Abstract
Pre-harvest sprouting, the germination of mature seeds on the mother plant under moist condition, is a serious problem in cereals. To investigate the effect of reduced abscisic acid (ABA) catabolism on germination in hexaploid wheat (Triticum aestivum L.), we cloned the wheat ABA 8′-hydroxyase gene which was highly expressed during seed development (TaABA8′OH1) and screened for mutations that lead to reduced ABA catabolism. In a screen for natural variation, one insertion mutation in exon 5 of TaABA8′OH1 on the D genome (TaABA8′OH1-D) was identified in Japanese cultivars including ‘Tamaizumi’. However, a single mutation in TaABA8′OH1-D had no clear effect on germination inhibition in double haploid lines. In a screen for a mutation, one deletion mutant lacking the entire TaABA8′OH1 on the A genome (TaABA8′OH1-A), TM1833, was identified from gamma-ray irradiation lines of ‘Tamaizumi’. TM1833 (a double mutant in TaABA8′OH1-A and TaABA8′OH1-D) showed lower TaABA8′OH1 expression, higher ABA content in embryos during seed development under field condition and lower germination than those in ‘Tamaizumi’ (a single mutant in TaABA8′OH1-D). These results indicate that reduced ABA catabolism through mutations in TaABA8′OH1 may be effective in germination inhibition in field-grown wheat.
Keywords: ABA 8′-hydroxylase, abscisic acid, dormancy, germination, Triticum aestivum L., pre-harvest sprouting, wheat
Introduction
Pre-harvest sprouting, the germination of mature seeds while still on the mother plant, occurs in wet or humid conditions before harvest and is one of the biggest problems in wheat production. Pre-harvest sprouting causes not only reduced grain yield, but also damages the quality of the end-product resulting in economic losses. Although pre-harvest sprouting could be prevented by a high level of dormancy at harvest, this limits replanting of newly harvested grain; therefore, breeding for an adequate level of dormancy (neither so low as to lead to pre-harvest sprouting nor so high as to lead to non-uniform germination at the time of the next sowing) is highly desirable.
It is known that a sesquiterpenoid phytohormone, abscisic acid (ABA), is strongly involved in regulation of seed germination (Bewley 1997, Fang and Chu 2008, Gubler et al. 2005, Kermode 2005, Walker-Simmons 1987). Catabolism of ABA plays an important role in reducing the ABA content in seeds during imbibition and thus in promoting germination (Nambara and Marion-Poll 2005). Hydroxylation of ABA at the 8′-position to produce 8′-hydroxy ABA, catalyzed by ABA 8′-hydroxylase (ABA8′OH), is considered to be the dominant pathway of ABA catabolism. Four genes encoding ABA8′OH (CYP707A1, CYP707A2, CYP707A3 and CYP707A4) have been reported in Arabidopsis (Kushiro et al. 2004, Saito et al. 2004). Single and double mutants of these genes contained a higher level of ABA and show reduced germination (Kushiro et al. 2004, Matakiadis et al. 2009, Okamoto et al. 2006). Two genes encoding ABA8′OH have been reported in barley (Millar et al. 2006). Expression analysis has shown that one of these genes (HvABA8′OH1) is highly expressed during seed germination, while the other gene is not (Millar et al. 2006). The increase in the expression of HvABA8′OH1 was followed by a rapid decrease in ABA content, associated with higher germinability (Chono et al. 2006). Transgenic grains introduced RNAi constructs derived from HvABA8′OH1 have shown reduced germination (Gubler et al. 2008). These results indicate that reduced ABA catabolism, which may be reflected in the increase or slower decrease of ABA content, leads to reduced germination.
Although common wheat (Triticum aestivum L.) is one of the most important crops, there have been few mutants with altered seed dormancy (Kawakami 1997, Rikiishi and Maekawa 2010, Schramm 2010, Warner et al. 2000). It is much more difficult to create mutants in polyploid species than in diploid species because it is necessary to simultaneously mutate at two or three loci in the polyploid. Common wheat is a hexaploid species with three genomes, A, B and D in general and contains triplicated homologues from each genome. ABA content is highest in developing seeds and declines as the seeds undergo maturation drying (Radley 1976, Walker-Simmons 1987). Environmental conditions, including temperature, rainfall and humidity surrounding mother plants, affect on the appearance of ABA content during seed development (King 1993, Radley 1976, Walker-Simmons and Sessing 1990, Wiedenhoeft et al. 1988). Although reduced ABA catabolism may play an important role to prevent germination under field condition in cereal crops, field studies on the regulation of ABA content and germinability during seed development are few. To investigate the effect of reduced ABA catabolism on germination in hexaploid wheat, we cloned the wheat homologue of HvABA8′OH1, TaABA8′OH1 and screened for mutations that lead to reduced ABA catabolism. The expression patterns of TaABA8′OH1 and changes in ABA content in a newly identified mutant were examined using field-grown plants.
Materials and Methods
Plant materials
Wheat cultivars used in this study were grown in an experimental field at the NARO Agricultural Research Center (NARC) in each wheat growing season. For genomic DNA isolation, the flag leaf of each variety was collected and kept at −80°C. For the germination test, gene expression, and ABA quantification analysis, spikes were tagged with its pollination date to provide the same developmental stage of seeds. Twenty seeds per spike (only 1st and 2nd florets) were collected from the center part of the freshly harvested tagged spike and subjected to the following experiments. To prepare non-dormant seeds, tagged spikes were harvested from the field 45 days after pollination (DAP) when the water content of seeds was 13%, and were kept at 22°C to dry for one month. Seeds (only 1st and 2nd florets) collected from dried spikes were stored at 4°C for one year.
Molecular cloning
Total RNA was prepared from the embryos of ‘Chinese Spring’ using a TRIZOL reagent (Invitrogen, CA, USA) according to the manufacturer’s instructions. Poly (A+) RNA was purified from total RNA using Dynabeads Oligo (dT)25 resin (Dynal Biotech, Oslo, Norway) and subjected to the following cDNA synthesis. cDNA synthesis and RACE reactions were performed using a Marathon cDNA amplification kit (Clontech, CA, USA) following the manufacturer’s instructions. To perform RACE reactions, gene-specific primers were designed from the sequence data of the EST clone (accession number CJ702265) on the database. 5′-RACE was performed with an A3 primer (Table 1) and adaptor primer provided by a Marathon cDNA amplification kit, and 3′-RACE was performed with an S1 primer (Table 1) and adaptor primer. The amplified PCR fragment was cloned into a pCR 4 TOPO vector (Invitrogen). DNA sequencing was performed using a Big Dye terminator cycle sequencing kit (Applied Biosystems, CA, USA) and an ABI PRISM 3100Avant Genetic Analyzer (Applied Biosystems). Sequence analysis was performed using DNASIS pro software (Hitachi Software Engineering Co., Tokyo, Japan). To confirm the sequence of a full-length cDNA, end-to-end PCR was done using an S5 primer and A4 primer (Table 1) with high-fidelity DNA polymerase (Pyrobest DNA polymerase, Takara Bio Inc., Shiga, Japan). The amplified PCR fragment was cloned into a pCR 4 Blunt-TOPO vector (Invitrogen) and sequenced.
Table 1.
A list of the primers used in this study
| Primer name | Sequence (5′→3′) |
|---|---|
| Primer sequences for TaABA8′OH1 | |
| A1 | CCGCTCTCGGACTTGGAGGTGGAC |
| A2 | CGTYGCCWCTATCGYGCYGTTGAT |
| A3 | CCGAAGATGGACAGCAATGCCACAT |
| A4 | GGGCCTCCTTTTGGCCCTTTGC |
| A5 | TTYCCTTTGGGAACGAGGAGGAG |
| A6 | GCCCACGTACGGCCACCCCAT |
| A7 | TTGTCGCCGAGGAACTTGACCATC |
| A8 | CACTTTCCAGCCCTTGGGAATCAG |
| A9 | ACTTCTTTGTGGCGCCGGATCTC |
| A10 | GGCATGAACGTGTTGGGCTTGG |
| A11 | GGCACGAGTGGGTCCCGTTCC |
| A12 | ATCTGTACTTGGTGGCGAGGTGGTG |
| S1 | AGGCCGTGGAGGACGTGGAGTACC |
| S2 | AAGCCCAACACGTTCATGCCGTTC |
| S3 | CAAGAACCCCAACGTCTTCTTCG |
| S4 | GKKCGTCGTCGGGCGGCGGC |
| S5 | GCCTTTCGTTGRCATGGCWGCTT |
| S6 | ATGTGGCATTGCTGTCCATCTTCGG |
| S7 | CTCCCTCGGCTCCTGGGAAGAC |
| S8 | GTGCTCACGTGGATGGTCAAGTTCC |
| dS1 | GACGCGCTCCTAACCGAACGAAC |
| dA1 | CGTCGCCTCTATCGCGCTGTTGAT |
| D-insert S1 | GATGCTCGTCCTCTGCCAGCGTAG |
| D-insert A1 | CTACTGCCGGATGCCACACAACAC |
| Primer sequences for expression analysis | |
| TaABA8′OH2_QRT_S1 | CAGGTGGGAGGTTGTTGGATCGAG |
| TaABA8′OH2_QRT_A1 | AAAGGGAGAAAAGAGCCCTCTAGC |
| 18SFw | GAATTGACGGAAGGGCACCACCAG |
| 18SRv | GGACATCTAAGGGCATCACAGACC |
To clone the TaABA8′OH1 genome, high-fidelity PCR was done using a gene-specific primer pair (S5 primer and A9 primer, Table 1) with genomic DNA isolated from the leaves of ‘Chinese Spring’ using the DNeasy Plant Mini kit (QIAGEN, CA, USA) as a template. The amplified PCR fragment was cloned and sequenced. For chromosomal assignment of each gene, nullisomic-tetrasomic lines of ‘Chinese Spring’ (Endo and Gill 1996), kindly provided by the National BioResources Project, Japan (NBRP-Wheat), were used. The primer sequences used in this study were listed in Table 1.
Screening for TaABA8′OH1 mutations
A total of 271 wheat genotypes (Supplemental Table 1), including commercial cultivars released in Japan (160 cultivars), Japanese breeding lines (42 cultivars), Japanese landraces (28 cultivars) and foreign cultivars introduced for breeding (41 cultivars) were grown in the 2004–2005 wheat growing season and subjected to screening for a natural variation in TaABA8′OH1s. To screen a mutation, the Japanese cultivar ‘Tamaizumi’ was used to make gamma-ray irradiated lines. In total, 126,000 seeds were irradiated with 200 Gy of gamma rays at the Institute of Radiation Breeding, National Institute of Agrobiological Science (NIAS; Hitachiohmiya, Ibaraki, Japan). M1 seeds were sown and gathered M2 seeds by single seed descent from each spike of M1 plants. In the 2007–2008 wheat growing season, 2,200 M2 seeds were sown and developed the first population of 2,074 M2 plants. In the 2009–2010 wheat growing season, 1,600 M2 seeds were sown and developed the second population of 1,349 M2 plants. In this study, a total of 3,423 M2 plants were obtained and subjected to a screen for mutation.
Genomic DNA was isolated from the flag leaf derived from each cultivar, breeding line and mutant lines using the DNeasy Plant 96 kit (QIAGEN). By using a gene-specific primer pair (S2 primer and A2 primer, Table 1), which were designed to amplify at around the stop codon, PCR was performed with Ex Taq DNA polymerase (Takara Bio Inc.), following the manufacturer’s instructions. The amplified fragments were subjected to polyacrylamide gel electrophoresis. To achieve the required efficiency for electrophoresis, the high-efficiency genome scanning system (Kawasaki and Murakami 2000) was adopted and a set of electrophoresis apparatus equipped with two sets of 24.5 × 26.5-cm glass plates, each accommodating a gel with 100 lanes, was used. Detection of the insertion mutation was performed by PCR using the D genome specific primer pair (dS1 primer and dA2 primer, Table 1) with EX taq DNA polymerase. The PCR conditions were as follows: 30 second denaturation at 98°C, then 35 cycles of 10 second at 98°C and 1 minute at 68°C. Amplified fragments were separated by electrophoresis on agarose gel.
Germination tests
To execute a germination test, doubled haploid (DH) lines derived from a cross between ‘Zenkoujikomugi’ (non-TaABA8′OH1-D mutant, a highly dormant Japanese red wheat cultivar) and ‘Tamaizumi’ (TaABA8′OH1-D mutant, a relatively dormant Japanese white wheat cultivar) were sown in the 2003–2004 wheat growing season. ‘Tamaizumi’ and its mutant (named TM1833, M4 generation) seeds were sown in the 2009–2010 wheat growing season. Thirty whole seeds were placed in a Petri dish (9-cm diameter) containing two Whatman No. 2 filter papers (8.5-cm diameter) and 6 ml of water. Dishes were sealed with Parafilm and placed in a growth cabinet. Germinated seeds (those showing the emergence of coleorhiza beyond the seed coat) were counted every 24 h and removed from the dishes for 7 days. Germination tests during seed development were performed in triplicate. The mean and standard error (SE) values of the germination percentage were calculated. The dry weight of seeds was measured after incubating 50 seeds at 135°C for 4 h. The water content (%) of seeds was estimated by (fresh weight − dry weight)/fresh weight. To examine ABA responsiveness during seed germination, ABA (2-cis, 4-trans-ABA; Sigma, MO, USA) was dissolved in DMSO to prepare a 10 mM stock solution. The stock solution was mixed to make an ABA solution at different concentrations, containing 0.1% of DMSO. Control solution was the same solution without ABA.
Quantitative reverse transcription-PCR (QRT-PCR)
To examine the effect of a deletion mutant on gene expression during seed maturation, the embryos of freshly harvested seeds of ‘Tamaizumi’ and TM1833 were isolated and immediately frozen in liquid nitrogen at 5-day-intervals from 20 to 60 DAP. Frozen embryos (about 90 embryos) were ground into powder in a mortar and pestle with liquid nitrogen. Powdered tissue was divided into two portions for ABA quantification analysis and gene expression analysis. For gene expression analysis, total RNA was prepared from embryos using TRIZOL reagent. First-strand cDNA derived from total RNA samples, which were treated with DNase I before cDNA synthesis, was prepared using the method described in our previous paper (Chono et al. 2006). Gene-specific primer pairs, capable of amplifying a 140 bp 3′ end fragment of TaABA8′OH1 and a 206 bp 3′ end fragment of TaABA8′OH2, respectively, were S2 primer and A1 primer and TaABA8′OH2_QRT_S1 primer and TaABA8′OH2_ QRT_A1 primer (Table 1). RT-PCR was performed using a 7500 Real-Time PCR System (Applied Biosystems) and a SYBR Premix EX Taq Kit (Takara Bio Inc.) with a gene-specific primer pair and an 18S primer pair (18SFw primer and 18SRv primer, Table 1) as an internal control, following the manufacturer’s instructions. The amplification efficiency of the primers was determined in accordance with the manufacturer’s instructions. Each sample was run in triplicate. The obtained data using the ΔΔ cycle threshold analysis method were analyzed by RQ study software (Applied Biosystems). Results shown are means ± SE from a single experiment performed in triplicate, representative of two independent experiments with similar results.
ABA quantification
The powdered embryos as mentioned in gene expression analysis were extracted by methanol. The methanol extract was mixed with 20% volume of water and then loaded onto a Bond Elut C18 (Varian, CA, USA) cartridge. This cartridge was washed with 5 ml of 80% methanol and the effluent was dried down in a centrifugal vacuum concentrator until approximately 40 μl of the liquid remained. Tris-buffered saline (25 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM MgCl2) was added to a final volume of 400 μl. These extracts were diluted 8-fold in Tris-buffered saline and subjected onto a Phytodetek ABA test kit (AGDIA Inc., IN, USA) following the manufacturer’s instructions. The color absorbance was detected with the SH-9000Lab microplate reader (Corona Electric Co., Ltd., Ibaraki, Japan) at 405 nm.
Results
Cloning of wheat ABA8′OH genes
To examine the role of ABA8′OH in wheat, we tried to isolate wheat HvABA8′OH1 homologues. When we started to clone wheat ABA8′OH homologues, only the EST clone (CJ702265) with a high sequence homology to HvABA8′OH1 was identified in the BLAST database. Then, we performed 5′-RACE and 3′-RACE reactions to obtain full-length cDNA sequences. Three full-length cDNAs from a cultivar ‘Chinese Spring’ were cloned and sequenced. Their sequences were highly similar to HvABA8′OH1 and nearly identical to the CJ702265 sequence. Next, we performed high-fidelity PCR using a gene-specific primer pair (S5A9, Fig. 1) with genomic DNA as a template and obtained three genomic sequences (Supplemental Fig. 1). These sequences were nearly identical to wheat ABA8′OH genes reported by Ji et al. (2011) and Zhang et al. (2009). Then, these three genes were named TaABA8′OH1s.
Fig. 1.
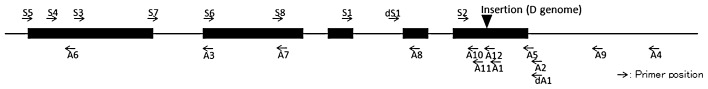
Structure of TaABA8′OH1. Black boxes indicate exons, and lines indicate introns and untranslated regions. White boxes indicate amplified PCR fragments of the S2A2 primer pair for TaABA8′OH1-A, TaABA8′OH1-B and TaABA8′OH1-D, respectively. Arrows indicate primer positions of TaABA8′OH1. Black arrowhead indicates the insertion mutation in exon 5 of TaABA8′OH1-D.
The sequences of TaABA8′OH1s have a small difference at around the stop codon (Supplemental Fig. 1). The primer pair, S2 primer and A2 primer (S2A2, Fig. 1), was designed to amplify the region including the sequence differences and was used to assign each PCR fragment derived from these genes to the A, B and D genomes, respectively. By using genomic DNA prepared from the nullisomic-tetrasomic lines of ‘Chinese Spring’ as a template, these three genes were assigned to A, B and D genomes in homoeologous group-6 chromosomes, respectively. As shown in Fig. 2, three PCR fragments derived from A, B and D genomes were separated on acrylamide gel. These three genes located on the chromosomes 6A, 6B and 6D were named TaABA8′OH1-A, TaABA8′OH1-B and TaABA8′OH1-D, respectively (GenBank accession numbers for genome and cDNA: AB714574 to AB714579).
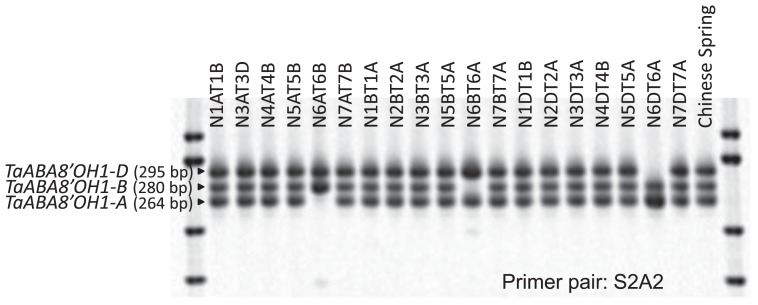
Fig. 2.
PCR fragments of TaABA8′OH1 with the S2A2 primer pair in nullisomic-tetrasomic lines of ‘Chinese Spring’. Total wheat genomic DNAs were isolated from the leaves of nullisomic-tetrasomic lines of ‘Chinese Spring’. The names of nullisomic-tetrasomic lines (for example, N1AT1B indicates nullisomic 1A and tetrasomic 1B) were shown on the top of the gel image. Three PCR fragments amplified using the S2A2 primer pair were separated on acrylamide gel. The three genes corresponding to these PCR fragments, assigned to the A, B and D genomes in homoeologous group-6 chromosomes, were named TaABA8′OH1-A, TaABA8′OH1-B and TaABA8′OH1-D, respectively.
Screen and characterization of a natural variation in TaABA8′OH1s
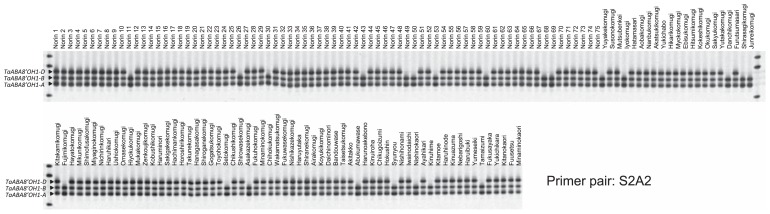
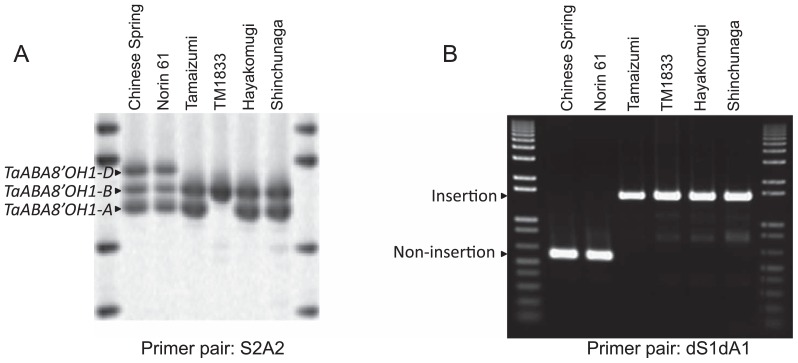
A total of 271 wheat genotypes (Supplemental Table 1), including commercial cultivars released in Japan (160 cultivars), Japanese breeding lines (42 cultivars), Japanese landraces (28 cultivars) and foreign cultivars (41 cultivars) were screened for a natural variation in TaABA8′OH1s, based on PCR amplification with the S2A2 primer pair. In 23 Japanese cultivars (‘Norin 11’, ‘Norin 26’, ‘Norin 30’, ‘Norin 43’, ‘Norin 49’, ‘Norin 50’, ‘Norin 53’, ‘Norin 60’, ‘Norin 68’, ‘Norin 69’, ‘Yuyakekomugi’, ‘Iyokomugi’, ‘Danchikomugi’, ‘Shirasagikomugi’, ‘Junreikomugi’, ‘Fujimikomugi’, ‘Setokomugi’, ‘Asakazekomugi’, ‘Abukumawase’, ‘Syunyou’, ‘Nishinokaori’, ‘Kinuhime’ and ‘Tamaizumi’) within 160 Japanese commercial cultivars, the PCR fragment derived from the D genome was not amplified (Fig. 3). These cultivars without TaABA8′OH1-D amplification were developed in the Kyusyu to Kanto area (Supplemental Table 1). The length of the PCR fragments derived from three genomes in Japanese commercial cultivars (e.g. ‘Norin 61’) was the same as ‘Chinese Spring’ (Fig. 4A). The absence of the fragment derived from the D genome was also identified in nine Japanese breeding lines in the Kyushu to Kanto area (Supplemental Table 1) and two Japanese landraces ‘Shinchunaga’ and ‘Hayakomugi’ (Fig. 4A and Supplemental Table 1). ‘Shinchunaga’ and ‘Hayakomugi’ were used as cross parents in early Japanese cultivars in the southern part of Japan.
Fig. 3.
PCR fragments of TaABA8′OH1 with the S2A2 primer pair in commercial cultivars released in Japan. Three PCR fragments amplified using the S2A2 primer pair were separated on acrylamide gel.
Fig. 4.
Screen for mutations of TaABA8′OH1. Using the S2A2 primer pair, the M2 population of gamma-ray treated ‘Tamaizumi’ was screened to detect a deletion mutation in TaABA8′OH. (A) PCR fragments amplified by the S2A2 primer pair. One mutant in TaABA8′OH1-A, TM1833, did not amplify the fragment derived from the A genome. (B) PCR fragments amplified by the dS1dA1 primer pair. The length of the fragment derived from the D genome in mutants (‘Shinchunaga’, ‘Tamaizumi’ and its mutant TM1833) was 966 bp longer than that in the normal type (‘Norin 61’).
To clarify the absence of the fragment derived from the D genome, ‘Tamaizumi’, ‘Shinchunaga’ and ‘Hayakomugi’ were further analyzed using the D genome-specific primer pair, dS1dA1 (Fig. 1 and Table 1). The extension time was increased from 30 seconds to 1 minute and then a fragment approximately 1 kbp longer than the fragment from ‘Norin 61’ and ‘Chinese Spring’ was amplified (Fig. 4B). The amplified fragment of ‘Shinchunaga’ was cloned and sequenced, and revealed that the fragment contains a 966-bp insertion in exon 5 of TaABA8′OH1-D of ‘Chinese Spring’ (Supplemental Fig. 2). A homology search revealed that the partial sequence of this insertion showed similarity to wheat transposon-like element Hikkoshi (AY376310). The insertion is predicted to create a premature stop codon upstream from the normal stop codon and lead to reduced ABA8′OH activity on the D genome due to changes in amino acids in the region relating to the recognition site of a substrate (Supplemental Fig. 3). A long PCR fragment using the dS1dA1 primer pair was detected in the cultivars and breeding lines of which the PCR fragment derived from the D genome using the S2A2 primer pair was absent (data not shown). The length of the dS1dA1 fragment was the same in these cultivars and breeding lines. In ‘Tamaizumi’ and ‘Hayakomugi’, the amplified fragment using dS1dA1 primer pair was cloned and sequenced and revealed that their sequences were identical to the sequence of ‘Shinchunaga’. These results indicated that these Japanese cultivars may have the same insertion sequence in exon 5 of TaABA8′OH1-D in ‘Shinchunaga’ and ‘Hayakomugi’.
Effect of an insertion mutation in TaABA8′OH1-D on germination in DH lines
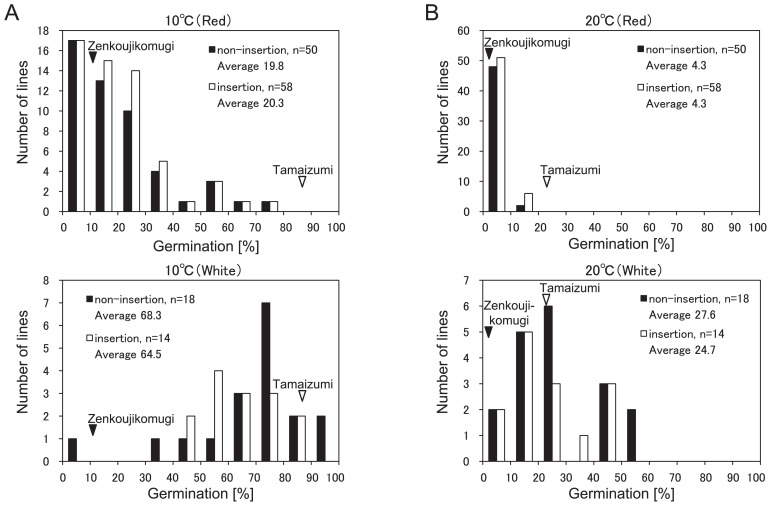
We examined the effect of an insertion mutation in TaABA8′OH1-D on germination using DH lines derived from ‘Zenkoujikomugi’ and ‘Tamaizumi’. The seeds of each DH line (DAP50) were imbibed at 10°C and 20°C for 7 days. In the parental cultivars, the germination percentage after 7 days of imbibition were 10.0% at 10°C and 3.3% at 20°C for ‘Zenkoujikomugi’ and 86.7% at 10°C and 23.3% at 20°C for ‘Tamaizumi’ (Fig. 5). The mean germination percentage of insertion TaABA8′OH1-D lines were 29.0% at 10°C and 8.3% at 20°C, while that of non-insertion TaABA8′OH1-D lines were 32.6% at 10°C and 10.5% at 20°C. There was no statistical significance in mean germination percentage between non-insertion and insertion lines at 10°C and 20°C. It is well known that the lines with red seed-coat color are generally more dormant than those with white seed-coat color (Flintham 2000, Himi et al. 2002, McCaig and Depauw 1992). Based on seed-coat color, 140 DH lines were separated into two groups. The group of white seed-coat color was more germinative than that of red seed-coat color at 10°C (Fig. 5A) and 20°C (Fig. 5B). There was no clear difference in the frequency distributions between the subgroups of insertion and non-insertion TaABA8′OH1-D lines at 10°C and 20°C. The result of the germination assay revealed that a single mutation in TaABA8′OH1-D had no clear effect on germination inhibition in the two groups divided by seed-coat color in DH lines, while the red seed color had a strong effect on germination inhibition.
Fig. 5.
Frequency distribution of germination percentages after 7 days of imbibition at 10°C and 20°C. The average of germination percentages for ‘Zenkoujikomugi’ and ‘Tamaizumi’ were indicated by a black and white arrowhead, respectively. The DH lines were imbibed at 10°C (A) and 20°C (B). The DH lines were separated by seed-coat color: upper figures were red seed-coat color, lower figures were white seed-coat color.
Screen of a mutation in TaABA8′OH1s
To make a double mutant in TaABA8′OH1s, a Japanese cultivar ‘Tamaizumi’, which carries an insertion mutation in TaABA8′OH1-D, was used to produce mutation lines by gamma-ray irradiation. We focused on the isolation of a mutant which have a deletion in TaABA8′OH1 resulting in a reduced ABA8′OH activity. A total of 3,423 M2 plants were screened using PCR with a S2A2 primer pair. One mutant in TaABA8′OH1-A, TM1833, did not amplify the fragment derived from the A genome (Fig. 4A). TM1833 was further analyzed to clarify the region of the mutation. Eleven primer pairs with the following enzyme digestion (S7A3/Alu I, S7A7/Alu I, S7A8/Mse I, S7A9/Kpn I, S6A8/Mse I, S8A8/Mse I, S1A8/Alu I, S1A10/Alu I + Mbo I, S1A11/Alu I + Mbo I, S1A12/Alu I + Mbo I and S1A1/Alu I + Mbo I) and two primer pairs (S2A2 and S2A5) were used to examine amplification of the fragment derived from the A genome. The position and sequence of the primer were shown in Fig. 1 and Table 1, respectively. No PCR fragment of TaABA8′OH1-A was amplified with the TM1833 genome as a template, while the accurate length of the PCR fragment was amplified with the ‘Tamaizumi’ genome (data not shown). A PCR fragment using S5A3 primer pair was cloned and sequenced and revealed that the fragment was derived from TaABA8′OH1-B and TaABA8′OH1-D in TM1833. These results indicated that the entire coding region of TaABA8′OH1-A was deleted in TM1833. Therefore, TM1833 was the double mutant of the deletion in TaABA8′OH1-A and the insertion in TaABA8′OH1-D.
Effect of a deletion mutation in TaABA8′OH1-A on germination in ‘Tamaizumi’ background
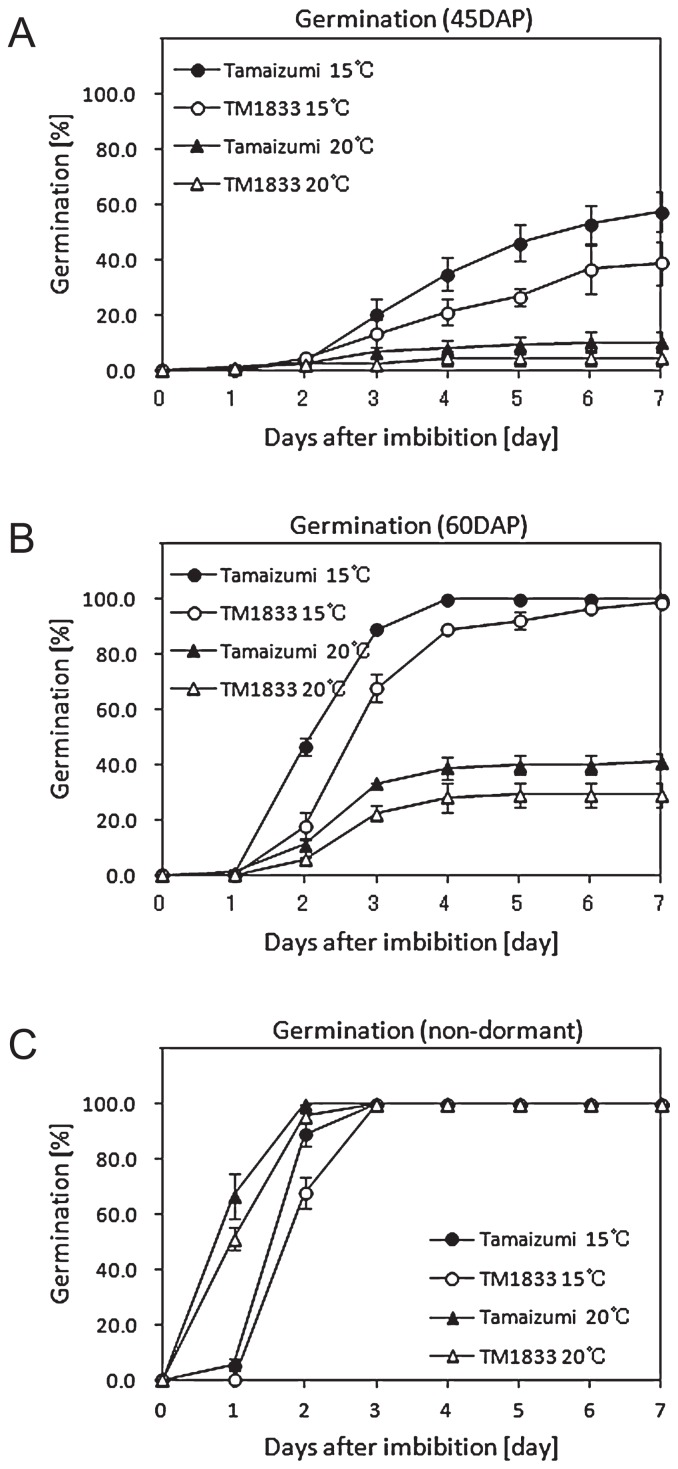
To investigate the effect of a deletion mutation in TaABA8′OH1-A on seed germination in field-grown wheat, the germination patterns of ‘Tamaizumi’ and TM1833 seeds were examined. ‘Tamaizumi’ and TM1833 seeds were freshly harvested at 45 and 60 DAP and imbibed at 15°C and 20°C without drying and storage.
At 45 DAP, ‘Tamaizumi’ seeds germinated gradually at 15°C and its germination percentage reached 57.4% at 7 days after the start of imbibition (Fig. 6A). The germination percentage of TM1833 seeds was lower than that of ‘Tamaizumi’ seeds at 15°C (Fig. 6A). In ‘Tamaizumi’ and TM1833 seeds, the germination percentage at 20°C was lower than that at 15°C.
Fig. 6.
Germination patterns of ‘Tamaizumi’ and TM1833 seeds at different seed maturing stages. ‘Tamaizumi’ (black) and TM1833 (white) seeds were freshly harvested at 45DAP (A) and 60DAP (B) and imbibed at 15°C (circle) and 20°C (triangle). Stored seeds (C) were also imbibed at 15°C and 20°C.
At 60 DAP, ‘Tamaizumi’ seeds germinated quickly and its germination percentage reached 100% within 4 days at 15°C (Fig. 6B). TM1833 seeds also germinated at 15°C, but the germination of TM1833 seeds was clearly delayed at the beginning of imbibition at 15°C (Fig. 6B). ‘Tamaizumi’ and TM1833 seeds showed 41.1% germination and 29.3% germination, respectively, after7 daysofimbibitionat 20°C (Fig. 6B). The germination percentage of TM1833 seeds was consistently and significantly lower than that of ‘Tamaizumi’ seeds for 7 days at 20°C.
To examine the germinability of non-dormant seeds in ‘Tamaizumi’ and TM1833, the stored seeds were imbibed at 15°C and 20°C. The germination percentage of non-dormant seeds at 20°C was higher than that at 15°C for 2 days (Fig. 6C). These seeds germinated poorly (‘Tamaizumi’) or not at all (TM1833) after 1 day of imbibition at 15°C and rapidly increased their germination percentage to 88.9% (‘Tamaizumi’) and 67.8% (TM1833) after 2 days (Fig. 6C). The non-dormant seeds of ‘Tamaizumi’ and TM1833 germinated efficiently after 1 day of imbibition at 20°C and showed 100 ± 0.0% germination (‘Tamaizumi’) and 95.6 ± 1.1% germination (TM1833) after 2 days (Fig. 6C). The germination percentage of TM1833 seeds was slightly lower than that of ‘Tamaizumi’ seeds for 2 days at 15°C and 20°C and then, both seeds were able to geminate to nearly 100% within 3 days.
Change in the expressions of ABA8′OH genes and ABA content during seed development
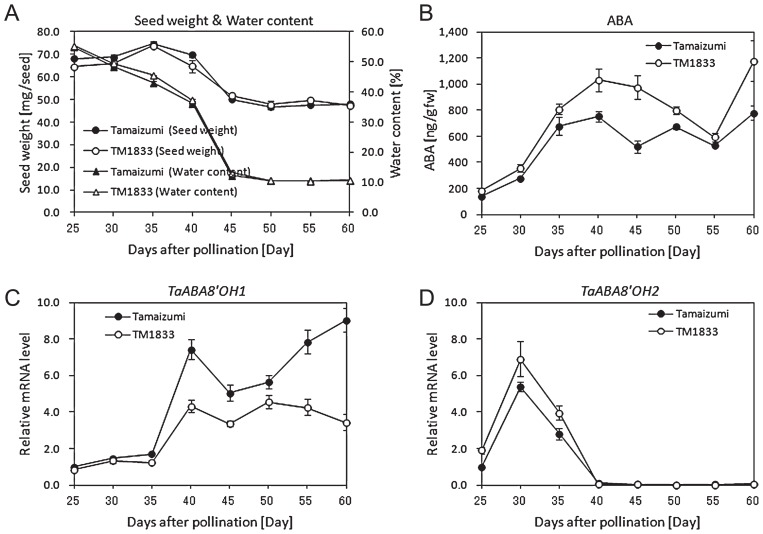
The patterns of gene expressions and ABA accumulation during seed development were examined. The seeds of ‘Tamaizumi’ and TM1833 were freshly harvested at 5-day-intervals from 25 to 60 DAP and subjected to gene expression and ABA quantification analysis. Patterns in the change in water content and seed weight were similar between ‘Tamaizumi’ and TM1833 (Fig. 7A).
Fig. 7.
Changes in seed growth, embryonic ABA content and expression levels of TaABA8′OHs in embryos during development of ‘Tamaizumi’ and TM1833 seeds. Seeds were collected from field-grown wheat at 5-day intervals. Seeds were used without artificial drying and storage. (A) Changes in seed weight and water content of seeds during seed development. Seed weight of ‘Tamaizumi’ (black circle) and TM1833 (white circle) seeds 5 days after imbibition and water content of ‘Tamaizumi’ (filled triangle) and TM1833 (open triangle) seeds are shown. (B) Changes in embryonic ABA contents during grain development. Endogenous ABA content in embryos from ‘Tamaizumi’ (black circle) and TM1833 (white circle) seeds is shown. (C, D) Changes in expression levels of TaABA8′OHs in embryos during seed development. The expression levels of TaABA8′OH1 (C) and TaABA8′OH2 (D) were analyzed by QRT-PCR. Results were normalized to the expression of 18S ribosomal RNA (internal control). Results from three independent experiments are shown with error bars (SE).
To examine gene expression of TaABA8′OH1, QRT-PCR was performed using the S2A1 primer pair (Fig. 1). The amplified fragment was derived from TaABA8′OH1-A and TaABA8′OH1-B in ‘Tamaizumi’ and from TaABA8′OH1-B in TM1833. The S2A1 primer pair spanning the insertion site of TaABA8′OH1-D did not produce any PCR fragment derived from the D genome in our PCR condition. The expression of the insertion type of TaABA8′OH1-D transcript was examined by using the insertion-specific primer pair (Table 1 and Supplemental Fig. 2) and revealed that the expression was detectable but quite low level in ‘Tamaizumi’ and ‘TM1833’ (data not shown).
In the embryos of ‘Tamaizumi’ and TM1833 seeds, TaABA8′OH1 mRNA were mainly expressed in the middle to latter stages of seed development (40 DAP to 60 DAP) (Fig. 7C). The expression level of TaABA8′OH1 mRNA in TM1833 was clearly lower than that in ‘Tamaizumi’. In addition to TaABA8′OH1, one more gene encoding ABA 8′-hydroxylase has been reported by Ji et al. (2011) and Nakamura et al. (2010). The second ABA8′OH gene, TaABA8′OH2, was mainly expressed in the early stage of seed development (25 DAP to 35 DAP), peaked at 30 DAP and the level of its expression was quite low during the middle to latter stages of grain development (Fig. 7D). While the expression level of TaABA8′OH2 in TM1833 was slightly higher than that in ‘Tamaizumi’, the expression pattern of TaABA8′OH2 was the same in ‘Tamaizumi’ and TM1833.
Quantification analysis revealed that the ABA content in the embryos of ‘Tamaizumi’ seeds increased gradually and peaked at 40 DAP (Fig. 7B). Although TM1833 and ‘Tamaizumi’ contained nearly the same level of ABA content in the early stage of seed development, TM1833 contained higher levels of ABA than Tamaizumi in the middle to latter stages of seed development (Fig. 7B). In TM1833, the lower expression of TaABA8′OH1 mRNA was consistent with the higher ABA content during grain development. In ‘Tamaizumi’ and TM1833, the increase in the level of ABA was observed at 60 DAP and the increase in TM1833 was larger than that in ‘Tamaizumi’.
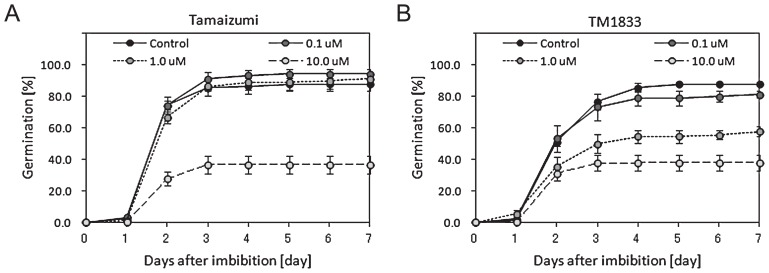
Effect of a deletion mutation in TaABA8′OH1-A on ABA responsiveness during seed germination in ‘Tamaizumi’ background
To further characterize the germination phenotype of TM1833, the response to an increased concentration of ABA during imbibition was examined using ‘Tamaizumi’ and TM1833 seeds at 50 and 72 DAP. ‘Tamaizumi’ and TM1833 seeds were imbibed in water with 0 (control), 0.1, 1.0 and 10 μM ABA at 20°C and germination was recorded every 24 hours for 7 days. At 50 DAP, less than 10% of ‘Tamaizumi’ and TM1833 seeds germinated during imbibition, and the germination inhibition in response to ABA was not clear (data not shown). ‘Tamaizumi’ and TM1833 seeds at 72 DAP were able to germinate to nearly 85% at 4 days in the control (Fig. 8). The germination of ‘Tamaizumi’ seeds was only inhibited in 10 μM ABA and the germination patterns of ‘Tamaizumi’ seeds in 0.1 and 1.0 μM ABA were similar to that in the control (Fig. 8A). Germination of TM1833 seeds was inhibited at low concentrations of ABA, at which germination of ‘Tamaizumi’ seeds was not inhibited (Fig. 8). The germination of TM1833 seeds was inhibited in 0.1, 1.0 and 10.0 μM ABA in a dose-dependent manner (Fig. 8B). TM1833 seeds showed a higher sensitivity to ABA than ‘Tamaizumi’ seeds. The non-dormant seeds of ‘Tamaizumi’ and TM1833 germinated quickly after the start of imbibition and the germination inhibition in response to ABA was detectable but weak (data not shown).
Fig. 8.
ABA responsiveness of ‘Tamaizumi’ and TM1833 seeds. Seeds at 72 DAP were imbibed in the dark at 20°C. The germination percentage of ‘Tamaizumi’ (A) and TM1833 (B) seeds during imbibition is shown. ‘Tamaizumi’ and TM1833 seeds were imbibed in water with 0, 0.1, 1.0 and 10 μM ABA at 20°C and germination was recorded every 24 hours for 7 days.
Discussion
In a screen for natural variation, one insertion mutation in TaABA8′OH1 on the D genome was identified in Japanese cultivars and breeding lines, which are localized in the Kyushu to Kanto area, but not in the Tohoku to Hokkaido area. The same mutation was identified in Japanese landraces ‘Shinchunaga’ and ‘Hayakomugi’ (Fig. 4), which were used as the cross parent of early wheat breeding in the southern part of Japan. Twenty-three cultivars carry the insertion mutation in TaABA8′OH1-D and all of these cultivars, except for ‘Norin 11’, were related to ‘Shinchunaga’ and/or ‘Hayakomugi’ in their pedigrees. The insertion mutation in TaABA8′OH1-D might be derived from ‘Shinchunaga’ and ‘Hayakomugi’, and localized in these areas. ‘Norin 11’ was selected from a cross between ‘Yushoki 347’ and ‘Hiroshimashipuree 3’ in 1924. These two landraces, at least the seeds stored in NICS and used in this study, carry non-insertion type of TaABA8′OH1-D (Supplemental Table 1). ‘Yushoki 347’ and ‘Hiroshimashipuree 3’ seeds were not provided by National Bio Resource Projest (NBRP) and NIAS Genebank in Japan. The same insertion mutation in TaABA8′OH1-D was not detected in 41 foreign cultivars we used (Supplemental Table 1). To investigate whether the same mutation is observed in other Japanese landraces and other foreign cultivars, more detail experiment to screen for a mutation in TaABA8′OH1-D may be needed.
The insertion mutation in exon 5 of TaABA8′OH1-D resulted in changes in amino acid residues which relate to the recognition site of a substrate (Werck-Reichhart 2000). The region of which amino acid residues were changed was highly conserved between ABA8′OHs in plants (Supplemental Fig. 3). Arabidopsis ABA8′OH mutant (cyp707a2-2), which carries a T-DNA insertion in this region, has shown reduced germination (Kushiro et al. 2004, Supplemental Fig. 3). In many cases, T-DNA insertion in the protein-coding region of gene reduces the transcript level of its target gene in Arabidopsis (Wang 2008). The expression analysis of TaABA8′OH1-D by using the insertion-specific primer pair revealed that the expression of the insertion type of TaABA8′OH1-D transcript was detectable but quite low level (data not shown). Therefore, the insertion mutation in TaABA8′OH1-D may reduced ABA8′OH activity via modified ABA8′OH protein and/or reduced TaABA8′OH transcript, resulting in reduced germination in wheat. As shown in Fig. 5, the effect of the insertion mutation in TaABA8′OH1-D on germination was examined using DH lines derived from a cross between ‘Zenkoujikomugi’ and ‘Tamaizumi’ and showed that the single mutation in TaABA8′OH1-D has no clear effect on germination inhibition in the DH lines. The reduced ABA8′OH activity on the D genome may be masked by the ABA8′OH activity on the A and B genomes. The genetic background of each cultivar may have some effect on germination. To investigate the effect of the single mutation in TaABA8′OH1-D on germination, more precise experiments using other DH lines may be needed.
Analysis of ‘Chinese Spring’ partial chromosome deletion lines (6DL-1, 6DL-2, 6DL-6, 6DL-7, 6DL-10, 6DL-11, 6DL-12 and 6DL-13), which were kindly provided by National BioResources Project, revealed that TaABA8′OH1-D was located between break points of 6DL-1 and 6DL-6 (Supplemental Fig. 4). Sourdille et al. (2004) reported physical-genetic map relationships in wheat and 7 molecular markers were located between break points of 6DL-1 and 6DL-6. One of these markers, Xcfd76, was the closest marker flanking the peak of the quantitative trait locus (QTL) for ABA responsiveness at seedling stage in wheat (Kobayashi et al. 2010). Kobayashi et al. (2010) reported that the QTL for ABA responsiveness was in the vicinity of the pre-harvest sprouting locus QPhs.cnl-6D.1 (Munkvold et al. 2009). These results suggest that ABA catabolism might have an effect on ABA sensitivity and/or pre-harvest sprouting, but there is no direct evidence for a relationship between ABA catabolism and these QTLs.
In a screen for a mutation in TaABA8′OH1, one deletion mutant in TaABA8′OH1-A named TM1833 was identified. In addition to the deletion mutation in TaABA8′OH1-A, TM1833 carries the insertion mutation in TaABA8′OH1-D derived from its parent cultivar ‘Tamaizumi’. In our field condition, TM1833 showed lower TaABA8′OH1 expression than that in ‘Tamaizumi’ during the middle to latter stages of seed development (Fig. 7C). Reduced expression of TaABA8′OH1 in TM1833 may have resulted in lower ABA8′OH activity and higher ABA content than those in ‘Tamaizumi’ (Fig. 7B). Increased ABA content would reduce germination in freshly harvested seeds (Fig. 6A, 6B). In Arabidopsis and barley, reduced ABA catabolism has resulted in high levels of ABA in seeds during imbibition and the inhibition of germination (Gubler et al. 2008, Kushiro et al. 2004, Matakiadis et al. 2009, Okamoto et al. 2006). These results indicate that a deletion mutation in TaABA8′OH1-A may be effective for reducing germination in a mutant TaABA8′OH1-D background. In hexaploid wheat, double or triple mutations in TaABA8′OH1 may be needed to reduce the activity of ABA8′OH during seed germination. We will investigate whether a single mutation in TaABA8′OH1-A can be effective for reducing germination in a normal TaABA8′OH1-D background.
The endogenous level of ABA is modulated by the precise balance between biosynthesis and catabolism. The oxidative cleavage of cis-epoxycarotenoids catalyzed by 9-cis-epoxycarotenoid dioxygenase (NCED) is the key regulatory step of ABA biosynthesis in higher plants (Nambara and Marion-Poll 2005). It has been shown previously that the expression of NCED gene is affected by environmental conditions (such as rainfall, high humidity and temperature) during grain development in barley (Chono et al. 2006). It has also been shown that the expression of NCED gene is induced by light during grain imbibition in barley and the light induction of NCED gene expression results in an increased ABA content (Gubler et al. 2008). Environmental conditions affect on the appearance of ABA peaks during seed development in wheat (King 1993, Radley 1976, Walker-Simmons and Sessing 1990, Wiedenhoeft et al. 1988). King (1993) has shown that during wheat seed maturation, ABA content in embryos is higher in seeds produced under wet conditions (90–100% relative humidity) than in seeds produced under dry conditions (35–40% relative humidity). In ‘Tamaizumi’ and TM1833, the endogenous ABA content was increased at 60 DAP in 2010 (Fig. 7B). The amount of precipitation from 58 to 59 DAP in 2010 was 7.0 mm, and the mean humidity was almost 85% (data not shown). The spikes were wet on these days and then dried in the field. The wetting of seeds during rainfall and/or the following drying might affect the expression of NCED gene and the embryonic ABA content in ‘Tamaizumi’ and TM1833 seeds thereafter. The increased ABA content in the latter stage of seed development might, in part, be explained by the light effect on germination inhibition during seed imbibition. The increase of endogenous ABA content in ‘Tamaizumi’ was smaller than that in TM1833. Difference in the expression level of TaABA8′OH1 might result in the different ABA accumulation in ‘Tamaizumi’ and TM1833.
Arabidopsis ABA8′OH mutant (cyp707A2) has shown an increased responsiveness to exogenously applied ABA in comparison with wild type (Okamoto et al. 2006). In TM1833, ABA8′OH activity may be suppressed through decreases in TaABA8′OH1 expression and then, the responsiveness of seeds to applied ABA may change. Alternatively, an increased ABA content during seed development may alter ABA responsiveness directly through changes in signalling gene expression. To clarify the mechanisms of increased ABA responsiveness in TM1833, more precise experiments relate to a complex network of ABA signaling pathways may be needed.
Gubler et al. (2008) has shown that decreased HvABA8′OH1 expression in RNAi experiments with transgenic barley results in reduced germination in freshly harvested grains, but its effect is small in after-ripened grains. They has also shown that the reduced HvABA8′OH1 expression in RNAi grains had only a small effect on after-ripening time compared to wild-type and null grains. A double mutant in TaABA8′OH1s showed reduced germination than those in a single mutant in TaABA8′OH1 in freshly harvested seeds (Fig. 6A, 6B), but had only a small effect on germination inhibition in non-dormant seeds (Fig. 6C). The manipulation of TaABA8′OH1 might provide an attractive opportunity to prevent pre-harvest sprouting in wheat, which may not restrain the replanting of newly harvested seeds in field.
Recently, Ji et al. (2011) reported that TaABA8′OH1 deletions affected drought tolerance at the reproductive stage. Drought treatment causes a loss in grain number in TaABA8′OH1 deletions, which accumulates as significantly higher spike ABA levels than that in the wild type. In our field conditions, TM1833 did not show lower grain numbers than those in ‘Tamaizumi’ (data not shown). Japan is located in an Asian monsoon climate region, which brings much rain to the country. In our field conditions, there were no big differences in agricultural characteristics, such as plant type, plant height, spike length, heading date, harvesting time, or fertility between ‘Tamaizumi’ and TM1833 (data not shown). To investigate the effect of mutation(s) in TaABA8′OH1 on grain numbers and other agricultural characteristics under field conditions in Japan, more precise experiments using ‘Tamaizumi’ and TM1833 may be required.
Supplementary Materials
Acknowledgements
We acknowledge Prof. T.R. Endo, Graduate School of Agriculture, Kyoto University, Kyoto, Japan for his kindness in giving us the experimental materials. We heartily thank Dr. E. Nambara and Dr. T. Kushiro for their helpful comments. We thank M. Nakamura, K. Yokoi, K. Suzuki, C. Amemiya, M. Someya, A. Yamase, Y. Maki and M. Yamada at NARO Institute of Crop Science for their technical assistance; members of the field cultivation team at National Agricultural Research Center for cultivation of experimental material; Dr. H. Hanai for his helpful comments and ongoing support. This work was supported in part by a “Development of innovative crops through the molecular analysis of useful genes” project grant from the National Agriculture and Food Research Organization.
Literature Cited
- Bewley, J.D. (1997) Seed germination and dormancy. Plant Cell 9: 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chono, M., Honda, I., Shinoda, S., Kushiro, T., Kamiya, Y., Nambara, E., Kawakami, N., Kaneko, S., and Watanabe, Y. (2006) Field studies on the regulation of abscisic acid content and germinability during grain development of barley: molecular and chemical analysis of pre-harvest sprouting. J. Exp. Bot. 57: 2421–2434 [DOI] [PubMed] [Google Scholar]
- Endo, T.R., and Gill, B.S. (1996) The deletion stocks of common wheat. J. Hered. 87: 295–307 [Google Scholar]
- Fang, J., and Chu, C. (2008) Abscisic acid and the pre-harvest sprouting in cereals. Plant Signal Behav. 3: 1046–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flintham, J.E. (2000) Different genetic components control coat-imposed and embryo-imposed dormancy in wheat. Seed Science Research 10: 43–50 [Google Scholar]
- Gubler, F., Millar, A.A., and Jacobsen, J.V. (2005) Dormancy release, ABA and pre-harvest sprouting. Curr. Opin. Plant Biol. 8: 183–187 [DOI] [PubMed] [Google Scholar]
- Gubler, F., Hughes, T., Waterhouse, P., and Jacobsen, J. (2008) Regulation of dormancy in barley by blue light and after-ripening: effects on abscisic acid and gibberellins metabolism. Plant Physiol. 147: 886–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himi, E., Mares, D.J., Yanagisawa, A., and Noda, K. (2002) Effect of grain colour gene (R) on grain dormancy and sensitivity of the embryo to abscisic acid (ABA) in wheat. J. Exp. Bot. 53: 1569–1574 [DOI] [PubMed] [Google Scholar]
- Ji, X., Dong, B., Shiran, B., Talbot, M.J., Edlington, J.E., Hughes, T., White, R.G., Gubler, F., and Dolferus, R. (2011) Control of abscisic acid catabolism and abscisic acid homeostasis is important for reproductive stage stress tolerance in cereals. Plant Physiol. 156: 647–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami, N., Miyake, Y., and Noda, K. (1997) ABA insensitivity and low ABA levels during seed development of non-dormant wheat mutants. J. Exp. Bot. 48: 1415–1421 [Google Scholar]
- Kawasaki, S., and Murakami, Y. (2000) Genome analysis of Lotus japonicas. J. Plant Res. 113: 497–506 [Google Scholar]
- Kermode, A.R. (2005) Role of abscisic acid in seed dormancy. J. Plant Growth Regul. 24: 319–344 [Google Scholar]
- King, R.W. (1993) Manipulation of grain dormancy in wheat. J. Exp. Bot. 44: 1059–1066 [Google Scholar]
- Kobayashi, F., Takumi, S., and Handa, H. (2010) Identification of quantitative trait loci for ABA responsiveness at the seedling stage associated with ABA-regulated gene expression in common wheat. Theor. Appl. Genet. 121: 629–641 [DOI] [PubMed] [Google Scholar]
- Kushiro, T., Okamoto, M., Nakabayashi, K., Yamagishi, K., Kitamura, S., Asami, T., Hirai, N., Koshiba, T., Kamiya, Y., and Nambara, E. (2004) The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J. 23: 1647–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matakiadis, T., Alboresi, A., Jikumaru, Y., Tatematsu, K., Pichon, O., Renou, J.P., Kamiya, Y., Nambara, E., and Truong, H.N. (2009) The Arabidopsis abscisic acid catabolic gene CYP707A2 plays a key role in nitrate control of seed dormancy. Plant Physiol. 149: 949–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaig, T.N., and Depauw, R.M. (1992) Breeding for preharvest sprouting tolerance in white-seed-coat spring wheat. Crop Sci. 32: 19–23 [Google Scholar]
- Millar, A.A., Jacobsen, J.V., Ross, J.J., Helliwell, C.A., Poole, A.T., Scofield, G., Reid, J.B., and Gubler, F. (2006) Seed dormancy and ABA metabolism in Arabidopsis and barley: the role of ABA 8′-hydroxylase. Plant J. 45: 942–954 [DOI] [PubMed] [Google Scholar]
- Munkvold, J.D., Tanaka, J., Benscher, D., and Sorrells, M.E. (2009) Mapping quantitative trait loci for pre-harvest sprouting resistance in white wheat. Theor. Appl. Genet. 119: 1223–1235 [DOI] [PubMed] [Google Scholar]
- Nakamura, S., Chono, M., Abe, F., and Miura, H. (2010) Mapping a diploid wheat abscisic acid 8′-hydroxylase homologue in the seed dormancy QTL region on chromosome 5Am. Euphytica 171: 111–120 [Google Scholar]
- Nambara, E., and Marion-Poll, A. (2005) Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56: 165–185 [DOI] [PubMed] [Google Scholar]
- Okamoto, M., Kuwahara, A., Seo, M., Kushiro, T., Asami, T., Hirai, N., Kamiya, Y., Koshiba, T., and Nambara, E. (2006) CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 141: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radley, M. (1976) The development of wheat grain in relation to endogenous growth substances. J. Exp. Bot. 27: 1009–1021 [Google Scholar]
- Rikiishi, K., and Maekawa, M. (2010) Characterization of a novel wheat (Triticum aestivum L.) mutant with reduced seed dormancy. J. Cereal Sci. 51: 292–298 [Google Scholar]
- Saito, S., Hirai, N., Matsumoto, C., Ohigashi, H., Ohta, D., Sakata, K., and Mizutani, M. (2004) Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 134: 1439–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm, E.C., Abellera, J.C., Strader, L.C., Campbell, K.G., and Steber, C.M. (2010) Isolation of ABA-responsive mutants in allohexaploid bread wheat (Triticum aestivum L.): drawing connections to grain dormancy, preharvest sprouting, and drought tolerance. Plant Sci. 179: 620–629 [Google Scholar]
- Sourdille, P., Singh, S., Cadalen, T., Brown-Guedira, G.L., Gay, G., Qi, L., Gill, B.S., Dufour, P., Murigneux, A., and Bernard, M. (2004) Microsatellite-based deletion bin system for the establishment of genetic-physical map relationships in wheat (Triticum aestivum L.). Funct. Integr. Genomics. 4: 12–25 [DOI] [PubMed] [Google Scholar]
- Walker-Simmons, M. (1987) ABA levels and sensitivity in developing wheat embryos of sprouting resistant and susceptible cultivars. Plant Physiol. 84: 61–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker-Simmons, M., and Sesing, J. (1990) Temperature effects on embryonic abscisic acid levels during development of wheat grain dormancy. J. Plant Growth Regul. 9: 51–56 [Google Scholar]
- Wang, Y.H. (2008) How effective is T-DNA insertional mutagenesis in Arabidopsis? J. Biochem. Tech. 1: 11–20 [Google Scholar]
- Warner, R.L., Kudrna, D.A., Spaeth, S.C., and Jones, S.S. (2000) Dormancy in white-grain mutants of Chinese Spring wheat (Triticum aestivum L.). Seed Sci. Res. 10: 51–60 [Google Scholar]
- Werck-Reichhart, D., Hehn, A., and Didierjean, L. (2000) Cytochromes P450 for engineering herbicide tolerance. Trends Plant Sci. 5: 116–123 [DOI] [PubMed] [Google Scholar]
- Wiedenhoeft, M.H., Chevalier, P., Walker-Simmons, M., and Ciha, A.J. (1988) Field studies on abscisic acid and embryonic germinability in winter wheat. Field Crops Res. 18: 271–278 [Google Scholar]
- Zhang, C.-L., He, X.-Y., He, Z.-H., Wang, L.-H., and Xia, X.-C. (2009) Cloning of TaCYP707A1 gene that encodes ABA 8′-hydroxylase in common wheat (Triticum aestivum L.). Agric. Sci. China 8: 902–909 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.