Abstract
Functional connectivity mapping with resting-state magnetic resonance imaging (MRI) has become an immensely powerful technique that provides insight into both normal cognitive function and disruptions linked to neurological disorders. Traditionally, connectivity is mapped using data from an entire scan (minutes), but it is well known that cognitive processes occur on much shorter time scales (seconds). Recent studies have demonstrated that the correlation between the blood oxygenation level-dependent (BOLD) MRI signal from different areas varies over time, motivating a further exploration of these fluctuations in apparent connectivity. However, it has also been shown that similar changes in correlation can arise when the timing relationships between voxels are randomized (Handwerker et al., 2012). In this work, we show that functional connectivity in the anesthetized rat exhibits dynamic properties that are similar to those previously observed in awake humans (Chang and Glover, 2010) and anesthetized monkeys (Hutchison et al., 2012). Sliding window correlation between BOLD time courses obtained from bilateral cortical and subcortical regions of interest results in periods of variable positive and negative correlation for most pairs of areas except homologous areas in opposite hemispheres, which exhibit a primarily positive correlation. A comparison with sliding window correlation of randomly matched time courses suggests that with the exception of homologous areas and sensorimotor connections, the dynamics cannot be distinguished from random fluctuations in correlation over time, supporting the idea that some of these dynamic patterns may be due to inherent properties of the signal rather than variations in neural coherence. Within the pairs of areas where the dynamics are most different from those of randomly matched time courses, ten common patterns of connectivity are identified, and their occurrence as a function of time is plotted for all animals. The observation of time-varying correlation in the rodent model will facilitate the future multimodal experiments needed to determine whether the changes in apparent connectivity are linked to underlying neural variability.
Key words: dynamic, functional connectivity, resting state MRI, rodent, sliding window correlation somatosensory cortex
Introduction
Functional connectivity mapping with resting-state magnetic resonance imaging (rsMRI) has become a popular and powerful technique for investigating the role of network activity in both normal brain function and neurological disorders. Until recently, functional connectivity was assumed to be stationary, with network connections calculated using data from the entire scan (5–10 min) (see, for example, Biswal et al., 1995; Cordes et al., 2000; Lowe et al., 1998). However, cognitive processes are known to occur on much shorter time scales, motivating a recent work that explores the dynamic properties of the spontaneous blood oxygenation level dependent (BOLD) signal fluctuations (Chang and Glover, 2010; Hutchison et al., 2012; Majeed et al., 2009, 2011).
One of the earliest studies examining the dynamics of the spontaneous BOLD signal demonstrated that the BOLD signals exhibit intrinsic spatiotemporal organization in both rats (Majeed et al., 2009) and humans (Majeed et al., 2011), showing that there are aspects of activity which are not captured by traditional analysis techniques that examine only “steady-state” relationships. Chang and Glover (2010) looked specifically at the relationship between the posterior cingulate cortex (PCC) and other brain areas, demonstrating that the correlation between these areas varied over time and that this temporal variability was greater than would be expected by chance. A recent report by Hutchison and colleagues demonstrates that similar dynamics occur in humans and in anesthetized monkeys. Since random thought processes are not expected to be present during anesthesia, their work suggests that these dynamics are not related to changing cognitive states (Hutchison et al., 2012). However, other physiological origins are not excluded, and the cause of the time-varying correlation remains unknown. A recent work by Handwerker and colleagues found that variations in correlation over time exhibit periodic properties and can be observed even when the timing relationships between voxels are randomized (Handwerker et al., 2012).
To further explore the properties and potential sources of fluctuations in apparent connectivity, we implement sliding window analysis in the rodent model of functional connectivity to examine a well-characterized network containing the bilateral sensorimotor cortex and the caudate putamen. The sensory and motor areas are strongly connected across hemispheres via the corpus callosum and exhibit high BOLD correlation when typical “steady-state” analysis methods are used (Pawela et al., 2008; Williams et al., 2010; Zhao et al., 2008). In contrast, the left and right caudate putamen (CP) are only indirectly connected but still exhibit BOLD-based functional connectivity (Williams et al., 2010; Zhao et al., 2008). The caudate also receives input from somatosensory cortex, but a signal from the two areas is usually not correlated in functional connectivity studies. The variety of anatomical and functional connectivity present in this network makes it an excellent setting in which to explore BOLD dynamics.
One of the challenges in dynamic analysis lies in determining the physiological significance of the time-varying relationships between areas. Two possible approaches suggest themselves: first, linking the dynamic patterns to an external measurement of behavior; and second, finding a direct neural analog of the changes in functional connectivity. The first of these possibilities is addressed in part by a recent paper which shows that in human subjects, network relationships within a short 12 second window before the onset of a psychomotor vigilance task predict performance on that task (Thompson et al., 2012), a finding that is consistent with previous work linking behavioral variability to functional connectivity and network activity calculated from entire scans (e.g., Fox et al., 2007; Kelly et al., 2008; Li et al., 2007). The second possibility is appealing in that it would directly link the BOLD signal to electrical measures of neural activity, but it is difficult to perform in humans. Electrical measurements in normal subjects are limited to noninvasive surface electrodes with poor spatial resolution and depth penetration, and although intracortical recordings can be obtained in special patient populations, they are constrained to areas of clinical interest and may reflect pathological function. One of the benefits of characterizing BOLD dynamics in the rat is that future studies combining simultaneous MRI and microelectrode recording can place the BOLD dynamics on a firm neural footing (Pan et al., 2010, 2011).
As a first step toward these multimodal experiments, we obtained resting-state MRI from a single coronal slice in the rat brain containing the sensorimotor cortex and the caudate putamen. Relationships between cortical and subcortical regions of interest were calculated using a traditional static approach and compared with sliding window correlation. The results show that all pairs of areas exhibit substantial variations in correlation, with correlation remaining mostly positive between homologous cortical areas but exhibiting periods of strong correlation and anti-correlation in other areas. The findings are consistent with the variations previously reported in humans and macaques, showing that changes in BOLD correlation over time are not limited to a particular species or network. However, as in the recent report by Handwerker and colleagues, a similar variability in correlation was observed in time courses that were randomly matched across scans, indicating that care should be taken when interpreting dynamics as indicative of neural processes.
Materials and Methods
All experiments were performed following guidelines set by the Institutional Animal Care and Use Committee (IACUC) of Emory University. Three scans each from four male Sprague–Dawley rats (200–300 g) were chosen from data acquired for another study (preparation described briefly below). These scans represent the best specimens collected for the study and were selected based on image quality, lack of motion, physiological stability, and a clear presentation of functional connectivity in somatosensory areas. The latter was determined by a preliminary analysis for each scan, indicating that cross-correlation based on a seed manually chosen in primary somatosensory cortex (SI) resulted in a typical pattern of localized bilateral correlation (Pawela et al., 2008; Williams et al., 2010; Zhao et al., 2008).
Animal preparation
Each rat was anesthetized with 2% isoflurane mixed with 1:1 oxygen and room air, and maintained under anesthesia for 2.5 h to replicate the time allotted for surgeries in our other studies. After this “wait period,” isoflurane was reduced to 1.5% for 30 additional minutes before discontinuing isoflurane and switching to a subcutaneous infusion of dexmedetomidine. Heart rate and blood oxygen saturation percentage were recorded with a pulse oximeter placed on the rear left paw. Body temperature was monitored with a rectal thermometer and maintained at approximately 37°C (±0.5°C) using an adjustable warm water pad. Respiratory rate was also monitored by using a pressure-sensitive pad placed under the rat's chest. The rat was then placed in the MRI cradle, and the head was secured with a bite bar and ear bars.
After setup was completed, the rat was given a subcutaneous bolus injection of 0.025 mg/kg dexmedetomidine (Dexdomitor, Pfizer, Karlsruhe, Germany). Five minutes after the bolus, isoflurane was discontinued, and 15 min later, a subcutaneous infusion of 0.05 mg/kg/h dexmedetomidine was initiated to maintain anesthesia for the duration of the experiment (Weber et al., 2006). Approximately 80 min after the initial dexmedetomidine bolus, the infusion dosage was increased to 0.15 mg/kg/h (3× initial infusion rate) for maintaining anesthetic depth, in accordance with the protocol established in (Pawela et al., 2009).
Image acquisition and processing
All images were acquired on a 20 cm bore 9.4 T Bruker BioSpec magnet interfaced to an AVANCE (Bruker, Billerica, MA) console. An actively decoupled imaging protocol was used, with a 7 cm volume coil for RF transmission and a 2 cm surface coil for signal reception. A FLASH image was acquired in three planes, and a single slice was positioned over the primary somatosensory cortex based on known anatomical markers. Manual shimming was conducted on this slice to obtain maximum signal to noise ratio (SNR) and spatial homogeneity. Each resting state scan was acquired using a single-shot gradient echo echo planar imaging sequence with the following parameters: Repetitions=1000, relaxation time=500 ms, echo time=15 ms, total scan time=8 min 20 sec, slice thickness=2 mm, field of view=2.56 cm×2.56 cm, matrix size=64×64. Approximately ten resting-state scans were acquired from a total of seven animals for the original study. For the analysis performed in this work, we used three resting-state scans each from the four best rats collected at the 2.5+h time point following the cessation of isoflurane, because we have previously found that functional connectivity measurements stabilize after that time. For the period when the scans were acquired for this analysis, the average heart rate was 300–310 bpm; respiratory rate was 70–80 breaths per minute; oxygenation was 98%–99%; and body temperature was 37°C–37.5°C.
All functional MRI data processing and analysis was performed using code written in MATLAB (MathWorks, Natick, MA). The time course from each voxel was linearly detrended, followed by finite impulse response band-pass filtering between 0.01–0.3 Hz based on previous work demonstrating correlation over a wide range of frequencies in the anesthetized rat (Magnuson et al., 2010; Majeed et al., 2009, 2011). Data points were removed from the beginning of each scan to reduce any transient effects of scanner instability, and again after filtering to reduce filter effects, leaving a total of 800 images for each scan.
“Static” functional connectivity analysis
For each rat, regions of interest (2×2 voxels) were manually selected in left and right primary somatosensory cortex (SI), secondary somatosensory cortex (SII), motor cortex (MI), and CP by comparison with an atlas (Paxinos and Watson, 1998) for a total of eight sites. The average time course from each region of interest (ROI) was calculated, and correlation and partial correlation were calculated pairwise between all nodes. Partial correlation between two ROIs controls for common inputs to the two from the remaining ROIs (i.e., calculates the correlation between the residuals after linear regression of the control ROIs) and should result in reduced sensitivity to external inputs.
Sliding window analysis
Using the same data, sliding window correlation was performed using window lengths of 25, 50, and 100 images (12.5, 25, or 50 sec). The longest of the windows is comparable to that previously used in the monkey (Hutchison et al., 2012). The relatively high temporal resolution of our scan provided increased sampling density and allowed us to examine shorter time windows than in previous studies.
For each window length, the correlation for each pair of areas was plotted as a function of time. The time courses were then segmented into strong positive (>0.4), moderate positive (0.2 to 0.4), weak (−0.2 to 0.2), moderate negative (−0.2 to −0.4), and strong negative (<−0.4) correlation. These thresholds were chosen based on a preliminary examination of the time courses of correlation, with the goal of simplifying analysis while preserving some information about dynamic range. States that were present for fewer than 10 images (5 s) were merged with the preceding state to minimize the effect of short-lived excursions to neighboring correlation ranges that are artificially exacerbated by the thresholding process. To quantify the stationarity of the correlation time courses, the number of transitions between states was determined for each scan from each rat, and the average and standard deviation were calculated for each pair of areas. For each scan and each pair of areas, the relative percentage of time spent in each state was determined, and averages were obtained for each pair of areas. Mirrored pairs (e.g., left SI to right MI and right SI to left MI; left SI to left MI and right SI to right MI; all pairs except those consisting of homologous areas in left and right hemispheres) were averaged for subsequent analysis to minimize comparisons and increase signal to noise, under the assumption that connectivity is bilaterally symmetric for this network. We also calculated the distribution of sliding window correlation coefficients for each pair of areas.
A control analysis was performed to determine whether results were due to inherent signal properties and/or preprocessing. Sliding window correlation was performed for the same areas but with time courses chosen randomly from different scans, so that any relationships between areas arise solely from the properties of the signals themselves rather than from underlying changes in neural coherence. A data set equal in size to the actual experimental data was derived from the randomized comparisons (i.e., 12 data points for each pair of areas), and the same procedures were followed to calculate the number of transitions and length of time spent in each state. The steady-state correlation and distribution of sliding window correlation coefficients were also calculated.
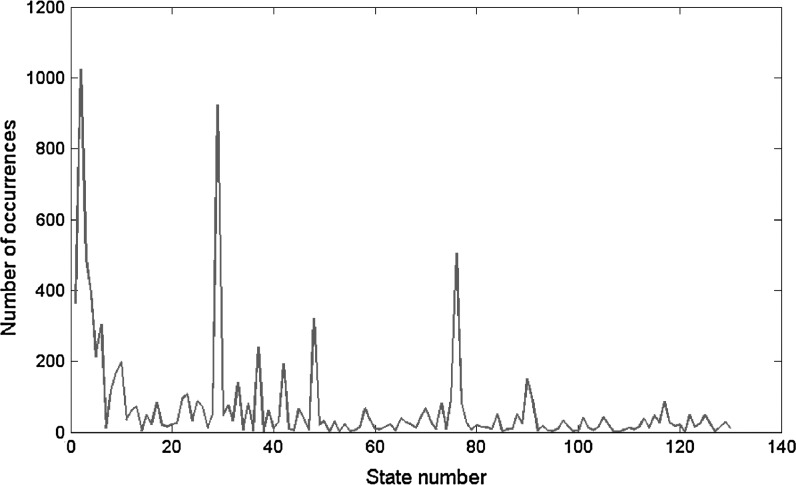
Real data from pairs of areas in which the duration of multiple states fell outside of one standard deviation from those observed in the randomly matched data were chosen for further analysis. The time courses were further simplified into three states (positive correlation (>0.2), no correlation, and negative correlation (<−0.2)) to facilitate analysis. This data reduction resulted in 729 possible combinations of the three states for the six pairs of sites, as compared with the 15,625 possible combinations for five states and six site pairs. Of these, one hundred and thirty unique combinations were detected in the concatenated time courses from all rats. The number of instances for each combination was tabulated, and the top ten patterns (>200 occurrences) were chosen for further examination. The amount of time spent in each of these patterns was plotted for the concatenated time courses from all rats.
Results
Correlation and partial correlation
Cross correlation and partial correlation were calculated between all ROIs and averaged over all scans from all rats (Table 1). Voxels with correlation values above 0.2 (corresponding to approximately p<0.001; p<0.03 with Bonferroni correction for multiple comparisons) were considered connected and are highlighted in the table. As expected, the strongest correlations occur between homologous areas in the left and right cortex. Left SII and right SII exhibit the highest correlation (0.56±0.18), while left and right CP have the weakest correlation of the homologous areas (0.25±0.13). These results are similar to those seen in previous works (Williams et al., 2010). Three other pairs of areas exhibit correlation above the threshold: left SI and left MI (0.22±0.2), right SI and right MI (0.37±0.16), and right MI and left SI (0.31±0.19). The mirror of the last pair, left MI and right SI, is more strongly correlated than the remaining pairs of areas but has a correlation below the threshold (0.16±0.14). Steady-state correlation values calculated from the randomly matched time courses ranged from−0.03 to 0.05, with standard deviations of 0.03 to 0.09.
Table 1.
The Average and Standard Deviation of Cross Correlation (Top Half of Table) and Partial Correlation (Bottom Half of Table) for All Pairs of Areas from All Scans
| LSII | LSI | LMI | LCP | RSII | RSI | RMI | RCP | |
|---|---|---|---|---|---|---|---|---|
| LSII | 1 | −0.01±0.06 | −0.1±0.12 | 0.07±0.06 | 0.56±0.18 | 0.02±0.08 | −0.06±0.05 | 0.03±0.01 |
| LSI | 0±0.05 | 1 | 0.22±0.2 | 0.02±0.05 | 0.05±0.11 | 0.49±0.12 | 0.31±0.19 | 0.05±0.04 |
| LMI | −0.06±0.11 | 0.13±0.12 | 1 | 0.02±0.11 | −0.06±0.12 | 0.16±0.14 | 0.36±0.17 | 0.05±0.12 |
| LCP | 0.06±0.1 | −0.02±0.05 | 0.03±0.07 | 1 | 0.05±0.08 | 0.02±0.03 | 0.04±0.04 | 0.25±0.13 |
| RSII | 0.56±0.18 | 0.02±0.06 | − 0.03±0.07 | − 0.01±0.11 | 1 | 0.06±0.12 | −0.01±0.14 | 0.05±0.08 |
| RSI | 0.03±0.06 | 0.38±0.1 | 0.01±0.11 | 0.01±0.03 | 0.02±0.08 | 1 | 0.37±0.16 | 0.03±0.05 |
| RMI | −0.01±0.05 | 0.1±0.14 | 0.29±0.11 | 0.01±0.02 | −0.01±0.09 | 0.26±0.12 | 1 | 0.04±0.12 |
| RCP | −0.03±0.05 | 0.03±0.04 | 0.05±0.07 | 0.23±0.13 | 0.05±0.07 | 0.01±0.05 | 0.01±0.11 | 1 |
Values for homologous areas in the left and right hemispheres are marked with dark gray cells, and nonhomologous pairs with values above 0.2 are highlighted in light gray. The dark boxes along the diagonal represent the correlation of an area with itself and are the dividing line between the upper and lower halves of the table.
Partial correlation, shown in the bottom half of the table, displays a similar pattern of connections, although the values are reduced and two of the nonhomologous pairs are no longer above the threshold. It is interesting to note that partial correlation for left and right SII and for left and right CP is nearly the same as the cross correlation, while the values for left and right SI and left and right MI are reduced. The areas where partial correlation values are reduced may receive more input from the other areas in this network than the areas where partial correlation is equivalent to cross correlation. This is consistent with our previous work which presents an intrinsic pattern of BOLD signal propagation from lateral to medial areas that suggests some level of a driving influence (Majeed et al., 2009, 2011).
Sliding window correlation
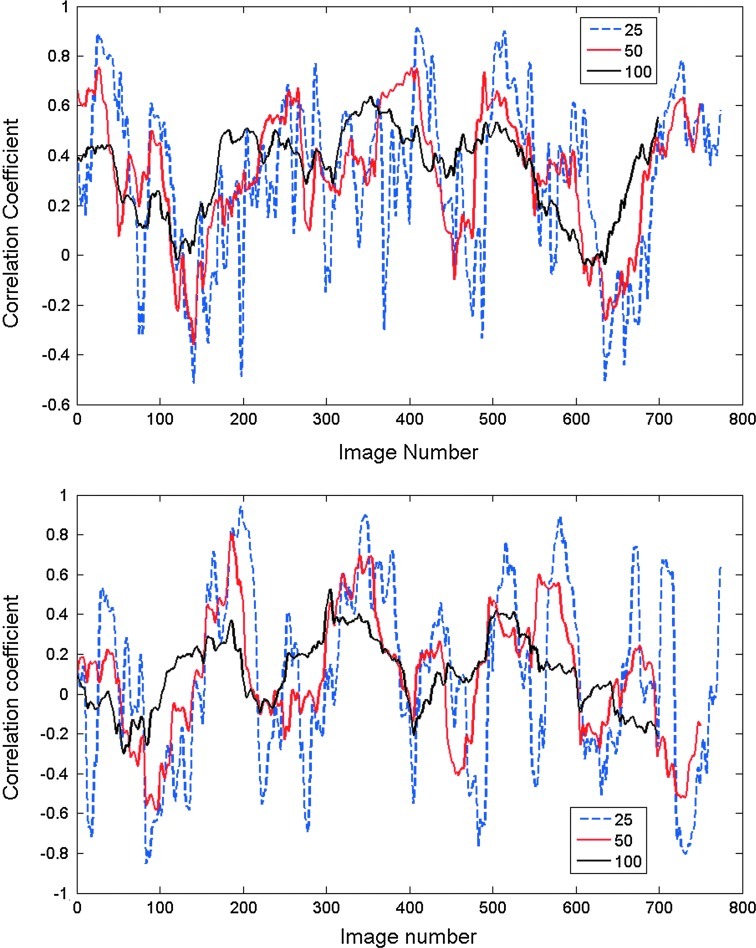
Correlation as a function of time was plotted for each scan using window lengths of 25, 50, or 100 images. All correlation time courses exhibited variance over time, with greater variance observed when shorter window lengths were used. These results are consistent with previous findings in the human (Chang and Glover, 2010) and monkey (Hutchison et al., 2012). In general, correlation time courses from homologous areas varied between no correlation and strong positive correlation, while time courses from other areas exhibited greater fluctuation from strong anticorrelation to strong positive correlation. Two examples from the first scan of the first rat are shown in Figure 1.
FIG. 1.
Sliding window correlation between left MI and right MI (top) and between left SII and left SI (bottom) for the first scan from the first rat. Correlation is plotted as a function of image number for window lengths of 25, 50, or 100 images (12.5, 25, or 50 sec). As window length increases, the variability in the correlation time course decreases. Correlation between left and right SI is mostly positive, while correlation between left SII and left SI is more variable and ranges from strongly negative to strongly positive, particularly for short window lengths.
Temporal segmentation
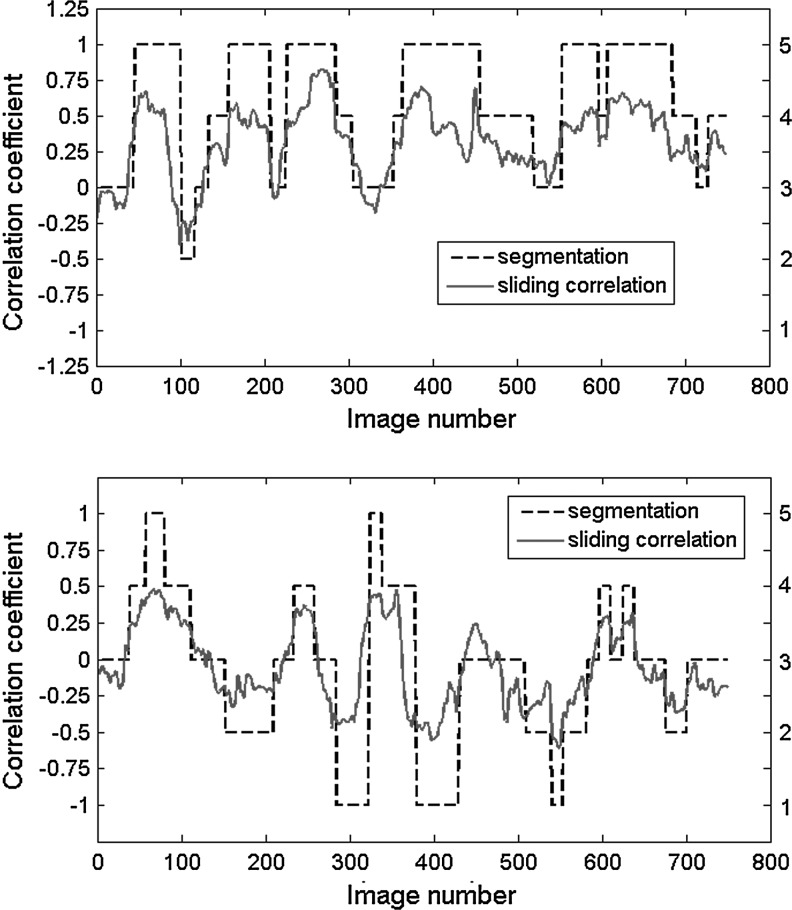
To further examine the dynamic aspects of connectivity, each correlation time course was segmented into five possible states: strong anticorrelation (<− 0.4), moderate anticorrelation (−0.2 to −0.4), weak or no correlation (−0.2 to 0.2), moderate correlation (0.2 to 0.4), and strong correlation (>0.4). Examples for left and right SI and for left SI and left CP are shown for a randomly chosen rat in Figure 2, using a window length of 50. As in Figure 1, the homologous SI areas exhibit mostly positive correlation, while the time course for left SI and CP ranges from strong anticorrelation to strong correlation. Similar segmented time courses were created for window lengths of 25 and 100.
FIG. 2.
Sliding window correlation (window length 50) for left and right SI (top) and left SI and left caudate putamen (CP) (bottom) for a randomly chosen rat. Each time course was segmented into 5 possible states (1=strong anticorrelation, 3=no correlation, 5=strong correlation), plotted along the right vertical axis. The time course for left and right SI falls mostly into the strong, moderate, or weak correlation categories, while the time course for left SI and CP ranges from strong anticorrelation to strong correlation.
Based on the segmented time courses, the number of transitions between states was calculated for each pair of areas and averaged across all rats. The results for a window length of 50 are reported in Table 2. Most pairs make 15–17 transitions over the course of the scan, with strongly correlated areas such as left and right SII making fewer transitions. This pattern was preserved for the other window lengths, although the number of transitions varied (15–18 for window length of 25; 7–10 for window length of 100; data not shown).
Table 2.
The Average and Standard Deviation for the Number of Transitions for All Scans (Window Length of 50)
| LSII | LSI | LMI | LCP | RSII | RSI | RMI | RCP | |
|---|---|---|---|---|---|---|---|---|
| LSII | 17.2±4 | 15.6±1.8 | 16.1±1.9 | 7.3±6.3 | 16.4±3.7 | 17.3±3.6 | 15.8±4.9 | |
| LSI | 15.1±5 | 16±4.6 | 15.9±2.8 | 11.6±4.4 | 15.3±5.4 | 16.3±2.1 | ||
| LMI | 17.8±3 | 17.5±2.1 | 16.3±2.3 | 13.5±4.1 | 16.1±2.5 | |||
| LCP | 16.3±3.1 | 17.5±3 | 17.7±2.1 | 16.2±5.2 | ||||
| RSII | 17.3±3.9 | 16±2.7 | 16.2±3.8 | |||||
| RSI | 13.3±5.4 | 17±3 | ||||||
| RMI | 16.7±2.9 | |||||||
| RCP |
Homologous areas are highlighted in light gray. Homologous cortical areas exhibit the lowest number of transitions, particularly in the strongly connected SII regions, while left and right caudate putamen (CP) have a higher number of transitions. Right SI and right MI also exhibit fewer transitions than most areas. A similar reduction in the number of transitions is observed in homologous cortical areas when window lengths of 25 or 100 are used (data not shown).
The segmented time courses and number of transitions were also calculated for the randomly matched data. For a window length of 25, the number of transitions was 16.3±0.9 for 25; for a window length of 50, 16.6±1; and for a window length of 100, 8±1.
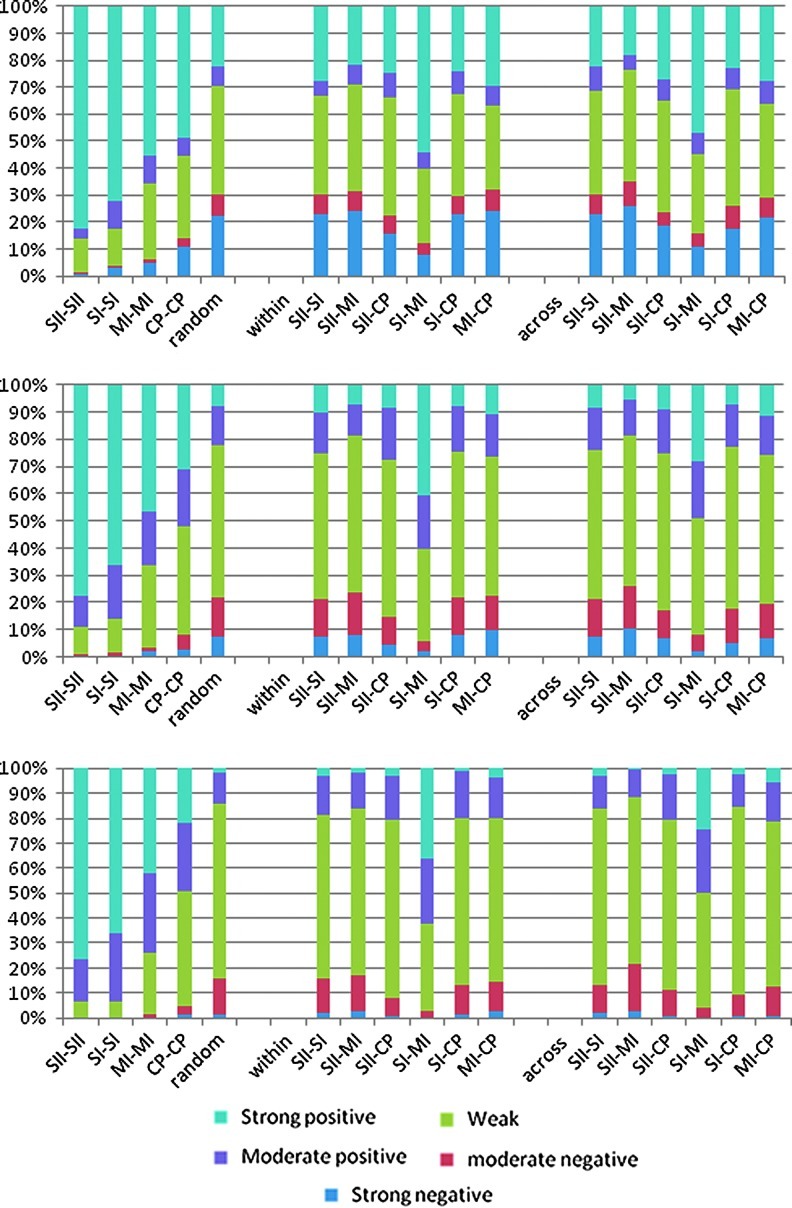
Based on the segmented time courses, the amount of time spent in each of the five states was calculated for each pair of areas and averaged across rats. Results for each of the three window lengths are shown in Figure 3. For most pairs of areas, the pattern is similar to that of random data, summarized in Table 3. In general, the amount of time spent in a state with weak to no relationship increases as window length increases, while the amount of time spent in a strongly correlated or anticorrelated state decreases (for example, ipsilateral SI-SII are uncorrelated 37% of the time when a window length of 25 is used; 53% for a length of 50; and 66% for a length of 100). The exceptions are the homologous pairs and within- and across-hemisphere connections between MI and SI, which exhibit increasingly positive correlation as window length increases. The relationships for all areas are consistent across window lengths and suggest that the apparent increase in variability observed with shorter windows is an artifact of the properties of the signal rather than an indication of increasing sensitivity to short-lived states. A window size of 50 was used for the remaining analysis.
FIG. 3.
Relative time spent in each state for each pair of areas, averaged over all scans, for window lengths of 25 (top), 50 (middle), and 100 (bottom). The first group of areas are bilateral homologues; the second are within-hemisphere pairs; and the third are cross-hemisphere pairs. Values for randomly matched data are shown next to the homologous pairs for comparison. In general, the amount of time spent in a state with weak to no relationship increases as window length increases, while the amount of time spent in a strongly correlated or anticorrelated state decreases. For most pairs of areas, the pattern is similar to that of random data. The exceptions are the homologous pairs and within- and across-hemisphere connections between MI and SI.
Table 3.
Average and Standard Deviation for the Relative Amount of Time Spent in Each State for Randomly Matched Data Analyzed with Three Different Window Lengths
| Window length | Strong anticorrelation | Moderate anticorrelation | Weak | Moderate correlation | Strong correlation |
|---|---|---|---|---|---|
| 25 | 22.5%±2.5% | 7.4%±1.7% | 40.7%±3.7% | 7.1%±1.4% | 22.2%±3% |
| 50 | 7.4%±1.9% | 14.4%±2.7% | 56.3%±3.5% | 14.4%±2.3% | 7.8%±1.9% |
| 100 | 1.6%±1% | 13.6%±2.5% | 69.2%±3.4% | 12.3%±2.8% | 1.5%±1% |
Correlation histograms
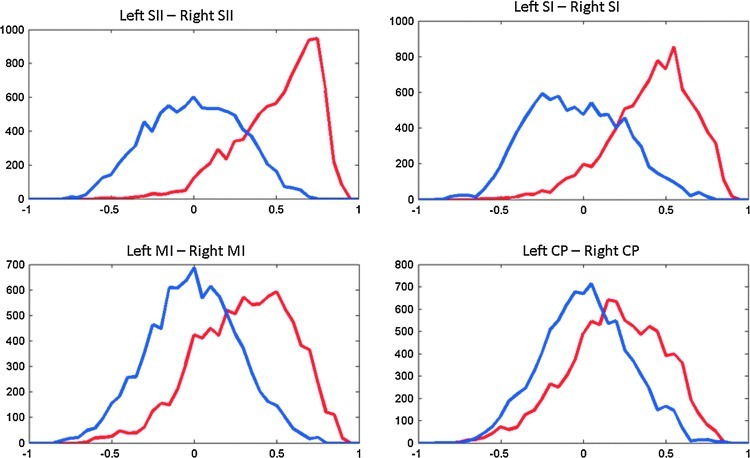
To more closely examine the distribution of correlation coefficients resulting from sliding window analysis, a histogram of the correlation coefficients for each pair of areas from both real and randomly matched data was calculated and is shown in Figure 4. As expected, the correlation histograms for real and randomly matched data are very similar for most pairs of areas. In the areas that exhibited differences from random on the state-length analysis, however, the histograms are also different. Especially for areas with very high correlation (left and right SII, for example), the histograms are skewed rather than simply shifted, which may indicate a difference in the underlying dynamics rather than a simple positive offset.
FIG. 4.
Histograms of correlation coefficients for homologous areas in left and right hemispheres after sliding window correlation with a window length of 50 images (all rats, all scans). Correlation value is plotted along the x axis, and number of instances is plotted along the y axis. Real data are shown in red, and results for randomly matched time courses are shown in blue. All histograms are skewed toward positive values, particularly for SII.
Occurrence of common states
For the remaining examination of network dynamics, only the six pairs of areas that exhibited correlation time courses that were clearly different from those of the randomly matched areas were retained. To further simplify analysis, the five correlation states were compressed to three: positive correlation (>0.2), weak correlation (−0.2 to 0.2), and negative correlation (<− 0.2). This resulted in a pattern of connectivity at each time point which was represented by a 6-element vector containing 1, 0, and −1 sec. For example, using the same ordering shown in Table 4, [1 1 1–1 0 0] would indicate that left and right SI, SII, and MI were positively correlated; left and right CP were negatively correlated; and both SI-MI pairs were uncorrelated at a given point in time. The time courses from all scans were concatenated, and a count of the number of occurrences for each unique pattern was obtained (Fig. 5). A total of 130 unique patterns were detected. We identified the ten most common patterns, all of which occurred at least 200 times throughout the concatenated time course. These patterns are shown in Table 4. The most common pattern consists of positive correlation for all area pairs (1024 occurrences), closely followed by positive correlation for all pairs other than CP (925 occurrences). No negative correlation was observed in the most common patterns. Left and right SI were positively connected in all common patterns, with the remaining areas showing varying degrees of stability. The greatest variability was observed in the within- and across-hemisphere SI-MI pairs.
Table 4.
The Ten Most Common Correlation Patterns Observed Using Sliding Window Correlation, Ranked by Occurrence
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| SII-SII | ||||||||||
| SI-SI | ||||||||||
| MI-MI | ||||||||||
| CP-CP | ||||||||||
| SI-MI within | ||||||||||
| SI-MI across | ||||||||||
| Total occurrences | 1024 | 925 | 506 | 487 | 391 | 365 | 321 | 303 | 242 | 214 |
A white cell indicates positive correlation; a black cell, weak or no correlation. No negative correlation was present in the most common patterns. Left and right SI are positively correlated in all patterns. The within-hemisphere and across-hemisphere SI-MI pairs exhibit the greatest variation.
FIG. 5.
Plot of the number of occurrences for each of the 130 unique states detected in the concatenated, segmented time series for all scans. The states are numbered in the order of detection. Ten states occurred more than 200 times and are examined in greater detail in the following table and figure.
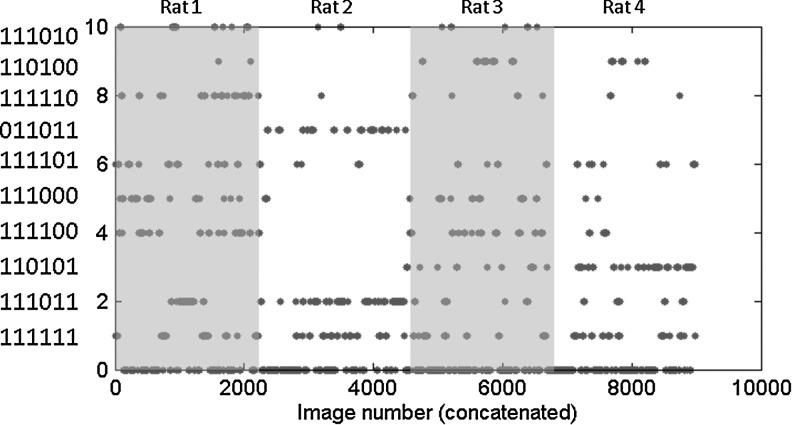
A plot of the pattern of correlation as a function of image number for the concatenated time series from all rats is given in Figure 6. The most common patterns are present in all rats, while some of the less common states are unevenly distributed. Patterns 1, 2, and 6 are spread across all rats; while patterns 3, 4, and 5 originate primarily from two rats; and pattern 7 is only observed in 1 rat.
FIG. 6.
Correlation pattern as a function of image number for the concatenated scans from all rats. State 0 includes all time points not assigned to one of the ten most common patterns, and the remaining patterns are ranked from 1 to 10 in order of the number of occurrences. The key along the vertical axis defines the state, where 1 is positive correlation and 0 is weak or no correlation, as described in Table 4. From left to right, the numbers represent left SII-right SII, left SI-right SI, left MI-right MI, left CP-right CP, within-hemisphere SI-MI, and across-hemisphere SI-MI. The highlighted areas on the graph correspond approximately to the division between the rats (3 scans each). The most common patterns are observed in all rats, while some of the less common states have a more inhomogeneous distribution.
Discussion
The time-varying correlation between sites in the rat somatosensory cortex and CP observed in this study is comparable in magnitude and timing to that previously observed in human subjects (Chang and Glover, 2010) and anesthetized macaques (Hutchison et al., 2012). The results of the current study provide confirmation that time-varying correlation between brain areas can be observed in multiple species and under different anesthetic conditions. Areas that are strongly correlated when examined with traditional “static” analysis exhibit fluctuations in connectivity but maintain primarily positive correlation. Other, less correlated areas exhibit correlation time courses that fluctuate between strong positive and strong negative values, similar to previous reports.
The findings of this study are in concordance with a work by Handwerker and colleagues, which warns against over-interpretation of the variations in correlation (Handwerker et al., 2012). While many pairs of areas exhibited variable periods of very strong positive and negative correlation, similar dynamics occurred when the time courses from one scan were randomly matched with time courses from another scan or another rat, suggesting that it is possible for the variability to arise from inherent properties of the postprocessed signal itself rather than the underlying biological processes. The lowpass filter used to reduce high-frequency noise results in strong autocorrelation within the signal, and the BOLD frequencies and propagation patterns are very similar across rats (Majeed et al., 2009). However, one would expect that the random selection of time courses from different scans would introduce a random phase to each pair of fluctuations, in contrast to the phase locking that one would expect from neurally-based changes. While strong correlation between time courses from different scans may be present at a particular time lag, all analysis was performed with zero lag and should, therefore, result in random phase.
In context of previous studies in humans and macaques, it should be noted that although the sliding window correlation time courses in our study appear similar to those previously reported, no direct comparison was performed. Chang and Glover showed that temporal variability in human subjects in some areas and at some time scales was significantly greater than would be expected by chance, and it is possible that the species or anesthetized condition used in the current study led to loss of meaningful dynamics. Both the work in humans and in macaques examined different networks than the sensorimotor regions characterized here. The default mode network, in particular, has been shown to exhibit varying degrees of correlation/anticorrelation with other brain areas (Fox et al., 2005; Kelly et al., 2008) and may be considered a better target for dynamic analysis. Additional support for meaningful dynamics in the default network comes from a recent study that links the variations in connectivity between the default mode network and the task-positive network in short time windows (∼12 sec) for performance on a vigilance task (Thompson et al., 2012), suggesting that the variability may have behavioral importance even if conscious cognition is not involved. However, the study by Handwerker et al. used the same seed region (PCC) as Chang and Glover, though different methods and parameters, and most of the variation in correlation was comparable to that obtained with time-randomized data. The ability to detect significant changes in connectivity between networks may prove highly sensitive to the methods and parameters used for analysis. The default mode is not yet well characterized in rats, although a candidate network has been described (Lu et al., 2012). Since our ultimate goal is to tie network dynamics to electrical signaling, we chose to work within the limitations of the rodent model.
At least two distinct types of dynamics have been reported in spontaneous BOLD fluctuations, one a quasi-periodic reproducible pattern possibly linked to large-scale modulatory signals (Majeed et al., 2009, 2011) and one a variation in the strength of the connection between areas over time (Chang and Glover, 2010; Hutchison et al., 2012). This article focuses on the second, but it is quite plausible that the two interact. Partial correlation resulted in a decrease of 15%–20% in the correlation coefficient as compared with cross correlation for connections between left and right SI and between left and right MI, while the connections between left and right SII were relatively unaffected. This is ideologically consistent with our previous findings that quasi-periodic waves often begin in SII and propagate along the cortex to SI and MI, and, thus, common inputs from SII may be responsible for a part of the correlation between the more medial areas. The interesting finding of periodic variations in connectivity (Handwerker et al., 2012) may be linked to this phenomenon. Cross correlation rather than partial correlation was performed for the dynamic analysis due to the loss of SNR inherent in the use of short windows, but this choice is likely to have only minor effects on the reported results.
The variation in correlation increases when shorter window lengths are used (Fig. 2). While shorter windows increase the high frequency information in the resulting correlation time course, the shorter windows also contain fewer samples, reducing the SNR of the measurement. We had hypothesized that the use of shorter windows would provide more sensitivity to short-lived changes in connectivity between two areas, but this idea is not supported by the data. For all window lengths, only 6 pairs of areas show differences from randomly matched data. We did not observe any areas that showed differences from randomly matched data at short window lengths nor at long window lengths. In addition to the effects of window length, the method used in this study for reducing the contribution of fluctuations near the selected threshold (merging states that last less than 5 sec with the previous state) also reduces the high-frequency information in the correlation time course. Of course, the hemodynamic response itself imposes an inherent lowpass filter on the information that can be obtained.
With these caveats in mind, our current work neither confirms nor rules out a neural basis for the variations in correlation over time. Similarity to randomly generated results does not guarantee that the relationship between two areas is actually random, and it is possible that genuine changes in neural activity occur on the same time scales as the random transitions. Conversely, the changes in correlation in areas that exhibited different temporal characteristics than the randomly matched data are not necessarily more meaningful than those of other areas. They may simply be created by adding the variation inherent to the signal to an existing baseline of positive correlation. However, an examination of the histogram of the correlation coefficients for each pair of areas shows that the shape of the histogram is skewed for highly correlated areas, not simply shifted (Fig. 4 and Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/brain). While many time courses exhibited substantial periods of anti-correlation, most of them could not be distinguished from randomly matched time courses based on the percentage of time they spent in each state, and no anticorrelation was observed within the most common states of the network. Some patterns seem to originate predominantly from a single rat (pattern 7), which may be due to a slight tilt or rotation of the image slice that brings more or less of an area into the slice. The same shift may explain why left MI and right SI are not as strongly correlated as the other SI-MI pairs. The fluctuations in correlation cannot be solely attributed to physiological noise or external sources, as similar patterns are present in the randomly matched data and the noise sources would not be expected to be time-locked across different scans or different animals.
Other limitations of the study are due to the data reduction strategies. The segmentation of the correlation time courses into 5 and finally into 3 states for each pair of areas discards large amounts of information and may cause more subtle effects to be missed. The thresholds for the segmentation were somewhat arbitrary and may not be ideal. Another possible approach would be to examine the temporal standard deviation of the correlation coefficients, to provide an estimate of variability without an arbitrary threshold. This was not implemented for this study, because we believed that the temporal duration of each state may be important, and that relatively minor changes in connectivity were less likely to be physiologically relevant. We also plan to perform further studies using pattern-detection algorithms on data from all pairs of areas to minimize the effects of data reduction. Finally, the use of anesthesia warrants extreme caution when extrapolating from these results to human studies. However, the results provide an impetus for comparing dynamics of interest in humans to randomly matched time courses to evaluate their potential significance. Ultimately, however, the difficulty lies in determining whether the dynamics are meaningful markers of underlying neural processes. The detection of time-varying connectivity in the anesthetized rodent will allow future studies using simultaneous imaging and multisite recording (Pan et al., 2010, 2011) to search for a neural basis of the dynamic connectivity. Our previous work has suggested that low-frequency coherence (delta and theta bands) may be most indicative of BOLD correlation (Pan et al., 2011), and, therefore, changes in this coherence between left and right SI, for example, would be a natural target for comparison with time-varying connectivity.
The possibility of using resting-state MRI to examine network dynamics is an exciting and potentially transformative development, but it remains to be seen whether the variations in BOLD connectivity can be decisively linked to neural changes or behavioral outcomes. The results of this study show that time-varying correlation can be detected in the rat but that in most cases, the properties of connectivity over time cannot be distinguished from that of randomly matched time courses. While these findings make it clear that caution is needed in interpreting changes in BOLD correlation, we remain optimistic that with the appropriate measures, some level of information about network dynamics can be extracted from resting-state scans. Multimodal studies will be essential in determining which properties are neurally or behaviorally meaningful.
Supplementary Material
Acknowledgments
The authors would like to thank Josh Grooms, for discussion and feedback on the article, and Dieter Jaeger, for discussions about network dynamics. This work was supported by NIH R21NS057718, NIH R21NS072810, and the Biomedical Imaging Technology Center (BITC).
Author Disclosure Statement
No competing financial interests exist.
References
- Biswal B. Yetkin FZ. Haughton VM. Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Chang C. Glover GH. Time-frequency dynamics of resting-state brain connectivity measured with fMRI. NeuroImage. 2010;50:81–98. doi: 10.1016/j.neuroimage.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes D. Haughton VM. Arfanakis K. Wendt GJ. Turski PA. Moritz CH. Quigley MA. Meyerand ME. Mapping functionally related regions of brain with functional connectivity MR imaging. AJNR Am J Neuroradiol. 2000;21:1636–1644. [PMC free article] [PubMed] [Google Scholar]
- Fox MD. Snyder AZ. Vincent JL. Corbetta M. Van Essen DC. Raichle ME. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci U S A. 2005;102:9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD. Snyder AZ. Vincent JL. Raichle ME. Intrinsic fluctuations within cortical systems account for intertrial variability in human behavior. Neuron. 2007;56:171–184. doi: 10.1016/j.neuron.2007.08.023. [DOI] [PubMed] [Google Scholar]
- Handwerker DA. Roopchansingh V. Gonzalez-Castillo J. Bandettini PA. Periodic changes in fMRI connectivity. NeuroImage. 2012;63:1712–1719. doi: 10.1016/j.neuroimage.2012.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM. Womelsdorf T. Gati JS. Everling S. Menon RS. Resting-state networks show dynamic functional connectivity in awake humans and anesthetized macaques. Hum Brain Mapp. 2012 doi: 10.1002/hbm.22058. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AM. Uddin LQ. Biswal BB. Castellanos FX. Milham MP. Competition between functional brain networks mediates behavioral variability. NeuroImage. 2008;39:527–537. doi: 10.1016/j.neuroimage.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Li CS. Yan P. Bergquist KL. Sinha R. Greater activation of the “default” brain regions predicts stop signal errors. NeuroImage. 2007;38:640–648. doi: 10.1016/j.neuroimage.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe MJ. Mock BJ. Sorenson JA. Functional connectivity in single and multislice echoplanar imaging using resting-state fluctuations. Neuroimage. 1998;7:119–132. doi: 10.1006/nimg.1997.0315. [DOI] [PubMed] [Google Scholar]
- Lu H. Zou Q. Gu H. Raichle ME. Stein EA. Yang Y. Rat brains also have a default mode network. Proc Natl Acad Sci U S A. 2012;109:3979–3984. doi: 10.1073/pnas.1200506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson M. Majeed W. Keilholz SD. Functional connectivity in BOLD and CBV weighted resting state fMRI in the rat brain. J Magn Reson Imag. 2010;32:584–592. doi: 10.1002/jmri.22295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeed W. Magnuson M. Hasenkamp W. Schwarb H. Schumacher EH. Barsalou L. Keilholz SD. Spatiotemporal dynamics of low frequency BOLD fluctuations in rats and humans. NeuroImage. 2011;54:1140–1150. doi: 10.1016/j.neuroimage.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeed W. Magnuson M. Keilholz SD. Spatiotemporal Dynamics of Low Frequency Fluctuations in BOLD fMRI of the Rat. J Magn Reson Imag. 2009;30:384–393. doi: 10.1002/jmri.21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W. Thompson G. Magnuson M. Majeed W. Jaeger D. Keilholz S. Simultaneous fMRI and electrophysiology in the rodent brain. J Visualized Exp. 2010;42:1901. doi: 10.3791/1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W. Thompson G. Magnuson M. Majeed W. Jaeger D. Keilholz S. Broad-band LFPs correlate with spontaneous fluctuations in fMRI signals in the rat somatosensory cortex under isoflurane anesthesia. Brain Connect. 2011;1:119–131. doi: 10.1089/brain.2011.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawela CP. Biswal BB. Cho YR. Kao DS. Li R. Jones SR. Schulte ML. Matloub HS. Hudetz AG. Hyde JS. Resting-state functional connectivity of the rat brain. Magn Reson Med. 2008;59:1021–1029. doi: 10.1002/mrm.21524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawela CP. Biswal BB. Hudetz AG. Schulte ML. Li R. Jones SR. Cho YR. Matloub HS. Hyde JS. A protocol for use of medetomidine anesthesia in rats for extended studies using task-induced BOLD contrast and resting-state functional connectivity. Neuroimage. 2009;46:1137–1147. doi: 10.1016/j.neuroimage.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G. Watson C. The Rat Brain in Stereotaxic Coordinates. San Diego: Academic Press; 1998. [Google Scholar]
- Thompson G. Magnuson M. Merritt M. Schwarb H. Pan W. McKinley A. Tripp L. Schumacher E. Keilholz S. Short time windows of correlation between large scale functional brain networks predict vigilance intra-individually and inter-individually. Hum Brain Mapp. 2012 doi: 10.1002/hbm.22140. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber R. Ramos-Cabrer P. Wiedermann D. van Camp N. Hoehn M. A fully noninvasive and robust experimental protocol for longitudinal fMRI studies in the rat. Neuroimage. 2006;29:1303–1310. doi: 10.1016/j.neuroimage.2005.08.028. [DOI] [PubMed] [Google Scholar]
- Williams KA. Magnuson M. Majeed W. LaConte SM. Peltier SJ. Hu X. Keilholz SD. Comparison of alpha-chloralose, medetomidine and isoflurane anesthesia for functional connectivity mapping in the rat. Magn Reson Imaging. 2010;28:995–1003. doi: 10.1016/j.mri.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F. Zhao T. Zhou L. Wu Q. Hu X. BOLD study of stimulation-induced neural activity and resting-state connectivity in medetomidine-sedated rat. NeuroImage. 2008;39:248–260. doi: 10.1016/j.neuroimage.2007.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.