Abstract
The mammalian immune system has evolved in the presence of microbes, both pathogenic and commensal. The consequences of microbial recognition by the host has led to the development of compensatory mechanisms by both the host and microbe to either resist or tolerate the existence of the other. In this review we discuss examples of this co-evolutionary relationship. Due to space considerations and for conceptual clarity, we have focused on detection of bacteria by the Toll-like receptor (TLR) family and highlight examples of bacterial strategies to evade, subvert and in some cases even utilize these receptors.
Introduction
TLRs are a family of membrane-spanning innate immune receptors that recognize ligands derived from bacteria, fungi, viruses, and parasites. Recognition of conserved microbial features by TLRs leads to a variety of downstream signals in immune cells, including pro-inflammatory cytokine production, costimulatory molecule upregulation, anti-microbial peptide secretion, and phagosomal maturation (1, 2). While individual TLRs typically recognize a specific class of microbial ligands, collectively this family of receptors can detect a broad range of microbes. Of the thirteen TLRs present in mammals, ligands have been identified for twelve: lipopolysaccharide (LPS) for TLR4, lipopeptides for TLR2/1 and TLR2/6 heterodimers, flagellin for TLR5, unmethylated CpG motifs in DNA for TLR9, profilin and Salmonella flagellin for TLR11 (3), and various forms of RNA for TLRs 3, 7, 8 and 13, with no known ligand currently identified for TLR 12 (4). Thus, multiple TLRs can potentially recognize bacteria, although individual TLRs will play a more dominant role for certain bacterial species or in certain contexts (e.g., recognition of nucleic acids released from degraded bacteria).
Signaling
All TLRs share a common modular structure: a leucine rich repeat (LRR)-containing ectodomain responsible for ligand-binding, a membrane spanning region, and a cytosolic signaling domain called the Toll-interleukin 1 receptor homology domain (TIR). Ligand binding induces recruitment of TIR domain-containing signaling adaptors that associate with the TLR via homotypic TIR:TIR interactions. All TLRs, with the exception of TLR3, recruit MyD88 at this initial step. MyD88 can associate with IL-1R-associated kinase (IRAK) members. Upon dissociation from the TIR complex, IRAK proteins interact with tumor necrosis factor receptor-associated factor 6 (TRAF6)to propagate TLR activation signals that include the activation of transcription factors NF-κB, AP-1 (mediated by JNK, p38 and ERK), and IRF5 (5). TLR3 and TLR4 can recruit a different adaptor, TIR-domain-containing adapter-inducing interferon-β (TRIF), that leads to the dimerization and activation of inhibitor of NF-κB kinase (IKKi) and TANK (TRAF–family member associated NF-κB activator)-binding kinase 1 (TBK1). Activated TBK1/IKKi phosphorylates the transcription factor, IRF3, inducing its nuclear translocation and subsequent transcription of interferon-related genes (6).
Function
TLR activation leads to production of pro-inflammatory cytokines, including TNFα, IL-12, and IL-6. These cytokines induce local inflammation, support the survival and expansion of B and T cells and activate natural killer (NK) cells. A subset of TLRs can also induce type I interferon production (IFNα/β) (4, 6). This family of cytokines can inhibit translation and/or induce apoptosis in host cells, thereby exposing intracellular bacteria to the extracellular environment and killing by other infiltrating immune cells. TLR signaling also leads to the upregulation of costimulatory molecules and MHC molecules presenting bacterial antigens. Costimulatory molecules include CD80, CD86 and CD40 and have the overall outcome of generating protective adaptive immune responses by the activation of antigen-specific T cells (7).
TLR activation can also induce cell-intrinsic antimicrobial activity. For example, TLR2 and TLR4 activation can recruit NADPH oxidase assembly as well as mitochondria relocalization to the bacteria-containing phagosome, leading to a burst of reactive oxygen and nitrogen species within this compartment (8–10). Evidence also suggests that TLR signaling can lead to a rapid acidification of the phagosome in which TLR signaling has occurred, likely through recruitment of vacuolar-ATPase subunits to the phagosomal membrane (11–14). Both of these activities increase the antimicrobial capacity of the phagosome, although some bacteria have actually co-opted these signals to regulate their virulence programs (discussed in greater detail below). Detection of microbial ligands by TLRs can also induce the expression and secretion of antimicrobial peptides (AMPs), such as beta-defensins, and cathelicidin, further supporting the role of TLR-mediated detection in cell-intrinsic antimicrobial activity (15–17).
Evasion and Subversion of Immunity by Bacteria
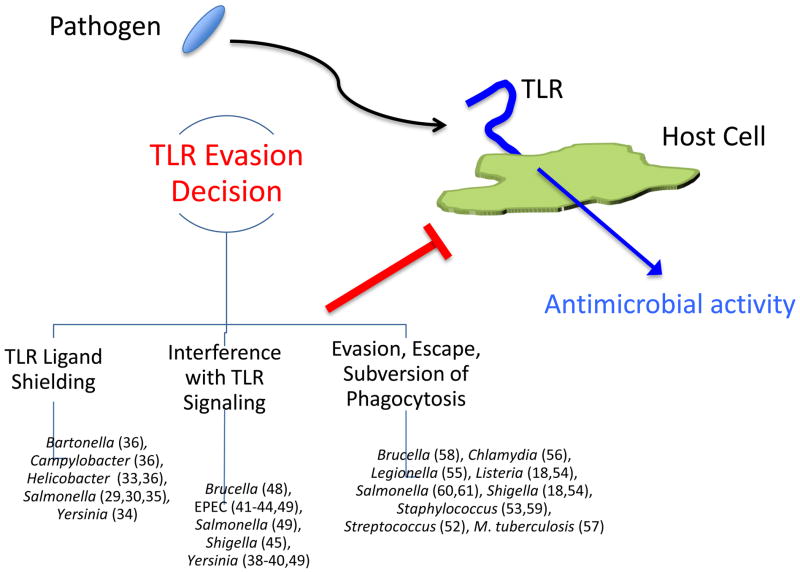
Because the specificity of TLRs are fixed in the germline and activation initiates the earliest aspects of the immune response to infection, these receptors have applied tremendous selective pressure on the virulence mechanisms of potential pathogens. Not surprisingly, pathogens have evolved a variety of strategies to survive despite recognition by the innate immune system. Here we identify and focus on three general themes, each representing disruption or interference at distinct stages of the host response (Figure 1): 1) evasion of host detection by shielding ligands, 2) interference with TLR signaling pathways, and 3) inhibiting, escaping, or subverting phagocytosis (18–20) (21). Our discussion of these strategies cannot be exhaustive; instead, we highlight key examples.
Figure 1. Strategies used by pathogens to evade TLR signaling.
Three common evasion strategies utilized by different pathogens are illustrated. Bacteria that employ these mechanisms (and relevant citations) are indicated beneath each strategy. See text for discussion.
Surface structure modification
The outer-membranes of gram-negative bacteria (including Salmonella and Yersinia species, for example) contain LPS, a potent activator of TLR4 (22–27). Activation of this TLR can lead to the production of pro-inflammatory cytokines, co-stimulatory molecule upregulation and secretion of type I IFN (1, 2, 5). These signals depend on the activation of NF-κB and MAP kinase pathways (1). Other surface proteins such as flagellin, a potent TLR5 ligand, can also lead to immune activation, albeit without the production of IFN, via similar pathways(28–30). It is therefore not surprising that bacteria have evolved mechanisms to make these surface structures less detectable by their corresponding TLRs.
LPS, crucial to the growth and survival of gram-negative bacteria, consists of a lipid A moiety conjugated to an O-linked polysaccharide. Lipid A is typically hexa-acylated, yet changes in the LPS structure (importantly the level of acylation) correlate with the ability for LPS-isolates from different bacteria to activate TLR4(31–34). For example, isolates of Helicobacter pylori, a human pathogen, have been shown to express penta-acylated LPS, making them less immunostimulatory (35). Further, some bacteria are capable of actively altering their LPS composition in order to make them less stimulatory during infection(31, 32, 36). For example, Salmonella is able to decrease the stimulatory capabilities of its LPS by expressing a lipid A deacetylase, PagL, that is specifically expressed during infection (via control of the PhoP/PhoQ two-component system) (37). A similar mechanism is used by the causative agent of bubonic plague, Yersinia pestis, which expresses a TLR4-stimulatory hexa-acylated lipid A component of its LPS during growth at 21–27 °C, but upon entry into humans or rodents (and an increase in temperature to 37 °C) expresses a non-stimulatory tetra-acylated form (36). Flagellin, much like the LPS of certain pathogenic bacteria (including isolates of Bartonella, Helicobacter, and Campylobacter), has also been shown to be less capable of stimulating TLR5. In these scenarios it was found that these pathogenic bacteria have acquired mutations within the N-terminal domain of their flagellin, specifically within regions responsible for activating TLR5 (28, 38). In turn, these mutations make bacteria less immunostimulatory and able to evade detection by the immune system during infection.
In his landmark paper, Janeway proposed that the targets of innate receptors must be highly conserved and difficult for pathogens to alter; otherwise, pathogens will quickly evolve to avoid detection (39). Based on the examples discussed above (as well as many others), pathogens are clearly capable of modifying TLR ligands to avoid detection. What prevents all pathogens from rapidly evolving away from the specificity of innate receptors? Certainly there are multiple answers to this intriguing question, but one explanation is that modification of the features targeted by TLRs reduces the overall fitness of the pathogen. For example, in the case of Yersinia, tetra-acylated lipid A may affect the integrity of the outer membrane or reduce the fitness of bacteria in non-human hosts (otherwise bacteria would only express the tetra-acylated form). Perhaps in rodents and humans this loss of fitness is countered by the avoidance of TLR4 recognition. A similar case is true for flagellin mutations. These mutations lead to reduced bacterial motility, unless coupled with secondary compensatory mutations that can rescue the motility defect in certain bacteria. Hence, the cost of reduced motility is overcome by the benefit afforded by survival via TLR5 evasion (28, 38).
Modulation of Intracellular Signaling Pathways
Instead of avoiding detection altogether, some bacteria have evolved to inhibit the signaling pathways downstream of TLR signaling. These mechanisms lead to the same overall outcome; inhibiting production of inflammatory cytokines as well as other anti-microbial activities that are initiated downstream of TLR activation (40–49). To do so, many different pathogens encode secretion systems that puncture host cell membranes and inject bacterially-encoded effector proteins that mimic or degrade members of TLR signaling pathways or directly interfere with normal signaling by covalently modifying signaling intermediates. For example, the Yersinia effector protein YopJ is an acetyltransferase that modifies key residues within MAP kinases and IKKβ, preventing their phosphorylation and inhibiting activation of the MAPK cascade and NF-κB(40–42). In another set of examples, enteropathogenic E. coli (EPEC) type 3 secretion system (T3SS) effector NleC cleaves RelA (p65), a subunit of NF-κB, and another EPEC effector, NleD, cleaves JNK to inhibit activation of AP-1(43–46). A similar bacterial strategy for targeting TLR signaling molecules can be found in an E3 ubiquitin ligase T3SS effector protein, IpaH9.8, encoded by the gastrointestinal pathogen Shigella flexenri. Upon injection into the cytosol, IpaH9.8 is able to bind to the IKK regulator, NEMO, and the ubiquitin adaptor protein ABIN-1, leading to ubiquitination and destruction of NEMO and inhibition of the NF-κB signaling pathway after TLR activation(47). Other forms of TLR signaling inhibition take place more proximally to the receptor. For example, Salmonella, Brucella, E. coli and certain Yersinia species encode different TIR-domain containing proteins that interfere with the homotypic TIR:TIR interactions between TLRs and their signaling adaptors (50, 51).
Inhibiting, Escaping or Subverting Phagocytosis
The nucleic acid sensing TLRs (TLR3, TLR7/8, and TLR9) are localized intracellulary and are recruited to phagosomes (4, 52), and surface localized TLRs, such as TLR4 and TLR2, can be internalized and sense bacterial products within the phagosome (53). Certain pathogens try to avoid phagocytosis by immune cells as a means to avoid this detection and the induction of antimicrobial mechanisms. Numerous virulence mechanisms resulting in inhibition of phagocytosis have been described, including inhibition of complement deposition on the bacterial cell surface, as is the case for Streptococcus pyogenes M protein, and shielding of the bacterium in fibrin clots via coagulase expression by Staphylococcus aureus (54, 55). Other classes of pathogens escape phagosomes and replicate within the cytosol. This strategy avoids detection by TLRs, but renders bacteria susceptible to cytosolic innate immune sensors. Both Shigella and Listeria monocytogenes utilize this virulence strategy (21, 56)—mutants that are unable to escape have a heightened production of pro-inflammatory cytokines, presumably due to increased activation of phagosomal TLRs.
Some bacteria survive within the phagosome despite its antimicrobial nature. This virulence strategy has several implications for the coevolutionary relationship with TLRs—those that survive within the phagosome must have a means of inhibiting the antimicrobial mechanisms induced by engaging these innate receptors. Salmonella, M. tuberculosis, Legionella, and Chlamydia are among those pathogens capable of surviving within phagosomes by preventing fusion with lysosomes (57–59). The process by which this occurs is different for each bacterial species, and in some cases greatly depends on recognizing features of the phagosome in order to induce virulence genes required for inhibiting the phagosomal maturation process or protecting the bacterium from antimicrobial onslaught. For example, in order to neutralize radicals that are produced via TLR-induced recruitment of NADPH oxidase to the phagosomal membrane, Brucella abortus and Staphylococcus aureus express superoxide dismutase and catalase (60, 61). Salmonella inhibits recruitment of NADPH oxidase via injection of effectors into the host cell (62). Several bacteria also alter their cell wall structure in order to make them less susceptible to antimicrobial peptides and other intra-phagosomal antimicrobial mechanisms induced by TLR activation (17, 32, 63–66). These critical virulence mechanisms rely on timely induction upon entry into host cells. The process by which this regulated expression occurs is a common theme for many intracellular bacteria and relies on the use of cues provided, in many cases, by the innate immune system. In the next section we focus on examples of bacteria coopting innate signals to coordinate expression of virulence genes for survival in diverse host environments.
Coopting Innate Immune Signals
Various phagosomal parameters induced upon TLR activation can be used by bacteria to identify their presence within host cells. A number of bacteria utilize phagosomal acidification for this purpose. For example, Salmonella requires TLR-dependent phagosomal acidification to coordinate expression of the SPI-2 T3SS, which is required for intracellular replication (12). Brucella suis also relies on acidification to induce virulence genes (67). Several cytosolic pathogens, including Listeria and Shigella, require phagosomal acidification to activate lysins and escape into the cytosol (56). M. tuberculosis utilizes signals associated with phagosomal maturation to regulate expression of efflux pumps that increase resistance to certain antibiotics (68). Also, the Salmonella two-component sensor PhoP/PhoQ, responsible for mediating anti-microbial-resistant LPS modifications, is induced by cationic antimicrobial peptides present in the phagosome upon activation (64).
These examples suggest that innate immune signals may represent common phagosomal features used by bacteria to coordinate virulence gene expression and raise an interesting point of discussion regarding why bacteria would become dependent on immunity-driven phagosomal signals to induce virulence mechanisms. While we can only speculate about “why” questions relating to host-pathogen interactions, an emerging theme of these relationships is that many of the cues leading to virulence gene induction are linked to innate immune signaling. Certainly all pathogens must regulate expression of their virulence genes, and a defining feature of a given pathogen may be which cues it uses to induce the genes required to transition between distinct niches. For example, to cause systemic infection after oral ingestion Salmonella must traverse the intestinal epithelium, encounter phagocytic immune cells (mainly macrophages and dendritic cells), survive and replicate within these cells, and eventually disseminate to systemic sites. This process relies on the expression of adhesion molecules, two different T3SSs, and a multitude of additional evasion mechanisms. Moreover, inappropriate induction of virulence genes can negatively impact each of these steps, and detection of the virulence factors themselves or the consequences of their action can activate additional innate immune pathways (69, 70). For these reasons, pathogens must quickly identify signals associated with these transitions between niches. Thus, while the consequences of innate immune activation may select the emergence of virulence strategies, these same features of the host response may best define the context in which these strategies are required.
Conclusion
Throughout this review we have focused on the interaction between pathogenic bacteria and TLRs, highlighting examples in which bacteria have attempted to evade TLR signaling by altering their surface structures, interfering with TLR signaling pathways, or escaping, inhibiting, or subverting phagocytosis. We have focused on examples of bacteria using innate immune signaling to regulate induction of virulence strategies. The examples and discussion presented herein speak to the evolution of pathogens with and in response to the innate immune system and highlight the ability of pathogens to evade or exploit host responses to enhance their virulence
Highlights.
Pathogens have evolved multiple mechanisms to evade TLR recognition and signaling
Alterations in bacterial surface structures make pathogens less immunostimulatory
Pathogens encode virulence factors that directly inhibit TLR signaling pathways
Inhibition, escape, or subversion of phagocytosis is a method to avoid TLR detection
Some pathogens coopt innate immune signals to initiate virulence
Acknowledgments
We apologize to any colleagues whose work was not cited due to space constraints. Research in the Barton laboratory related to the topic of this review is supported by the NIH (AI095587, AI06332) and a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease Award (to GMB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–84. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 2.Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327:291–5. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mathur R, Oh H, Zhang D, Park SG, Seo J, Koblansky A, Hayden MS, Ghosh S. A mouse model of salmonella typhi infection. Cell. 2012;151:590–602. doi: 10.1016/j.cell.2012.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annual review of immunology. 2011;29:185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- 5.Fitzgerald KA, Rowe DC, Barnes BJ, Caffrey DR, Visintin A, Latz E, Monks B, Pitha PM, Golenbock DT. LPS-TLR4 Signaling to IRF-3/7 and NF-{kappa}B Involves the Toll Adapters TRAM and TRIF. J Exp Med. 2003;198:1043–55. doi: 10.1084/jem.20031023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–81. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Van Gool SW, Vandenberghe P, de Boer M, Ceuppens JL. CD80, CD86 and CD40 provide accessory signals in a multiple-step T-cell activation model. Immunol Rev. 1996;153:47–83. doi: 10.1111/j.1600-065x.1996.tb00920.x. [DOI] [PubMed] [Google Scholar]
- 8.Underhill DM, Ozinsky A. Phagocytosis of microbes: complexity in action. Annu Rev Immunol. 2002;20:825–52. doi: 10.1146/annurev.immunol.20.103001.114744. [DOI] [PubMed] [Google Scholar]
- 9.Lambeth JD. NOX enzymes and the biology of reactive oxygen. Nat Rev Immunol. 2004;4:181–9. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- 10.West AP, Brodsky IE, Rahner C, Woo DK, Erdjument-Bromage H, Tempst P, Walsh MC, Choi Y, Shadel GS, Ghosh S. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011;472:476–80. doi: 10.1038/nature09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blander JM, Medzhitov R. Regulation of phagosome maturation by signals from toll-like receptors. Science. 2004;304:1014–8. doi: 10.1126/science.1096158. [DOI] [PubMed] [Google Scholar]
- 12.Arpaia N, Godec J, Lau L, Sivick KE, McLaughlin LM, Jones MB, Dracheva T, Peterson SN, Monack DM, Barton GM. TLR signaling is required for Salmonella typhimurium virulence. Cell. 2011;144:675–88. doi: 10.1016/j.cell.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blander JM, Medzhitov R. Toll-dependent selection of microbial antigens for presentation by dendritic cells. Nature. 2006;440:808–12. doi: 10.1038/nature04596. [DOI] [PubMed] [Google Scholar]
- 14.Trombetta ES, Ebersold M, Garrett W, Pypaert M, Mellman I. Activation of lysosomal function during dendritic cell maturation. Science. 2003;299:1400–3. doi: 10.1126/science.1080106. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez-Martinez S, Cancino-Diaz ME, Cancino-Diaz JC. Expression of CRAMP via PGN-TLR-2 and of alpha-defensin-3 via CpG-ODN-TLR-9 in corneal fibroblasts. Br J Ophthalmol. 2006;90:378–82. doi: 10.1136/bjo.2005.082289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Redfern RL, Reins RY, McDermott AM. Toll-like receptor activation modulates antimicrobial peptide expression by ocular surface cells. Exp Eye Res. 2011;92:209–20. doi: 10.1016/j.exer.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tauszig S, Jouanguy E, Hoffmann JA, Imler JL. Toll-related receptors and the control of antimicrobial peptide expression in Drosophila. Proc Natl Acad Sci USA. 2000;97:10520–5. doi: 10.1073/pnas.180130797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ernst JD. Bacterial inhibition of phagocytosis. Cell Microbiol. 2000;2:379–86. doi: 10.1046/j.1462-5822.2000.00075.x. [DOI] [PubMed] [Google Scholar]
- 19.Visser LG, Annema A, van Furth R. Role of Yops in inhibition of phagocytosis and killing of opsonized Yersinia enterocolitica by human granulocytes. Infect Immun. 1995;63:2570–5. doi: 10.1128/iai.63.7.2570-2575.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosqvist R, Bolin I, Wolf-Watz H. Inhibition of phagocytosis in Yersinia pseudotuberculosis: a virulence plasmid-encoded ability involving the Yop2b protein. Infect Immun. 1988;56:2139–43. doi: 10.1128/iai.56.8.2139-2143.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goebel W, Kuhn M. Bacterial replication in the host cell cytosol. Curr Opin Microbiol. 2000;3:49–53. doi: 10.1016/s1369-5274(99)00050-8. [DOI] [PubMed] [Google Scholar]
- 22.O’Brien AD, Rosenstreich DL, Scher I, Campbell GH, MacDermott RP, Formal SB. Genetic control of susceptibility to Salmonella typhimurium in mice: role of the LPS gene. J Immunol. 1980;124:20–4. [PubMed] [Google Scholar]
- 23.Poltorak A, He X, Smirnova I, Liu MY, Huffel CV, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–8. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 24.Qureshi ST, Lariviere L, Leveque G, Clermont S, Moore KJ, Gros P, Malo D. Endotoxin-tolerant mice have mutations in Toll-like receptor 4 (Tlr4) J Exp Med. 1999;189:615–25. doi: 10.1084/jem.189.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawai T, Takeuchi O, Fujita T, Inoue J, Muhlradt PF, Sato S, Hoshino K, Akira S. Lipopolysaccharide stimulates the MyD88-independent pathway and results in activation of IFN-regulatory factor 3 and the expression of a subset of lipopolysaccharide-inducible genes. J Immunol. 2001;167:5887–94. doi: 10.4049/jimmunol.167.10.5887. [DOI] [PubMed] [Google Scholar]
- 26.Ahmad-Nejad P, Hacker H, Rutz M, Bauer S, Vabulas RM, Wagner H. Bacterial CpG-DNA and lipopolysaccharides activate Toll-like receptors at distinct cellular compartments. Eur J Immunol. 2002;32:1958–68. doi: 10.1002/1521-4141(200207)32:7<1958::AID-IMMU1958>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 27.Royle MC, Tötemeyer S, Alldridge LC, Maskell DJ, Bryant CE. Stimulation of Toll-like receptor 4 by lipopolysaccharide during cellular invasion by live Salmonella typhimurium is a critical but not exclusive event leading to macrophage responses. J Immunol. 2003;170:5445–54. doi: 10.4049/jimmunol.170.11.5445. [DOI] [PubMed] [Google Scholar]
- 28.Smith KD, Andersen-Nissen E, Hayashi F, Strobe K, Bergman MA, Barrett SL, Cookson BT, Aderem A. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat Immunol. 2003;4:1247–53. doi: 10.1038/ni1011. [DOI] [PubMed] [Google Scholar]
- 29.Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, Eng JK, Akira S, Underhill DM, Aderem A. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 30.Stecher B, Hapfelmeier S, Müller C, Kremer M, Stallmach T, Hardt W-D. Flagella and chemotaxis are required for efficient induction of Salmonella enterica serovar Typhimurium colitis in streptomycin-pretreated mice. Infect Immun. 2004;72:4138–50. doi: 10.1128/IAI.72.7.4138-4150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo L, Lim KB, Gunn JS, Bainbridge B, Darveau RP, Hackett M, Miller SI. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science. 1997;276:250–3. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- 32.Guo L, Lim KB, Poduje CM, Daniel M, Gunn JS, Hackett M, Miller SI. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell. 1998;95:189–98. doi: 10.1016/s0092-8674(00)81750-x. [DOI] [PubMed] [Google Scholar]
- 33.Okusawa T, Fujita M, Nakamura J, Into T, Yasuda M, Yoshimura A, Hara Y, Hasebe A, Golenbock DT, Morita M, Kuroki Y, Ogawa T, Shibata K. Relationship between structures and biological activities of mycoplasmal diacylated lipopeptides and their recognition by toll-like receptors 2 and 6. Infect Immun. 2004;72:1657–65. doi: 10.1128/IAI.72.3.1657-1665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohlson MB, Fluhr K, Birmingham CL, Brumell JH, Miller SI. SseJ deacylase activity by Salmonella enterica serovar Typhimurium promotes virulence in mice. Infect Immun. 2005;73:6249–59. doi: 10.1128/IAI.73.10.6249-6259.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tran AX, Stead CM, Trent MS. Remodeling of Helicobacter pylori lipopolysaccharide. J Endotoxin Res. 2005;11:161–6. doi: 10.1179/096805105X37349. [DOI] [PubMed] [Google Scholar]
- 36.Robinson RT, Khader SA, Locksley RM, Lien E, Smiley ST, Cooper AM. Yersinia pestis evades TLR4-dependent induction of IL-12(p40)2 by dendritic cells and subsequent cell migration. J Immunol. 2008;181:5560–7. doi: 10.4049/jimmunol.181.8.5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawasaki K, Ernst RK, Miller SI. 3-O-deacylation of lipid A by PagL, a PhoP/PhoQ-regulated deacylase of Salmonella typhimurium, modulates signaling through Toll-like receptor 4. J Biol Chem. 2004;279:20044–8. doi: 10.1074/jbc.M401275200. [DOI] [PubMed] [Google Scholar]
- **38.Andersen-Nissen E, Smith KD, Strobe KL, Barrett SL, Cookson BT, Logan SM, Aderem A. Evasion of Toll-like receptor 5 by flagellated bacteria. Proc Natl Acad Sci U S A. 2005;102:9247–52. doi: 10.1073/pnas.0502040102. The authors show that flagellin from α and ε Proteobacteria have mutations in their flagellin at the percise site recognized by TLR5. The same mutation, in Samonella flagellin, makes Salmonella unable to stimulate TLR5 but also non-motile. α and ε Proteobacteria have secondary compensatory mutations in their flgellin to maintain motility. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janeway CA., Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 40.Mukherjee S, Orth K. In vitro signaling by MAPK and NFkappaB pathways inhibited by Yersinia YopJ. Methods Enzymol. 2008;438:343–53. doi: 10.1016/S0076-6879(07)38024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *41.Sweet CR, Conlon J, Golenbock DT, Goguen J, Silverman N. YopJ targets TRAF proteins to inhibit TLR-mediated NF-kappaB, MAPK and IRF3 signal transduction. Cell Microbiol. 2007;9:2700–15. doi: 10.1111/j.1462-5822.2007.00990.x. The data presented within this paper suggest that YopJ acts at the level of TRAF3 and TRAF6 to remove activating K63 ubiquitin conjugates. YopJ, therefore, inhibits TLR-dependent MAPK and NF-κB signaling. [DOI] [PubMed] [Google Scholar]
- 42.Yoon S, Liu Z, Eyobo Y, Orth K. Yersinia effector YopJ inhibits yeast MAPK signaling pathways by an evolutionarily conserved mechanism. J Biol Chem. 2003;278:2131–5. doi: 10.1074/jbc.M209905200. [DOI] [PubMed] [Google Scholar]
- 43.Muhlen S, Ruchaud-Sparagano MH, Kenny B. Proteasome-independent degradation of canonical NFkappaB complex components by the NleC protein of pathogenic Escherichia coli. J Biol Chem. 2011;286:5100–7. doi: 10.1074/jbc.M110.172254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *44.Pearson JS, Riedmaier P, Marches O, Frankel G, Hartland EL. A type III effector protease NleC from enteropathogenic Escherichia coli targets NF-kappaB for degradation. Mol Microbiol. 2011;80:219–30. doi: 10.1111/j.1365-2958.2011.07568.x. The authors identify NleC from EPEC as being a type III effector protease that specifcially degrades NF-κB components p65, c-Rel and p50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sham HP, Shames SR, Croxen MA, Ma C, Chan JM, Khan MA, Wickham ME, Deng W, Finlay BB, Vallance BA. Attaching and effacing bacterial effector NleC suppresses epithelial inflammatory responses by inhibiting NF-kappaB and p38 mitogen-activated protein kinase activation. Infect Immun. 2011;79:3552–62. doi: 10.1128/IAI.05033-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yen H, Ooka T, Iguchi A, Hayashi T, Sugimoto N, Tobe T. NleC, a type III secretion protease, compromises NF-kappaB activation by targeting p65/RelA. PLoS Pathog. 2010;6:e1001231. doi: 10.1371/journal.ppat.1001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **47.Ashida H, Kim M, Schmidt-Supprian M, Ma A, Ogawa M, Sasakawa C. A bacterial E3 ubiquitin ligase IpaH9.8 targets NEMO/IKKgamma to dampen the host NF-kappaB-mediated inflammatory response. Nat Cell Biol. 2010;12:66–73. 1–9. doi: 10.1038/ncb2006. This report identifies IpaH9.8 from Shigella as being responsible for targeting NEMO/IKKgamma for degradation to dampen NF-κB signaling. IpaH9.8 interacts with ABIN-1, acting as an E3 ligase to induce polyubiquitylation of NEMO and subsequent proteosomal degradation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim DW, Lenzen G, Page AL, Legrain P, Sansonetti PJ, Parsot C. The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc Natl Acad Sci U S A. 2005;102:14046–51. doi: 10.1073/pnas.0504466102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krachler AM, Woolery AR, Orth K. Manipulation of kinase signaling by bacterial pathogens. J Cell Biol. 2011;195:1083–92. doi: 10.1083/jcb.201107132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *50.Radhakrishnan GK, Yu Q, Harms JS, Splitter GA. Brucella TIR Domain-containing Protein Mimics Properties of the Toll-like Receptor Adaptor Protein TIRAP. J Biol Chem. 2009;284:9892–8. doi: 10.1074/jbc.M805458200. The authors show that TcpB, a Brucella-encoded TIR-containing protein, mimics that activity of TIRAP and can inhibit TLR4 signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Newman RM, Salunkhe P, Godzik A, Reed JC. Identification and characterization of a novel bacterial virulence factor that shares homology with mammalian Toll/interleukin-1 receptor family proteins. Infect Immun. 2006;74:594–601. doi: 10.1128/IAI.74.1.594-601.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barton GM, Kagan JC. A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat Rev Immunol. 2009;9:535–42. doi: 10.1038/nri2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat Immunol. 2008;9:361–8. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carlsson F, Sandin C, Lindahl G. Human fibrinogen bound to Streptococcus pyogenes M protein inhibits complement deposition via the classical pathway. Mol Microbiol. 2005;56:28–39. doi: 10.1111/j.1365-2958.2005.04527.x. [DOI] [PubMed] [Google Scholar]
- 55.Cheng AG, McAdow M, Kim HK, Bae T, Missiakas DM, Schneewind O. Contribution of coagulases towards Staphylococcus aureus disease and protective immunity. PLoS Pathog. 2010;6:e1001036. doi: 10.1371/journal.ppat.1001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ray K, Marteyn B, Sansonetti PJ, Tang CM. Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat Rev Microbiol. 2009;7:333–40. doi: 10.1038/nrmicro2112. [DOI] [PubMed] [Google Scholar]
- 57.de Felipe KS, Glover RT, Charpentier X, Anderson OR, Reyes M, Pericone CD, Shuman HA. Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS Pathog. 2008;4:e1000117. doi: 10.1371/journal.ppat.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scidmore MA, Fischer ER, Hackstadt T. Restricted fusion of Chlamydia trachomatis vesicles with endocytic compartments during the initial stages of infection. Infect Immun. 2003;71:973–84. doi: 10.1128/IAI.71.2.973-984.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Armstrong JA, Hart PD. Phagosome-lysosome interactions in cultured macrophages infected with virulent tubercle bacilli. Reversal of the usual nonfusion pattern and observations on bacterial survival. J Exp Med. 1975;142:1–16. doi: 10.1084/jem.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gee JM, Valderas MW, Kovach ME, Grippe VK, Robertson GT, Ng WL, Richardson JM, Winkler ME, Roop RM., 2nd The Brucella abortus Cu, Zn superoxide dismutase is required for optimal resistance to oxidative killing by murine macrophages and wild-type virulence in experimentally infected mice. Infect Immun. 2005;73:2873–80. doi: 10.1128/IAI.73.5.2873-2880.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Das D, Saha SS, Bishayi B. Intracellular survival of Staphylococcus aureus: correlating production of catalase and superoxide dismutase with levels of inflammatory cytokines. Inflamm Res. 2008;57:340–9. doi: 10.1007/s00011-007-7206-z. [DOI] [PubMed] [Google Scholar]
- 62.Vazquez-Torres A, Xu Y, Jones-Carson J, Holden DW, Lucia SM, Dinauer MC, Mastroeni P, Fang FC. Salmonella Pathogenicity Island 2-Dependent Evasion of the Phagocyte NADPH Oxidase. Science. 2000;287:1655–8. doi: 10.1126/science.287.5458.1655. [DOI] [PubMed] [Google Scholar]
- 63.Brodsky IE, Ghori N, Falkow S, Monack D. Mig-14 is an inner membrane-associated protein that promotes Salmonella typhimurium resistance to CRAMP, survival within activated macrophages and persistent infection. Mol Microbiol. 2005;55:954–72. doi: 10.1111/j.1365-2958.2004.04444.x. [DOI] [PubMed] [Google Scholar]
- **64.Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005;122:461–72. doi: 10.1016/j.cell.2005.05.030. This report shows an interesting example of a bacterium detecting a phagosomal innate immune signal to initiate virulence mechansims. The PhoP/PhoQ two-component system is reported to recognize antimicrobial peptides for activation of cell surface modfications in Salmonella. [DOI] [PubMed] [Google Scholar]
- 65.Gunn JS, Miller SI. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J Bacteriol. 1996;178:6857–64. doi: 10.1128/jb.178.23.6857-6864.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prost L, Miller S. The Salmonellae PhoQ sensor: mechanisms of detection of phagosome signals. Cellular Microbiology. 2008;10:576–82. doi: 10.1111/j.1462-5822.2007.01111.x. [DOI] [PubMed] [Google Scholar]
- 67.Boschiroli ML, Ouahrani-Bettache S, Foulongne V, Michaux-Charachon S, Bourg G, Allardet-Servent A, Cazevieille C, Liautard JP, Ramuz M, O’Callaghan D. The Brucella suis virB operon is induced intracellularly in macrophages. Proc Natl Acad Sci U S A. 2002;99:1544–9. doi: 10.1073/pnas.032514299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *68.Adams KN, Takaki K, Connolly LE, Wiedenhoft H, Winglee K, Humbert O, Edelstein PH, Cosma CL, Ramakrishnan L. Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell. 2011;145:39–53. doi: 10.1016/j.cell.2011.02.022. The authors report that Mycobacteria upregulate efflux pumps that make them resistant to antibiotic killing. Efflux pump expression is shown to be induced upon entry and replication in macrophage phagosomes, suggesting a link between the detection of innate immune signals and the induction of virulence mechanisms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Waterman SR, Holden DW. Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell Microbiol. 2003;5:501–11. doi: 10.1046/j.1462-5822.2003.00294.x. [DOI] [PubMed] [Google Scholar]
- 70.Bäumler AJ, Tsolis RM, Heffron F. The lpf fimbrial operon mediates adhesion of Salmonella typhimurium to murine Peyer’s patches. Proc Natl Acad Sci USA. 1996;93:279–83. doi: 10.1073/pnas.93.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]