Abstract
GRAIN SIZE 3 (GS3) is a cloned gene that is related to seed length. Here we report the discovery of new deletion alleles at the GS3 locus, each of which confer short seed. We selected ten short seeded cultivars from a collection of 282 diverse cultivars. Sequence analysis across the GS3 gene in these ten cultivars identified three novel alleles and a known allele that contain several independent deletion(s) in the fifth exon of GS. These independent deletion variants each resulted in a frameshift mutation that caused a premature stop codon, and they were functionally similar to one another. Each coded for a truncated gene product that behaved as an incomplete dominant allele and conferred a short seeded phenotype. Haplotype analysis of these sequence variants indicated that two of the variants were of japonica origin, and two were from indica. Transformation experiments demonstrated that one of the deletion alleles of GS3 decrease the cell number in the upper epidermis of the glume, resulting in a significant reduction in seed length. The multiple and independent origins of these short seeded alleles indicate that farmers and early breeders imposed artificial selection favoring short seeds.
Keywords: seed length, frameshift mutation, haplotype analysis, diversity, cell number
Introduction
Rice (Oryza sativa L.) is the most important staple crop for billions of people worldwide. Given rapid population and income growth in many rice-eating parts of the world, the breeding of high-yielding and high-quality rice cultivars is critical to meet growing demand (Khush 1999). Rice seed shape is a breeding target because it directly affects grain weight, which is an important component of grain yield. In addition, rice seed shape is an important determinant of consumer preference around the world. Long, slender grains are preferred by many consumers in India, Pakistan, Thailand, China and the United States, while consumers in Japan, South Korea and Sri Lanka prefer short, bold grained varieties (Juliano and Villareal 1993, Unnevehr et al. 1992).
There is greater variation for seed length found among cultivated varieties than in wild rice, probably due to human selection (Takano-Kai et al. 2009). Preferences for different seed sizes and shapes are dependent on how rice is cooked, processed and consumed. For example, increasing the amount of rice bran available for extraction of rice oil can be most easily accomplished by reducing the size of the rice seed, which increases the surface area per volume of brown rice. There is evidence that breeders have selected for short seed size as well as large seed size in rice (Mikami et al. 2004).
Many genes are known to control seed size and several independent studies based on inter- and intraspecific crosses of rice have previously identified quantitative trait loci (QTLs) associated with seed length (Li et al. 2004, Redoña and Mackill 1998, Tan et al. 2000, Tsunematsu et al. 1995), some of which have been identified and characterized. GRAIN SIZE3 (GS3) encodes a protein with several conserved domains, including a phosphatidylethanolamine-binding protein (PEBP)-like domain, a transmembrane region (TM), a putative tumour necrosis factor receptor/nerve growth factor receptor (TNFR/NGFR) family domain and a von Willebrand factor type C (VWFC) domain (Fan et al. 2006, Takano-Kai et al. 2009). GRAIN WEIGHT2 (GW2) encodes an unknown RING-type protein with E3 ubiquitin ligase activity (Song et al. 2007). The identical gene of qSW5 and GW5 has no apparent homolog in the database but was shown to interact with the polyubiquitin-proteasome pathway to regulate cell division during seed development (Shomura et al. 2008, Weng et al. 2008).
Among the genes known to regulate seed length, GS3 is an interesting case because its mutants both positively and negatively regulate seed length. GS3 consists of five exons. The wild type allele of GS3 results in a medium seed phenotype. A C-to-A nonsense mutation in the second exon of GS3 results in a long seeded phenotype (Fan et al. 2006, Takano-Kai et al. 2009). A 1-bp deletion in the fifth exon of GS3, leading to a frameshift mutation at the C terminus, produced a short seeded phenotype due to a truncated protein that lacked the TNFR/NGFR and VWFC domains (Mao et al. 2010).
Here we report the identification of novel, incomplete dominant alleles at GS3 that all confer short seeds. The variants were identified based on sequencing across the GS3 locus in ten short seeded cultivars, each having seed length <6.5 mm. These alleles represent similar functional mutations, suggesting that the fifth exon of the GS3 gene is a hotspot for mutation and has been the target of selection by many independent groups of humans during the evolution of O. sativa. We also examined sequence haplotypes across the GS3 gene in short seeded cultivars to identify the subpopulation origin of these alleles.
Materials and Methods
Plant materials used in survey for short seeded cultivars
We surveyed seed size in 281 diverse cultivars maintained in the Plant Breeding Laboratory, Faculty of Agriculture, Kyushu University (Supplemental Table 1). Eighty one of these cultivars overlapped with the 235 cultivars of O. sativa previously investigated for grain size (Takano-Kai et al. 2009). Included in this collection was the short seeded rice mutant line, H343, maintained in Hokkaido University. H343 carries the MINUTE (Mi) locus for short seeds that was previously mapped near GS3 on chromosome 3 (Fraker et al. 2004, Takamure et al. 1991, Takeda and Saito 1977). Seed length in the 282 rice strains was measured using a caliper (KORI Dial Caliper, KORI SEIKI, Tokyo, Japan).
Sequence analysis of GS3
Approximately 6 kbp of genomic sequence across the GS3 gene was generated from the ten short grained strains. This region contained 1122-bp upstream from the start codon and 119-bp downstream from the stop codon. Three pairs of PCR primers were designed to amplify overlapping regions within the gene, and internal primers within each amplicon were designed to sequence the PCR products. PCR reactions to generate the sequencing template were performed in 25 μl of reaction mixture containing 1× KOD-Plus PCR buffer, 1 mM MgSO4, 200 μM of each dNTP, 0.2 μM of each primer, 1 unit of KOD-Plus DNA polymerase (TOYOBO, Osaka, Japan) and approximately 25 ng of template DNA in a GeneAmp PCR 9700 system (Applied Biosystems, Foster City, CA, USA). The PCR program used was 95°C for 2 min, followed by 35 cycles of 98°C for 30 s, 60°C for 30 s and 68°C for 5 min. The PCR products were sequenced with the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA, USA) using the ABI 3130X genetic analyzer. Sequences were assembled and aligned using the Sequencher program (Gene Codes, Ann Arbor, MI).
Plant materials for genetic analysis
A medium seeded temperate japonica cultivar, Taichung 65 (T65), a short seeded aromatic cultivar, JC73-4, an F1 and an F2 population derived from a cross between T65 and JC73-4 were genetically analyzed. The seed lengths of the T65, JC73-4, F1 and F2 population were measured by the method described above.
DNA extraction and genetic analysis
Genomic DNA for the simple sequence repeat (SSR) analysis was extracted from freeze-dried leaf samples using a modified potassium acetate-SDS method (Dellaporta et al. 1983). The PCR reactions were performed in 15 μl of reaction mixture containing 50 mM KC1, 10 mM Tris-HCl (pH 9.0), 1.5 mM MgCl2, 200 μl of each dNTP, 0.2 μM of each primer, 0.75 units Taq polymerase (TaKaRa, Otsu, Japan) and approximately 25 ng template DNA in a GeneAmp PCR 9700 system (Applied Biosystems, Foster City, CA, USA). The PCR program used was 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, 55°C for 30 s and 72°C for 30 s. PCR products were run in 4% agarose gels (Agarose HT; Amresco Inc., Solon, OH, USA) in 0.5× TBE buffer.
Vector construction and plant transformation
To generate the GS3 construct used to introduce the 320-bp deletion into the medium seeded cultivar, Nipponbare, a construct used previously by Takano-Kai et al. (2009) was used as the mutagenesis template. The primers used for the mutagenesis were GCAGCAGCAGCTCGGCCTCTTGAGGTTGAA and GCGCGTACTTAATTAGCTCAAGATTAGTTCTCGATGATGATC, each containing a modified phosphoric acid at the 5′ end. PCR reactions for mutagenesis were performed in 50 μl of reaction mixture containing 1× KOD-Plus PCR buffer, 1 mM MgSO4, 200 μM each dNTP, 0.2 μM each primer, 1 unit of KOD-Plus DNA polymerase and approximately 1 μg template construct in a GeneAmp PCR 9700 system. The PCR program used was 95°C for 2 min, followed by 20 cycles of 98°C for 10 s and 68°C for 17 min. The PCR product was re-circularized using a DNA ligation kit, Ligation High (TOYOBO, Osaka, Japan) and was introduced into competent bacteria Escherichia coli DH5α (TOYOBO). Positive colonies were identified by PCR screening using the two primer sets TCCCACAAAACCATCAACTTGTTA and CAAGCAGAGCAAGGCAATCAA and GGGAATAGGGATTCCACACG and AATGGTGATGCCCAGCTTTT. The presence of the 320-bp deletion was confirmed by DNA sequencing. This construct (referred to here as the GS3-d construct) (Supplemental Fig. 1) was introduced into Nipponbare by Agrobacterium-mediated transformation (Toki 1997).
GS3 from Nipponbare was used as a control (referred to here as the GS3 construct) (Supplemental Fig. 1). In addition, the RNA interference (RNAi) trigger region was amplified by PCR with the primers CACCCGCGAGATCGGATTCCTC and AGCGACACGGACTCTTCGTT and then inserted into the pENTR/D-TOPO cloning vector (Invitrogen, Carlsbad, CA, USA) to yield an entry vector. The RNA silencing vector was produced by an LR Clonase-catalysed reaction (Invitrogen) between the entry vector and pANDA (Miki and Shimamoto 2004, referred to here as the GS3-p construct). The GS3-p construct (Supplemental Fig. 1) was also used as a control. These two constructs, GS3 and GS3-p, were also introduced into Nipponbare by Agrobacterium-mediated transformation.
For measurement of seed length of the transformants, five seeds from each of five T0 plants were used.
Measurement of cell number and cell size in the upper epidermis of the rice glume
Mature seeds were coated with gold–palladium and viewed under a scanning electron microscope (SEM; JSM-5200, Jeol, Tokyo, Japan). Longitudinal surface images of each seed were recorded as sequential JPEG files. The sequential images of each seed were then merged into a single image using Photoshop software. Longitudinal rows of tubercles were observed on the outer surface of the rice hull. These tubercles are formed by accumulation of silicic acid in the cell walls of the upper epidermal cell (Takeoka 1976). The cell number of upper epidermis in the longitudinal tubercles was counted from the apex to the base of the seed in transgenic plants. The cell number of upper epidermal cells of each transgenic line was calculated from the mean of three values from each of five plants. In addition, the length of upper epidermal cells of the transgenic plants was measured by determining the length between the tubercles in the middle of the glume. The cell size of upper epidermal cells of the transgenic lines was calculated from the mean of 120 values per seed from five independent seeds. The number and length of cells in the palea and lemma were obtained using the ImageJ software (http://rsb.info.nih.gov/ij/).
Results
Sequence analysis of GS3 in short seeded cultivars
A diverse collection of 282 strains of O. sativa (Supplemental Table 1) was evaluated for seed size. The seed length of the 282 strains varied from 4.99 mm to 10.52 mm. In our previous study, we surveyed the sequence variant of the GS3 locus among 54 rice strains and no functional nucleotide polymorphism was found except for a nonsense mutation in the second exon, which results in a long seeded phenotype (Takano-Kai et al. 2009). All of the 54 strains showed the seed length of 6.56–10.67 mm. Therefore, ten out of 282 strains (3.5%) that had a seed length <6.5 mm (Fig. 1A and Table 1) were subsequently subjected for sequence analysis of GS3.
Fig. 1.
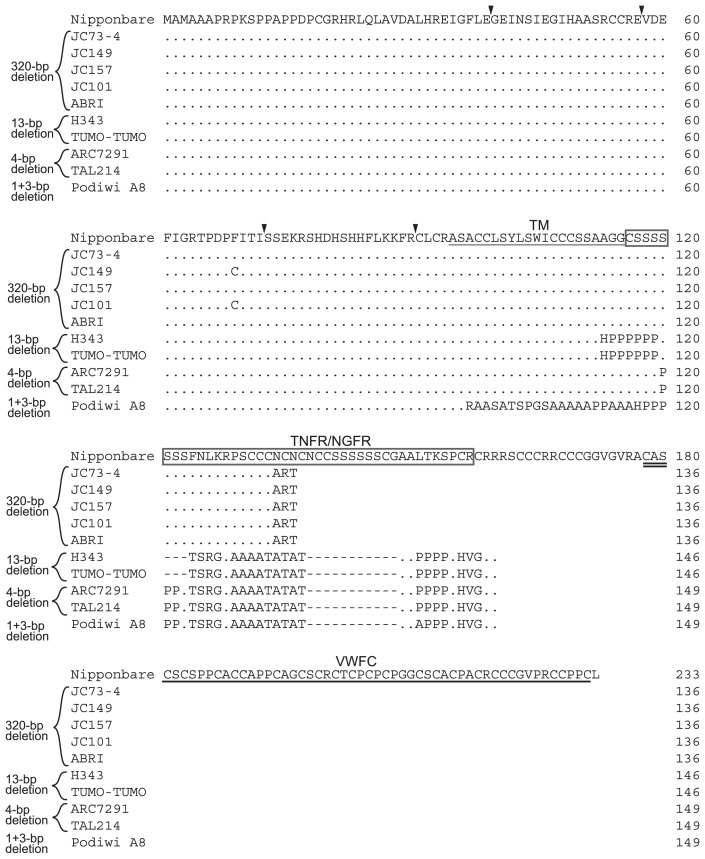
Seed length comparison of the selected short seeded cultivars and sequence analysis of GS3 in the short seeded cultivars. (A) The seeds of Nipponbare and short seeded cultivars. From left to right, Nipponbare, ARC7291, Podiwi A8, JC149, JC157, JC101, TAL214, Tumo-Tumo, JC73-4, ABRI and H343. Bar is 6.5 mm. (B) The nucleotide sequences from the fifth exon of Nipponbare GS3 are shown. The red boxes indicate the sites of deletion. Blue characters indicate the stop codon. The numbers indicate the nucleotide numbers of Nipponbare GS3.
Table 1.
Plant material used for the sequence analysis of GS3
| Accession name | Origin of Country | Seed length (mm ± SD) |
|---|---|---|
| JC73-4 | India | 5.92 ± 0.14 |
| JC149 | India | 6.26 ± 0.18 |
| JC157 | India | 6.14 ± 0.27 |
| JC101 | India | 6.00 ± 0.07 |
| ABRI | Bhutan | 5.66 ± 0.15 |
| Tumo-Tumo | Malaysia | 5.94 ± 0.18 |
| ARC7291 | India | 6.46 ± 0.05 |
| TAL214 | Taiwan | 5.97 ± 0.15 |
| Podiwi A8 | Sri lanka | 6.43 ± 0.16 |
| H343 | Japan | 4.99 ± 0.01 |
A total of 78 SNPs and 26 indels relative to Nipponbare were identified among the selected 10 strains in the sequence analysis of GS3. Of these mutations, only deletion(s) in the fifth exon of GS3 were observed in all 10 strains. JC73-4, JC149, JC157, JC101 and ABRI showed a 320-bp deletion 5064-bp downstream from the start codon (referred to as the 5064-bp site); H343 and Tumo-Tumo had a 13-bp deletion at the 5000-bp site; ARC7291 and TAL214 had a 4-bp deletion at the 5021-bp site (similar to the allele found in Chuan 7 by Mao et al. 2010); Podiwi A8 had a 1-bp deletion at the 4953-bp site and a 3-bp deletion at the 5016-bp site (l + 3-bp deletions, Fig. 1B).
Deletion of the fifth exon alleles in GS3 causes a frameshift mutation resulting in a premature stop codon
The 320-bp, 13-bp, 4-bp and 1 + 3-bp deletions of GS3 each cause a frameshift that generates a premature stop codon (Fig. 2). The deduced amino acid sequence of the 320-bp deletion allele of GS3 (JC73-4, JC149, JC157, JC101 and ABRI) indicated that the product lacked a part of the TNFR/NGFR domain and the entire VWFC domain. The deduced amino acid sequence of the 13-bp deletion allele (H343 and Tumo-Tumo) indicated that the product lacked all of the TNFR/NGFR and VWFC domains. Although the three residues of the TM of the 13-bp deletion allele were changed, the domain prediction database (http://www.ebi.ac.uk/Tools/pfa/iprscan/) found that the product had a TM. The deduced amino acid sequence of the 4-bp deletion allele of GS3 (ARC7291 and TAL214) revealed that the product also lacked all of the TNFR/NGFR and VWFC domains, and these sequences were the same as that of Chuan 7 (Mao et al. 2010). The deduced amino acid sequence of the 1 + 3-bp deletion allele of GS3 (Podiwi A8) also lacked the TM, three domains of TNFR/NGFR and VWFC, and encoded a different C-terminus compared to the GS3 locus of Nipponbare.
Fig. 2.
Alignment of the deduced amino acid sequence of GS3 of Nipponbare and the 10 short seeded cultivars. The single solid underline indicates the transmembrane region (TM), the double underlines indicate the TNFR/NGFR family cysteine-rich domain (TNFR/NGFR) and the boxes indicate the von Willebrand factor type C (VWFC) domain. The triangles indicate the exon/exon boundaries of GS3. Dots indicate the same residues as Nipponbare sequence and hyphens indicate the deletions. Numbers at right side indicate the number of residues.
Mao et al. (2010) demonstrated that the functional short-seeded allele in the variety Chuan 7 encoded a gene product that lacked the TNFR/NGFR and VWFC domains. Therefore, since all of the GS3 variants found in this study produce GS3 products lacking the normal TNFR/NGFR and VWFC domains, we conclude that this is the reason they confer short seeded phenotypes.
Origin of deletion alleles by haplotype analysis
To investigate the origin of the deletions in the 5th exon of GS3, we examined GS3 sequence haplotypes in the 10 short seeded varieties. A total of 104 SNPs/indels were identified in the 10 short seeded strains relative to Nipponbare. Sixty one of these SNPs/indels were identical to those used for haplotype analysis in a previous study (Takano-Kai et al. 2009) and they were used to determine the origin of the haplotypes found in the 10 short seeded cultivars in this study.
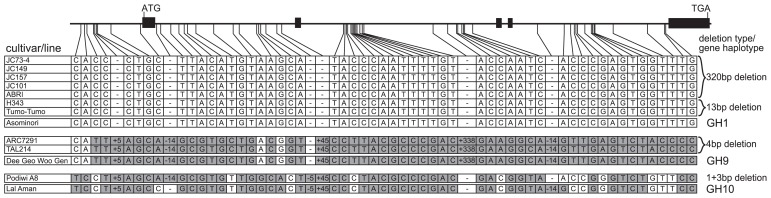
The GS3 gene haplotypes from the 10 short seeded cultivars were classified into three groups. The first gene haplotype group included cultivars carrying the 320-bp deletion (JC73-4, JC149, JC157, JC101 and ABRI) and those carrying the 13-bp deletion (H343 and Tumo-Tumo). This gene haplotype group corresponded to the previously identified gene haplotype group 1 (GH1) that was reported to be of japonica origin (Takano-Kai et al. 2009). The second gene haplotype included cultivars carrying the 4-bp deletion (ARC7291 and TAL214) and corresponded to GH9 that was previously reported to be of indica origin. The last gene haplotype included the cultivar carrying the 1 + 3 deletion (Podiwi A8) and was most similar to GH10, previously reported to be of indica origin (Fig 3). These results demonstrated that the 320-bp and 13-bp deletions occurred in a japonica-like ancestor and that the 4-bp and 1 + 3-bp deletions occurred in an indica-like ancestor.
Fig. 3.
Haplotype variation of GS3 in short seeded cultivars. The gene model for GS3 containing five exons and comprising approximately 6.2 kb is shown horizontally along the top. SNP positions with GS3 are connected by lines to the table below. Columns indicate the distribution of polymorphisms at each SNP/indel positions. Numbers indicate the size of an indel relative to Nipponbare. Asominori is one of cultivars belonging to japonica haplotype GH1, Dee Geo Woo Gen is one of cultivars belonging to indica haplotype GH9 and Lal Aman is a cultivar belonging to indica haplotype GH10 (Takano-Kai et al. 2009). White indicates the common GH1 and grey indicates a variant SNP allele.
Analysis of the JC73-4 with 320-bp deletion allele
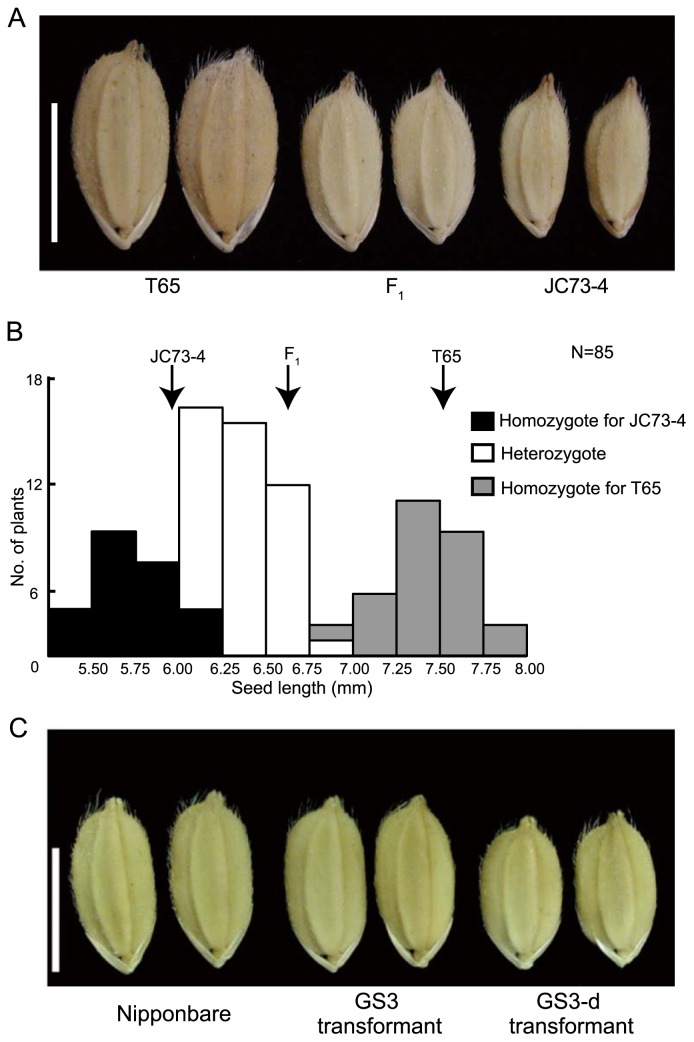
We performed genetic analysis of JC73-4 with the 320-bp deletion allele. The F1 and F2 populations derived from a cross between T65 and JC73-4 were used. T65 with wild-type allele of GS3 showed medium seed length and the F1 plants showed an intermediate seed length (6.62 ± 0.11 mm) relative to the two parents (Fig. 4A). The seed length of the F2 population showed a trimodal frequency distribution (Fig. 4B). The 85 plants in the F2 population were divided into three groups: the short seed group (seed length: 5.41–5.93 mm), the intermediate seed group (seed length: 6.01–6.86 mm) and the medium seed group (seed length: 6.92–7.77 mm); 17 plants had short seeds, 43 plants had intermediate seeds and 25 plants had medium seeds. This was consistent with a 1 : 2 : 1 segregation ratio (chi-square = 1.518, P = 0.468), indicating that short seed length was governed by a single, incompletely dominant allele coming from JC73-4. We designed PCR primers to detect the 320-bp deletion and genotyped each individual of the F2 population. All T65 homozygous plants had medium seeds, the heterozygous plants and three JC73-4 homozygotes had intermediate seeds and 17 of the JC73-4 homozygotes had short seeds (Fig. 4B). Thus, seed length co-segregated with the 320-bp deletion in the GS3 gene.
Fig. 4.
Genetic analysis of seed length in JC73-4 and transformation experiment for GS3 including the 320-bp deletion. (A) The seeds of T65, F1 and JC73-4. Bar is 5 mm. (B) Frequency distribution of the seed length in the F2 population, classified by the genotypes of a marker to detect the 320-bp deletion. (C) The seeds of Nipponbare, GS3 transformants and GS3-d transformants. Bar is 5 mm.
Next, we performed a set of transgenic experiments to obtain direct evidence that the 320-bp deletion of GS3 caused the short seeded phenotype. Using site-directed mutagenesis, a plasmid construct (GS3-d construct) was generated from the Nipponbare sequence containing the 320-bp deletion. Nipponbare GS3 (GS3 construct) was used as a control (Supplemental Fig. 1). Each construct was transfected into Nipponbare.
The seed length of the regenerated transgenic plants transformed with the GS3-d construct (GS3-d transformant, 6.43 ± 0.12 mm) was significantly shorter than that of the control (GS3 transformant, 7.12 ± 0.14 mm) (Fig. 4C). These results indicate that the 320-bp deletion allele is a functional form of GS3 that causes the short seeded phenotype. The culm length of GS3-d transformant was shorter than that of GS3-transformant (data not shown).
GS3 decreases cell number in the upper epidermis of the glume
To investigate the effect of GS3 on seeds at the cellular level, we measured the total number of cells and mean cell length in the upper epidermis of the glume using transgenic plants.
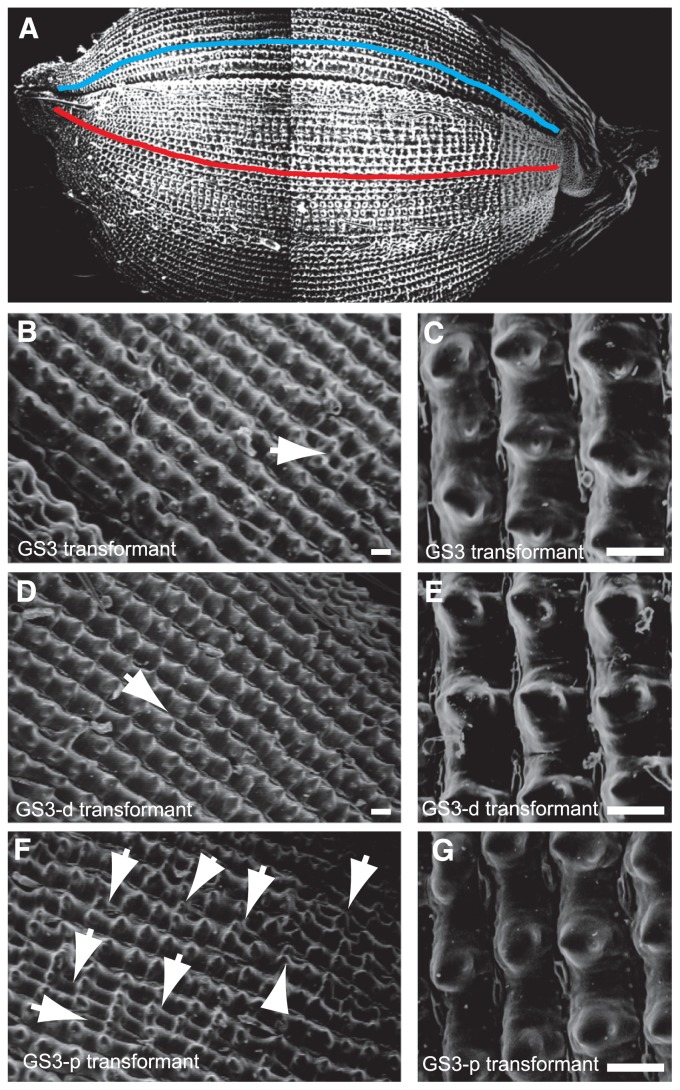
We measured the cell number and cell size in the upper epidermis of the lemma and palea of GS3 transformants, GS3-d transformants and GS3-p transformants using SEM (Fig. 5). Although the cell size of the GS3-d transformants was smaller than that of the GS3 and GS3-p transformants, no significant difference was observed between the cell sizes of the GS3 and GS3-p transformants in the lemma or the palea. Conversely, significant differences were observed in the cell numbers of the GS3, GS3-d and GS3-p transformants in both the lemma and palea (Table 2); namely, the 320-bp deletion decreased the cell number, while a knockdown construct utilizing RNAi increased the cell number. These results indicate that GS3 controls cell number in the upper epidermis of the glume.
Fig. 5.
Scanning electron micrographs of the upper epidermis in the transgenic plants. (A) The three sequential images of a seed were merged into a single image. The cell numbers of the upper epidermis were counted in the longitudinal tubercles that are formed from the apex to the base of a seed. Red and blue lines indicate a row of longitudinal tubercles at the lemma and palea, respectively. (B, C) The upper epidermis in the GS3 transformant. (D, E) The upper epidermis in the GS3-d transformant. (F, G) The upper epidermis in the GS3-p transformant. Arrows indicate the irregular thin rows. Bars are 50 μm.
Table 2.
Cell number and size of the upper epidermis of transgenic lines
| Line | Palea | Lemma | ||
|---|---|---|---|---|
|
|
|
|||
| Cell number | Cell size (μm ± SE) | Cell number | Cell size (μm ± SE) | |
| GS3 transformant | 92.17 ± 1.93a | 79.71 ± 1.98a | 82.86 ± 1.81a | 95.97 ± 1.95a |
| GS3-d transformant | 72.33 ± 1.85b | 66.66 ± 1.33b | 71.40 ± 1.45b | 80.96 ± 1.72b |
| GS3-p transformant | 109.00 ± 1.14c | 84.30 ± 1.41a | 98.31 ± 1.65c | 92.45 ± 1.31a |
Multiple means comparisons based on Tukey-Kramer Honestly Significant Difference (HSD) test. Levels not connected by the same letter are significantly different.
The surface of the outer glume of rice has rows of stripes along the longitudinal axis of the rice seed. The GS3-p transformant glume had more irregular, thinner rows of stripes than GS3 and GS3-d transformant glumes (Fig. 5). The loss of function of GS3 leads to an increase in irregular cells.
Discussion
GS3 has multiple alleles conferring the short seeded phenotype
In this study, we evaluated the seed size of 282 diverse rice strains and identified three novel GS3 alleles with independent deletions that all map to the fifth exon of the gene (320-bp, 13-bp and 1 + 3-bp deletion). We also identified a variety carrying a fourth deletion (4-bp deletion) that had been previously shown to be a determinant of short seed size in rice (Mao et al. 2010). Using genetic and transgenic analysis, we demonstrated that the 320-bp deletion causes a short seeded phenotype. The other two alleles discovered here (13-bp, 1 + 3-bp deletions) caused premature stops and were found to give rise to identical GS3 amino acid sequences, with gene products that were identical to those generated by the 4-bp deletion allele and 320-bp deletion allele found in Chuan 7 (Mao et al. 2010) and in JC73-4 (this study), respectively. From these results, we conclude that diverse deletion alleles of GS3 are all responsible for the short seeded phenotype.
The short seeded alleles of GS3 have both japonica and indica origins
We showed that the 320-bp and 13-bp deletions in the fifth exon of GS3 occurred in a japonica-like ancestor, and the 4-bp and 1 + 3-bp deletions occurred in an indica-like ancestor. Thus, we conclude that multiple GS3 haplotypes conferring short seeds occurred independently in the japonica and indica groups. This is in contrast to the mutational history of the long seeded phenotype where a unique, nonsense mutation occurred in the second exon of GS3 in the japonica gene pool or in a japonica-like ancestor and was disseminated into the indica gene pool via introgression during the process of rice domestication (Takano-Kai et al. 2009).
The short or small seeded phenotype of rice was occasionally a target of artificial selection in some restricted areas. The short seeded cultivars JC73-4, JC149, JC157 and JC101 are aromatic cultivars collected from Orissa, India by Oka (1957). Although aromatic rice which is popular as a premium rice in world markets has long seeds and grains, the majority of indigenous Indian aromatic rice varieties have short or medium sized seeds (Singh et al. 2000). Aromatic, short grained rice in India is widely used by local consumers for making kheer (sweet rice) for religious and festive occasions (Rani et al. 2006).
The function of GS3 in rice seed
GS3 has previously been shown to regulate stigma length and is involved in stigma exsertion in rice. A nonsense mutation in the second exon of GS3 results in an increase in cell number, resulting in stigma elongation (Takano-Kai et al. 2011). In this study, we demonstrated that, in addition to controlling stigma length, GS3 controls cell number in the upper epidermis of the glume. Several genes that regulate cell number have been isolated, and one of them is DENSE AND ERECT PANICLE1 (DEP1), a gene involved in rice grain yield (Huang et al. 2009, Taguchi-Shiobara et al. 2011, Wang et al. 2009, Zhou et al. 2009). DEP1 is reported to carry a previously unknown PEBP-like domain protein sharing some homology with the N terminus of GS3, and three VWFC domains in the C-terminus (Huang et al. 2009, Wang et al. 2009). The dominant allele at the DEP1 locus (dep1) is a gain-of-function mutation that truncates at the C-terminus of DEP1. The effect of dep1 is to enhance meristematic activity, resulting in reduced length of the inflorescence internode, an increase in the number of grains per panicle, and a consequential increase in grain yield (Huang et al. 2009). The domain structure of dep1 is very similar to that of the 320-bp, 13-bp, 4-bp and 1 + 3-bp deletion alleles of GS3 identified in this study, because the amino acid sequence of dep1 lacks two VWFC domains in the C-terminus. Recently, an atypical heterotrimeric G-protein γ-subunit (AGG3) was identified in Arabidopsis thaliana (Chakravorty et al. 2011). AGG3 contains a γ-subunit in its N terminus followed by a putative transmembrane domain and a C-terminal cysteine-rich region. The γ-subunit of AGG3 interacts with the Arabipopsis G-protein β-subunit (Chakravorty et al. 2011) and regulates organ size and shape in A. thaliana (Li et al. 2012). GS3 and DEP1 in rice are homologues of AGG3 (Chakravorty et al. 2011). Botella (2012) speculates that GS3 and DEP1 are G-protein γ-subunits and interact with rice G-protein β-subunit, resulting in the inhibition of seed expansion and enhancement of meristematic activity, respectively. Moreover, Botella (2012) proposes that the cysteine-rich regions (VWFC domain) of GS3 and DEP1 have an inhibitory effect in the βγ dimer signaling of GS3 and DEP1, therefore the lack of the cysteine-rich region results in increased signaling by βγ dimer. These interpretations are consistent with the protein structures and phenotypes of the deletion alleles of GS3 identified in this study. Although it remains unclear how GS3 and DEP1 regulate cell number, elucidating that GS3 and DEP1 function as rice heterotrimeric G-proteins may help clarify the regulatory mechanism of cell division and grain development.
Supplementary Materials
Acknowledgments
This study was partly supported by a grant from the Programme for Promotion of Basic and Applied Researches for innovations in Bio-oriented industry. We are grateful the Biotron Application Center at Kyushu University, for use of the closed greenhouse to grow the transgenic plants.
Literature Cited
- Botella, J.R. (2012) Can heterotrimeric G proteins help to feed the world? Trends Plant Sci. 17: 563–568 [DOI] [PubMed] [Google Scholar]
- Chakravorty, D., Trusov, Y., Zhang, W., Acharya, B.R., Sheahan, M.B., McCurdy, D.W., Assmann, S.M., and Botella, J.R. (2011) An atypical heterotrimeric G-protein γ-subunit is involved in guard cell K+-channel regulation and morphological development in Arabidopsis thaliana. Plant J. 67: 840–851 [DOI] [PubMed] [Google Scholar]
- Dellaporta, S.L., Wood, J., and Hicks, J.B. (1983) A plant DNA minipreparation: version II. Plant Mol. Biol. Rep. 1: 19–21 [Google Scholar]
- Fan, C., Xing, Y., Mao, H., Lu, T., Han, B., Xu, C., Li, X., and Zhang, Q. (2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative trans-membrane protein. Theor. Appl. Genet. 112: 1164–1171 [DOI] [PubMed] [Google Scholar]
- Fraker, E., Li, J., and McCouch, S. (2004) SSR mapping of Minute, a major gene for grain size, on rice chromosome 3. Rice Genet. Newsl. 21: 21–23 [Google Scholar]
- Huang, X., Qian, Q., Liu, Z., Sun, H., He, S., Luo, D., Xia, G., Chu, C., Li, J., and Fu, X. (2009) Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 41: 494–497 [DOI] [PubMed] [Google Scholar]
- Juliano, B.O., and Villareal, C.P. (1993) Grain quality evaluation of world rices. International Rice Research Institute, Manila, Philippines [Google Scholar]
- Khush, G.S. (1999) Green revolution: preparing for the 21st century. Genome 42: 646–655 [PubMed] [Google Scholar]
- Li, J., Xiao, J., Grandillo, S., Jiang, L., Wan, Y., Deng, Q., Yuan, L., and McCouch, S. (2004) QTL detection for rice grain quality traits using an interspecific backcross population derived from cultivated Asian (O. sativa L.) and African (O. glaberrima S) rice. Genome 47: 697–704 [DOI] [PubMed] [Google Scholar]
- Li, S., Liu, Y., Zheng, L., Chen, L., Li, N., Corke, F., Lu, Y., Fu, X., Zhu, Z., and Bevan, M.W.et al. (2012) The plant-specific G protein γ subunit AGG3 influences organ size and shape in Arabidopsis thaliana. New Phytol. 194: 690–703 [DOI] [PubMed] [Google Scholar]
- Mao, H., Sun, S., Yao, J., Wang, C., Yu, S., Xu, C., Li, X., and Zhang, Q. (2010) Linking differential domain function of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. USA 107: 19579–19584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami, T., Takadate, M., Yokoyama, H., Kawamura, Y., Kobayashi, W., Tateyama, M., Maeda, K., Nakahori, T., and Oyamada, Z. (2004) A new rice variety ‘Tsubuyuki’. Bull. Aomori Agric. Forest. Res. Cent. 40: 39–56 [Google Scholar]
- Miki, D., and Shimamoto, K. (2004) Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 45: 490–495 [DOI] [PubMed] [Google Scholar]
- Oka, H.I. (1957) Report of study tour to India for collection of rice, 1957. In: Morishima, H. (ed.) Reports of the Study-Tours for Investigation of Wild and Cultivated Rice Species Part F. pp. 1–34 [Google Scholar]
- Rani, N.S., Pandey, M.K., Prasad, G.S.V., and Sudharshan, I. (2006) Historical significance, grain quality features and precision breeding for improvement of export quality basmati varieties in India. Indian J. Crop Sci. 1: 29–41 [Google Scholar]
- Redona, E.D., and Mackill, D.J. (1998) Quantitative trait locus analysis for rice panicle and grain characteristics. Theor. Appl. Genet. 96: 957–963 [Google Scholar]
- Shomura, A., Izawa, T., Ebana, K., Ebitani, T., Kanegae, H., Konishi, S., and Yano, M. (2008) Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 40: 1023–1028 [DOI] [PubMed] [Google Scholar]
- Singh, R.K., Singh, U.S., and Khush, G.S. (2000) Aromatic rices. Oxford & IBH Pub. Co. Pvt. Ltd., New Delhi, India [Google Scholar]
- Song, X.J., Huang, W., Shi, M., Zhu, M.Z., and Lin, H.X. (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39: 623–630 [DOI] [PubMed] [Google Scholar]
- Taguchi-Shiobara, F., Kawagoe, Y., Kato, H., Onodera, H., Tagiri, A., Hara, N., Miyao, A., Hirochika, H., Kitano, H., and Yano, M.et al. (2011) A loss-of-function mutation ofrice DENSE PANICLE 1 causes semi-dwarfness and slightly increased number of spikelets. Breed. Sci. 61: 17–25 [Google Scholar]
- Takamure, I., Yamamoto, T., and Kinoshita, T. (1991) Determination of loci for Mi (Minute grain) and Lk-f (Fusayoshi long grain) on the 3rd linkage group in rice. Jpn. J. Breed. 41(Suppl. 1): 322–323 [Google Scholar]
- Takano-Kai, N., Jiang, H., Kubo, T., Sweeney, M., Matsumoto, T., Kanamori, H., Padhukasahasram, B., Bustamante, C., Yoshimura, A., and Doi, K.et al. (2009) Evolutionary history of GS3, a gene conferring grain length in rice. Genetics 182: 1323–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano-Kai, N., and Doi, K. andYoshimura, A. (2011) GS3 participates in stigma exsertion as well as seed length in rice. Breed. Sci. 61: 244–250 [Google Scholar]
- Takeda, K., and Saito, K. (1977) The inheritance and character expression of the minute gene derived from a rice genetic tester “Minute”. Bull. Fac. Agric. Hirosaki Univ. 27: 1–29 [Google Scholar]
- Takeoka, Y. (1976) Histogenesis of lemma in japonica paddy rice. Proc. Crop Sci. Soc. Jpn. 45: 569–581 [Google Scholar]
- Tan, Y.F., Xing, Y.Z., Li, J.X., Yu, S.B., Xu, C.G., and Zhang, Q.F. (2000) Genetic bases of appearance quality of rice grains in Shanyou 63, an elite rice hybrid. Theor. Appl. Genet. 101: 823–829 [DOI] [PubMed] [Google Scholar]
- Toki, S. (1997) Rapid and efficient Agrobacterium-mediated transformation in rice. PlantMol. Biol. Rep. 15: 16–21 [Google Scholar]
- Tsunematsu, H., Yoshimura, A., Yano, M., and Iwata, N. (1995) Quantitative trait locus analysis using recombinant inbred lines and restriction fragment length polymorphism markers in rice. In: Khushu, G.S. (ed.) Rice Genet. Ill, International Rice Research Institute, Manila, Philippines, pp. 619–623 [Google Scholar]
- Unnevehr, L.J., Duff, B., and Juliano, O. (1992) Consumer demand for rice grain quality. International Rice Research Institute, Manila, Philippines [Google Scholar]
- Wang, J., Nakazaki, T., Chen, S., Chen, W., Saito, H., Tsukiyama, T., Okumoto, Y., Xu, Z., and Tanisaka, T. (2009) Identification and characterization of the erect-pose panicle gene EP conferring high grain yield in rice (Oryza sativa L.). Theor. Appl. Genet. 119: 85–91 [DOI] [PubMed] [Google Scholar]
- Weng, J., Gu, S., Wan, X., Gao, H., Guo, T., Su, N., Lei, C., Zhang, X., Cheng, Z., and Guo, X.et al. (2008) Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 18: 1199–1209 [DOI] [PubMed] [Google Scholar]
- Zhou, Y., Zhu, J., Li, Z., Yi, C., Liu, J., Zhang, H., Tang, S., Gu, M., and Liang, G. (2009) Deletion in a quantitative trait gene qPE9-l associated with panicle erectness improves plant architecture during rice domestication. Genetics 183: 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.