-155, and miR-17-92 are important regulators of Th1 responses.
Much less is known about the role of individual miRNAs regulating Th2 responses, particularly the intrinsic control of Th2 differentiation. Although miR-155-deficient T cells are prone to produce Th2-type cytokines (IL-10, IL-4) (74), detailed studies beyond miRNA expression profiling are largely missing. In addition, relatively little is known about how individual miRNAs regulate Th17 cells. miR-155 promotes Th17 production, but it remains unclear how this miRNA affects Th17 differentiation and/or function (76). miR-326 was reported to be required for Th17 cell development and important for the development of EAE in mice and possibly MS in humans (92). However, the findings were challenged by other studies that either did not find differential expression of miR-326 in samples from MS patients or that were unable to detect miR-326 in mouse or human Th17 cells (39, 41, 93). Of note though, miR-326 was detected in brain lesions from MS patients using laser capture microscopy to isolate diseased tissue (94). Thus, further research on the role of miR-326 in MS and Th17 cells is certainly warranted to resolve the current controversy (95, 96). Another miRNA, miR-301a, is enriched in murine Th17 cells (97). Pharmacological miR-301a inhibition using chemical compounds called antagomiRs led to decreased IL-17A and signal transducer and activator of transcription 3 (Stat3) phosphorylation, and a miR-301a mimic increased CCR6 expression. This effect is likely mediated through direct repression of the Stat3 inhibitor PIAS3. Transfection of CD4+ T cells with miR-301a mimic worsened and antagomiR treatment suppressed EAE in an adoptive transfer model, suggesting that miR-301a controls Th17 function.
miR-182 expression is increased in Th1, Th2, and Th17 cells, probably through stimulation of the TCR in combination with IL-2 (72). STAT5 bound the miR-183-miR-96-miR-182 locus, and STAT5 inhibition led to reduced miR-182 expression. Thus, it was proposed that IL-2 induces miR-182 in the late phase of clonal expansion where miR-182 represses FOXO1 expression. Supporting this notion, pharmacological inhibition of miR-182 led to mildly decreased in vitro proliferation, and adoptive transfer of cells treated with the inhibitor transferred slightly milder arthritis. However, genetic models are required to thoroughly assess the role of miR-182 in different Th subsets, because results obtained by the use of antagomiRs and other pharmacological miRNA inhibitors can reportedly differ significantly from genetic miRNA ablation (43, 98).
Regulatory T cells
A number of miRNAs (miR-10a, miR-21, miR-146a, and miR-155) are enriched in Treg compared to Tconv cells (39, 43, 44, 78, 79). However, all individual miRNAs that were ablated in vivo were largely dispensable for Treg function under homeostatic conditions (43, 77). In contrast, when immune homeostasis is challenged by an immune response or immune reconstitution, miRNA functions have been revealed. For instance, miR-155 is an important regulator of Treg homeostasis in a competitive setting (78). Facilitated IL-2 signaling likely contributes to the dependence of Tregs on miR-155, although in vitro the defect can be overcome by increased IL-2 concentrations. miR-146a is critical for Treg function in a high stress setting (79), but its role under homeostatic conditions has not yet been reported. miR-10a displays an intriguingly specific expression pattern in Tregs, yet ablation of miR-10a was largely dispensable for Treg generation under homeostatic conditions and for Treg function in an adoptive transfer colitis model (43). However, using a lineage-tracing system, we found that miR-10a expression correlated with the most stable Tregs and is lower in exFoxP3 cells (i.e. those T cells that have lost FoxP3 expression following activation of the locus) (31, 43). Furthermore, miR-10a inhibitors (antagomiRs) destabilized FoxP3 expression in vitro. In contrast, miR-10a-deficient mice did not display reduced FoxP3 expression in Tregs. Further investigation is needed to address this discrepancy. It is possible that antagomiR-10a-mediated FoxP3 downregulation is due to off-target effects, that other miRNAs such as miR-10b compensate for genetic miR-10a deficiency in vivo, or that specific challenges such as Treg activation or inflammatory signals are needed for FoxP3 downregulation. Nevertheless, the expression pattern and in vitro results together with the finding that RA, known to stabilize Tregs (69), induces miR-10a in T cells suggest that miR-10a might be a stabilizing factor for Tregs in general or a Treg subset, for instance in the gut. In support of this hypothesis, it has been reported that miR-10a directly represses BCL-6, restraining the ability of in vitro generated iTregs to convert to Tfh cells (44). Whether this is also true for thymically derived Tregs remains to be assessed. In comparison to all Tregs, miR-10a expression was strongly enriched in a subset of Tregs expressing PD-1 and CXCR5, designated T-follicular regulatory (Tfr) cells (44). Thus, the rather low miR-10a expression in polyclonal Tregs could underestimate the expression in Tfr (43) and would suggest that miR-10a is particularly important for this subset of Tregs. Therefore, future in vivo studies need to address if miR-10a-deficient Tregs have a specific defect in Tfr development, function, or stability. The conundrum that remains, however, is why Tfr cells express high levels of miR-10a and BCL-6 (which is crucial for Tfr development) (99) if miR-10a represses BCL-6 (44). One possibility is that BCL-6 avoids targeting by miR-10a through alternative polyadenylation. The only computationally predicted miR-10a binding site in the BCL-6 3′UTR is indeed located in the distal part (7). Alternatively, miR-10a might limit BCL-6, whose expression would otherwise be even higher and could therefore inhibit BLIMP-1 expression, which seems to be responsible for IL-10 production in Tfr (99). Thus, the molecular regulation of these seemingly paradoxical differentiation programs awaits further investigation.
In addition to miRNAs enriched in Tregs, we recently investigated the role of miRNAs induced by CD28, which is a critical molecule for Treg homeostasis (100). miR-17-92 was induced in a CD28-dependent manner in Tconv and Treg cells (De Kouchkovsky, Bluestone, and Jeker, manuscript submitted). However, ablation of miR-17-92 in all T cells (47) or specifically in Tregs (De Kouchkovsky, Bluestone, and Jeker, manuscript submitted) did not affect Treg numbers or function, similar to what was observed for miRNAs enriched in Tregs. In contrast, inducing EAE resulted in very severe disease, where miR-17-92-deficient Tregs could hardly control disease (De Kouchkovsky, Bluestone, and Jeker, manuscript submitted). Polyclonal Treg numbers during priming were normal and only modestly reduced in the CNS at peak disease. Although overexpression of miRNAs from the miR-17-92 promotes T-cell proliferation (26, 91), Treg proliferation was surprisingly intact despite miR-17-92-deficiency. Interestingly, one week post priming, antigen-specific Tregs were significantly reduced, whereas tetramer negative cells were present in normal numbers. Thus, miR-17-92 is critical for the accumulation of antigen-specific Tregs. This is likely due to the fact that the antigen-specific Tregs become activated and adapt to the challenges (stress) through proliferation, differentiation, and migration. Since T-cell costimulation is a key process that equips T cells with properties to do so by changing the metabolism and increasing survival signals, we conclude that miR-17-92 is an important part of the CD28 costimulatory pathway. Of note, miR-17-92 inhibits the generation of TGFβ-induced iTregs in vitro (47). This may be in part because miR-17-92 targets the TGFβ receptor II and likely reduces sensitivity to TGFβ. Alternatively, this could reflect that miR-17-92 is induced by costimulatory signals because strong CD28 signals inhibit FoxP3 induction in Tconv cells (69).
The function of miR-17-92 was not limited to accumulation of antigen-specific Tregs though. In the inflamed CNS, both antigen-specific and polyclonal Treg numbers were only marginally reduced, but miR-17-92-deficient Tregs displayed significantly reduced frequencies of IL-10-producing Tregs as compared to miR-17-92-sufficient Tregs (De Kouchkovsky, Bluestone, and Jeker, manuscript submitted). This was most pronounced in mice with severe disease and among antigen-specific Tregs. In this regard, miR-17-92 overexpression did not affect polyclonal Treg numbers but increased Tfr frequencies, while miR-17-92 heterozygosity resulted in reduced Tfr frequencies but normal polyclonal Treg numbers (Baumjohann, Ansel, and Jeker, manuscript submitted). Thus, the dose of miR-17-92 correlated with Tfr frequencies, suggesting that miR-17-92 controls the differentiation of Treg precursors into IL-10-producing effector and follicular Tregs. Thus, IL-2 might preferentially induce B-lymphocyte-induced maturation protein-1 (BLIMP-1), which is required for IL-10 production, in Treg and Tfr cells, which express high levels of CD25 compared to Tfh cells, which downregulate CD25 because IL-2 signaling inhibits Tfh cell generation (101–103). Collectively, these results suggest that miR-17-92 might be required for the differentiation into BLIMP-1-expressing IL-10-producing effector Tregs in general.
T-follicular helper cells
Tfh cell differentiation is initiated shortly after priming of naive T cells by dendritic cells, which induces BCL-6 (104), a major transcription factor required for Tfh differentiation (105–107). BCL-6 regulates a gene expression program including upregulation and downregulation of chemokine receptors that allow the differentiating cell to migrate to GCs where Tfh cells interact with B cells (108). Thus, T-cell activation, micro-anatomic positioning, interactions with different types of APCs and the cytokine milieu are tightly controlled parameters determining Tfh differentiation (109). However, despite substantial recent advances including the discovery that BCL-6 is a major driver of Tfh differentiation (105–107), the molecular networks controlling Tfh differentiation are incompletely understood. The only report on miRNAs in Tfh differentiation suggested that BCL-6 represses the miR-17-92 cluster, which in turn represses CXCR5 expression (106).
Treg and Tfh cells share a number of similarities: (i) CD28 is not only critical for Treg homeostasis (110) but also Tfh cell generation (111), (ii) Tfh and Treg subsets (specifically IL-10-producing effector Tregs) depend on inducible costimulator (ICOS) and IFN regulatory factor 4 (IRF4) for IL-10 production, (iii) the signature cytokine of both cell types, IL-21 and IL-10, respectively, are both STAT3 signaling cytokines, and (iv) Tregs can turn into Tfh cells under certain conditions (112). Based on this interconnectivity, we hypothesized that miR-17-92 may not only be required for effector Treg formation but also Tfh differentiation and/or function. Immunization of mice with protein or infection with lymphocytic choriomeningitis virus (LCMV) demonstrated that miR-17-92 was required for Tfh accumulation (Baumjohann, Ansel, and Jeker, manuscript submitted). T-cell-specific miR-17-92 ablation not only led to reduced Tfh cell but also GC B-cell numbers and a reduced antibody response. Using protein immunization and an adoptive transfer model, we demonstrated that the observed defect was T-cell-intrinsic. In addition, as described above for Tregs, miR-17-92 was largely dispensable for proliferation but led to reduced BCL-6, CXCR5, and IL-21 production. Thus, miR-17-92 is an important regulator of Tfh differentiation.
Transgenic mice overexpressing miR-17-92 in all T and B cells display increased T-cell proliferation and (auto)antibody production (91). Since Tfh cells are major drivers of the GC response, it is possible that the miR-17-92 transgene increased Tfh generation. In support of this notion, CD4cre-mediated miR-17-92 overexpression in T cells led to increased Tfh cells in Peyer’s patches, a site of natural constant microbial stimulation and GC formation (Baumjohann, Ansel, and Jeker, manuscript submitted). Although at first sight contradictory to Yu et al., our findings are not necessarily in conflict to the previously reported findings (106). It is possible that the different experimental approaches (overexpression in a B-cell line versus loss of function in T cells) explain the seemingly contradictory results. Alternatively, dynamic miR-17-92 expression might explain the differences.
We found that PTEN, a known inhibitor of Tfh cell generation (113), was a functionally relevant miR-17-92 target during the first divisions after priming of naive T cells. However, the functional importance of PTEN repression was limited to the early phase of Tfh differentiation. Furthermore, genome-wide gene expression analysis demonstrated that miR-17-92 regulated dozens of genes during Tfh differentiation (Baumjohann, Ansel, and Jeker, manuscript submitted). Thus, miR-17-92 is a key parameter shaping the genetic network of Tfh cells.
One important component of T-cell differentiation is repression of genes (2). In fact, BCL-6 is a transcriptional repressor that is expressed in all T cells shortly after activation (104, 114). Early after initiating the Tfh program, the cells express cytokines associated with Th1, Th2, and Th17 cells (111), yet BCL-6 represses the key transcription factors TBET (encoded by Tbx21), GATA3, and RORc, which drive development of Th1, Th2, and Th17 cells, respectively (111, 115). Thus, BCL-6 represses transcription factors that might interfere with Tfh differentiation. However, repression is incomplete, and Tfh cells retain substantial potential to differentiate into other Th subsets (115). A current model of Tfh differentiation therefore includes substantial flexibility during differentiation (114). Since miRNAs are generally repressors, future studies need to decipher which genes are directly repressed by miR-17-92 during Tfh differentiation.
In summary, miR-17-92 deficiency strongly impaired Tfh differentiation at multiple checkpoints. Early during Tfh differentiation, miR-17-92 repressed the Tfh inhibitor PTEN. Other pathways are very likely also regulated but remain to be investigated.
Complexities in determining the functional roles of miRNAs and their targets
The demonstration of dramatic functional activities of miRNAs has been complicated due to a number of intrinsic miRNA attributes. First, individual miRNAs are redundant, since multiple related miRNA family members act on a similar set of target genes. Thus, effective genetic knockout approaches can require the combination of multiple loss-of-function alleles to observe substantial biological changes (116, 117). In addition, even the deletion of entire miRNA families is often tolerated in the absence of stress signals or ‘perturbagens’ (83, 118–120). Moreover, characterization of target genes regulated by individual miRNAs is particularly demanding. miRNAs generally regulate hundreds of targets with often less than 50% repression (65, 121). Therefore, separating real regulation from technical artifacts or biologic fluctuation is challenging. Since mRNA destabilization is the predominant mechanism of miRNA function in mammals, mRNA arrays, which are readily available to many researchers, can be used as a proxy to determine genetic networks regulated by miRNAs, but they do not discriminate direct from indirect targets (54, 55). Bioinformatic algorithms can help to predict direct targets, and sophisticated proteomics approaches allow genome-wide quantification of miRNA regulation of the proteome (54, 65, 121). Finally, immunoprecipitation-based biochemical assays allow direct genome-wide miRNA target site mapping (54, 68, 122). However, none of these approaches can determine the functional importance of individual miRNA:target interactions. Approaches for functional validation of a target include the demonstration that a miRNA knockout phenocopies transgenic expression of its target and vice versa that miRNA overexpression phenocopies (genetic) inhibition of its target genes. This approach has been used to demonstrate functional importance of miR-150-mediated targeting of c-Myb in B (123) and NK cells (124) and miR-155-mediated SOCS-1 repression in Tregs (78). Importantly, the phenotypes were sensitive to gene dosage, validating that indeed subtle changes in gene expression are functionally relevant. However, it would be premature to infer from these examples that in general a few targets are key, while others are dispensable. For instance, while transgenic miR-17-92 overexpression leads to repression of PTEN and BIM in T cells, genetic Pten/Bim compound mutants only partially phenocopied miR-17-92 overexpression, arguing for additional functionally relevant miR-17-92 targets (91). In addition, genetic rescue approaches have been used to ‘revert’ a phenotype observed in the absence of a given miRNA (79). The underlying assumption is that the loss of miRNA function leads to derepression of its target, which can be neutralized by genetically reducing (heterozygous) or removing (homozygous) the target gene. However, although Pten is a well-documented direct target of miR-17-92, our own genetic rescue experiments suggest that miR-17-92-mediated Pten repression on its own may play a more limited role in Treg and Tfh cells than what could be anticipated based on the literature (de Kouchkovsky, Bluestone, and Jeker, manuscript submitted; Baumjohann, Ansel, and Jeker, manuscript submitted). Finally, the most direct and arguably gold standard method to rigorously test the contribution of a specific miRNA binding site in a specific gene is to generate a knockin mutation of the miRNA binding site. Strikingly, a single miR-155 binding site in the activation-induced cytidine deaminase (AID) gene regulates its mRNA stability (125). Myc-Igh translocations were increased but less than in miR-155-deficient B cells, suggesting that additional miR-155 target genes must be functionally relevant.
These results collectively suggest that minor dysregulation of multiple miRNA targets functionally cooperate to culminate in a phenotypic abnormality in the absence of miRNA repression. We propose that the emphasis to understand miRNA function should be on target networks rather than individual miRNA targets (126, 127). Individual miRNA targets may perhaps dominate in a certain cellular context, but in isolation, they are unlikely to account for an entire phenotype. Although 3′UTR assays are useful to confirm bioinformatic predictions, extrapolations about functional importance from such assays need to be made cautiously. This is particularly true if the 3′UTR assays were done in cell lines rather than the primary cell of interest. Without a doubt, functional validation of miRNA:target gene interactions will be complex and tedious. Given these complexities, interdisciplinary approaches will likely become highly relevant. Although novel approaches such as targeted proteomics are promising (128), new methods to functionally validate multiple targets and combinations thereof need to be developed. The scientific community needs to find ways of dealing with such datasets, and we need to try to avoid over interpretation and perhaps more importantly, exaggerated expectations. After all, we do not expect the networks of genes bound and regulated by individual transcription factors to be individually and functionally validated in a single study. Why would we demand this for miRNAs?
miRNAs set thresholds
Contrary to the prevailing view that miRNAs are mainly fine-tuners of gene expression, single cell analysis shows that on a per cell basis miRNAs can be very effective gene repressors (129). A single miRNA binding site in the 3′UTR of a reporter gene suppresses that gene very effectively. Importantly, repression at low target abundance is more effective than repression at high target expression, leading to a non-linear threshold effect (129). Increasing numbers of miR-20 binding sites (which is part of the miR-17-92 cluster) in the target mRNA ‘sharpened’ the threshold: at high reporter gene expression, a twofold repression was achieved independent of whether the reporter contained one or seven miRNA binding sites. In contrast, at low target levels one miRNA binding site resulted in a twofold repression but seven binding sites resulted in a 10-fold repression (129). The gain in repressive activity was higher when increasing the number of binding sites from one to four than going from four to seven. Thus, miRNAs can set thresholds, particularly if the target gene contains multiple miRNA-binding sites and is expressed at low levels. Since such repression allows to prevent gene expression, this kind of miRNA function has been referred to as binary off-switch (6). In certain situations the same miRNA:target pair could result in an off-switch at low target abundance but fine-tuning activity when the target reaches higher abundance, i.e. represent a threshold-linear effect (129).
During T-cell differentiation, STAT proteins are important in shaping the active enhancer landscape guiding gene expression, which points to cytokines as sensors of environmental cues directing T-cell differentiation (130). As outlined above, after initial activation of naive T cells, the early stages of Th subsets retain substantial flexibility (114). This transitional differentiation stage is likely susceptible to environmental fluctuations. Coherent feed-forward loops are one way to increase robustness of a genetic network by suppressing unintentional responses to environmental changes (130, 131). Setting a miRNA-mediated threshold is another way for a cell to filter real signals from noise (118). Once an intended signal becomes strong enough, the threshold is passed, and a rapid response results (Fig. 4). In contrast, fluctuations below the threshold or insufficient triggering will be buffered without consequences on the protein output or cellular level. Intriguingly, computational models suggest that optimal attenuation of fluctuations is achieved with modest repression (132). Thus, select miRNAs likely represent a safety feature to prevent responses to stochastic triggering of cytokine or other cell surface receptors, for example, and therefore are important for cell fate decisions and maintenance of cellular identity.
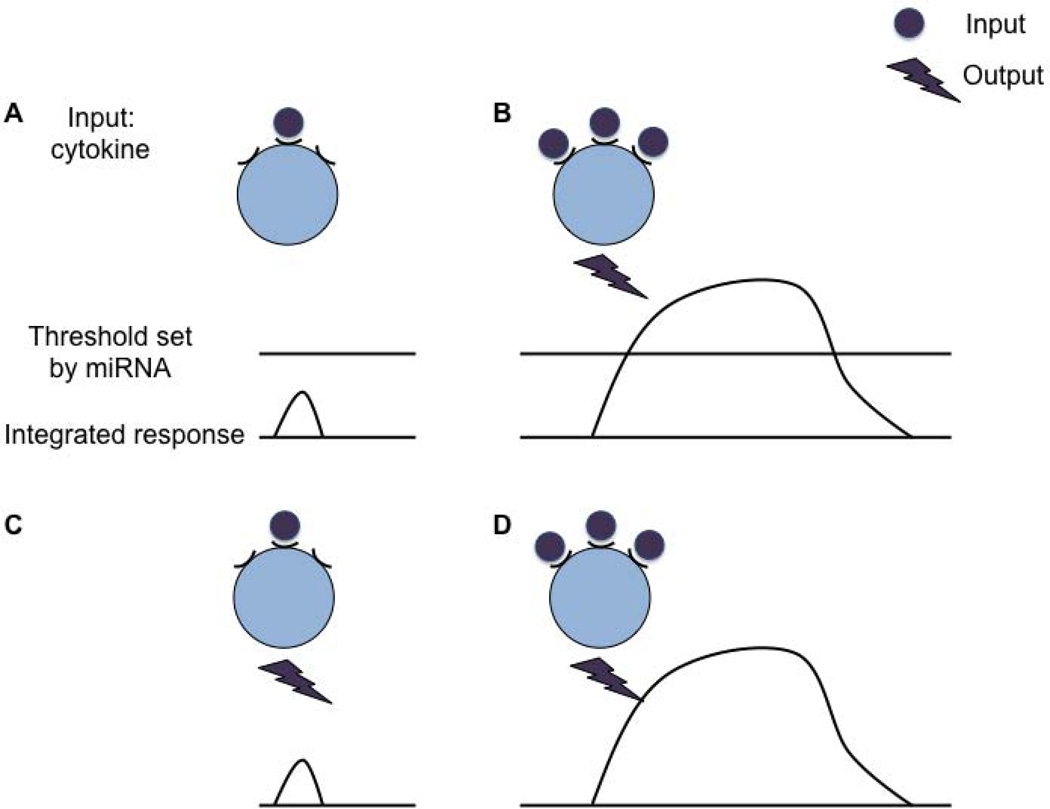
Fig. 4. miRNAs repress stochastic gene expression by setting a threshold.
miRNAs can set genetic thresholds. (A) Random interaction of a cytokine with its receptor results in a subthreshold integrated response, which does not pass the threshold and therefore the cell does not react. (B) Continued or stronger cytokine signals amplify the integrated response which leads to an output once the threshold is passed. (C) In the absence of miRNA regulation, there is no threshold and therefore random cytokine signals lead to a cellular response. (D) In the absence of a miRNA the cell responds to continued or stronger cytokine signals. The response can be the same or intensified.
Illustrations of miRNA-mediated thresholding have been reported in T cells: miR-29 is sufficient to repress the characteristic IFNγ hyperproduction in miRNA-deficient T cells (26). Among many genes regulated by miR-29, two notable ones are Tbx21 and Eomes, both of which are known to induce IFNγ production. Importantly, CD4+ T cells physiologically express very low levels of EOMES. However, inhibition of miR-29 in wildtype CD4+ T cells led to increased EOMES expression, demonstrating that miR-29 normally prevents EOMES expression.
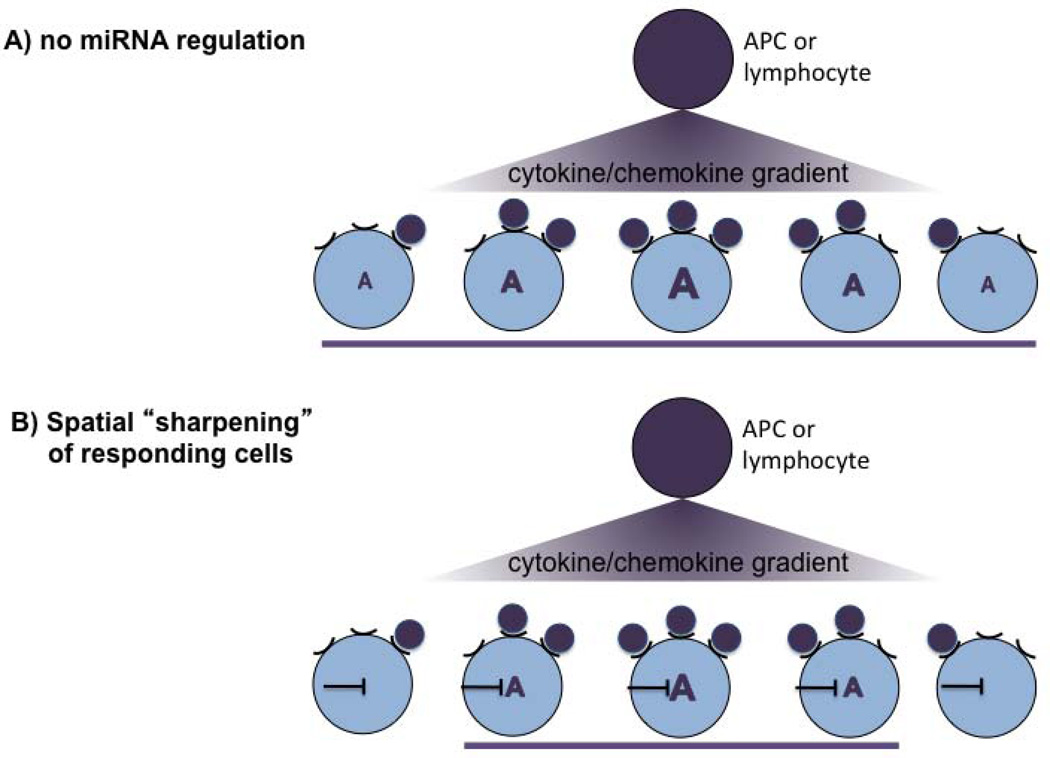
Intriguingly, miRNAs often target genes expressed in their neighboring cells, suggesting that they spatially sharpen gene expression by repressing unwanted genes (58). Translated to the immune system, thresholding is likely important to spatially constrain responsiveness of cells to a given cytokine or chemokine gradient for instance in lymph nodes where positioning in a highly defined microenvironment is critical for Tfh development and GC reactions (Fig. 5). In support of this hypothesis, absence of miR-17-92 led to an eightfold increase in FoxP3-expressing iTregs in response to low TGFβ concentrations, whereas the increase was only twofold with higher concentrations (47). The difference between wildtype and miR-17-92 heterozygous cells was more pronounced than the difference between heterozygous and homozygous miR-17-92-deficient cells, illustrating how highly sensitive the repression is. Collectively, these observations suggest that the role of miR-17-92 in this system is mainly to suppress TGFβ signaling in cells that are exposed to low TGFβ concentrations. The repression is neither linear nor absolute, however, giving the immune system flexibility.
Fig. 5. Spatial sharpening of cells responding to a cytokine/chemokine gradient.
A model how a miRNA-mediated threshold can spatially sharpen responsivness to a chemical gradient. (A) In the absence of a miRNA, all cells that receive a cytokine/chemokine signal respond with gene expression ‘A’. The strength of the response is dose-dependent and correlates with the cytokine/chemokine concentration in the gradient. (B) Presence of a miRNA reduces responsiveness of cells receiving very low signals. This leads to spatial sharpening of the responding cells (purple bar). Thus, although the cell will be exposed to the cytokine and expresses its receptor, it will not react to it. This potentially allows to ‘sharpen’ the functional consequences of a cytokine/chemokine gradient in a given microenvironment without altering the cytokine/chemokine gradient itself.
Cooperative activity of miRNAs potentiates gene regulation
The finding that in a controlled experimental system multiple binding sites are exponentially more effective at repressing its target gene suggests that cooperative binding by several miRNAs will be non-linearly more effective. As an example, PTEN and Bim, two important genes for immune homeostasis (91), contain high numbers of predicted miRNA-binding sites, suggesting significant regulation by miRNAs. Furthermore, two miRNAs expressed in T cells, miR-21 and miR-155, on their own have a small effect on repression of Hip2. In contrast, deletion of both binding sites in the distal 3′UTR of Hip2 restored the expression to levels of the short 3′UTR (46). This demonstrates that interplay between 3′UTR length and cooperative targeting of multiple miRNAs shapes gene expression in T cells.
Private versus public genetic programs regulated by a particular miRNA?
It is currently not clear if a given miRNA has a defined repertoire of targets in each cell type or if the network of targets is cell-type dependent. Distinct networks could be achieved by differential accessibility of the 3′UTR and/or differential alternative polyadenylation between cell types. In addition, gene expression strength likely is important (129). Of note, CD25 is a direct non-canonical miR-17-92 target (68). miR-17-92-deficient Tregs do not display increased CD25 mean fluorescence intensity (MFI) compared to control Tregs (de Kouchkovsky, Bluestone, and Jeker, manuscript submitted), whereas miR-17-92-deficient Tfh have increased CD25 MFI compared to controls (Baumjohann, Ansel, and Jeker, manuscript submitted). This discrepancy could be a result of CD25 expression close to saturation in Tregs (where miRNAs are less efficient), whereas in Tfh cells CD25 levels need to be lower.
Concluding remarks
Regulatory RNAs and in particular miRNAs are emerging as key regulators of T-cell differentiation and function. miRNAs are involved in many aspects of the immune system including differentiation, cytokine signaling, cytokine production, and cellular migration. A fascinating world has opened up begging for more investigation into the intricate regulation of immune function by regulatory RNAs. Studying RNA-mediated gene regulation will continue to reveal the beauty of nature but should ultimately form the basis for a new generation of diagnostics and therapeutics.
Table 1.
Summary of the functions individual miRNAs play in T cells
| miRNA | Function | Reference |
|---|---|---|
| miR-10a | Associated with stable Tregs | (43) |
| Restraining ability of iTreg to convert to Tfh | (44) | |
| miR-29ab | Potent repression of IFN-γ (CD4) | (26) |
| Repression of IFN-γ | (88) | |
| Repression of IFN-γ in a negative feedback loop | (73) | |
| miR-146a | Promotes Treg function in high stress setting and suppresses IFN-γ production in CD4+ effector T cells | (79) |
| Limits T-cell numbers | (80) | |
| Limits CD4+ and CD8+ T-cell activation and numbers by negatively regulating NF-κB pathway in negative feedback loop | (81) | |
| miR-155 | Pleiotropic effects on multiple cell types including regulation of T-helper differentiation, germinal center reaction, and cytokine production | (74) |
| Pleiotropic effects on multiple cell types including B, T, and dendritic cells. Regulation of immune response and cytokine production | (75) | |
| Promotes competitive fitness of Tregs | (78) | |
| Promotes thymic Treg development but is largely dispensable for Treg function | (77) | |
| Cell-intrinsically required for Th1 and Th17 responses | (76) | |
| CD8+ T memory cell formation | (84) | |
| Intrinsic requirement for normal CD8+ T-cell responses to LCMV and L. monocytogenes | (85) | |
| miR-17-92 | Promotes T-cell expansion | (91) |
| Promotes CD4+ T-cell expansion and Th1 function. Inhibits TGFβ-induced iTreg generation | (47) | |
| CD8+ T-cell memory formation (promotes short-lived terminal effector CD8+ T cells, inhibits memory CD8+ T cells | (86) | |
| Required for accumulation of antigen-specific Treg and IL-10 production | de Kouchkovsky et al., manuscript submitted | |
| Regulation of Tfh differentiation and function | Baumjohann et al., manuscript submitted | |
| miR-182 | Induced by IL-2, promotes clonal expansion | (72) |
| miR-301a | Promotes Th17 generation and function | (97) |
| miR-326 | Promotes Th17 generation and function in mice. Association of elevated miR-326 expression in blood with multiple sclerosis | (92) |
Acknowledgements
We thank members of the Bluestone laboratory and of the “UCSF miRNA in lymphocyte group” for discussions. This work was supported by the NIH grants P01 AI35297, U19 AI056388 (to J.A.B.), P30 DK63720 (for core support), and a Scholar Award from the Juvenile Diabetes Research Foundation (to J.A.B.). L.T.J. was supported by the Swiss Foundation for Grants in Biology and Medicine (SFGBM)/SNSF (PASMP3-124274/1) and NIH grant P30 DK63720.
Footnotes
The authors have no conflicts of interest to declare.
References
- 1.O'Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations. Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharp PA. The centrality of RNA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunham I, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suh N, et al. MicroRNA function is globally suppressed in mouse oocytes and early embryos. Current Biol. 2010;20:271–277. doi: 10.1016/j.cub.2009.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma J, et al. MicroRNA activity is suppressed in mouse oocytes. Current Biol. 2010;20:265–270. doi: 10.1016/j.cub.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiang HR, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 13.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Molecular Cell. 2010;38:323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 15.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cifuentes D, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patrick DM, et al. Defective erythroid differentiation in miR-451 mutant mice mediated by 14-3-3zeta. Genes Dev. 2010;24:1614–1619. doi: 10.1101/gad.1942810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cobb BS, et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cobb BS, et al. A role for Dicer in immune regulation. J Exp Med. 2006;203:2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang N, Bevan MJ. Dicer controls CD8+ T-cell activation, migration, and survival. Proc Natl Acad Sci USA. 2010;107:21629–21634. doi: 10.1073/pnas.1016299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ravi A, et al. Proliferation and tumorigenesis of a murine sarcoma cell line in the absence of DICER1. Cancer Cell. 2012;21:848–855. doi: 10.1016/j.ccr.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steiner DF, et al. MicroRNA-29 regulates T-box transcription factors and interferon-gamma production in helper T cells. Immunity. 2011;35:169–181. doi: 10.1016/j.immuni.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chong MM, Rasmussen JP, Rundensky AY, Littman DR. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J Exp Med. 2008;205:2005–2017. doi: 10.1084/jem.20081219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liston A, Lu LF, O'Carroll D, Tarakhovsky A, Rudensky AY. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J Exp Med. 2008 doi: 10.1084/jem.20081062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou X, et al. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med. 2008;205:1993–2004. doi: 10.1084/jem.20080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou X, et al. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nature Immunol. 2009;10:1000–1007. doi: 10.1038/ni.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chatterjee S, Grosshans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009;461:546–549. doi: 10.1038/nature08349. [DOI] [PubMed] [Google Scholar]
- 33.Grosshans H, Chatterjee S. MicroRNases and the regulated degradation of mature animal miRNAs. Adv Esxp Med Biol. 2010;700:140–155. [PubMed] [Google Scholar]
- 34.Ruegger S, Grosshans H. MicroRNA turnover: when how, and why. Trends Biochem Sci. 2012;37:436–446. doi: 10.1016/j.tibs.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 35.Thomas MF, et al. Eri1 regulates microRNA homeostasis and mouse lymphocyte development and antiviral function. Blood. 2012;120:130–142. doi: 10.1182/blood-2011-11-394072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prosser HM, Koike-Yusa H, Cooper JD, Law FC, Bradley A. A resource of vectors and ES cells for targeted deletion of microRNAs in mice. Nat Biotechnol. 2011;29:840–845. doi: 10.1038/nbt.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park CY, et al. A resource for the conditional ablation of microRNAs in the mouse. Cell Reports. 2012;1:385–391. doi: 10.1016/j.celrep.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuchen S, et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity. 2010;32:828–839. doi: 10.1016/j.immuni.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monticelli S, et al. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 2005;6:R71. doi: 10.1186/gb-2005-6-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rossi RL, et al. Distinct microRNA signatures in human lymphocyte subsets and enforcement of the naive state in CD4+ T cells by the microRNA miR-125b. Nat Immunol. 2011;12:796–803. doi: 10.1038/ni.2057. [DOI] [PubMed] [Google Scholar]
- 42.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 43.Jeker LT, et al. MicroRNA 10a Marks Regulatory T Cells. PLoS ONE. 2012;7:e36684. doi: 10.1371/journal.pone.0036684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi H, et al. TGF-beta and retinoic acid induce the microRNA miR-10a, which targets Bcl-6 and constrains the plasticity of helper T cells. Nat Immunol. 2012;13:587–595. doi: 10.1038/ni.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barski A, et al. Chromatin poises miRNA- and protein-coding genes for expression. Genome Res. 2009;19:1742–1751. doi: 10.1101/gr.090951.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang S, et al. Molecular dissection of the miR-17-92 cluster's critical dual roles in promoting Th1 responses and preventing inducible Treg differentiation. Blood. 2011;118:5487–5497. doi: 10.1182/blood-2011-05-355644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu H, et al. miRNA profiling of naive, effector and memory CD8 T cells. PLoS ONE. 2007;2:e1020. doi: 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haasch D, et al. T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene, BIC. Cell Immunol. 2002;217:78–86. doi: 10.1016/s0008-8749(02)00506-3. [DOI] [PubMed] [Google Scholar]
- 50.Bronevetsky Y, et al. T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J Exp Med. 2013;210:417–432. doi: 10.1084/jem.20111717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 52.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 53.O'Carroll D, et al. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev. 2007;21:1999–2004. doi: 10.1101/gad.1565607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hendrickson DG, et al. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bethune J, Artus-Revel CG, Filipowicz W. Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep. 2012;13:716–723. doi: 10.1038/embor.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3'UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 59.Martin G, Gruber AR, Keller W, Zavolan M. Genome-wide analysis of pre-mRNA 3' end processing reveals a decisive role of human cleavage factor I in the regulation of 3' UTR length. Cell Reports. 2012;1:753–763. doi: 10.1016/j.celrep.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 60.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Y, et al. The miR-17-92 cluster expands multipotent hematopoietic progenitors whereas imbalanced expression of its individual oncogenic miRNAs promotes leukemia in mice. Blood. 2012;119:4486–4498. doi: 10.1182/blood-2011-09-378687. [DOI] [PubMed] [Google Scholar]
- 62.Olive V, et al. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mu P, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Farh KK, et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 65.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ziegelbauer JM, Sullivan CS, Ganem D. Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat Genet. 2009;41:130–134. doi: 10.1038/ng.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Loeb GB, et al. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol Cell. 2012;48:760–770. doi: 10.1016/j.molcel.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Benson MJ, Pino-Lagos K, Rosemblatt M, Noelle RJ. All-trans retinoic acid mediates enhanced T reg cell growth, differentiation, and gut homing in the face of high levels of co-stimulation. J Exp Med. 2007;204:1765–1774. doi: 10.1084/jem.20070719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol Cell. 2010;39:373–384. doi: 10.1016/j.molcel.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stittrich AB, et al. The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nat Immunol. 2010;11:1057–1062. doi: 10.1038/ni.1945. [DOI] [PubMed] [Google Scholar]
- 73.Smith KM, et al. miR-29ab1 deficiency identifies a negative feedback loop controlling Th1 bias that is dysregulated in multiple sclerosis. J Immunol. 2012;189:1567–1576. doi: 10.4049/jimmunol.1103171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thai TH, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 75.Rodriguez A, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.O'Connell RM, et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kohlhaas S, Garden OA, Scudamore C, Turner M, Okkenhaug K, Vigorito E. Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells. J Immunol. 2009;182:2578–2582. doi: 10.4049/jimmunol.0803162. [DOI] [PubMed] [Google Scholar]
- 78.Lu LF, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lu LF, et al. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142:914–929. doi: 10.1016/j.cell.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boldin MP, et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med. 2011;208:1189–1201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang L, et al. miR-146a controls the resolution of T cell responses in mice. J Exp Med. 2012;209:1655–1670. doi: 10.1084/jem.20112218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jeker LT, Bluestone JA. Small RNA regulators of T cell-mediated autoimmunity. J Clin Immunol. 2010;30:347–357. doi: 10.1007/s10875-010-9392-7. [DOI] [PubMed] [Google Scholar]
- 84.Tsai CY, Allie SR, Zhang W, Usherwood EJ. MiR-155 affects antiviral effector and effector memory CD8 T cell differentiation. J Virol. 2013;87:2348–2351. doi: 10.1128/JVI.01742-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lind EF, Elford AR, Ohashi PS. Micro-RNA 155 is required for optimal CD8+ T cell responses to acute viral and intracellular bacterial challenges. J Immunol. 2013;190:1210–1216. doi: 10.4049/jimmunol.1202700. [DOI] [PubMed] [Google Scholar]
- 86.Wu T, et al. Temporal expression of microRNA cluster miR-17-92 regulates effector and memory CD8+ T-cell differentiation. Proc Natl Acad Sci USA. 2012;109:9965–9970. doi: 10.1073/pnas.1207327109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huffaker TB, et al. Epistasis between MicroRNAs 155 and 146a during T cell-mediated antitumor immunity. Cell Reports. 2012;2:1697–1709. doi: 10.1016/j.celrep.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ma F, et al. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nat Immunol. 2011;12:861–869. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- 89.Papadopoulou AS, et al. The thymic epithelial microRNA network elevates the threshold for infection-associated thymic involution via miR-29a mediated suppression of the IFN-alpha receptor. Nat Immunol. 2012;13:181–187. doi: 10.1038/ni.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McClymont SA, et al. Plasticity of human regulatory T cells in healthy subjects and patients with type 1 diabetes. J Immunol. 2011;186:3918–3926. doi: 10.4049/jimmunol.1003099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xiao C, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Du C, et al. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10:1252–1259. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- 93.Cox MB, et al. MicroRNAs miR-17 and miR-20a inhibit T cell activation genes and are under-expressed in MS whole blood. PLoS ONE. 2010;5:e12132. doi: 10.1371/journal.pone.0012132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Junker A, et al. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain. 2009;132:3342–3352. doi: 10.1093/brain/awp300. [DOI] [PubMed] [Google Scholar]
- 95.Tufekci KU, Oner MG, Genc S, Genc K. MicroRNAs and multiple sclerosis. Autoimmune Dis. 2011;2011:807426. doi: 10.4061/2011/807426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Junker A, Hohlfeld R, Meinl E. The emerging role of microRNAs in multiple sclerosis. Nat Rev Neurol. 2011;7:56–59. doi: 10.1038/nrneurol.2010.179. [DOI] [PubMed] [Google Scholar]
- 97.Mycko MP, Cichalewska M, Machlanska A, Cwiklinska H, Mariasiewicz M, Selmaj KW. MicroRNA-301a regulation of a T-helper 17 immune response controls autoimmune demyelination. Proc Natl Acad Sci USA. 2012;109:E1248–E1257. doi: 10.1073/pnas.1114325109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Patrick DM, et al. Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J Clin Invest. 2010;120:3912–3916. doi: 10.1172/JCI43604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Linterman MA, et al. Foxp3+ follicular regulatory T cells control the germinal center response. Nat Med. 2011;17:975–982. doi: 10.1038/nm.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Salomon B, et al. B7/CD28 costimulation is essential for the homeostasis of the CD4+CD25+ immunoregulatory T cells that control autoimmune diabetes. Immunity. 2000;12:431–440. doi: 10.1016/s1074-7613(00)80195-8. [DOI] [PubMed] [Google Scholar]
- 101.Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. STAT5 is a potent negative regulator of TFH cell differentiation. J Exp Med. 2012;209:243–250. doi: 10.1084/jem.20111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nurieva RI, et al. STAT5 protein negatively regulates T follicular helper (Tfh) cell generation and function. J Biol Chem. 2012;287:11234–11239. doi: 10.1074/jbc.M111.324046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ballesteros-Tato A, et al. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity. 2012;36:847–856. doi: 10.1016/j.immuni.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Baumjohann D, Okada T, Ansel KM. Cutting Edge: Distinct waves of BCL6 expression during T follicular helper cell development. J Immunol. 2011;187:2089–2092. doi: 10.4049/jimmunol.1101393. [DOI] [PubMed] [Google Scholar]
- 105.Johnston RJ, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yu D, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 107.Nurieva RI, et al. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vinuesa CG, Cyster JG. How T cells earn the follicular rite of passage. Immunity. 2011;35:671–680. doi: 10.1016/j.immuni.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 109.Crotty S. Follicular helper CD4 T cells (TFH) Annu Re Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 110.Bour-Jordan H, Esensten JH, Martinez-Llordella M, Penaranda C, Stumpf M, Bluestone JA. Intrinsic and extrinsic control of peripheral T-cell tolerance by costimulatory molecules of the CD28/ B7 family. Immunol Rev. 2011;241:180–205. doi: 10.1111/j.1600-065X.2011.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Linterman MA, Liston A, Vinuesa CG. T-follicular helper cell differentiation and the co-option of this pathway by non-helper cells. Immunol Rev. 2012;247:143–159. doi: 10.1111/j.1600-065X.2012.01121.x. [DOI] [PubMed] [Google Scholar]
- 112.Tsuji M, et al. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer's patches. Science. 2009;323:1488–1492. doi: 10.1126/science.1169152. [DOI] [PubMed] [Google Scholar]
- 113.Rolf J, et al. Phosphoinositide 3-kinase activity in T cells regulates the magnitude of the germinal center reaction. J Immunol. 2010;185:4042–4052. doi: 10.4049/jimmunol.1001730. [DOI] [PubMed] [Google Scholar]
- 114.Ma CS, Deenick EK, Batten M, Tangye SG. The origins, function, and regulation of T follicular helper cells. J Exp Med. 2012;209:1241–1253. doi: 10.1084/jem.20120994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Luthje K, et al. The development and fate of follicular helper T cells defined by an IL-21 reporter mouse. Nat Immunol. 2012;13:491–498. doi: 10.1038/ni.2261. [DOI] [PubMed] [Google Scholar]
- 116.Park CY, Choi YS, McManus MT. Analysis of microRNA knockouts in mice. Hum Mol Genet. 2010;19:R169–R175. doi: 10.1093/hmg/ddq367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ventura A, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Alvarez-Saavedra E, Horvitz HR. Many families of C. elegans microRNAs are not essential for development or viability. Curr Biol. 2010;20:367–373. doi: 10.1016/j.cub.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 122.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xiao C, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 124.Bezman NA, Chakraborty T, Bender T, Lanier LL. miR-150 regulates the development of NK and iNKT cells. J Esp Med. 2011;208:2717–2731. doi: 10.1084/jem.20111386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dorsett Y, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Subramanyam D, et al. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol. 2011;29:443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chou J, Lin JH, Brenot A, Kim JW, Provot S, Werb Z. GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA-29b expression. Nat Cell Biol. 2012;15:201–213. doi: 10.1038/ncb2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jovanovic M, et al. A quantitative targeted proteomics approach to validate predicted microRNA targets in C. elegans. Nat Methods. 2010;7:837–842. doi: 10.1038/nmeth.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, van Oudenaarden A. MicroRNAs can generate thresholds in target gene expression. Nat Genet. 2011;43:854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Vahedi G, et al. STATs Shape the Active Enhancer Landscape of T Cell Populations. Cell. 2012;151:981–993. doi: 10.1016/j.cell.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- 132.Osella M, Bosia C, Cora D, Caselle M. The role of incoherent microRNA-mediated feedforward loops in noise buffering. PLoS Comput Biol. 2011;7:e1001101. doi: 10.1371/journal.pcbi.1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]