Abstract
Rationale
Changes in the cortical expression of small non-coding microRNA (miRNA) have been observed in postmortem analysis of psychotic disorders. Antipsychotic drugs (APDs) are the most effective treatment option for these disorders and have been associated with changes in gene expression. MicroRNA regulate numerous genes involved in brain development and function. It is therefore plausible to question whether miRNA expression is also altered and hence whether they take part in the neuroleptic mechanism of action.
Objectives
We sought to investigate whether treatment with APDs induces changes in miRNA expression and query the functional implications of such changes. Furthermore, we investigated the possible functional interplay of miRNA–gene regulatory interactions.
Method
High-throughput miRNA profiling of the whole brain of C57BL/6 mice treated with haloperidol, olanzapine or clozapine for 7 days was performed. Functional analysis was conducted on the putative targets of altered microRNA. Significant miRNA–gene regulatory interactions were evaluated by the integration of genome-wide mRNA expression analysis using the Bayesian networks with splitting–averaging strategy and functional analysis conducted.
Results
Small subsets of miRNA were altered with each treatment with potential neurologically relevant influence. Metabolic pathways were enriched in olanzapine and clozapine treatments, possibly associated with their weight gain side effects. Neurologically and metabolically relevant miRNA–gene interaction networks were identified in the olanzapine treatment group.
Conclusion
This study is the first to suggest a role for miRNA in the mechanism of APD action and the metabolic side effects of the atypical ADPs, and adds support for their consideration in pharmacogenomics.
Electronic supplementary material
The online version of this article (doi:10.1007/s00213-012-2939-y) contains supplementary material, which is available to authorized users.
Keywords: Antipsychotic drugs, MicroRNA, Schizophrenia, Olanzapine, Clozapine, Haloperidol, Pharmacogenomics
Introduction
Psychotic disorders such as schizophrenia and bipolar disorder are severely debilitating mental illnesses. While antipsychotic drugs (APDs) are the most effective treatment option, the precise mechanisms of their action are not fully understood. There are two classes of APDs—first generation (typical) and second generation (atypical). Both classes are known to induce their therapeutic effects, relieving positive psychotic symptoms, by antagonism of dopamine receptors, particularly D2 receptors (Seeman et al. 1975). Haloperidol is a typical APD displaying highly potent dopamine receptor antagonism, allowing for a lower effective dose. It is especially effective in treating positive symptoms, particularly acute hallucinations and delusions, as well as states of aggression, impulsivity and excitement (Joy et al. 2006). Additional to treating psychotic symptoms, atypical APDs are also effective in relieving negative symptoms and improving cognition (Arranz and de Leon 2007). Their binding profiles show lower affinity for dopamine receptors than typical APDs and greater affinity to serotonergic, histaminergic, muscarinic and adrenergic receptors (Miyamoto et al. 2005). It is thought that the ability to reduce negative symptoms and improve cognition may be largely due to antagonism of serotonin (5-HT) 2A and 1A receptors (Meltzer and Sumiyoshi 2008). Clozapine was the first approved atypical APD useful in treating drug-resistant patients (Kane et al. 1988). Olanzapine is another atypical APD derived from clozapine with slight differences in receptor binding affinities (Bymaster et al. 1996; Duggan et al. 2005).
All APDs are associated with serious side effects. The most concerning side effects of typical APDs are the ‘acute extrapyramidal symptoms’ (EPS) or movement disorders, such as Parkinsonism and dyskinesia (Settle and Ayd 1983). Haloperidol is associated with high EPS, the result of potent antagonism of dopamine D2 receptors, whereas atypical APDs only weakly bind dopamine D2 receptors and dissociate before EPS is triggered (Kapur and Seeman 2000). Blockade of other neurotransmitter receptors is thought to underlie the alternate spectrum of side effects caused by atypical APDs. Clozapine is known to induce weight gain, and this is particularly so for the structurally related olanzapine (Kane et al. 1988; Duggan et al. 2005). Affinity for the serotonergic receptors, especially 5-HT2C and to a slightly lesser extent 5-HT1A, has been most strongly implicated in the weight gain effects of atypical APDs thus far (Reynolds et al. 2006).
Numerous gene expression studies have been performed on APD-treated animals to help refine our understanding of the intracellular consequences as well as mechanisms of their action and side effects. In accordance with expectation, these studies broadly reveal altered genes associated with neurotransmission, signal transduction and synaptic plasticity (Chong et al. 2002; Duncan et al. 2008; Fatemi et al. 2006; Girgenti et al. 2010; Kontkanen et al. 2002a; MacDonald et al. 2005). These studies, however, also identified changes in a host of other pathways not predicted by alteration of neurotransmission and associated signal transduction pathways. This suggests that a range of mechanisms are involved in antipsychotic activity and associated side effects. One influence that is not usually observed in conventional mRNA expression arrays is the change in level of mature microRNA (miRNA). These small non-coding RNAs could have a profound influence on global gene expression as they are believed to regulate around two thirds of the mammalian genome (Friedman et al. 2009). These molecules are thought to be particularly important for brain development and function (Giraldez et al. 2005; Krichevsky et al. 2003; Sempere et al. 2004). These studies implicate a range of miRNA alterations in the postmortem cerebral cortex and, through bioinformatic and expression analysis, suggest that these molecules play an important role in the neuropathology of psychosis (Beveridge and Cairns 2012). As most individuals experiencing psychotic illness are exposed to APDs, these molecules may represent both an important intermediate target of drug action and the point of influence leading to the disease-associated changes. Perkins et al. (2007) considered changes in miRNA expression in the prefrontal cortex of haloperidol-treated rats as a control for human miRNA expression profiling in schizophrenia and schizoaffective subjects. Their study reported three miRNA with altered expression—miR-199a, miR-128a and miR-128b, none of which correlated with the disease-related changes. Similarly, Zhou et al. (2009) examined miRNA expression in the hippocampus of rats treated with the mood stabilisers lithium and sodium valproate, as treatments for bipolar disorder, and identified several dysregulated miRNA with gene targets involved in neurogenesis and brain signalling pathways, some of which are bipolar disorder candidate genes.
In the current study, we investigated changes in miRNA expression associated with haloperidol, clozapine and olanzapine treatment in the mouse brain. We also compared the predicted miRNA target genes with gene expression profiles derived from the same treatments and identified significant miRNA–mRNA interactions associated with olanzapine, which may provide further insight into its metabolic side effects.
Methods
Antipsychotic drug treatment
A total of 76 male C57BL/6 mice, 8–10 weeks of age, were treated with either clozapine (10 mg/kg) (Sigma-Aldrich; Sydney, Australia), olanzapine (10 mg/kg) (Eli Lily; Sydney, Australia), haloperidol (1 mg/kg) (Sigma-Aldrich; Sydney, Australia) or 0.9 % saline (10 ml/kg) as a control, daily for 7 days as described in Duncan et al. (2008). Dosages were deemed consistent with previous APD animal treatment studies (Arnaiz et al. 1999; Emamian et al. 2004; Kontkanen et al. 2002b; Schreiber et al. 1999; Simosky et al. 2003). Dosage trials were also conducted for the atypicals, in which drug serum concentrations were measured with increasing doses over a 7-day period and compared to the human therapeutic dose range (Duncan et al. 2008; Online resource 1). The doses administered did not lead to any overt behavioural changes in wild-type mice.
The focus on an intermediate 7-day treatment period was driven by previous research that shows that the greatest improvement in psychotic symptoms is seen in the first 2 weeks of treatment as compared to the changes seen in subsequent weeks of chronic treatment, disproving the traditional ‘delayed onset’ hypothesis of APD action (Agid et al. 2006; Li et al. 2007; Raedler et al. 2007).
Total RNA extraction from mouse whole brain
Mouse whole brain was removed and homogenised in TRIzol reagent solution (Invitrogen; Carlsbad, CA, USA), and total RNA extracted, purified and quality assessed as described previously (Duncan et al. 2008). Whole brain was used on the basis of providing an unbiased approach to expression profiling of the effects of drugs with a mechanism of action that is highly unspecific in terms of both receptor targets and sites, and that provide treatment for psychotic disorders which are not confined to a particular region of the brain. The use of whole brain, as opposed to a specific brain region, therefore allowed us to focus on the most globally and significantly altered molecules, while avoiding dissection artefact associated with region-specific studies in the small mammalian brain.
Principles of laboratory animal care were followed. For whole brain tissue collection, animal treatment studies were approved by the Garvan Institute of Medical Research/St. Vincent’s Hospital Animal Experimentation Ethics Committee (NSW, Australia, AEEC # 03/09). Animal treatment studies for region-specific tissue collection were approved by the University of New South Wales Animal Care and Ethics Committee (ACEC # 07/22A).
MicroRNA microarray hybridisation
MicroRNA expression profiling was performed using the commercial bead-based miRNA microarray platform and mouse miRNA panel developed by Illumina (Sunnyvale, CA, USA). The mouse miRNA panel contains assays for 380 annotated mouse miRNA sequences from Sanger miRBase version 9.1 (Chen et al. 2008). A total of 45 high-quality total mouse RNA samples were selected for microarray profiling, 12 of these from the clozapine treatment group and 11 each from the olanzapine, haloperidol and saline treatment groups. Total RNA was amplified, labelled and hybridised to the miRNA beadarray matrix according to the manufacturer’s instructions.
MicroRNA microarray data normalisation and analysis
Microarray expression data were background subtracted in the BeadStudio software package (Illumina, version 3.0; Sunnyvale, CA, USA) and normalised to the geometric mean of sno-142 and sno-234. These snoRNA were selected for endogenous normalisation as they displayed the most stable expression across the control cohort according to geNorm analysis (Vandesompele et al. 2002). Non-expressed miRNA (<2 background fluorescence) were filtered from the analysis. Differential expression analysis was executed by the Significance Analysis of Microarrays statistical analysis program (full academic version 2.23) (Tusher et al. 2001) (http://www-stat.stanford.edu/~tibs/SAM/), using a two-class unpaired Wilcoxon test of unlogged data and 5,000 permutations. Significantly altered miRNA were identified as those with q value <5 (FDR < 5 %).
Quantitative real-time reverse-transcription PCR validation
Differentially expressed miRNA were validated by quantitative real-time RT-PCR (qPCR) similarly to that described previously (Santarelli et al. 2011). A standard curve was produced using serial dilutions of pooled cDNA in order to evaluate reaction efficiencies. Relative miRNA expression was analysed using the relative quantitation method, in which the ratio of miRNA expression to the geometric mean of sno-142, sno-202 and sno-234 was calculated and efficiency corrected. These snoRNA displayed stable expression across the control cohort according to geNorm analysis (Vandesompele et al. 2002). Outliers were identified as values outside the mean ± 3SD and were removed. The statistical significance of the differential expression between saline and treatment groups was assessed by multivariate analysis of variance.
Target gene predictions and pathway analysis
Potential gene targets of the differentially expressed miRNA were predicted using the miRanda algorithm. The stringency of this method was increased by filtering out predicted targets with less than three putative miRNA binding sites, whether for the same or different miRNA. Pathway analysis was then performed on this target list using Ingenuity Systems Pathway Analysis (IPA) software’s (Ingenuity Systems; Redwood City, CA, USA) core analysis functions. Metabolic analysis was also performed on the target lists by comparing each list to a list of genes with roles in metabolic processes and diseases and performing an additional core analysis.
Gene microarray hybridisation
From each treatment, 16 total RNA samples were selected and split into eight samples to form two pooled samples. These pooled samples were prepared for hybridisation to Affymetrix GeneChip Mouse Genome 430v2.0 arrays (Santa Clara, CA, USA) as described in Duncan et al. (2008) and according to the manufacturer’s instructions. Briefly, cDNA was generated from total RNA and transcribed to form biotinylated cRNA probes, which were then purified and fragmented for hybridisation to the microarrays. The probes were hybridised to Affymetrix GeneChip Test3 microarrays prior to the mouse genome microarrays for quality control assessment, using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin.
Integration analysis of microRNA and gene array data
The fluorescent signal intensities of the Affymetrix GeneChip arrays were determined using GeneChip Operating Software v1.2 (Affymetrix) and normalised to the mean intensity of all probe sets. Differentially expressed mRNA and miRNA were identified by the Bayesian empirical linear model with R. mRNA with fold change >2 and p value <0.05, and with fold change >1.3 and p value <0.05, were selected. The corresponding gene list was filtered for predicted targets of the selected miRNA. Potential miRNA–mRNA regulatory modules for each treatment group were identified using the Bayesian network with splitting–averaging strategy (BN-SA) (Liu et al. 2009). Briefly, BN-SA reports a p value that denotes the strength of dependency of the mRNA expression on the miRNA and a correlation between mRNA and miRNA expression. Those modules of p value <0.05 and reported high confidence were selected, and pathway analysis of the mRNA was performed using the core analysis feature within IPA.
Results
Significantly altered microRNA in antipsychotic drug-treated mice
High-throughput miRNA expression analysis of each APD treatment group compared to the saline treatment control group revealed several miRNA with significantly (FDR < 5 %) altered expression in each of the treatment groups. A total of six miRNA were shown to be downregulated by haloperidol and five downregulated by olanzapine, whereas five miRNA were shown to be upregulated by clozapine. Unsupervised hierarchical clustering of the microarray expression data illustrates a clean split in haloperidol and control samples (Fig. 1a) and olanzapine and control samples (Fig. 1b), and a moderate split between clozapine and control samples (Fig. 1c).
Fig. 1.
Changes in miRNA expression after treatment with APDs. a–c Hierarchical clustering of significantly dysregulated miRNA microarray expression in APD-treated mouse brain (log transformed, median centered and uncentered correlation, average linkage clustering; Cluster 3.0) (Eisen et al. 1998). Blue low expression, yellow high expression. Images produced with Java TreeView (version 1.1.1) (Saldanha 2004). a Haloperidol and saline treatment groups. b Olanzapine and saline treatment groups. c Clozapine and saline treatment groups. d Microarray expression and qPCR expression of selected miRNA from olanzapine and haloperidol treatment groups. Bars represent fold change in expression + SEM (APD treated vs. saline)
Microarray validation was performed by qPCR on a selection of miRNA from each treatment group (Fig. 1d). For the miRNA associated with the olanzapine treatment group, we were able to confirm significant differential expression of mmu-miR-193, with a fold change of 0.65 (p = 0.02). For the haloperidol treatment group, mmu-miR-434-5p and mmu-miR-22 were also significant by qPCR with a fold change of 0.37 (p = 0.0004) and 0.48 (p = 0.007), respectively. All miRNA tested in these two groups followed the trend for downregulation as reported by the microarray analysis. Validation of the clozapine treatment group was conflicting, with miR-329 and miR-342-5p displaying significant downregulation by qPCR as opposed to upregulation on the microarrays.
MicroRNA target pathways implicated in APD mechanism and side effects
A total of 1,392 genes were identified as potential targets of the haloperidol-altered miRNA-1462 for olanzapine and 1060 for clozapine. Interestingly, each list of predicted targets included a range of serotonin, glutamate, GABA receptors, as well as neurexins, synapsins and brain-derived neurotrophic factor (BDNF). Notably, HTR2C appeared as a predicted target of miRNA associated with clozapine and, similarly, HTR1A with olanzapine. Predicted targets that we have previously found to be altered by microarray analysis (Duncan et al. 2008) are annotated in Online resource 1.
Pathway analysis was performed on the predicted miRNA target genes compiled for each APD treatment group using IPA. A biological network was considered significant if the network score was ≥20. Networks relevant to the neurobiology of APDs, containing various schizophrenia candidate genes, showed significant enrichment of target genes, including: (1) nervous system development and function, cell-to-cell signalling and interaction, and genetic disorder (n = 28, score = 24) in the haloperidol treatment group; (2) cellular function and maintenance, molecular transport and cell-to-cell signalling and interaction (n = 31, score = 28) in the olanzapine treatment group; and (3) developmental disorder, genetic disorder and neurological disease (n = 33, score = 38) in the clozapine treatment group. Metabolic terms appeared in a significant clozapine network, including cellular movement, carbohydrate metabolism and cellular growth and proliferation (n = 28, score = 27).
Numerous canonical pathways and biological functions relevant to APD treatment were also identified in the IPA core analysis (Table 1). In the haloperidol and olanzapine analyses, nervous system development and function is significantly represented, which included neurogenesis (haloperidol: n = 66, p value = 1.20E−05; olanzapine: n = 79, p value = 5.11E−08; see Online resources 2 and 3). All APD analyses reported significant enrichment of axonal guidance signalling. IPA also identified classes of diseases and disorders in which focus genes are known to be involved, including neurological disease, psychological disorders and developmental disorders (Table 1).
Table 1.
Top diseases, functions and pathways enriched for predicted gene targets of miRNA altered with APD treatment. (IPA core analysis; N = number of focus genes in function/term)
| Haloperidol | Olanzapine | Clozapine | ||||||
|---|---|---|---|---|---|---|---|---|
| Diseases and disorders | p value range | N | Diseases and disorders | p value range | N | Diseases and disorders | p value range | N |
| Neurological disease | 5.88E−08 to 1.06E−02 | 90 | Cancer | 1.55E−05 to 2.33E−02 | 134 | Cancer | 3.00E−05 to 2.32E−02 | 105 |
| Psychological disorders | 2.17E07 to 6.14E03 | 22 | Haematological disease | 2.66E−05 to 2.33E−02 | 29 | Cardiovascular disease | 3.56E−05 to 2.73E−03 | 4 |
| Developmental disorder | 1.08E−06 to 1.25E−02 | 79 | Genetic disorder | 2.84E−05 to 1.52E−02 | 8 | Developmental disorder | 1.43E−04 to 1.72E−02 | 20 |
| Cancer | 3.00E−06 to 1.39E−02 | 120 | Developmental disorder | 2.94E−05 to 1.65E−02 | 76 | Genetic disorder | 1.43E−04 to 7.16E−03 | 30 |
| Haematological disease | 2.57E−04 to 6.69E−03 | 24 | Neurological disease | 1.99E−04 to 2.07E−02 | 90 | Endocrine system disorders | 2.91E−04 to 7.91E−03 | 20 |
| Molecular and cellular functions | Molecular and cellular functions | Molecular and cellular functions | ||||||
| Gene expression | 2.22E−12 to 1.43E−02 | 151 | Gene expression | 2.03E−10 to 2.42E−02 | 158 | Gene expression | 2.52E−14 to 2.46E−02 | 128 |
| Cellular assembly and organization | 2.43E−08 to 1.44E−02 | 143 | Post-translational modification | 4.40E−08 to 2.33E−02 | 84 | Cellular assembly and organization | 7.51E−10 to 2.46E−02 | 96 |
| Protein synthesis | 2.43E−08 to 1.25E−02 | 125 | Cellular assembly and organization | 2.85E−07 to 2.01E−02 | 130 | Cellular function and maintenance | 7.51E−10 to 2.35E−02 | 83 |
| Cellular development | 7.96E−07 to 1.26E−02 | 92 | Cellular function and maintenance | 2.85E−07 to 2.33E−02 | 115 | Cell death | 4.28E−07 to 2.13E−02 | 118 |
| Cellular movement | 1.04E−06 to 1.44E−02 | 179 | Cellular development | 1.97E−06 to 2.34E−02 | 210 | Cellular growth and proliferation | 1.63E−06 to 2.41E−02 | 147 |
| Physiological system development and function | Physiological system development and function | Physiological system development and function | ||||||
| Tissue development | 2.02E−10 to 1.44E−02 | 230 | Tissue development | 5.87E−09 to 2.33E−02 | 229 | Tissue development | 1.38E−09 to 2.46E−02 | 195 |
| Embryonic development | 9.18E−08 to 1.46E−02 | 162 | Nervous system development and function | 5.11E−08 to 2.33E−02 | 176 | Embryonic development | 5.94E−09 to 2.43E−02 | 142 |
| Organ development | 9.18E−08 to 1.44E−02 | 131 | Organismal development | 4.63E−07 to 2.24E−02 | 210 | Organ development | 5.94E−09 to 2.43E−02 | 131 |
| Organismal development | 9.18E−08 to 1.46E−02 | 195 | Embryonic development | 1.66E−05 to 2.20E−02 | 161 | Organismal development | 5.94E−09 to 2.43E−02 | 160 |
| Nervous system development and function | 3.74E−07 to 1.44E−02 | 163 | Organ development | 1.66E−05 to 2.20E−02 | 131 | Cardiovascular system development and function | 8.04E−06 to 2.20E−02 | 55 |
| Canonical pathways | p value | Ratio | Canonical pathways | p value | Ratio | Canonical pathways | p value | Ratio |
| Axonal guidance signalling | 5.85E−08 | 54/398 | TGF-β signalling | 8.40E−05 | 17/85 | Reelin signalling in neurons | 1.87E−04 | 13/80 |
| Ephrin receptor signalling | 3.44E−05 | 26/183 | BMP signalling pathway | 1.50E−04 | 15/73 | Axonal guidance signalling | 3.65E−04 | 36/398 |
| B Cell receptor signalling | 1.42E−04 | 22/144 | Regulation of IL-2 expression in activated and anergic T lymphocytes | 1.55E−04 | 16/83 | Role of NFAT in cardiac hypertrophy | 4.46E−04 | 21/188 |
| BMP signalling pathway | 1.95E−04 | 14/73 | Glucocorticoid receptor signalling | 1.57E−04 | 35/263 | Factors promoting cardiogenesis in vertebrates | 1.85E−03 | 12/89 |
| RhoA signalling | 2.43E−04 | 18/108 | PPARα/RXRα activation | 3.10E−04 | 25/169 | Calcium signalling | 2.36E−03 | 18/177 |
Weight gain/metabolically relevant pathways, glucocorticoid receptor signalling and PPARα/RXRα activation were reported in the olanzapine group (Table 1). Further inspection of the full functional lists also shows significant weight gain-related terms in the clozapine treatment group (Online resource 4), including insulin resistance (n = 15, p value = 2.91E−04) and obesity (n = 23, p value = 6.97E−04), and a lack of metabolic terms in the haloperidol treatment group. The ‘metabolic filtered’ core analysis of the olanzapine and clozapine groups reported several relevant significant diseases and disorders (Table 2). In the olanzapine analysis, nutritional disease appeared in the top five diseases and disorders, which consisted of nutritional disorder (p value = 3.11E−12), obesity (p value = 8.81E−10) and weight gain (p value = 2.04E−05), whilst lipid metabolism and carbohydrate metabolism appeared in the top five molecular and cellular functions. In the clozapine analysis, metabolic disease appeared in the top five diseases and disorders, containing metabolic disorder (p value = 1.17E−10) and glucose metabolism disorder (p value = 8.21E−10), and in the top five molecular and cellular functions appeared lipid metabolism. Additional data are given in Online resources 3 and 4.
Table 2.
Top diseases, functions and pathways involving predicted metabolically involved gene targets of miRNA altered with olanzapine and clozapine treatment. (IPA core analyses–metabolic filter; N = number of focus genes in function/term)
| Olanzapine | Clozapine | ||||
|---|---|---|---|---|---|
| Diseases and disorders | p value range | N | Diseases and disorders | p value range | N |
| Developmental disorder | 4.98E−19 to 7.21E−05 | 34 | Developmental disorder | 1.54E−13 to 3.19E−04 | 29 |
| Neurological disease | 1.24E−17 to 1.44E−04 | 40 | Gastrointestinal disease | 2.07E−12 to 9.19E−04 | 26 |
| Cancer | 7.67E−16 to 1.86E−04 | 44 | Neurological disease | 8.22E−12 to 1.05E−03 | 32 |
| Genetic disorder | 4.25E−13 to 8.81E−10 | 34 | Genetic disorder | 2.08E−11 to 1.07E−03 | 33 |
| Nutritional disease | 3.11E−12 to 2.04E−05 | 20 | Metabolic disease | 1.17E−10 to 1.13E−03 | 21 |
| Molecular and cellular functions | Molecular and cellular functions | ||||
| Cellular growth and proliferation | 3.30E−22 to 1.85E−04 | 63 | Cell death | 1.86E−16 to 1.06E−03 | 50 |
| Cellular development | 2.75E−21 to 1.85E−04 | 66 | Cellular development | 1.68E−14 to 1.05E−03 | 62 |
| Lipid metabolism | 1.03E−17 to 1.65E−04 | 46 | Cellular movement | 7.71E−14 to 1.05E−03 | 38 |
| Small molecule biochemistry | 1.03E−17 to 1.85E−04 | 56 | Lipid metabolism | 3.97E−13 to 1.07E−03 | 47 |
| Carbohydrate metabolism | 7.48E−17 to 1.85E−04 | 32 | Small molecule biochemistry | 3.97E−13 to 1.07E−03 | 61 |
| Physiological system development and function | Physiological system development and function | ||||
| Organismal development | 1.64E−24 to 1.85E−04 | 72 | Tissue development | 3.72E−15 to 1.07E−03 | 60 |
| Tissue development | 5.88E−24 to 1.86E−04 | 68 | Tissue morphology | 1.22E−13 to 1.05E−03 | 45 |
| Tissue morphology | 1.92E−22 to 1.85E−04 | 57 | Organismal development | 1.38E−13 to 1.07E−03 | 56 |
| Embryonic development | 1.44E−21 to 1.85E−04 | 56 | Embryonic development | 2.51E−13 to 1.07E−03 | 45 |
| Organ development | 1.33E−21 to 1.85E−04 | 50 | Organ development | 2.51E−13 to 1.05E−03 | 40 |
| Canonical pathways | p value | Ratio | Canonical pathways | p value | Ratio |
| Glucocorticoid receptor signalling | 1.13E−12 | 18/263 | Reelin signalling in neurons | 3.15E−07 | 8/80 |
| Role of NFAT in cardiac hypertrophy | 1.45E−08 | 12/185 | Role of tissue factor in cancer | 3.40E−07 | 9/109 |
| FGF signalling | 1.81E−08 | 9/84 | Myc mediated apoptosis signalling | 6.49E−07 | 7/60 |
| Role of JAK family kinases in IL-6-type cytokine signalling | 3.39E−08 | 6/26 | Ovarian cancer signalling | 1.44E−06 | 9/133 |
| PPARα/RXRα activation | 6.66E−08 | 11/169 | Renal Cell carcinoma signalling | 1.56E−06 | 7/68 |
Integration of APD-associated gene and miRNA expression
As a strategy to further explore miRNA–mRNA interactions that may play a role in the mechanisms of APD action, we also performed integration analysis of miRNA and mRNA expression data in each 7-day APD treatment. Several miRNA–mRNA regulatory modules were identified in each APD treatment using the BN-SA method; however, the networks identified in the haloperidol and clozapine groups were not significant. In regard to olanzapine, both negatively and positively correlated interactions were identified (Fig. 2).
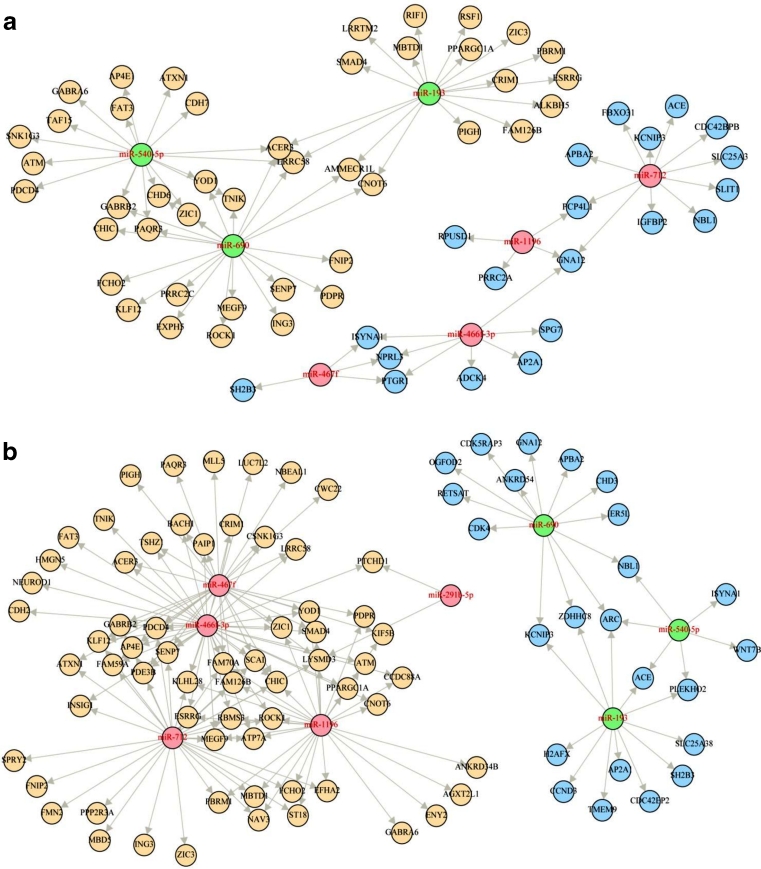
Fig. 2.
Potential miRNA–mRNA regulatory networks identified in the olanzapine treatment group. Predicted miRNA target genes identified as significantly differentially expressed were identified. The strength of the potential miRNA–mRNA interactions was assessed in regard to correlation of miRNA to mRNA expression values using the BN-SA method. Those interactions reported with strong correlation with a high confidence comment are pictured here. a Negatively correlated interactions. b Positively correlated interactions. (pink upregulated miRNA, green downregulated miRNA, blue downregulated mRNA, yellow upregulated mRNA)
Pathway analysis on the combined list of mRNA identified in the olanzapine networks was performed using IPA (Table 3). One significant, interesting network was reported in the top five networks that included neurological disease and behaviour (n = 14, score = 19). Associations with highly relevant diseases/disorders were also reported: neurological disease and developmental disorder, as well as nervous system development and function and axonal guidance signalling (Table 2). As in the predictive analysis, a metabolically relevant network was also reported: lipid metabolism, molecular transport, and small molecule biochemistry (n = 21, score = 34). Metabolic disease was also reported in the top ten diseases/disorders. Full results are given in Online resource 5.
Table 3.
Top diseases, functions and pathways involving predicted gene targets of miRNA altered with olanzapine treatment—integration analysis of miRNA and gene expression. (IPA core analyses; N = number of focus genes in function/term)
| Olanzapine | ||
|---|---|---|
| Diseases and disorders | p value range | N |
| Neurological disease | 1.76E−05 to 4.71E−02 | 24 |
| Developmental disorder | 9.81E−05 to 4.01E−02 | 21 |
| Cancer | 3.23E−04 to 4.87E−02 | 27 |
| Organismal injury and abnormalities | 9.62E−04 to 4.87E−02 | 10 |
| Haematological disease | 1.10E−03 to 3.78E−02 | 10 |
| Molecular and cellular functions | ||
| Cellular assembly and organization | 2.92E−04 to 4.87E−02 | 26 |
| Cell death | 7.52E−04 to 4.87E−02 | 14 |
| Gene expression | 1.37E−03 to 4.44E−02 | 25 |
| Cellular compromise | 1.42E−03 to 4.87E−02 | 10 |
| Cellular growth and proliferation | 1.57E−03 to 4.24E−02 | 27 |
| Physiological system development and function | ||
| Organismal functions | 6.53E−05 to 2.95E−02 | 13 |
| Tissue development | 4.98E−04 to 4.87E−02 | 47 |
| Nervous system development and function | 5.67E−04 to 4.87E−02 | 39 |
| Tissue morphology | 5.67E−04 to 4.75E−02 | 21 |
| Embryonic development | 5.90E−04 to 4.87E−02 | 34 |
| Canonical pathways | p value | Ratio |
| IL-8 signalling | 2.62E−04 | 8/172 |
| Gα12/13 signalling | 9.59E−04 | 6/117 |
| Axonal guidance signalling | 1.36E−03 | 11/398 |
| TR/RXR activation | 1.49E−03 | 5/87 |
| VEGF signalling | 1.57E−03 | 5/89 |
Discussion
Since the inception of high-throughput genomics, genome-wide approaches to gene expression analysis have been employed for both human samples and animal models to provide a survey of gene activity in relation to postmortem neuropathology and the role of antipsychotic medication. With significant changes in schizophrenia-associated gene expression, a number of research groups, including our own, have investigated and identified dysregulation of miRNA expression (reviewed in Beveridge and Cairns 2012). In this study, we wanted to determine how influential APDs are on the expression of miRNA and if the changes have functional significance to the mechanism of action and side effects. To this end, we performed miRNA expression profiling and integration analysis with gene expression data in mouse brain following treatment with haloperidol, olanzapine or clozapine.
We observed significant changes in the expression of small sets of miRNA in each APD treatment group by microarray analysis. These observations broadly contrast with our previously reported analysis of miRNA expression in Brodmann Area (BA) 9 and 46 of the dorsolateral prefrontal cortex (DLPFC), and BA22 of the superior temporal gyrus, which consistently displayed schizophrenia-associated elevation of miRNA expression (Beveridge et al. 2008, 2010; Santarelli et al. 2011). We observed downregulation of miRNA after haloperidol and olanzapine treatment, which may suggest that normalisation of miRNA expression levels may be another mechanism of APD action. For example, miR-193, miR-223 and miR-544, shown to be downregulated by olanzapine treatment, were all previously observed to be elevated in the DLPFC in schizophrenia (Beveridge et al. 2010; Kim et al. 2010; Santarelli et al. 2011), suggesting that olanzapine may help to counter the disease-associated dysregulation of these miRNA. Similarly, in BA22, we reported upregulation of miR-339—observed here to be downregulated after haloperidol treatment. On the other hand, we also observed changes after APD treatment following the same direction as those observed in the brain in schizophrenia and bipolar disorder. For example, Moreau et al. (2011) reported downregulation of miR-22 and miR-339 in both disorders—found here to be downregulated after haloperidol treatment. In the case of clozapine, the five differentially expressed miRNA were upregulated, although validation of miR-329 and miR-342-5p by qPCR displayed an opposite trend. We have previously reported upregulation of miR-31 in BA9 in schizophrenia—observed here as upregulated in mouse brain after clozapine treatment. We have also reported downregulation of miR-31 and miR-342-5p in peripheral blood mononuclear cells in schizophrenia (Gardiner et al. 2011)—observed here to be altered after clozapine treatment. The remainder of miRNA found to be altered after APD treatment have not yet been reported in schizophrenia or bipolar disorder expression profiling studies. Other neuroleptic treatment studies have reported changes in the expression of other miRNA. For example, Perkins et al. (2007) observed upregulation of miR-199a, miR-128a and miR-128b in rodents after haloperidol treatment. Zhou et al. (2009) reported large sets of miRNA altered after treatment of the mood stabilisers lithium and sodium valproate, predominantly downregulated similar to our results. Downregulation of miR-410 after haloperidol treatment was similarly reported by Zhou et al. (2009) to be downregulated after sodium valproate treatment. Those findings in which changes in miRNA expression are observed in the same direction in the disorder and after APD treatment may suggest that some schizophrenia-associated changes in miRNA expression may be related to treatments. Nonetheless, our findings support that these compounds clearly have an influence on miRNA expression that is distinct from the underlying disorder and may even have significance to their mechanism of action and side effects.
In order to explore the biological influence of these APD-associated miRNA, we performed gene target predictions and functional/pathway analysis on target lists for each drug treatment group. Predicted targets included a range of known APD neurotransmitter receptor target genes, such as serotonin, glutamate and GABA receptors, which would suggest that miRNA are involved in the modulation of these receptors. N-Methyl-d-aspartate receptors (NMDAR) GRIN1 and GRIN2B were predicted targets of haloperidol- and clozapine-altered miRNA, respectively. NMDAR antagonism has been shown to induce positive, negative and cognitive symptoms in schizophrenia (Lahti et al. 2001), and both haloperidol and clozapine have demonstrated the ability to attenuate the effects of different NMDAR antagonists but do not directly bind NMDARs (Miyamoto et al. 2012). Hence, our findings may suggest that modulation of miRNA expression by these APDs may be part of the missing link. The serotonin receptors HTR2C and HTR1A were predicted targets of clozapine- and olanzapine-altered miRNA, respectively. Antagonism of these receptors has been attributed to the ability of atypical APDs to treat both negative symptoms and cognitive impairment in schizophrenia patients (Miyamoto et al. 2012). Other target genes with relevance to synaptic function included presynaptic neurexins and BDNF. Functional analysis of these genes identified associations with several highly relevant neuronal functions, pathways and diseases, further supporting a role for miRNA in the neuroleptic mechanism of action.
APD-associated changes in miRNA may also play a role in mediating some of the side effects experienced by patients. In this respect, it is interesting that HTR2C is thought to be involved in weight gain associated with atypicals (De Luca et al. 2007; Mulder et al. 2007). While this is likely to be a consequence of direct ligand receptor interactions, the gene was also predicted to be a target of the clozapine-associated miRNAs miR-31, miR-337-5p and miR-1198. Similarly, HTR1A, associated with modulation of appetite (Yadav et al. 2009), was a predicted target of the olanzapine-associated miRNAs miR-223 and miR-544. The glucocorticoid receptor signalling pathway was also reported for miRNA associated with the olanzapine treatment group. Inhibition of this pathway in rodents has been shown to counter the weight gain resulting from olanzapine treatment (Beebe et al. 2006). We also observed enrichment of target genes in the PPARα/RXRα activation pathway, which is reported to be associated with weight gain via fatty acid storage and glucose homeostasis (Evans et al. 2004). In view of these observations, we performed a metabolically filtered core analysis in IPA and found striking similarities between the atypicals clozapine and olanzapine consistent with well-reported side effects of the atypical APDs, while the haloperidol analysis showed no significant metabolic results. Interestingly, the analysis reported significant enrichment for lipid metabolism for both atypicals and carbohydrate metabolism in the olanzapine group. In further support for a metabolic influence from these APD-regulated miRNA, we observed enrichment of relevant diseases and disorders, such as nutritional disease, obesity and weight gain in the olanzapine group, and metabolic disorder and glucose metabolism disorder for the clozapine group. These terms are well reported in treatment with these atypical APDs (Reynolds and Kirk 2010) and suggest that miRNA may play a role in their metabolic side effects.
While the former analyses were of a predictive investigation, we also performed an integration analysis of miRNA and mRNA expression data on the olanzapine samples, in which we identified several significant interactions that collectively formed an extensive network. By further characterising the genes involved in this network, we again observed significant enrichment of relevant pathways, including modulation of axonal guidance signalling, and nervous system development and function. Significantly, lipid metabolism and metabolic disease were observed and further supported the predicted metabolic influence of the olanzapine-associated miRNA. This integration analysis served to provide a more stringent, functional approach to investigating the implications of miRNA-associated changes with direct reference to treatment-induced changes in gene expression that occur in the same tissue.
Our investigation served to observe the changes in miRNA expression at the baseline level, without the additional variable of psychosis-associated changes. The APD doses used in our study lie within the optimal dose ranges for reversing the hyperactivity exhibited by the chakragati mouse, reported to be the best model for psychosis to date (Dawe et al. 2010). While relevance and significance of animal models of psychosis is still a subject of considerable debate, further insight into the putative antipsychotic influence of the miRNA changes occurring after APD treatment may be achieved by a follow-up study in one of these models of psychosis. This would enable a comparison of the APD-associated changes in miRNA expression, observed here in the wild type, to be compared to those observed before and after rescue of the ‘psychotic phenotype’ the presence of pharmacologically relevant levels of APDs.
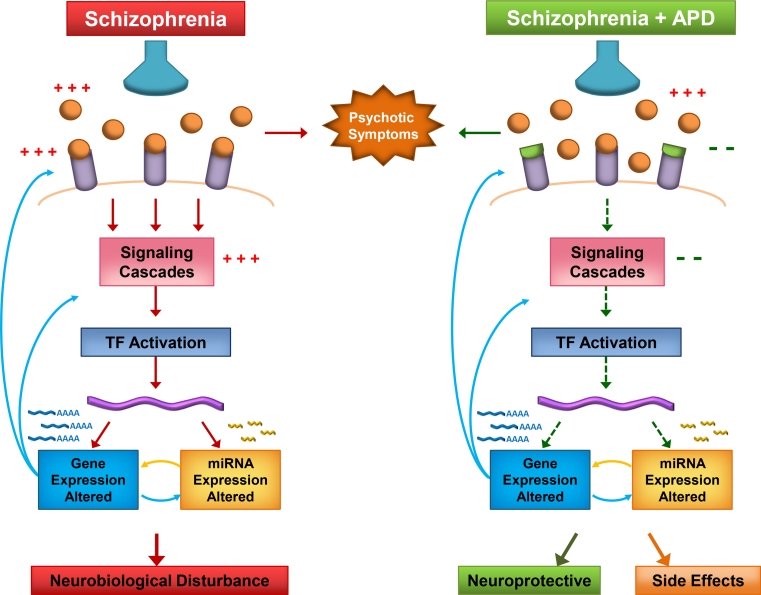
In conclusion, our investigation provides insight into the potential involvement of miRNA in the mechanisms of APD action. Functional annotation of predicted target genes and direct interaction analysis using both mRNA and miRNA expression analysis revealed pathways with significance to both the neuroleptic mechanism and side effects, particularly the metabolic effects associated with atypical APDs. Our findings may hence suggest a model that conforms to the growing focus of miRNA in the field of pharmacogenomics (Shomron 2010), in which miRNA play a significant role in mediating antipsychotic effects via regulation of drug targets and/or downstream signalling pathways (Fig. 3). The continual identification of changes in miRNA in diseases and after drug treatment is placing newfound importance of these molecules in terms of drug enhancement and new drug development (Rukov and Shomron 2011). This suggests that miRNA heterogeneity is a likely underlying factor in the complexity of psychosis and will play a role in the varied responses to APD efficacy and side effects.
Fig. 3.
The potential involvement of miRNA in the underlying mechanisms of APD therapeutic action or side effects. Neurotransmitters (orange balls) are released by presynaptic receptors (blue funnels) and bind to their postsynaptic receptors (purple tubes) to induce signalling cascades. Transcription factor activation of DNA (purple waves) leads to altered miRNA and gene expression, including miRNA biogenesis genes. Mature miRNA modulate gene expression leading to further upstream changes and ultimately to changes in neurobiological processes. In schizophrenia, neurotransmitter receptor binding and signalling cascades are enhanced (three plus signs), resulting in the presence of psychotic symptoms. Recent findings support enhancement of miRNA biogenesis, with miRNA dysregulation leading to changes in gene expression, which results in neurobiological disturbances. APDs (green cups) block postsynaptic neurotransmitter receptors, relieving psychotic symptoms. Signalling cascades are tuned down (two minus signs), and opposing changes in miRNA and gene expression occur. The end result is modulation of pathways and processes that are neuroprotective, as well as those that, when altered, may be involved in the manifestation of side effects
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOC 248 kb)
(XLS 176 kb)
(XLS 214 kb)
(XLS 203 kb)
(XLS 81 kb)
Acknowledgments
This study was supported by the Schizophrenia Research Institute, utilizing infrastructure funding from the NSW Ministry of Health and an M.C. Ainsworth Research Fellowship in Epigenetics (MC); a NARSAD Young Investigator Award; a Hunter Medical Research Institute project grant; an NHMRC project grant 631057; the Neurobehavioral Genetics Unit; and NSW Department of Health. The animal experiments performed comply with the current Australian laws. The authors declare no biomedical financial interests or potential conflicts of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
References
- Agid O, Seeman P, Kapur S. The “delayed onset” of antipsychotic action—an idea whose time has come and gone. J Psychiatry Neurosci. 2006;31:93–100. [PMC free article] [PubMed] [Google Scholar]
- Arnaiz SL, Coronel MF, Boveris A. Nitric oxide, superoxide, and hydrogen peroxide production in brain mitochondria after haloperidol treatment. Nitric Oxide Biol Chem Off J Nitric Oxide Soc. 1999;3:235–243. doi: 10.1006/niox.1999.0229. [DOI] [PubMed] [Google Scholar]
- Arranz MJ, de Leon J. Pharmacogenetics and pharmacogenomics of schizophrenia: a review of last decade of research. Mol Psychiatry. 2007;12:707–747. doi: 10.1038/sj.mp.4002009. [DOI] [PubMed] [Google Scholar]
- Beebe KL, Block T, Debattista C, Blasey C, Belanoff JK. The efficacy of mifepristone in the reduction and prevention of olanzapine-induced weight gain in rats. Behav Brain Res. 2006;171:225–229. doi: 10.1016/j.bbr.2006.03.039. [DOI] [PubMed] [Google Scholar]
- Beveridge NJ, Cairns MJ. MicroRNA dysregulation in schizophrenia. Neurobiol Dis. 2012;46:263–271. doi: 10.1016/j.nbd.2011.12.029. [DOI] [PubMed] [Google Scholar]
- Beveridge NJ, Tooney PA, Carroll AP, Gardiner E, Bowden N, Scott RJ, Tran N, Dedova I, Cairns MJ. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum Mol Genet. 2008;17:1156–1168. doi: 10.1093/hmg/ddn005. [DOI] [PubMed] [Google Scholar]
- Beveridge NJ, Gardiner E, Carroll AP, Tooney PA, Cairns MJ. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol Psychiatry. 2010;15:1–14. doi: 10.1038/mp.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bymaster FP, Calligaro DO, Falcone JF, Marsh RD, Moore NA, Tye NC, Seeman P, Wong DT. Radioreceptor binding profile of the atypical antipsychotic olanzapine. Neuropsychopharmacology. 1996;14:87–96. doi: 10.1016/0893-133X(94)00129-N. [DOI] [PubMed] [Google Scholar]
- Chen J, Lozach J, Garcia EW, Barnes B, Luo S, Mikoulitch I, Zhou L, Schroth G, Fan JB. Highly sensitive and specific microRNA expression profiling using BeadArray technology. Nucleic Acids Res. 2008;36:e87. doi: 10.1093/nar/gkn387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong VZ, Young LT, Mishra RK. cDNA array reveals differential gene expression following chronic neuroleptic administration: implications of synapsin II in haloperidol treatment. J Neurochem. 2002;82:1533–1539. doi: 10.1046/j.1471-4159.2002.01104.x. [DOI] [PubMed] [Google Scholar]
- Dawe GS, Nagarajah R, Albert R, Casey DE, Gross KW, Ratty AK. Antipsychotic drugs dose-dependently suppress the spontaneous hyperactivity of the chakragati mouse. Neuroscience. 2010;171:162–172. doi: 10.1016/j.neuroscience.2010.08.061. [DOI] [PubMed] [Google Scholar]
- De Luca V, Mueller DJ, de Bartolomeis A, Kennedy JL. Association of the HTR2C gene and antipsychotic induced weight gain: a meta-analysis. Int J Neuropsychopharmacol. 2007;10:697–704. doi: 10.1017/S1461145707007547. [DOI] [PubMed] [Google Scholar]
- Duggan L, Fenton M, Rathbone J, Dardennes R, El-Dosoky A, Indran S (2005) Olanzapine for schizophrenia. Cochrane Database Syst Rev: CD001359 [DOI] [PubMed]
- Duncan CE, Chetcuti AF, Schofield PR. Coregulation of genes in the mouse brain following treatment with clozapine, haloperidol, or olanzapine implicates altered potassium channel subunit expression in the mechanism of antipsychotic drug action. Psychiatr Genet. 2008;18:226–239. doi: 10.1097/YPG.0b013e3283053019. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emamian ES, Hall D, Birnbaum MJ, Karayiorgou M, Gogos JA. Convergent evidence for impaired AKT1-GSK3beta signaling in schizophrenia. Nat Genet. 2004;36:131–137. doi: 10.1038/ng1296. [DOI] [PubMed] [Google Scholar]
- Evans RM, Barish GD, Wang YX. PPARs and the complex journey to obesity. Nature Med. 2004;10:355–361. doi: 10.1038/nm1025. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Reutiman TJ, Folsom TD, Bell C, Nos L, Fried P, Pearce DA, Singh S, Siderovski DP, Willard FS, Fukuda M. Chronic olanzapine treatment causes differential expression of genes in frontal cortex of rats as revealed by DNA microarray technique. Neuropsychopharmacology. 2006;31:1888–1899. doi: 10.1038/sj.npp.1301002. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner E, Beveridge NJ, Wu JQ, Carr V, Scott RJ, Tooney PA, Cairns MJ. Imprinted DLK1-DIO3 region of 14q32 defines a schizophrenia-associated miRNA signature in peripheral blood mononuclear cells. Mol Psychiatry. 2012;17:827–840. doi: 10.1038/mp.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Girgenti MJ, Nisenbaum LK, Bymaster F, Terwilliger R, Duman RS, Newton SS. Antipsychotic-induced gene regulation in multiple brain regions. J Neurochem. 2010;113:175–187. doi: 10.1111/j.1471-4159.2010.06585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joy CB, Adams CE, Lawrie SM (2006) Haloperidol versus placebo for schizophrenia. Cochrane Database Syst Rev: CD003082 [DOI] [PubMed]
- Kane J, Honigfeld G, Singer J, Meltzer H. Clozapine for the treatment-resistant schizophrenic. A double-blind comparison with chlorpromazine. Arch Gen Psychiatry. 1988;45:789–796. doi: 10.1001/archpsyc.1988.01800330013001. [DOI] [PubMed] [Google Scholar]
- Kapur S, Seeman P. Antipsychotic agents differ in how fast they come off the dopamine D2 receptors. Implications for atypical antipsychotic action. J Psychiatry Neurosci. 2000;25:161–166. [PMC free article] [PubMed] [Google Scholar]
- Kim AH, Reimers M, Maher B, Williamson V, McMichael O, McClay JL, van den Oord EJ, Riley BP, Kendler KS, Vladimirov VI. MicroRNA expression profiling in the prefrontal cortex of individuals affected with schizophrenia and bipolar disorders. Schizophr Res. 2010;124:183–191. doi: 10.1016/j.schres.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontkanen O, Lakso M, Wong G, Castren E. Chronic antipsychotic drug treatment induces long-lasting expression of fos and jun family genes and activator protein 1 complex in the rat prefrontal cortex. Neuropsychopharmacology. 2002;27:152–162. doi: 10.1016/S0893-133X(02)00289-0. [DOI] [PubMed] [Google Scholar]
- Kontkanen O, Toronen P, Lakso M, Wong G, Castren E. Antipsychotic drug treatment induces differential gene expression in the rat cortex. J Neurochem. 2002;83:1043–1053. doi: 10.1046/j.1471-4159.2002.01213.x. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahti AC, Weiler MA, Tamara Michaelidis BA, Parwani A, Tamminga CA. Effects of ketamine in normal and schizophrenic volunteers. Neuropsychopharmacol Off Publi Am Coll Neuropsychopharmacol. 2001;25:455–467. doi: 10.1016/S0893-133X(01)00243-3. [DOI] [PubMed] [Google Scholar]
- Li M, Fletcher PJ, Kapur S. Time course of the antipsychotic effect and the underlying behavioral mechanisms. Neuropsychopharmacology. 2007;32:263–272. doi: 10.1038/sj.npp.1301110. [DOI] [PubMed] [Google Scholar]
- Liu B, Li J, Tsykin A, Liu L, Gaur AB, Goodall GJ. Exploring complex miRNA-mRNA interactions with Bayesian networks by splitting-averaging strategy. BMC Bioinforma. 2009;10:408. doi: 10.1186/1471-2105-10-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald ML, Eaton ME, Dudman JT, Konradi C. Antipsychotic drugs elevate mRNA levels of presynaptic proteins in the frontal cortex of the rat. Biol Psychiatry. 2005;57:1041–1051. doi: 10.1016/j.biopsych.2005.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meltzer HY, Sumiyoshi T. Does stimulation of 5-HT(1A) receptors improve cognition in schizophrenia? Behav Brain Res. 2008;195:98–102. doi: 10.1016/j.bbr.2008.05.016. [DOI] [PubMed] [Google Scholar]
- Miyamoto S, Duncan GE, Marx CE, Lieberman JA. Treatments for schizophrenia: a critical review of pharmacology and mechanisms of action of antipsychotic drugs. Mol Psychiatry. 2005;10:79–104. doi: 10.1038/sj.mp.4001556. [DOI] [PubMed] [Google Scholar]
- Miyamoto S, Miyake N, Jarskog LF, Fleischhacker WW, Lieberman JA. Pharmacological treatment of schizophrenia: a critical review of the pharmacology and clinical effects of current and future therapeutic agents. Mol Psychiatry. 2012;17:1206–1227. doi: 10.1038/mp.2012.47. [DOI] [PubMed] [Google Scholar]
- Moreau MP, Bruse SE, David-Rus R, Buyske S, Brzustowicz LM. Altered microRNA expression profiles in postmortem brain samples from individuals with schizophrenia and bipolar disorder. Biol Psychiatry. 2011;69:188–193. doi: 10.1016/j.biopsych.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder H, Franke B, van der Beek AA, Arends J, Wilmink FW, Egberts AC, Scheffer H. The association between HTR2C polymorphisms and obesity in psychiatric patients using antipsychotics: a cross-sectional study. Pharmacogenomics J. 2007;7:318–324. doi: 10.1038/sj.tpj.6500422. [DOI] [PubMed] [Google Scholar]
- Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, Parker JS, Jin J, Hammond SM. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8:R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raedler TJ, Schreiner A, Naber D, Wiedemann K. Early onset of treatment effects with oral risperidone. BMC Psychiatry. 2007;7:4. doi: 10.1186/1471-244X-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds GP, Kirk SL. Metabolic side effects of antipsychotic drug treatment—pharmacological mechanisms. Pharmacol Ther. 2010;125:169–179. doi: 10.1016/j.pharmthera.2009.10.010. [DOI] [PubMed] [Google Scholar]
- Reynolds GP, Arranz B, Templeman LA, Fertuzinhos S, San L. Effect of 5-HT1A receptor gene polymorphism on negative and depressive symptom response to antipsychotic treatment of drug-naive psychotic patients. Am J Psychiatry. 2006;163:1826–1829. doi: 10.1176/appi.ajp.163.10.1826. [DOI] [PubMed] [Google Scholar]
- Rukov JL, Shomron N. MicroRNA pharmacogenomics: post-transcriptional regulation of drug response. Trends Mol Med. 2011;17:412–423. doi: 10.1016/j.molmed.2011.04.003. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Santarelli DM, Beveridge NJ, Tooney PA, Cairns MJ. Upregulation of dicer and microRNA expression in the dorsolateral prefrontal cortex Brodmann area 46 in schizophrenia. Biol Psychiatry. 2011;69:180–187. doi: 10.1016/j.biopsych.2010.09.030. [DOI] [PubMed] [Google Scholar]
- Schreiber S, Getslev V, Backer MM, Weizman R, Pick CG. The atypical neuroleptics clozapine and olanzapine differ regarding their antinociceptive mechanisms and potency. Pharmacol Biochem Behav. 1999;64:75–80. doi: 10.1016/S0091-3057(99)00107-0. [DOI] [PubMed] [Google Scholar]
- Seeman P, Chau-Wong M, Tedesco J, Wong K. Brain receptors for antipsychotic drugs and dopamine: direct binding assays. Proc Natl Acad Sci USA. 1975;72:4376–4380. doi: 10.1073/pnas.72.11.4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settle EC Jr, Ayd FJ Jr (1983) Haloperidol: a quarter century of experience. J Clin Psychiatry 44:440–448 [PubMed]
- Shomron N. MicroRNAs and pharmacogenomics. Pharmacogenomics. 2010;11:629–632. doi: 10.2217/pgs.10.26. [DOI] [PubMed] [Google Scholar]
- Simosky JK, Stevens KE, Adler LE, Freedman R. Clozapine improves deficient inhibitory auditory processing in DBA/2 mice, via a nicotinic cholinergic mechanism. Psychopharmacology. 2003;165:386–396. doi: 10.1007/s00213-002-1285-x. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034 [DOI] [PMC free article] [PubMed]
- Yadav VK, Oury F, Suda N, Liu ZW, Gao XB, Confavreux C, Klemenhagen KC, Tanaka KF, Gingrich JA, Guo XE, Tecott LH, Mann JJ, Hen R, Horvath TL, Karsenty G. A serotonin-dependent mechanism explains the leptin regulation of bone mass, appetite, and energy expenditure. Cell. 2009;138:976–989. doi: 10.1016/j.cell.2009.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Yuan P, Wang Y, Hunsberger JG, Elkahloun A, Wei Y, Damschroder-Williams P, Du J, Chen G, Manji HK. Evidence for selective microRNAs and their effectors as common long-term targets for the actions of mood stabilizers. Neuropsychopharmacology. 2009;34:1395–1405. doi: 10.1038/npp.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 248 kb)
(XLS 176 kb)
(XLS 214 kb)
(XLS 203 kb)
(XLS 81 kb)