Abstract
Insulators help in organizing the eukaryotic genomes into physically and functionally autonomous regions through the formation of chromatin loops. Recent findings in Drosophila and vertebrates suggest that insulators anchor multiple loci through long-distance interactions which may be mechanistically linked to insulator function. Important to such processes in Drosophila is CP190, a common co-factor of insulator complexes. CP190 is also known to associate with the nuclear matrix, components of the RNAi machinery, active promoters and borders of the repressive chromatin domains. Although CP190 plays a pivotal role in insulator function in Drosophila, vertebrates lack a probable functional equivalent of CP190 and employ CTCF as the major factor to carry out insulator function/chromatin looping. In this review, we discuss the emerging role of CP190 in tethering genome, specifically in the perspective of insulator function in Drosophila. Future studies aiming genome-wide role of CP190 in chromatin looping is likely to give important insights into the mechanism of genome organization.
Keywords: Insulators, CP190, long-range interactions, chromatin organization
Introduction
One of the fundamental questions in nuclear biology is how eukaryotes package their relatively large genome in the tiny confines of the nucleus. Studies over the past many years have strongly supported the idea that eukaryotic genomes are organized into a series of structurally and functionally independent domains of chromatin through inter and intra-chromosomal interactions among various regulatory regions.1-4 The establishment as well as the maintenance of such domains involve the action of cis-regulatory elements referred to as “insulators” or “boundaries.” Chromatin insulators were first discovered in Drosophila melanogaster and subsequently in a variety of organisms including yeast, mosquito, Xenopus, chicken, mice and humans suggesting their widespread importance in genome organization.5-11 These elements exert their effects by preventing inappropriate cross-talk between genomic regions such as enhancers and promoters and active and silent states of the chromatin (Fig. 1). Insulators have been identified using transgenic assays in which they block enhancer-promoter interactions when present between the two (hence, referred to as enhancer-blockers) and/or prevent the spreading of the silencing effects of the heterochromatin (referred to as barriers).8,12,13 While some of the characterized insulators have been shown to act primarily as barriers to heterochromatin, others may possess both enhancer-blocking and barrier activity.14 For their function, insulators depend on specific proteins that associate with these elements. Although the presence of GAGA factor (GAF) has been reported recently,15 CTCF remains the major protein that mediates the insulator function in vertebrates.16 However, in Drosophila, multiple insulator elements have been characterized that vary widely in their DNA sequences and in the proteins that bind to them. Among the best studied elements are the scs and scs’ from the hsp70 heat shock locus, gypsy from the gypsy retrotransposon, SF1 from the Antennapedia complex and Mcp, Fab-7 and Fab-8 from the bithorax complex.11,13,17-20 While scs and scs’ use Zest-white-5 (Zw5) and Boundary Element Associated Factor (BEAF), respectively, as their main DNA binding proteins, the gypsy insulator binds to Suppressor of Hairy-wing [Su(Hw)], Fab-7 and SF1 bind to GAGA factor (GAF) and Fab-8 binds to the Drosophila homolog of the vertebrate CTCF (dCTCF).20-24 The function of these insulators often depends on the insulator co-factor, CP190 through protein-protein interactions.25-27 Recent studies have suggested that insulator proteins such as Su(Hw), BEAF, and dCTCF bind specific DNA sequences and recruit CP190 and Mod(mdg4), which through homotypic and heterotypic protein-protein interactions bridge contacts between distant genomic region.28 In this review, we will largely focus our discussion on the role of CP190 as a key player in insulator function and genome organization.
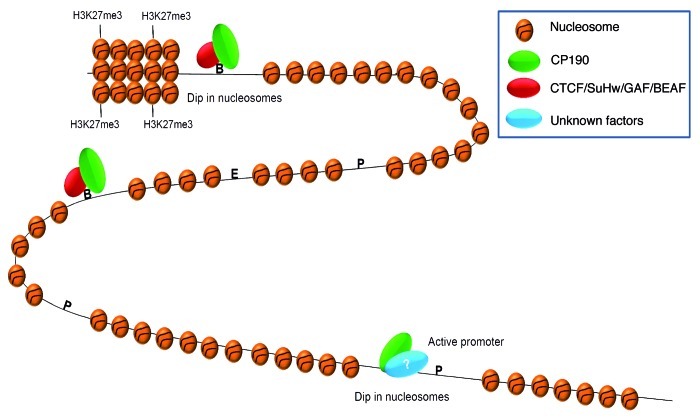
Figure 1. CP190 and the global organization of chromatin. CP190 acts as a mediator for dCTCF and Su(Hw) class of insulators by blocking enhancer-promoter communication. CP190 binds to active promoters which inversely correlate with nucleosome occupancy by unknown mechanisms. This trend is also seen at the borders of H3K27me3. Moreover, CP190 has many binding sites throughout the genome that do not overlap with any of the known insulator binding proteins. P, promoter; E, enhancer; B, boundary; ?, indicates an unknown protein partner.
The Centrosomal Protein 190 (CP190)
CP190 (for Centrosomal Protein of 190 kDa) is a protein of 1,096 amino acids with a predicted molecular weight of 121 kDa and an apparent molecular weight of about 190 kDa. The protein contains an N-terminal BTB/POZ (Broad-complex, Tramtrack and Bric-abrac/Poxvirus and Zinc Finger) domain; an aspartic-acid rich D-domain; three C2H2 zinc finger motifs; and a C-terminal E-rich domain (Fig. 2). Apart from these motifs, CP190 also contains a centrosomal targeting domain (CENT) for its localization to centrosomes during mitosis.29 The BTB/POZ, the aspartic-acid rich (D-rich) and the C-terminal glutamic-acid rich (E-rich) domains are essential for its association with insulator subclasses and insulator function. The E-rich region is important for the disassociation of CP190 from the chromosome during heat-shock, which may provide a mechanism for regulating insulator function.30
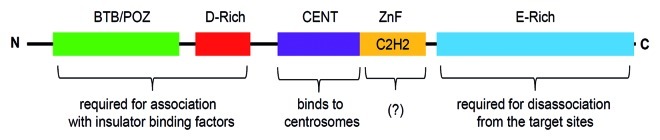
Figure 2. Schematic of the full-length Centrosomal Protein 190 (1,096 amino acid residues). CP190 contains BTB/POZ domain at the N-terminus, D-rich, CENT and zinc-finger (Zn) domains in the center and an E-rich domain at the C-terminus. Function of each domain is described in the text.
CP190 was originally identified in Drosophila melanogaster using a monoclonal anti-centrosomal antibody and was subsequently used to select the CP190 gene from a λgt11 expression library.31,32 Like other Drosophila insulator proteins (dCTCF and GAF being the exception) CP190 appears to be restricted to insects.15,33 Although CP190 was initially identified and characterized as a result of its association with centrosomes and microtubules, later studies showed it to be localized in the nucleus and bind to specific sites on polytene chromosomes, suggesting its role in the interphase nuclei.34 Early biochemical studies also suggest that CP190 is a component of nuclear matrix.35 Depleting CP190 in culture cells does not significantly interfere with centrosomes and microtubule organization, cell division or with cell viability. However, flies that are homozygous mutant for CP190 die at the late pupal stages, suggesting that it is essential for fly development.36 CP190 does not bind directly to DNA, however, it is crucial for the insulator function of Su(Hw) dependent gypsy and dCTCF dependent Fab-8 insulators.25,26 CP190 has also been shown to associate with other subclasses of insulator complexes such a BEAF-32.37,38
Genomic Distribution of CP190
Earlier studies on the localization of CP190 have shown it to be associated with centrosomes throughout the nuclear division cycle in syncytial Drosophila embryos. However, after the cellularization of the embryo, CP190 is exclusively found in nucleus during the interphase.32,35,39 Using indirect immunofluorescence staining of polytene chromosomes from salivary glands, CP190 was found at a number of sites along the entire length of the chromosomes localizing to band/interband boundaries.34 These early observations indicated the function of CP190 beyond centrosomes. A variety of non-histone proteins have been shown to have a structural/or regulatory role in chromatin. For example, the Polycomb group (PcG) proteins maintain the compact and transcriptionally repressive state of the homeotic genes during development.40,41 In contrast, the proteins of the trithorax group, maintain an open state of chromatin. GAF, a product of trithorax-like (Trl) gene is a zinc-finger protein that associates with a large number of chromosome loci and its mutation leads to enhancement of position effect variegation, suggesting its role in organizing chromatin structure.42,43 Similarly, Su(Hw) and dCTCF are also zinc-finger proteins which have been found to localize to a number of sites in the genome especially at the boundaries between bands and interbands and are important for insulator function and organization of chromatin.44 These studies tempt us to think that CP190, which has a global genomic distribution like other insulator factors, may be another protein with a larger role in organization of chromatin structure.25,26,38 Early studies using a genetic screen for dominant enhancers of mod(mdg4) identified CP190 as a third component of the gypsy insulator-complex.26 CP190 was found to co-localize extensively with Mod(mdg4)2.2 and the Su(Hw) proteins at endogenous insulator sites and at the borders of bands/interbands on the polytene chromosomes. Subsequently, CP190 was also shown to occupy a number of dCTCF target sites, when analyzed on polytene chromosomes.25 These results indicated that, although Su(Hw) and dCTCF have diverse binding specificity, they share co-factors.
Recent genome-wide ChIP-chip data revealed an extensive overlap of CP190 with the target sites of four major insulator factors; dCTCF, Su(Hw), BEAF and GAF.37,38 Although dCTCF and Su(Hw) target sites do not overlap, CP190 largely overlaps with both these insulator proteins and with around 80% of the GAF sites.45 Additionally, around 80% of the Stromalin (vertebrate Cohesin counterpart in Drosophila) sites and most of the binding sites of dCTCF also overlap with CP190. This analysis found that although many sites of the insulator subclasses overlap, a subset show cell-type specific localization.38 Similar results were obtained when dCTCF target sites were compared in S2 and Kc cells, where again a subset of dCTCF target sites showed cell-type specificity.37 These observations indicate that different subclasses of insulators organize genome in a cell-type specific manner which may be responsible for differential gene expression. However, More recently, using quantitative genome-wide analysis of insulator protein binding, Schwartz et al., found that there is no distinction between individual CP190 sites and sites co-bound by Su(Hw)+CP190 and dCTCF+CP190 in S2 and BG3 cells as well as in whole embryos suggesting that co-binding of CP190 is not a product of tissue specific regulation.46 Moreover, around 80% robust CP190 binding sites overlap with dCTCF, Su(Hw) or BEAF-32. Interestingly, they also found a number of sites in the genome where CP190 binds independently of Su(Hw), dCTCF and BEAF-32. It was also found that recruitment of CP190 and BEAF to co-bound sites is independent of each other as knockdown of either BEAF or CP190 do not effect each other’s recruitment. This study also found that majority of the insulator binding sites that showed enhancer-blocking activity in transgenic assays belong to those bound exclusively by CP190 or co-bound by BEAF suggesting that these sites may represent the major group of robust insulator elements in Drosophila.
CP190 Affects the Structure of Chromatin
The organization of nucleosomes and their chemical and compositional modifications play a key role in regulation of gene expression.47 Recently, lot of attention has focused on nucleosome organization and the factors that determine this organization. Underlying DNA sequences, the binding of a transcription factor or chromatin remodelling machinery can influence position of nucleosomes.48 Repositioning and partial or complete disruption of nucleosomes may allow DNA binding proteins to access their regulatory elements.49 Given the role of insulators in genome organization, it becomes interesting to look if they have any effects on the organization/re-organization of the nucleosomes. Previous work has shown that mammalian CTCF binding sites are associated with positioned nucleosomes.50,51 In Drosophila, boundary regions have also been associated with reduced nucleosome occupancy across the bithorax complex.52 These results are intriguing and point out a key role of insulator elements and their associated proteins in nucleosome organization. CP190 has been shown to bind promoters of active genes and such promoters are depleted of nucleosomes45 (Fig. 1) Target sites which are occupied by both dCTCF and CP190 show lack of H3 and therefore, loss of nucleosomes. In contrast, sites exclusively bound by dCTCF do not show any changes in the nucleosome occupancy. When CP190 is depleted, H3 levels are increased at these sites, suggesting that the loss of nucleosomes is due to CP190.45 The inverse-correlation between CP190 binding and nucleosome occupancy is also seen at the borders of H3K27me3 domains. Previously, several domains of H3K27me3 were identified in the Drosophila genome.53 Many of these domains show double occupancy for dCTCF/CP190 at the borders.45 These borders have reduced levels of H3K27me3 and also loss in H3 binding. Using CP190 and dCTCF mutants, the loss of H3 and decrease in H3K27me3 at the borders of repressive domains was found to be dependent on CP190 and not on dCTCF. Similar results were obtained by Negre et al. when they analyzed binding sites of insulator associated proteins including CP190 and found that these regions have low nucleosome density and high histone replacement or displacement.37 Taken together, these results strongly suggest a possible role of CP190 in chromatin/nucleosome organization and raise the possibility of barrier like function for CP190 bound regions.
CP190 is a Common Component of Multiple Insulator Complexes
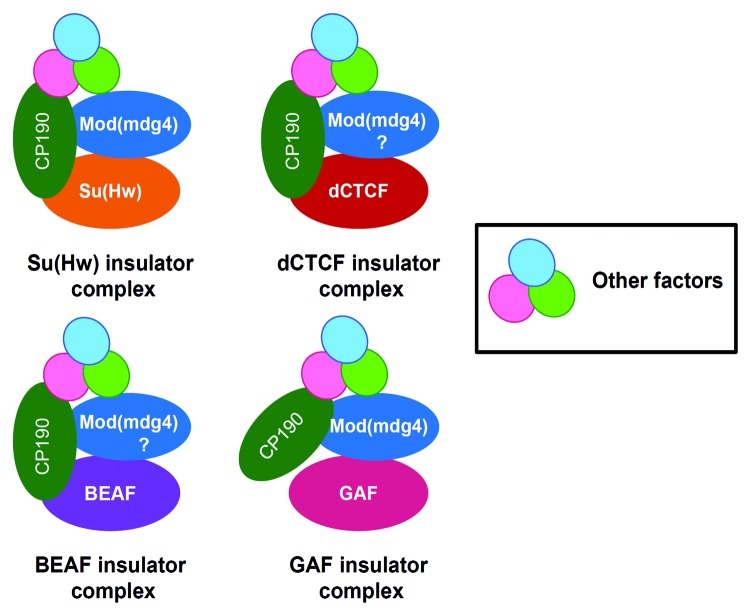
Unlike vertebrates, which appear to employ only a single factor CTCF for insulator activity, Drosophila possesses at least five classes of insulators defined by their DNA binding proteins. These include the Su(Hw), dCTCF, Zw5, BEAF and GAF.54 Genetic studies and genome-wide analysis have shown that, at least four of the insulator factors, Su(Hw), dCTCF, BEAF and GAF share a common component, the CP190 protein, although, in case of GAF it has not been conclusively proven (Fig. 3).44,25,45,38 CP190 directly interacts with components of the insulator complexes. For example, co-immunoprecipitation and yeast two-hybrid assays revealed that, CP190 physically interacts with Su(Hw) and Mod(mdg4)2.2.26 Furthermore, it was shown that CP190 is required for gypsy insulator function. Similarly, using a combination of co-immunoprecipitation, DNA FISH and affinity chromatography, two individual studies showed that CP190 interacts with dCTCF and is essential for the insulator function of Fab-8 insulator.25,44 Using a transgenic enhancer-blocking assay, CP190 was also shown to be required for the boundary activity of HS1 insulator in the Drosophila bithorax complex.55 Genome-wide ChIP-chip analysis has also revealed extensive overlap of CP190 with BEAF sites.38 These results suggest that most of the insulator factors recruit CP190 as a co-factor and, therefore, may utilize a common mechanism for their insulator function. However, a recently identified insulator, Wari at the 3` end of the white gene lacks binding sites for any of the known insulator factors but is bound by CP190.56 This report raises the possibility that, in some cases, CP190 may be sufficient to mediate the insulator function. This is supported by a recent study in which many individual CP190 sites were found across the Drosophila genome that act as strong enhancer blockers in transgenic assay.46 Alternatively, CP190 may be recruited to such sites by yet to be identified DNA binding factors.
Figure 3. CP190 is a common co-factor for four classes of insulators in Drosophila. Each class of insulator is represented by the DNA binding factor dCTCF, Su(Hw), GAF and BEAF, color coded differently to indicate insulator subclasses. CP190 is shared by all the insulator classes, although in case of GAF, its functional relevance is yet to be determined. CP190 is also found to associate with factors like Rm62, dTopors, Ago2, implicated in insulator function. In addition to CP190, insulator subclasses may also share Mod(mdg4). Furthermore, all the above classes of insulators may also share other co-factors that are yet to be identified.
Immunolocalization studies has shown that Su(Hw), Mod(mdg4)2.2, dCTCF, and CP190 colocalize and form nuclear speckles termed insulator bodies.26,57 It has been suggested that these structures are formed as a result of interaction among individual insulator-containing complexes located at distant genomic locations. However, other studies have argued against the clustering of distinct insulator DNA sequences within an insulator body and instead shown that these structures are aggregates of insulator proteins much like the promyelocytic leukemia nuclear bodies (PML-NB).58 Whether insulator bodies are functional insulator complexes or depots of aggregated insulator proteins is debatable, however, it is now established that CP190 is crucial to the formation of insulator bodies. The first evidence to support that CP190 is a key player in the formation of insulator bodies came from fluorescence microscopy where it was shown that CP190 co-localizes to Su(Hw) and dCTCF containing insulator bodies.44,57 The Su(Hw) and the dCTCF insulator complexes are present throughout the polytene chromosomes at the transition of bands/interbands, however they do not co-localize, suggesting that they bind to different region in the chromatin. Interestingly, both the factors come together to form distinct loci in the diploid nucleus. This interaction between individual sites of Su(Hw) and dCTCF has been shown to be mediated by CP190.25,44 Mutations in dCTCF or mod(mdg4) do not effect each other’s presence or CP190 at the insulator bodies. But interestingly, when CP190 is mutated, the formation of insulator bodies by dCTCF and Su(Hw) is disrupted. More recently, Golovnin and colleagues, showed that depletion of CP190 using RNAi results in a diffused distribution of both Su(Hw) and Mod(mdg4)-67.2 proteins and disruption of the insulator body formation.58,59 Interestingly, reduction of either Mod(mdg4)-67.2 or Su(Hw) did not affect the localization of CP190 to insulator bodies suggesting that CP190 can independently form insulator bodies. Additionally, the authors also showed that the SUMO is required for the assembly of insulator proteins into insulator bodies. Taken together, these results suggest that CP190 plays a critical role in the organization of insulator bodies by recruiting Su(Hw) or dCTCF to insulator bodies. Once assembled, insulator bodies can then act as warehouse of insulator proteins that may transiently interact with the chromatin fiber and detach from the insulator bodies via desumoylation.59 Apart from being a major component of insulator complexes, CP190 has been shown to interact with dTopors and Ago2 both of which are important for insulator function of Su(Hw) and Fab-8 insulator, respectively60,61. These studies suggest that dTopors and Ago2 may be a subset of other proteins with which CP190 interacts to carry out insulator function.
“Loopers” of the Genome: An Emerging View of Insulator Function
Recent data emerged from studies employing high resolution techniques such as 3D-FISH and Chromosome Conformation Capture assay (3C) and its variants, support the idea that insulators directly and physically interact with each other or with other regulatory elements such as promoters, enhancers and silencers, to form chromatin loops.62,63 Depending upon the nature and context in which such interactions take place, the target loci could either be relocated to a transcription factory, resulting in transcriptional activation or to a Polycomb group (PcG) body, causing transcriptional repression (Fig. 4).64 One of the earliest evidence of the existence of insulator-mediated long-range interaction in Drosophila came from studies on the scs and scs’ insulators of the heat shock locus. 3C assay showed that Zw5 bound scs and BEAF bound scs’ physically interact with each other.65 Similar interactions have been observed between two insertions of the gypsy in a Su(Hw) dependent manner.57 GAF binding insulators such as Fab-7 or Mcp and dCTCF/CP190 binding Fab-8 have been shown to mediate long-range interaction in the bithorax complex.66,67 It has been found that Abdominal-B (Abd-B) and Antennapedia (Antp) genes, which are located far away (~10Mb) in chromosome 3R colocalize in nuclei of cells in which both genes are repressed. However, when Fab-7 or Mcp is deleted their colocalization is reduced suggesting that these two insulators play an important role in mediating their interaction.66 Similarly, using reporter assay, Fab-7 and Fab-8 have been shown to interact with a CTCF site in the Abd-B promoter.67 Several studies have shown that Polycomb target sites interact over long distances, and in certain cases, such interactions are mediated by insulators.66,68,69 For example, a single copy of gypsy insulator restricts a PRE (polycomb response element) from interacting with a distal promoter, while two copies of gypsy bring a PRE to a downstream target gene to mediate its repression.70 Similarly, the interaction of Mcp and Fab-7 elements, which contain both PRE and insulator activity, has been shown to depend on the underlying insulator activity of these elements.71 These studies suggest insulators to be the chief factors mediating long-range interactions among diverse regulatory elements in Drosophila.
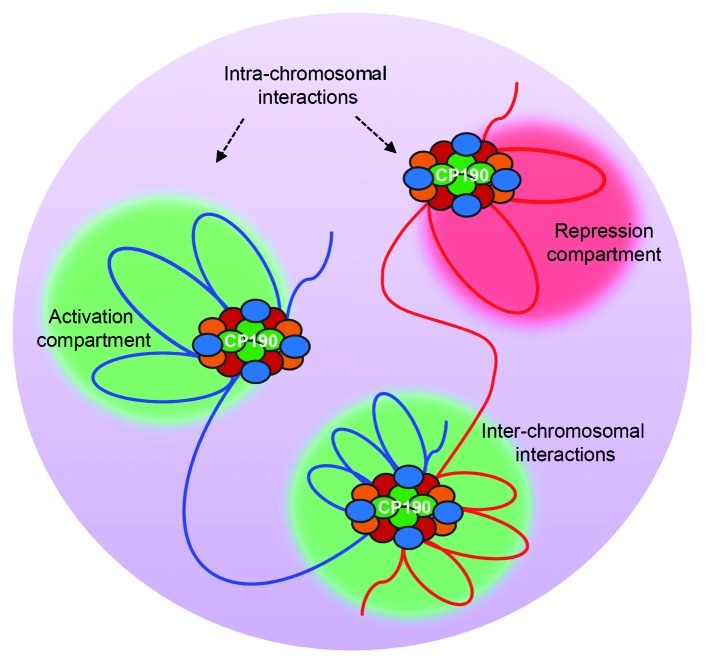
Figure 4. CP190 facilitates chromatin looping interactions. Insulator complexes of which CP190 is a key component, mediate intra- or inter-chromosomal interactions. Such looping interactions are thought to result in relocation of target loci to a repression compartment [such as Polycomb group (PcG) bodies] or an activation compartment (transcription factories). In the schematic, CP190 is shown as green ovals. Red, orange and blue ovals represent other insulator associated proteins. Red and blue lines indicate regions of two different chromosomes.
While the involvement of major insulator factors in long-range interactions has been shown previously, recent observation point to an important role of CP190 in mediating such interaction.2,72 It is proposed that CP190 is recruited to different insulator sites either by itself or by insulator binding proteins which then act as a common adaptor that mediates interaction among different insulator sites (Fig. 4).53,38 For example, Moshkovich and coworkers showed that chromosomal looping in the Abd-B locus is dependent on CP190.61 When CP190 is depleted in S2 cells, long-range interactions/loop formation among regulatory elements in the Abd-B locus is impaired. Further evidence for the role of CP190 in promoting chromatin looping has come from a recent study involving a CTCF dependent insulator at the Eip75B locus.72 CP190 was shown to stabilize loop formation necessary for restricting transcriptional activation to ecdysone regulated genes. More recently, Hou et al. found that domain boundaries which are enriched for insulator factors dCTCF, BEAF and CP190 interact more frequently which may facilitate clustering of active and silent genes to transcription factories and Polycomb (Pc) bodies respectively.2
It is interesting to note that CP190, which is critical for insulator function and chromatin looping in Drosophila, has not been conserved in vertebrates. Instead vertebrate CTCF co-operates with cohesin which creates or stabilizes chromatin loops by physically linking different CTCF-binding sites on the same or different chromosomes.73-76 Thus, it is tempting to propose that cohesin might play a role equivalent to that of CP190 in Drosophila. It has been observed that upon heat shock CP190 disassociates from the chromatin or insulator complexes while the localization of DNA binding proteins is not affected.59,72 Similarly, depletion of cohesin results in disruption of chromatin looping and changes in expression of genes under CTCF control, without affecting the expression or binding of CTCF.77 These results suggest that recruitment of CP190 and cohesin may function as a regulatory mechanism of controlling insulator activity in Drosophila and vertebrates, respectively. Another interesting connection between CP190 and cohesin is their interaction with DEAD-box RNA helicases and the regulation of insulator activity. CP190 physically interacts with RNA helicase Rm62 in an RNA dependent manner to negatively regulate insulator function of the gypsy insulator.78 Accordingly, reduction in the levels of Rm62 restores the insulator activity of the gypsy insulator. Similarly, RNA dependent interactions have also been observed between RNA helicase p68 (the vertebrate counterpart of Rm62) and cohesin.79 p68 interacts with cohesin to positively regulate insulator activity of Igf2/H19 ICR which upon depletion of p68 is compromised.79 Although the interaction between CP190 and Rm62 in Drosophila and that of p68 and cohesin in vertebrates have opposing effects, the conservation of this interaction is remarkable. Overall, the view emerging from these studies suggest that the central function of insulators may be to create or stabilize chromosome interactions and that CP190 plays a key role in mediating such interactions in Drosophila. However, in vertebrates, it appears that cohesin may have replaced CP190 to mediate long-range interactions.
Concluding Remarks
It is clear that CP190 acts as a common anchor protein of different insulator classes and plays a wide role in genome organization through the formation of chromatin loops. Several genome wide studies have shown that CP190 co-localizes with all the major insulator binding proteins.2,37,38 However, there are a number of sites in the Drosophila genome where CP190 does not co-localize with any of the known insulator proteins. Whether these represent CP190 target sites to which it binds on its own, as suggested in a recent study by Schwartz et al.,46 or sites where CP190 is recruited by DNA binding factors that are yet to be identified. In either case, it is clear that CP190 has broad range of genomic loci under its regulatory influence. Another feature of CP190 is its correlation with nucleosome occupancy. At active promoters and borders of repressive chromatin domains, CP190 negatively correlates with nucleosome occupancy suggesting that it may directly disrupt the nucleosome assembly or recruit histone modifiers/and or chromatin remodelers.45
Insulators mediate long-range interactions between distant genomic regions and, interestingly, such regions also include Polycomb target sites.71,80 It is tempting to speculate that CP190, which is involved in insulator mediated looping, may have a role in targeting such sites to polycomb bodies.61,72 Furthermore, given its global genomic distribution and the availability of high resolution 3C technique, it will also be important to study the role of CP190 in long-distance interactions on a genome-wide scale. Finally, while a global regulator of chromatin structure is expected to be conserved across the species, vertebrates appear to lack a CP190 counterpart. It remains to be seen if CTCF acquired different partners in vertebrates such as cohesin or a functional homolog of CP190 exists that remains to be identified.
Acknowledgments
The authors thank Rainer Renkawitz for critical reading of the manuscript. Research in Y.S.S and R.K.M. laboratories is supported by the Department of Biotechnology (DBT) and Counsel of Scientific and Industrial Research (CSIR), India. S.H.A acknowledges fellowships from ICMR, India and DAAD, Germany. We also thank Navneet K Matharu for helpful discussions. The authors declare no conflict of interest.
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/23389
References
- 1.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hou C, Li L, Qin ZS, Corces VG. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol Cell. 2012;48:471–84. doi: 10.1016/j.molcel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–72. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 5.Ahanger SH, Srinivasan A, Vasanthi D, Shouche YS, Mishra RK. Conserved boundary elements from the Hox complex of mosquito, Anopheles gambiae. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks1178. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–14. doi: 10.1016/0092-8674(93)80052-G. [DOI] [PubMed] [Google Scholar]
- 7.Donze D, Adams CR, Rine J, Kamakaka RT. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geyer PK, Corces VG. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 1992;6:1865–73. doi: 10.1101/gad.6.10.1865. [DOI] [PubMed] [Google Scholar]
- 9.Li Q, Stamatoyannopoulos G. Hypersensitive site 5 of the human beta locus control region functions as a chromatin insulator. Blood. 1994;84:1399–401. [PubMed] [Google Scholar]
- 10.Robinett CC, O’Connor A, Dunaway M. The repeat organizer, a specialized insulator element within the intergenic spacer of the Xenopus rRNA genes. Mol Cell Biol. 1997;17:2866–75. doi: 10.1128/mcb.17.5.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Udvardy A, Maine E, Schedl P. The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J Mol Biol. 1985;185:341–58. doi: 10.1016/0022-2836(85)90408-5. [DOI] [PubMed] [Google Scholar]
- 12.Kellum R, Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64:941–50. doi: 10.1016/0092-8674(91)90318-S. [DOI] [PubMed] [Google Scholar]
- 13.Kellum R, Schedl P. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol Cell Biol. 1992;12:2424–31. doi: 10.1128/mcb.12.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.West AG, Gaszner M, Felsenfeld G. Insulators: many functions, many mechanisms. Genes Dev. 2002;16:271–88. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- 15.Matharu NK, Hussain T, Sankaranarayanan R, Mishra RK. Vertebrate homologue of Drosophila GAGA factor. J Mol Biol. 2010;400:434–47. doi: 10.1016/j.jmb.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 16.Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr Opin Genet Dev. 2007;17:400–7. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gdula DA, Gerasimova TI, Corces VG. Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila. Proc Natl Acad Sci U S A. 1996;93:9378–83. doi: 10.1073/pnas.93.18.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belozerov VE, Majumder P, Shen P, Cai HN. A novel boundary element may facilitate independent gene regulation in the Antennapedia complex of Drosophila. EMBO J. 2003;22:3113–21. doi: 10.1093/emboj/cdg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagstrom K, Muller M, Schedl P. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 1996;10:3202–15. doi: 10.1101/gad.10.24.3202. [DOI] [PubMed] [Google Scholar]
- 20.Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith ST, et al. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 2005;6:165–70. doi: 10.1038/sj.embor.7400334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaszner M, Vazquez J, Schedl P. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev. 1999;13:2098–107. doi: 10.1101/gad.13.16.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hart CM, Zhao K, Laemmli UK. The scs’ boundary element: characterization of boundary element-associated factors. Mol Cell Biol. 1997;17:999–1009. doi: 10.1128/mcb.17.2.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerasimova TI, Gdula DA, Gerasimov DV, Simonova O, Corces VG. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell. 1995;82:587–97. doi: 10.1016/0092-8674(95)90031-4. [DOI] [PubMed] [Google Scholar]
- 24.Schweinsberg S, Hagstrom K, Gohl D, Schedl P, Kumar RP, Mishra R, et al. The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics. 2004;168:1371–84. doi: 10.1534/genetics.104.029561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohan M, Bartkuhn M, Herold M, Philippen A, Heinl N, Bardenhagen I, et al. The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J. 2007;26:4203–14. doi: 10.1038/sj.emboj.7601851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pai CY, Lei EP, Ghosh D, Corces VG. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol Cell. 2004;16:737–48. doi: 10.1016/j.molcel.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 27.Ghosh D, Gerasimova TI, Corces VG. Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. EMBO J. 2001;20:2518–27. doi: 10.1093/emboj/20.10.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, Corces VG. Insulators, long-range interactions, and genome function. Curr Opin Genet Dev. 2012;22:86–92. doi: 10.1016/j.gde.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oegema K, Whitfield WG, Alberts B. The cell cycle-dependent localization of the CP190 centrosomal protein is determined by the coordinate action of two separable domains. J Cell Biol. 1995;131:1261–73. doi: 10.1083/jcb.131.5.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliver D, Sheehan B, South H, Akbari O, Pai CY. The chromosomal association/dissociation of the chromatin insulator protein Cp190 of Drosophila melanogaster is mediated by the BTB/POZ domain and two acidic regions. BMC Cell Biol. 2010;11:101. doi: 10.1186/1471-2121-11-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frasch M, Glover DM, Saumweber H. Nuclear antigens follow different pathways into daughter nuclei during mitosis in early Drosophila embryos. J Cell Sci. 1986;82:155–72. doi: 10.1242/jcs.82.1.155. [DOI] [PubMed] [Google Scholar]
- 32.Whitfield WG, Millar SE, Saumweber H, Frasch M, Glover DM. Cloning of a gene encoding an antigen associated with the centrosome in Drosophila. J Cell Sci. 1988;89:467–80. doi: 10.1242/jcs.89.4.467. [DOI] [PubMed] [Google Scholar]
- 33.Schoborg TA, Labrador M. The phylogenetic distribution of non-CTCF insulator proteins is limited to insects and reveals that BEAF-32 is Drosophila lineage specific. J Mol Evol. 2010;70:74–84. doi: 10.1007/s00239-009-9310-x. [DOI] [PubMed] [Google Scholar]
- 34.Whitfield WG, Chaplin MA, Oegema K, Parry H, Glover DM. The 190 kDa centrosome-associated protein of Drosophila melanogaster contains four zinc finger motifs and binds to specific sites on polytene chromosomes. J Cell Sci. 1995;108:3377–87. doi: 10.1242/jcs.108.11.3377. [DOI] [PubMed] [Google Scholar]
- 35.Oegema K, Marshall WF, Sedat JW, Alberts BM. Two proteins that cycle asynchronously between centrosomes and nuclear structures: Drosophila CP60 and CP190. J Cell Sci. 1997;110:1573–83. doi: 10.1242/jcs.110.14.1573. [DOI] [PubMed] [Google Scholar]
- 36.Butcher RD, Chodagam S, Basto R, Wakefield JG, Henderson DS, Raff JW, et al. The Drosophila centrosome-associated protein CP190 is essential for viability but not for cell division. J Cell Sci. 2004;117:1191–9. doi: 10.1242/jcs.00979. [DOI] [PubMed] [Google Scholar]
- 37.Nègre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bushey AM, Ramos E, Corces VG. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 2009;23:1338–50. doi: 10.1101/gad.1798209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Callaini G, Riparbelli MG. Centriole and centrosome cycle in the early Drosophila embryo. J Cell Sci. 1990;97:539–43. doi: 10.1242/jcs.97.3.539. [DOI] [PubMed] [Google Scholar]
- 40.Franke A, DeCamillis M, Zink D, Cheng N, Brock HW, Paro R. Polycomb and polyhomeotic are constituents of a multimeric protein complex in chromatin of Drosophila melanogaster. EMBO J. 1992;11:2941–50. doi: 10.1002/j.1460-2075.1992.tb05364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Messmer SFA, Paro R. Polycomb and polyhomeotic are constitutents of a multimeric protein complex in chromatin of Drosophila melanogaster. Genes Dev. 1992;6:1241–54. doi: 10.1101/gad.6.7.1241. [DOI] [PubMed] [Google Scholar]
- 42.Farkas G, Gausz J, Galloni M, Reuter G, Gyurkovics H, Karch F. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature. 1994;371:806–8. doi: 10.1038/371806a0. [DOI] [PubMed] [Google Scholar]
- 43.Tsukiyama T, Becker PB, Wu C. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature. 1994;367:525–32. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- 44.Gerasimova TI, Lei EP, Bushey AM, Corces VG. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol Cell. 2007;28:761–72. doi: 10.1016/j.molcel.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartkuhn M, Straub T, Herold M, Herrmann M, Rathke C, Saumweber H, et al. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J. 2009;28:877–88. doi: 10.1038/emboj.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwartz YB, Linder-Basso D, Kharchenko PV, Tolstorukov MY, Kim M, Li HB, et al. Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res. 2012;22:2188–98. doi: 10.1101/gr.138156.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10:161–72. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Segal E, Widom J. From DNA sequence to transcriptional behaviour: a quantitative approach. Nat Rev Genet. 2009;10:443–56. doi: 10.1038/nrg2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Z, Pugh BF. Genomic organization of H2Av containing nucleosomes in Drosophila heterochromatin. PLoS One. 2011;6:e20511. doi: 10.1371/journal.pone.0020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fu Y, Sinha M, Peterson CL, Weng Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008;4:e1000138. doi: 10.1371/journal.pgen.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mito Y, Henikoff JG, Henikoff S. Histone replacement marks the boundaries of cis-regulatory domains. Science. 2007;315:1408–11. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- 53.Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, et al. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–5. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- 54.Maeda RK, Karch F. Making connections: boundaries and insulators in Drosophila. Curr Opin Genet Dev. 2007;17:394–9. doi: 10.1016/j.gde.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 55.Pérez-Lluch S, Cuartero S, Azorín F, Espinàs ML. Characterization of new regulatory elements within the Drosophila bithorax complex. Nucleic Acids Res. 2008;36:6926–33. doi: 10.1093/nar/gkn818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Erokhin M, Parshikov A, Georgiev P, Chetverina D. E(y)2/Sus1 is required for blocking PRE silencing by the Wari insulator in Drosophila melanogaster. Chromosoma. 2010;119:243–53. doi: 10.1007/s00412-009-0253-1. [DOI] [PubMed] [Google Scholar]
- 57.Gerasimova TI, Byrd K, Corces VG. A chromatin insulator determines the nuclear localization of DNA. Mol Cell. 2000;6:1025–35. doi: 10.1016/S1097-2765(00)00101-5. [DOI] [PubMed] [Google Scholar]
- 58.Golovnin A, Melnikova L, Volkov I, Kostuchenko M, Galkin AV, Georgiev P. ‘Insulator bodies’ are aggregates of proteins but not of insulators. EMBO Rep. 2008;9:440–5. doi: 10.1038/embor.2008.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Golovnin A, Volkov I, Georgiev P. SUMO conjugation is required for the assembly of Drosophila Su(Hw) and Mod(mdg4) into insulator bodies that facilitate insulator complex formation. J Cell Sci. 2012;125:2064–74. doi: 10.1242/jcs.100172. [DOI] [PubMed] [Google Scholar]
- 60.Capelson M, Corces VG. The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol Cell. 2005;20:105–16. doi: 10.1016/j.molcel.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 61.Moshkovich N, Nisha P, Boyle PJ, Thompson BA, Dale RK, Lei EP. RNAi-independent role for Argonaute2 in CTCF/CP190 chromatin insulator function. Genes Dev. 2011;25:1686–701. doi: 10.1101/gad.16651211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Steensel B, Dekker J. Genomics tools for unraveling chromosome architecture. Nat Biotechnol. 2010;28:1089–95. doi: 10.1038/nbt.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Erokhin M, Davydova A, Kyrchanova O, Parshikov A, Georgiev P, Chetverina D. Insulators form gene loops by interacting with promoters in Drosophila. Development. 2011;138:4097–106. doi: 10.1242/dev.062836. [DOI] [PubMed] [Google Scholar]
- 64.Pirrotta V, Li HB. A view of nuclear Polycomb bodies. Curr Opin Genet Dev. 2012;22:101–9. doi: 10.1016/j.gde.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blanton J, Gaszner M, Schedl P. Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev. 2003;17:664–75. doi: 10.1101/gad.1052003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, Bonnet J, et al. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell. 2011;144:214–26. doi: 10.1016/j.cell.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 67.Kyrchanova O, Toshchakov S, Podstreshnaya Y, Parshikov A, Georgiev P. Functional interaction between the Fab-7 and Fab-8 boundaries and the upstream promoter region in the Drosophila Abd-B gene. Mol Cell Biol. 2008;28:4188–95. doi: 10.1128/MCB.00229-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Muller M, Hagstrom K, Gyurkovics H, Pirrotta V, Schedl P. The mcp element from the Drosophila melanogaster bithorax complex mediates long-distance regulatory interactions. Genetics. 1999;153:1333–56. doi: 10.1093/genetics/153.3.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vazquez J, Müller M, Pirrotta V, Sedat JW. The Mcp element mediates stable long-range chromosome-chromosome interactions in Drosophila. Mol Biol Cell. 2006;17:2158–65. doi: 10.1091/mbc.E06-01-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Comet I, Schuettengruber B, Sexton T, Cavalli G. A chromatin insulator driving three-dimensional Polycomb response element (PRE) contacts and Polycomb association with the chromatin fiber. Proc Natl Acad Sci U S A. 2011;108:2294–9. doi: 10.1073/pnas.1002059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li HB, Müller M, Bahechar IA, Kyrchanova O, Ohno K, Georgiev P, et al. Insulators, not Polycomb response elements, are required for long-range interactions between Polycomb targets in Drosophila melanogaster. Mol Cell Biol. 2011;31:616–25. doi: 10.1128/MCB.00849-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wood AM, Van Bortle K, Ramos E, Takenaka N, Rohrbaugh M, Jones BC, et al. Regulation of chromatin organization and inducible gene expression by a Drosophila insulator. Mol Cell. 2011;44:29–38. doi: 10.1016/j.molcel.2011.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–33. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 74.Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, et al. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci U S A. 2008;105:8309–14. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–66. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 77.Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, et al. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009;5:e1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lei EP, Corces VG. RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat Genet. 2006;38:936–41. doi: 10.1038/ng1850. [DOI] [PubMed] [Google Scholar]
- 79.Yao H, Brick K, Evrard Y, Xiao T, Camerini-Otero RD, Felsenfeld G. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 2010;24:2543–55. doi: 10.1101/gad.1967810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bartkuhn M, Renkawitz R. Long range chromatin interactions involved in gene regulation. Biochim Biophys Acta. 2008;1783:2161–6. doi: 10.1016/j.bbamcr.2008.07.011. [DOI] [PubMed] [Google Scholar]