Abstract
Actinomycetes growing on acidified starch-casein agar seeded with suspensions of litter and mineral soil from a spruce forest were provisionally assigned to the genus Nocardia based upon colonial properties. Representative isolates were found to grow optimally at pH 5.5, have chemotaxonomic and morphological features consistent with their assignment to the genus Nocardia and formed two closely related subclades in the Nocardia 16S rRNA gene tree. DNA:DNA relatedness assays showed that representatives of the subclades belong to a single genomic species. The isolates were distantly associated with their nearest phylogenetic neighbour, the type strain of Nocardia kruczakiae, and were distinguished readily from the latter based on phenotypic properties. On the basis of these data it is proposed that the isolates merit recognition as a new species, Nocardia aciditolerans sp. nov. The type strain is isolate CSCA68T (=KACC 17155T = NCIMB 14829T = DSM 45801T).
Electronic supplementary material
The online version of this article (doi:10.1007/s10482-013-9887-3) contains supplementary material, which is available to authorized users.
Keywords: Nocardia aciditolerans, Polyphasic taxonomy, Spruce forest soil, Actinomycetes
Introduction
The genus Nocardia is a member of the family Nocardiaceae of the order Corynebacteriales within the class Actinobacteria (Goodfellow et al. 2012). The genus is well defined and presently encompasses 86 species with validly published names (http://www.bacteria.cict.fr/qr/nocardiae.html) which form a monophyletic clade within the evolutionary radiation occupied by genera classified in the order Corynebacteriales (Goodfellow and Jones 2012). The taxonomic status of most of these species is supported by an abundance of genotypic and phenotypic data (Goodfellow and Maldonado 2012) although it is evident that the genus is underspeciated (Wang et al. 1999; Maldonado et al. 2000; Roth et al. 2003). Many of the recently described Nocardia species were isolated from clinical material and considered to be opportunistic pathogens of humans and animals (Brown-Elliott et al. 2006; Goodfellow and Maldonado 2012). Saprophytic nocardiae have received less attention though they are widely distributed in natural habitats, notably soil, where they have a role in the turnover of organic matter (Orchard 1979, 1981). There is evidence that some species synthesize bioactive compounds of potential industrial value, as exemplified by Nocardia iowensis (Lamm et al. 2009).
During the characterisation of acidiphilic and aciditolerant actinomycetes isolated from a coniferous forest soil, a number of strains were found to have colonial properties typical of nocardiae. In the present study, the taxonomic status of representatives of these strains was determined by using a polyphasic approach. The resultant data show that the isolates belong to a new Nocardia species, for which we propose the name Nocardia aciditolerans sp. nov.
Materials and methods
Sampling site
Environmental samples were taken from the litter and mineral horizons of a pure stand of Picea sitchensis Carriere (Sitka spruce) at the southern end of Hamsterley Forest, County Durham, UK (National Grid Reference NZ 066292) in October, 2011. The trees were planted in 1929 with individual trees about 3 metres apart. The site was flat with no herbaceous or moss understory. The spruce needles formed a well defined horizon which was divided into three layers; the L layer consisted of about 4 cm of intact needles, the F layer of 4 cm of partially decomposed but recognisable needles and the H layer of a black-brown amorphous mass of decomposed needles, humic material and other organic matter, approximately 3.5 cm deep. The mean pH, moisture and organic matter contents of the three litter layers were 4.9, 4.0 and 3.8; 49, 68 and 32 % and 98, 95 and 93 %, respectively. The litter layers overlay the mineral soil which consisted of an A1 horizon (mean pH 3.9) about 7 cm thick, below which was the C horizon of parent millstone grit (mean pH 4.1); the moisture and organic matter contents of the horizons were 35 and 25 %, and 20 and 14 %, respectively.
Organisms, maintenance and culture conditions
Acidiphilic and aciditolerant filamentous actinomycetes were sought from the litter and mineral horizons of the spruce forest soil using a standard dilution plate procedure (Goodfellow et al. 1967). Serial dilutions of the five environmental samples were spread over the surfaces of starch-casein medium (Kűster and Williams 1964) with agar (SCA) and gellan gum (GG) as gelling agents; the media were supplemented with cycloheximide and nystatin (each at 50 μg ml−1) and adjusted to pH 4.5 with 1 N HCl. Aliquots (100 μl) of each dilution prepared from the environmental suspensions were spread over the surfaces of the selective isolation plates which had been dried for 15 min prior to inoculation, as recommended by Vickers and Williams (1987). The inoculated plates, four per dilution, were incubated at 28 °C for 4 weeks. In addition to typical streptomycete-like colonies, 130 small compact colonies covered with white aerial hyphae were detected on isolation plates seeded with suspensions from the H, A1, and C layers. These Nocardia-like colonies were subcultured from starch-casein media prepared with each of the gelling agents and found to grow optimally on starch-casein agar at pH 5.5, but poorly at pH 4.5 and 7.0. Seventeen of the isolates were randomly selected for further study, namely strains HGG34, HGG71, HGG12n, HSCA8, and HSCA46 from the H layer; A1GG7, A1GG56, A1GG10n, A1SCA40, A1SCA48 and A1SCA1n from the A1 horizon; and CGG42, CGG62, CGG22n, CSCA51, CSCA58 and CSCA68T from the C horizon. These strains were maintained as hyphal fragments in glycerol (20 %, v/v) at −80 °C and on glucose-yeast extract agar (GYEA; Gordon and Mihm 1962) slopes adjusted to pH 5.5.
Biomass for the molecular systematic and most of the chemosystematic studies was prepared by growing the representative strains in shake flasks of GYE broth (pH 5.5) at 150 revolutions per minute at 28 °C for 3 weeks. Cells were harvested by centrifugation and washed twice in distilled water; biomass for the chemotaxonomic analyses was freeze dried and that for the molecular systematic work stored at −20 °C. Biomass for the cellular fatty acid analysis carried out on isolate CSCA68T was harvested from modified Bennett’s broth (Jones 1949), adjusted to pH 5.5, after incubation at 28 °C for 7 days.
Phylogenetic analyses
Genomic DNA was extracted from the representative isolates using a GenElute™ Bacterial Genomic Kit (Sigma), according to the instructions of the manufacturer, albeit with lysozyme at 45 mg ml−1 and incubation overnight at 37 °C. The 16S rRNA genes were amplified using the universal primers p27f and p1525r (Lane 1991) under the following conditions: 1 μl DNA template (final concentration 100 ng ml−1), 5 μl 10× DNA polymerase buffer (Bioline), 3 μl MgCl2 (50 mM stock solution, Bioline), 1.6 μl of UTPs (12.5 mM stock mixture, Bioline), 1 μl of each primer (10 μM stock solution) and 1 μl polymerase (5 U, Bioline). The amplified products were separated by electrophoresis, purified with an ExoSap-IT kit (USB Corporation, Ohio, USA) and directly sequenced using a Dye Deoxy Terminator Cycle Sequencing Kit (Applied Biosystems), as described by Chun and Goodfellow (1995).
Nearly complete 16S rRNA gene sequences of the isolates (~1,400 nucleotides [nt]) were compared with corresponding sequences of the most closely related type strains using the EzTaxon server (Kim et al. 2012). Phylogenetic trees based on the aligned sequences were inferred using the maximum-likelihood (Felsenstein 1981) maximum parsimony (Fitch 1971) and neighbour-joining (Saitou and Nei 1987) tree-making algorithms drawn from the MEGA 5 (Tamura et al. 2011) and PHYML (Guindon and Gascuel 2003) software packages. An evolutionary distance matrix was generated using the distance model of Jukes and Cantor (1969). Confidence values of branches of the phylogenetic tree were determined in a bootstrap analysis based on 1,000 resampling of the neighbour-joining dataset (Felsenstein 1985) using the MEGA 5 software. The tree was rooted using the rRNA gene sequence of Nocardia acidivorans GW4-1778T (accession number AM40 2972). Similarly, an expanded neighbour-joining tree was generated based on almost complete 16S rRNA gene sequences of the isolates and corresponding sequences of the type strains of Nocardia species, including those of the two acidotolerant taxa, Nocardia jiangxiensis and Nocardia miyunensis (Cui et al. 2005).
Chemotaxonomy
Isolates HGG71, A1GG7, A1SCA48, CGG62, CGG22n, CSCA51 and CSCA68T were examined for chemical markers known to be of value in nocardial systematics (Goodfellow and Maldonado 2012). Standard chromatographic procedures were used to determine the isomers of diaminopimelic acid (Staneck and Roberts 1974), mycolic acids (Minnikin et al. 1975) and whole-organism sugars (Hasegawa et al. 1983), using appropriate controls. Isolates CSCA68T and A1SCA48 were examined for the presence of diagnostic menaquinones (Collins 1985) and polar lipids (Minnikin et al. 1984) with Nocardia brasiliensis strain NBRC 14402T (ATCC 19296T) as control. Cellular fatty acids of isolate CSCA68T were extracted, methylated and determined by gas chromatography (Hewlitt Packard instrument 6890) and analysed using the standard Sherlock Microbial Identification (MIDI) system, version 5 (Sasser 1990). The G+C mol% of the DNA of isolates A1SCA48 and CSCA68T were determined following the procedure described by Gonzalez and Saiz-Jimenez (2002).
DNA:DNA relatedness
The DNA:DNA relatedness value (∆Tm) between isolates CSCA68T and A1SCA48 were determined using a fluorimetric method (Gonzalez and Saiz-Jimenez 2005), the optimal temperature for renaturation (Tm) was calculated using the equation Tor −0.51 (% GC) + 41. The melting temperature (Tm) at which 50 % of the initial double stranded DNA denatured into single-stranded DNA for isolate CSCA68T and the isolates CSCA68T: A1SCA 48 hybrid DNA preparations was compared and the difference (∆Tm) calculated.
Cultural, morphological and staining properties
All of the isolates were examined for cultural and morphological features following growth on standard media at 28 °C for 3 weeks. Cultural properties were investigated using tryptone-yeast extract, yeast extract-malt extract, oatmeal, inorganic-salts starch, glucose-asparagine, peptone-yeast extract-iron and tyrosine agars (International Streptomyces Project media 1–7 respectively; Shirling and Gottlieb 1966). Gram- and alcohol-acid fast staining were carried out following growth on glucose-yeast extract agar for 14 days at 28 °C using Hucker’s (Gerhardt 1981) and Ziehl-Neelsen (Gordon 1967) methods. The morphological properties of mycelia taken from GYEA plates were observed under a light microscope, after Gram staining.
Phenotypic tests
All of the isolates were examined for a broad range of phenotypic properties known to be of value in nocardial systematics (Goodfellow 1971; Isik et al. 1999; Goodfellow and Maldonado 2012) with incubation at 28 °C for 3 weeks. The pH, temperature and sodium chloride tolerance tests were carried out using GYEA as the basal medium (Gordon and Mihm 1957).
Results
16S rRNA gene sequencing and DNA:DNA relatedness studies
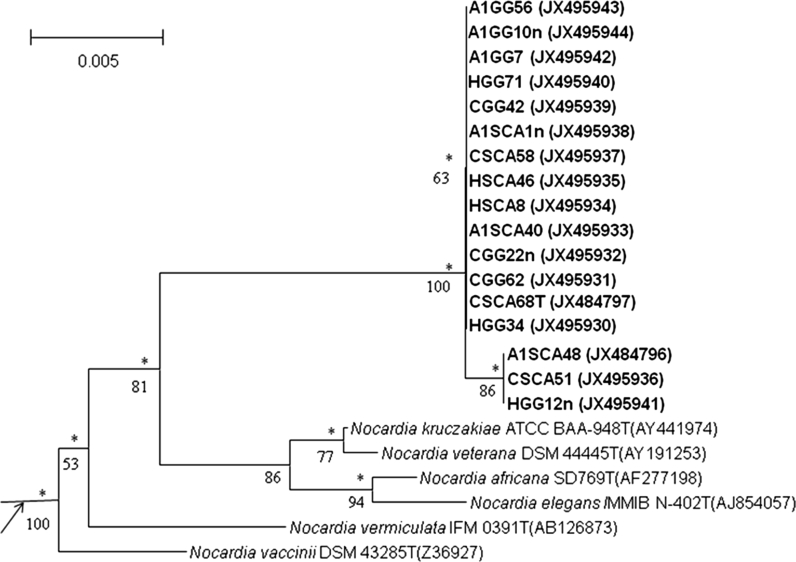
Near complete 16S rRNA gene sequences for strains A1SCA48, CSCA68T and HGG34, CGG62, CGG22n, A1SCA40, HSCA8, HSCA46, CSCA51, CSCA58, A1SCA1n, CGG42, HGG71, HGG12n, A1GG7, A1GG56, A1GG10n (GenBank accession numbers: JX484796–JX484797 and JX495930–JX495944, respectively) were determined. The sequences of all of 17 isolates formed a well-defined clade in the Nocardia 16S rRNA gene tree, a result underpinned by each of the tree-making algorithms and by a 100 % bootstrap value (Fig. 1). It is evident from the Figure that they formed two subclades, the larger of which included isolate CSCA68T. Members of the two subclades shared a 16S rRNA gene similarity of between 99.4 and 99.8 %, values equivalent to between 2 and 7 nt differences. The isolates were most closely related to the type strain of Nocardia kruczakiae sharing 16S rRNA gene similarities with the latter within the range 97.8–98.2 %, values that corresponded to between 25 and 30 nt differences at between 1,392 and 1,434 locations. The isolates were also distantly associated with the type strains of Nocardia africana (96.7–97.0 %), Nocardia elegans (97.4–97.7 % similarity) and Nocardia veterana (97.8–98.1 % similarity). It is apparent from the expanded neighbour-joining tree (Online supplementary Fig. 1) that the isolates are sharply separated from the type strains of N. jiangxiensis and N. miyunensis which form a clade supported by a 77 % bootstrap value. It is also apparent from this Figure that the clade encompassing the isolates is deep rooted but well within the Nocardia tree though the isolates are not closely related to any of the validly named Nocardia species.
Fig. 1.
Neighbour-joining tree based on nearly complete 16S rRNA gene sequences showing relationships between the isolates and between them and the most closely related Nocardia species. Numbers at the nodes indicate the levels of bootstrap support based on a neighbour-joining analyses of 1,000 re-sampled datasets, only values above 50 % are given. Asterisks indicate the branches of the tree that were also recovered using the maximum-likelihood and maximum-parsimony tree-making algorithms. T type strain. Bar 0.005 substitutions per nucleotide position.The root position of the tree was obtained using Nocardia acidivorans GW4-1778T as outgroup
The ∆Tm between isolate CSCA68T g DNA and isolates CSCA68T: A1SCA48 hybrid DNA was 2 °C, a result that corresponds to a DNA:DNA similarity value of 80 % (Gonzalez and Saiz-Jimenez 2005), a value well above the 70 % cut off point recommended for assigning strains to the same genomic species (Wayne et al. 1987).
Chemotaxonomic, cultural, morphological and phenotypic characteristics
Whole-organism hydrolysates of seven representative isolates were rich in meso-diaminopimelic acid (meso-A2pm), contained major amounts of arabinose and galactose with a trace of ribose and mycolic acids that co-chromatographed with those of N. brasiliensis NBRC 14402T (ATCC 19296T). Isolates CSCA68T and A1SCA48 were found to possess hexahydrogenated menaquinones with eight isoprene units where the last two isoprene units were cyclized (MK8 [H4 cyclo]) as the predominant isoprenologue. The strains were determined to contain diphosphatidylglycerol (DPG); phosphatidylethanolamine (PE), phosphatidylinositol (PI) and phosphatidylinositol mannosides (PIMS) as major polar lipids and a trace of phosphatidylglycerol (PG), as exemplified in Fig. 2. The major cellular fatty acids of isolate CSCA68T were identified as C16:0 (46.3 %), C18:1 ω9c (13.5 %) and 10-methyl C18:0 (tuberculostearic acid; 11.8 %). The genomic DNA G + C contents of isolates A1SCA48 and CSCA68T were determined to be the same, namely 71.3 mol%.
Fig. 2.
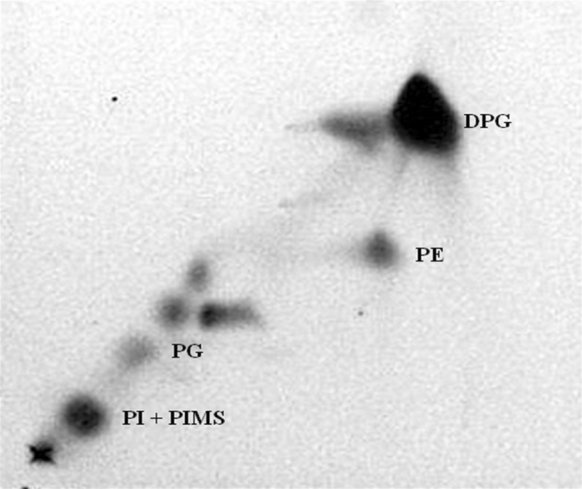
Two-dimensional thin-layer chromatography of polar lipids of isolate CSCA68T stained with molybdenum blue spray (Sigma). Chloroform: methanol: water (32.5: 12.5: 2.0) was used in the first direction and chloroform: acetic acid: methanol: water (40: 7.5: 6: 2) in the second direction. DPG diphosphatidylglycerol, PE phosphatidylethanolamine, PI phosphatidylinositol, PG phosphatidylglycerol and PIMS phosphatidylinositol mannosides
All of the isolates were observed to form an extensively branched substrate mycelium which carried light gray aerial hyphae on ISP7 agar following incubation at 28 °C for 3 weeks. The isolates were found to grow well on glycerol-asparagine, tyrosine, tryptone-yeast extract and yeast extract-malt extract agars (ISP media 5, 7, 1 and 2, respectively), poorly on oatmeal agar (ISP medium 3) and either poorly or not at all on peptone-yeast extract-iron agar (ISP medium 6) and inorganic salt starch agar (ISP medium 4). Sparse to moderate amounts of white to gray aerial hyphae were observed following growth on glycerol-asparagine and tyrosine agars. Isolates A1GG7, A1GG10n, CSCA68T and HSCA46 were observed to produce brown to orange pigments on tyrosine agar. It can be seen from Table 1 that the isolates share many phenotypic traits; grow on a broad range of compounds as sole carbon sources, are relatively inactive in the biochemical and degradative tests, and do not grow at 40 °C, at either pH 4.0 or 8.0 or in the presence of 3 % w/v sodium chloride.
Table 1.
Phenotypic properties of the aciditolerant isolates
| Characteristics | A1SCA48 | CSCA51 | CGG62 | HGG71 | A1SCA1n | HSCA8 | CSCA68T | HGG34 | A1SCA40 |
|---|---|---|---|---|---|---|---|---|---|
| Sole carbon sources 1 % (w/v) | |||||||||
| Adonitol | – | + | – | – | – | + | – | – | – |
| Amygdalin | – | – | + | – | – | – | + | + | – |
| l-Arabinose | – | + | – | – | – | + | + | – | + |
| d-Fructose | – | ++ | ++ | – | – | ++ | ++ | ++ | – |
| Glycogen | – | – | + | – | – | – | – | – | – |
| Inulin | – | – | + | – | – | + | + | + | – |
| d-Raffinose | – | + | + | – | – | – | + | + | – |
| d-Sorbitol | – | + | – | – | – | + | + | – | – |
| d-Trehalose | – | ++ | ++ | – | ++ | ++ | ++ | ++ | – |
| Xylitol | – | – | – | – | – | + | + | – | – |
| Sole carbon sources 0.1 % (v/w) | |||||||||
| Sodium succinate | – | + | + | + | – | + | + | + | + |
| Sole carbon & nitrogen sources | |||||||||
| Acetamide | + | + | + | – | – | + | + | + | + |
| l-Alanine | – | ++ | ++ | ++ | – | ++ | ++ | ++ | ++ |
| l-Asparagine | – | +++ | +++ | +++ | +++ | +++ | +++ | +++ | – |
| l-Phenylalanine | ++ | ++ | – | – | ++ | ++ | – | – | – |
| Uric acid | + | + | – | – | – | – | – | – | – |
| Characteristics | CSCA58 | CGG22n | HSCA46 | CGG42 | HGG12n | A1GG56 | A1GG10n | A1GG7 |
|---|---|---|---|---|---|---|---|---|
| Sole carbon sources 1 % (w/v) | ||||||||
| Adonitol | – | – | – | + | + | – | – | – |
| Amygdalin | – | – | + | + | + | + | – | – |
| l-Arabinose | – | – | – | + | – | – | – | – |
| d-Fructose | – | ++ | ++ | ++ | ++ | ++ | – | ++ |
| Glycogen | – | + | – | + | + | – | – | – |
| Inulin | – | – | + | + | + | + | – | – |
| d-Raffinose | – | – | + | + | – | – | – | – |
| d-Sorbitol | – | – | – | + | – | – | – | – |
| d-Trehalose | – | – | ++ | ++ | ++ | ++ | – | ++ |
| Xylitol | – | – | – | – | – | – | – | – |
| Sole carbon sources 0.1 % (v/w) | ||||||||
| Sodium succinate | + | + | + | + | + | + | + | + |
| Sole carbon & nitrogen sources | ||||||||
| Acetamide | – | – | + | + | + | – | + | + |
| l-Alanine | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| l-Asparagine | – | +++ | +++ | +++ | +++ | – | – | – |
| l-Phenylalanine | – | – | – | ++ | ++ | – | ++ | – |
| Uric acid | + | – | – | + | + | + | + | + |
All isolates were positive for: nitrate reduction; hydrogen sulphide production; degradation of Tween 60; growth on meso-erythritol, d-galactose, d-glucose, meso-inositol, d-mannitol, d-mannose, d-melezitose, d-melibiose, d-ribose and d-xylose (1 %, w/v) and sodium fumarate (0.1 %, w/v) as sole carbon sources; growth on l-proline, l-serine and l-valine (0.1 %, w/v) as sole carbon and nitrogen sources, and at 10–37 °C, pH 5–7 and in the presence of 1 % w/v sodium chloride
All isolates were negative for: aesculin, allantoin and arbutin hydrolysis; degradation of adenine, casein, elastin, guanine, hypoxanthine, Tweens 20, 40 and 80, l-tyrosine, uric acid, xanthine and xylan; growth on d-cellobiose, dulcitol, d-lactose, d-maltose, l-rhamnose, d-salicin and d-sucrose (1 %, w/v) and ethanol, iso-amyl alcohol and 1,2—propanediol and n-propanol (1 %, v/v), and acetate, adipate, benzoate, butyrate, citrate, malonate, oxalate and l- (+)-tartrate (sodium salts at 0.1 %, w/v) as sole carbon sources, l-aspartic acid, ethanolamine, gelatin, l-glutamic acid and urea as sole carbon and nitrogen sources (0.1 %, w/v) and growth at 40 °C, pH 4 and 8, and in the presence of 3 %, w/v sodium chloride
Variable results were shown for: adonitol, amygdalin, l-arabinose, d-fructose, glycogen, inulin, d-raffinose, d-sorbitol, d-trehalose and xylitol (1 %, w/v) and sodium succinate (0.1 %, w/v) as sole carbon sources and acetmide, d-alanine, l-asparagine, l-phenylalanine and uric acid as sole carbon and nitrogen sources (0.1 %, w/v)
+ positive, − negative
A range of phenotypic properties can be weighted to distinguish the isolates from their nearest phylogenetic neighbours, including the type strain of N. kruczakiae (Table 2).
Table 2.
Phenotypic properties that distinguish the 17 representatives isolates from the type strains of their closest phylogenetic neighbours
| Characteristic | Isolates |
N. africana
DSM 44491T |
N. elegans
DSM 44890T |
N. kruczakiae
DSM 44887T |
N. veterana
DSM 44445T |
|---|---|---|---|---|---|
| Aesculin hydrolysis | – | – | + | + | – |
| Arbutin hydrolysis | – | – | – | + | + |
| Casein degradation | – | + | – | – | – |
| Growth on sole carbon sources (1 %, w/v) | |||||
| meso-Erythritol | + | – | – | – | – |
| d-Galactose | + | – | – | – | – |
| meso-Inositol | + | – | – | – | + |
| d-Maltose | – | – | – | + | – |
| d-Mannitol | + | – | – | – | – |
| l-Rhamnose | + | – | – | – | + |
| d-Xylose | + | – | – | – | – |
| Growth at: | |||||
| 10 °C | + | – | – | – | – |
| 45 °C | – | + | + | + | + |
All of the strains grew on d-glucose as a sole carbon source (1 %, w/v) but were unable to degrade adenine, hypoxanthine, tyrosine or xanthine or to grow on d-cellobiose, d-sucrose (each at 1 %, w/v) or sodium acetate (0.1 %, w/v) as sole carbon sources
Data on the type strains were taken from Goodfellow and Maldonado (2012)
+ positive, − negative
Discussion
All of the representative aciditolerant strains isolated from the litter and mineral horizons of the spruce forest soil at Hamsterley Forest gave chemotaxonomic and morphological characteristics of the genus Nocardia (Goodfellow and Maldonado 2012). They were observed to form an extensively branched substrate mycelium that fragmented into rod-like elements and produced whole-cell hydrolysates rich in meso-A2pm, arabinose and galactose (wall chemotype IV sensu Lechevalier and Lechevalier 1970). Isolate CSCA68T was found to contain major proportions of straight-chain, saturated, unsaturated and tuberculostearic fatty acids, MK-8 (H4ω-cyclo) as the predominant isoprenologue, major amounts of DPG, PE, PI and PIMS (phospholipid type 2 after Lechevalier et al. 1977), mycolic acids with the same Rf value as those of N. brasiliensis strain NBRC 14402T (ATCC 19296T) and DNA G+C values within the range 64–72 mol%.
The aciditolerant isolates formed a distinct phyletic branch in the Nocardia 16S rRNA gene tree sharing low 16S rRNA gene similarities with their closest neighbours, results consistent with their assignment to a separate species (Goodfellow and Maldonado 2012). Their phylogenetic relatives include their closest phylogenetic neighbour, the type strain of N. kruczakiae, a respiratory pathogen originally isolated from an immunocompromised patient (Conville et al. 2004). The isolates shared even lower 16S rRNA gene similarities with N. jiangxiensis 43401T (CGMCC 4.1905T) and N. miyunensis 117T (CGMCC 4.1904T), aciditolerant strains isolated from acidic soils on an acidified selective isolation medium (Cui et al. 2005). It seems likely from these results that aciditolerant nocardiae may form an integral part of actinobacterial communities in acidic soils. The isolates can also be distinguished from the type strains of N. miyunensis and N. jiangxiensis for, unlike the latter, they grow at 10 °C but not on d-cellobiose, d-lactose, d-maltose, l-rhamnose, d-sucrose or sodium citrate as sole carbon sources nor do they hydrolyse aesculin.
The DNA:DNA homology assay showed that the representatives of the two closely related 16S rRNA gene subclades, namely strains CSCA68T and A1SCA48 belong to the same genomic species. The results of the phenotypic tests were in line with this finding as the isolates from each of the subclades shared a diverse range of phenotypic properties. All of the representatives isolates can be separated from their nearest phylogenetic neighbours, the type strains of N. africana, N. elegans, N. kruczakiae and N. veterana using a combination of phenotypic tests found to be of value in nocardial systematics (Goodfellow and Maldonado 2012). Thus, the isolates, unlike N. kruczakiae DSM 44887T, grew at 10 °C, but not at 45 °C, and used meso-erythritol, d-galactose, meso-inositol, d-mannitol, l-rhamnose and d-xylose as sole carbon sources but were unable to hydrolyse aesculin or arbutin. It is interesting that the four type strains, all of which were isolated from clinical material, grew well at 45 °C (Conville et al. 2004).
It can be concluded from the genotypic and phenotypic data that the isolates form a distinct taxon that can be distinguished from all validly named Nocardia species, including N. jiangxiensis and N. miyunensis, the only aciditolerant Nocardia species validly described to date. The isolates are therefore considered to represent a novel Nocardia species, for which the name proposed is N. aciditolerans.
Description of N. aciditolerans sp. nov.
Nocardia aciditolerans N.L. n. acidum (from L. adj. acidus, sour; L. pres. part. tolerans; N.L. past. adj. aciditolerans, acid tolerating).
Aerobic, Gram-positive, acid-alcohol-fast, aciditolerant actinomycetes that form an extensively branched, grayish yellow substrate mycelium which fragments into irregular rod- and coccoid-like elements after 14 days at 28 °C on glucose-yeast extract agar. Forms an extensively branched substrate mycelium bearing white or light gray aerial hyphae on glycerol—asparagine and tyrosine agars, respectively. Good growth also occurs on acidified tryptone-yeast extract and yeast extract-malt extract agars. Grows from pH 4.5–7.0 (optimally at pH 5.5), from 10 to 30 °C (optimally at 28 °C) and in the presence of 1 % but not 3 % w/v sodium chloride. Additional phenotypic properties are cited in the text or in Table 1. The cellular fatty acid profile of the type strain contains major proportions of C16:0, C18:1 ω9c, 10-methyl C18:0; lower proportions of C14:0, C16:1 ω9c, C17:1 ω8c, C17:1 ω5c, C17:0 and methyl C17:0, and traces of C12:0, C13:0, C15:1 ω8c, C15:1 ω5c, C15:0 30H, C18:3 ω6c and C18:0. The DNA G+C content of the type strain is 71.3 mol%.
The type strain, CSCA 68T (=KACC 17155T = NCIMB 14829T = DSM 45801T), was isolated from the C mineral horizon at Hamsterley Forest, County Durham, UK.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1 Neighbour-joining tree based on nearly complete 16S rRNA gene sequences (1292-1510 nt) showing relationships between the isolates and validly published Nocardia species. T, type strain. Bar, 0.005 substitutions per nucleotide position (PDF 25 kb)
Acknowledgments
Patrycja Golinska was supported by a research fellowship from Nicolaus Copernicus University (Toruń, Poland) “Enhancing Educational Potential of Nicolaus Copernicus University in the Disciplines of Mathematical and Natural Sciences” (Project No. POKL.04.01.01-00-081/10). The authors are indebted to Dr. Byung-Yong Kim for carrying out the fatty acid analysis on the type strain of N. aciditolerans.
References
- Brown-Elliott BA, Brown JM, Conville PS, Wallace RJ., Jr Clinical and laboratory features of the Nocardia spp. based on current molecular taxonomy. Clin Microbiol Rev. 2006;19:259–282. doi: 10.1128/CMR.19.2.259-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun J, Goodfellow M. A phylogenetic analysis of the genus Nocardia with 16S rRNA gene sequences. Int J Syst Bacteriol. 1995;45:240–245. doi: 10.1099/00207713-45-2-240. [DOI] [PubMed] [Google Scholar]
- Collins MD. Isoprenoid quinone analysis in bacterial classification and identification. In: Goodfellow M, Minnikin DE, editors. Chemical methods in bacterial systematics. London: Academic Press; 1985. pp. 267–287. [Google Scholar]
- Conville PS, Brown JM, Steigerwalt AG, Lee JW, Anderson VL, Fishbain JT, Holland SM, Vitebsky FG. Nocardia kruczakiae sp. nov., a pathogen of immunocompromised patients and a member of the “N. nova complex”. J Clin Microbiol. 2004;42:5139–5145. doi: 10.1128/JCM.42.11.5139-5145.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q, Wang L, Huang Y, Liu Z, Goodfellow M. Nocardia jiangxiensis sp. nov. and Nocardia miyunensis sp. nov., isolated from acidic soils. Int J Syst Evol Microbiol. 2005;56:1921–1925. doi: 10.1099/ijs.0.63644-0. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: maximum-likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- Fitch WM. Towards defining the course of evolution: minimum change for a specific tree topology. Syst Zool. 1971;20:406–416. doi: 10.2307/2412116. [DOI] [Google Scholar]
- Gerhardt P. Manual of methods for general bacteriology. Washington, DC: American Society for Microbiology; 1981. [Google Scholar]
- Gonzalez JM, Saiz-Jimenez C. A fluorimetric method for the estimation of G+C mol% content in microorganisms by thermal denaturation temperature. Environ Microbiol. 2002;4:770–773. doi: 10.1046/j.1462-2920.2002.00362.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez JM, Saiz-Jimenez C. A simple fluorimetric method for the estimation of DNA–DNA relatedness between closely related microorganisms by thermal denaturation temperatures. Extremophiles. 2005;9:75–79. doi: 10.1007/s00792-004-0417-0. [DOI] [PubMed] [Google Scholar]
- Goodfellow M. Numerical taxonomy of some nocardioform bacteria. J Gen Microbiol. 1971;69:33–80. doi: 10.1099/00221287-69-1-33. [DOI] [PubMed] [Google Scholar]
- Goodfellow M, Jones AL. Order V Corynebacteriales ord. nov. In: Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Suzuki K-I, Ludwig W, Whitman WB, editors. Bergey’s manual of systematic bacteriology, the actinobacteria, vol 5, part B. 2. New York: Springer; 2012. pp. 235–243. [Google Scholar]
- Goodfellow M, Maldonado LA. Genus I. Nocardia Trevisan 1889. In: Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Suzuki K-I, Ludwig W, Whitman WB, editors. Bergey’s manual of systematic bacteriology, the Actinobacteria, vol 5, part B. 2. New York: Springer; 2012. pp. 376–419. [Google Scholar]
- Goodfellow M, Hill IR, Gray TRG. Bacteria in a pine forest soil. In: Gray TRG, Parkinson D, editors. The ecology of soil bacteria. Liverpool: University Press; 1967. pp. 500–515. [Google Scholar]
- Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Suzuki K-I, Ludwig W, Whitman WB, editors. Bergey’s manual of systematic bacteriology, the Actinobacteria part A. 2. New York: Springer; 2012. pp. 1–1034. [Google Scholar]
- Gordon RE. The taxonomy of soil bacteria. In: Gray TRG, Parkinson D, editors. The ecology of soil bacteria. Liverpool: Liverpool University Press; 1967. pp. 293–321. [Google Scholar]
- Gordon RE, Mihm JM. A comparative study of some strains received as nocardiae. J Bacteriol. 1957;73:15–27. doi: 10.1128/jb.73.1.15-27.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon RE, Mihm JM. Identification of Nocardia caviae (Erikson) nov.com. Ann NY Acad Sci USA. 1962;98:628–636. doi: 10.1111/j.1749-6632.1962.tb30585.x. [DOI] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hasegawa T, Takizawa M, Tanida S. A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol. 1983;29:319–322. doi: 10.2323/jgam.29.319. [DOI] [Google Scholar]
- Isik K, Chun J, Hah YC, Goodfellow M. Nocardia salmonicida nom. rev., a fish pathogen. Int J Syst Bacteriol. 1999;49:833–837. doi: 10.1099/00207713-49-2-833. [DOI] [PubMed] [Google Scholar]
- Jones KL. Fresh isolates of actinomycetes in which the presence of sporogenous aerial mycelium is a fluctuating characteristic. J Bacteriol. 1949;57:141–145. doi: 10.1128/jb.57.2.141-145.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, editor. Mammalian protein metabolism. New York: Academic Press; 1969. pp. 21–132. [Google Scholar]
- Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012;62:716–721. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
- Kűster E, Williams ST. Selection of media for isolation of streptomycetes. Nature. 1964;202:928–929. doi: 10.1038/202928a0. [DOI] [PubMed] [Google Scholar]
- Lamm AS, Khare A, Conville P, Lau PCK, Bergeron H, Rosazza JPN. Nocardia iowensis sp. nov., an organism rich in biocatalytically important enzymes and nitric oxide synthase. Int J Syst Evol Microbiol. 2009;59:2408–2414. doi: 10.1099/ijs.0.007427-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow W, editors. Nucleic acid techniques in bacterial systematics. Chichester: Wiley; 1991. pp. 115–148. [Google Scholar]
- Lechevalier MP, Lechevalier HA. Chemical composition as a criterion in the classification of aerobic actinomycetes. Int J Syst Bacteriol. 1970;20:425–443. doi: 10.1099/00207713-20-4-435. [DOI] [Google Scholar]
- Lechevalier MP, De Biévre C, Lechevalier M. Chemotaxonomy of aerobic actinomycetes: phospholipid composition. Biochem Syst Ecol. 1977;5:249–260. doi: 10.1016/0305-1978(77)90021-7. [DOI] [Google Scholar]
- Maldonado L, Hookey JV, Ward AC, Goodfellow M. The Nocardia salmonicida clade, including descriptions of Nocardia cummidelens sp. nov., Nocardia fluminea sp. nov. and Nocardia soli sp. nov. Antonie Van Leeuwenhoek. 2000;78:367–377. doi: 10.1023/A:1010230632040. [DOI] [PubMed] [Google Scholar]
- Minnikin DE, Alshamaony L, Goodfellow M. Differentiation of Mycobacterium, Nocardia and related taxa by thin layer chromatographic analysis of whole organism methanolysates. J Gen Microbiol. 1975;88:200–204. doi: 10.1099/00221287-88-1-200. [DOI] [PubMed] [Google Scholar]
- Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods. 1984;2:233–241. doi: 10.1016/0167-7012(84)90018-6. [DOI] [Google Scholar]
- Orchard VA. Effect of sewage sludge additions on Nocardia in soil. Soil Biol Biochem. 1979;11:217–220. doi: 10.1016/0038-0717(79)90064-6. [DOI] [Google Scholar]
- Orchard VA. The ecology of Nocardia and related taxa. Zentralbl Bakteriol Supp. 1981;11:167–180. [Google Scholar]
- Roth A, Andrees S, Kroppenstedt RM, Harmsen D, Mauch H. Phylogeny of the genus Nocardia based on reassessed 16S rRNA gene sequences reveals underspeciation and division of strains classified as Nocardia asteroides into three established species and two unnamed taxons. J Clin Microbiol. 2003;41:851–856. doi: 10.1128/JCM.41.2.851-856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for constructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101, MIDI Inc. Newark
- Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species. Int J Syst Bacteriol. 1966;16:313–340. doi: 10.1099/00207713-16-3-313. [DOI] [Google Scholar]
- Staneck JL, Roberts GD. Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol. 1974;28:226–231. doi: 10.1128/am.28.2.226-231.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. Mega 5: molecular evolutionary genetics analysis using maximum likelihood evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers JC, Williams ST. An assessment of plate inoculation procedures for the enumeration and isolation of soil streptomycetes. Microbios Lett. 1987;35:113–117. [Google Scholar]
- Wang Y, Zhang ZS, Ruan JS, Wang YM, Ali SM. Investigation of actinomycete diversity in the tropical rainforests of Singapore. J Ind Microbiol Biotechnol. 1999;23:178–187. doi: 10.1038/sj.jim.2900723. [DOI] [Google Scholar]
- Wayne LG, Brenner DJ, Colwell RR, et al. International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol. 1987;37:463–464. doi: 10.1099/00207713-37-4-463. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1 Neighbour-joining tree based on nearly complete 16S rRNA gene sequences (1292-1510 nt) showing relationships between the isolates and validly published Nocardia species. T, type strain. Bar, 0.005 substitutions per nucleotide position (PDF 25 kb)