Abstract
The process of transcription termination is essential to proper expression of bacterial genes and, in many cases, to the regulation of bacterial gene expression. Two types of bacterial transcriptional terminators are known to control gene expression. Intrinsic terminators dissociate transcription complexes without the assistance of auxiliary factors. Rho-dependent terminators are sites of dissociation mediated by an RNA helicase called Rho. Despite decades of study, the molecular mechanisms of both intrinsic and Rho-dependent termination remain uncertain in key details. Most knowledge is based on the study of a small number of model terminators. The extent of sequence diversity among functional terminators and the extent of mechanistic variation as a function of sequence diversity are largely unknown. In this review, we consider the current state of knowledge about bacterial termination mechanisms and the relationship between terminator sequence and steps in the termination mechanism.
Keywords: RNA polymerase, intrinsic termination, Rho-dependent termination, polarity, NusG
Introduction
The mechanism of transcription termination by bacterial RNA polymerase (RNAP) has been a focus of gene expression studies since the earliest days of bacterial molecular genetics. Although the basic framework of both intrinsic and factor-dependent terminations (Rho-independent and Rho-dependent terminations, respectively) has been known for many years, the detailed molecular mechanisms by which terminators destabilize and dissociate the elongating transcription complex [elongation complex (EC)] have remained elusive. This uncertainty about the nature and order of molecular contacts and rearrangements is all the more remarkable given the detailed structural knowledge on ECs now available. Complicating this uncertainty about fundamental mechanisms, especially for intrinsic termination, has been the need to annotate termination sites in bacterial genomes computationally.1–11 Although vitally important to parse the burgeoning collection of bacterial genome sequences, computational methods are limited by our knowledge on what constitutes a validated terminator. Multiple models for the molecular basis of termination have been posited and substantiated to varying degrees,12–16 making this an appropriate time to review bacterial termination mechanisms.
We will focus only on intrinsic and Rho-dependent termination, which are thought to be the principal mechanisms by which bacterial transcription units are defined and by which bacterial gene expression is regulated during transcript elongation. We will consider first the molecular determinants of EC stability, since reduction in EC stability is the principal requirement for terminating transcription. We will then describe the determinants and mechanisms of intrinsic and Rho-dependent termination.
EC Stability and the Definition of Termination
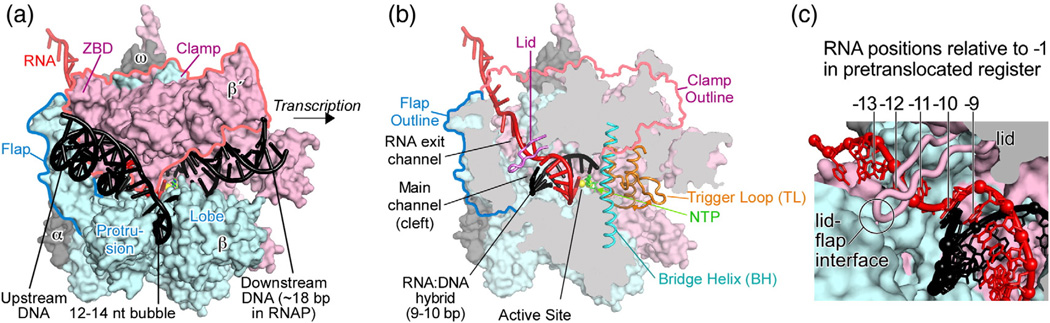
The structure of bacterial ECs and the molecular determinants of their exceptional stability are relatively well understood. Once RNAP has escaped the initiation phase of transcription, it maintains a canonical set of contacts to RNA and DNA that allow it to transcribe > 104 bp without dissociation. These include contacts to ~18 bp of not-yet-transcribed duplex DNA downstream of the active site, an 8- to 10-bp RNA:DNA hybrid within a 12- to 14 -nt transcription bubble, and ~5 nt of single-stranded RNA in an RNA exit channel (Fig. 1).20,21 The exit channel is separated from the main channel, which holds the hybrid, by the lid domain. The lid covers the RNA −11 nt, which is held in a shallow pocket by weak exit-channel contacts (Fig. 1c).17 The DNA duplex reforms as the template strand exits the main channel, but it does not make strong RNAP contacts (Fig. 1a); single-molecule fluorescence resonance energy transfer (FRET) in yeast RNAPII ECs places it near the clamp domain,22 and footprinting experiments confirm that upstream DNA is not protected by RNAP.23,24 Substrate nucleotide triphosphates are thought to enter the active site through a secondary channel that is separated from the main cleft by the bridge helix. Rapid nucleotide addition requires folding of the trigger loop (TL) into antiparallel trigger helices that pack against the bridge helix in a three-helix bundle and contact the bound nucleotide triphosphate substrate. Downstream DNA, the RNA:DNA hybrid, and the exiting RNA are surrounded by semi-mobile RNAP domains called the clamp, protrusion, and lobe (Fig. 1a).
Fig. 1.
Structure of the EC. (a) Model of EC based on a Thermus thermophilus EC crystal structure (Protein Data Bank ID 2o5i).17 (b) Cutaway view of EC model showing locations of open and closed conformations of the TL (Protein Data Bank IDs 1iw7 and 2o5j, respectively)18,19 and the location of the lid separating the RNA exit and main channels. (c) Close-up view of the RNA exit channel with the flap tip removed and the nucleotides numbered for an EC in the pretranslocated register.
In vitro, ECs containing Escherichia coli RNAP are stabile to 1 M NaCl or up to 65 °C.25–27 This remarkable stability is attributable principally to RNAP contacts to the RNA:DNA hybrid,27–29 but significant contributions are also made by contacts to the downstream DNA duplex and exiting RNA.20,26
Termination occurs when these contacts are sufficiently destabilized that the rate of EC inactivation and eventual dissociation becomes significant relative to the rate at which the next nucleotide is added to the growing RNA transcript.30 This branched kinetic mechanism has several important consequences. First, the rate of transcript elongation defines a kinetic window within which termination at any given DNA position is possible. For this reason, transcriptional pausing is thought to be the first step in a termination pathway and a prerequisite to efficient termination. Second, true termination requires dissociation of the EC, with release of RNA and DNA from RNAP. Some paused ECs become arrested on DNA (i.e., transcriptionally inactivated without dissociating), via either backtracking of the RNA and DNA chains through RNAP or other processes; true termination requires dissociation of the EC. Finally, the kinetic branch between elongation and termination may occur prior to actual EC dissociation. Formation of an inactivated EC that eventually dissociates, rather than direct EC dissociation, may compete with elongation,31 but the structure of the inactivated EC remains unresolved.12,32–34 Thus, termination may occur in at least three steps: an initial pause, formation of a termination intermediate, and dissociation of the EC.
The Intrinsic Termination Signal and Steps in the Pathway of Intrinsic Termination
Intrinsic termination, sometimes called Rho-independent termination, refers to dissociation of the EC caused solely by interactions of DNA and RNA with RNAP without the assistance of auxiliary transcription regulators. In E. coli and many other bacteria, intrinsic terminators are found at the end of operons where they form mRNA 3′-ends and also between, within, or upstream from genes where they can regulate transcription via attenuation. Intrinsic terminators exhibit canonical common features and characteristics, but vary in sequence, termination efficiency, and mechanism.
The intrinsic termination signal
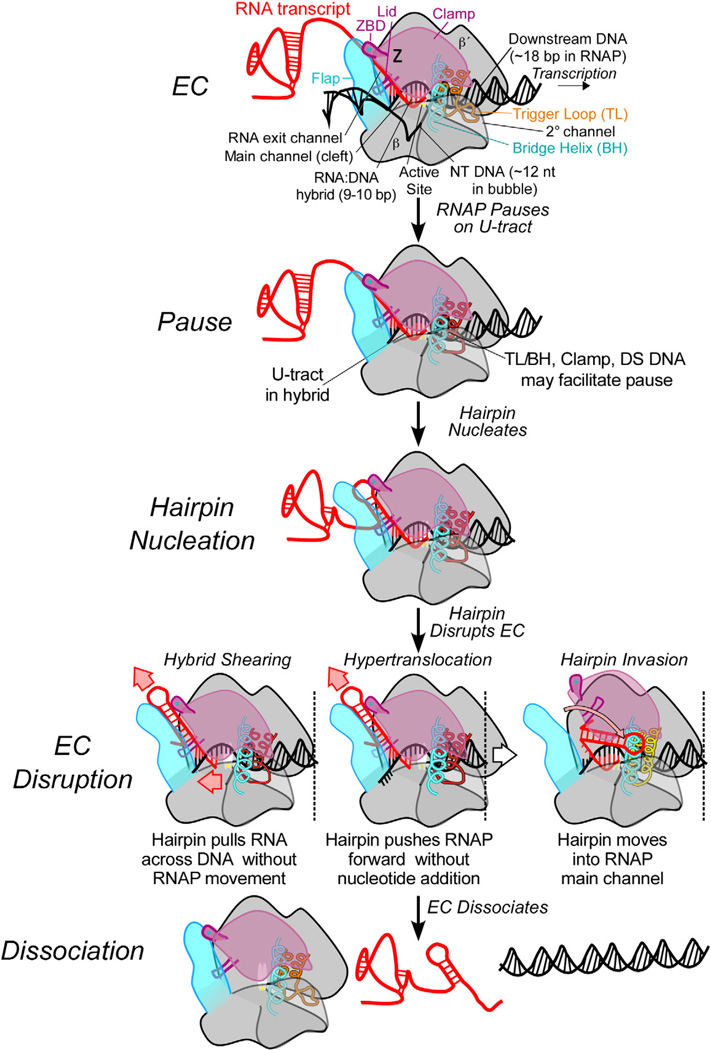
An intrinsic terminator is characterized by a GC-rich dyad repeat followed by a stretch of Ts in the nontemplate DNA strand that, when transcribed into RNA, forms a GC-rich stem–loop structure (or hairpin) followed by a 7- to 8 -nt U-rich tract in the RNA:DNA hybrid.1,35 The downstream DNA sequence also plays a role at some intrinsic terminators.12,36,37 An A-tract is sometimes present upstream from the hairpin, but this typically reflects encoding of a bidirectional intrinsic terminator; addition of an A-tract does not enhance sense-strand termination.38 The terminator hairpin and the U-tract appear to be universal features of intrinsic terminators, but differences in exact sequence and structure cause variations in the efficiency of termination and in mechanism.14 The intrinsic termination signal causes dissociation of ECs in discrete steps whose order and function are now relatively well understood: (1) a transcriptional pause, (2) hairpin nucleation, (3) EC disruption by hairpin completion, and (4) EC dissociation (Fig. 2).
Fig. 2.
Mechanisms of intrinsic termination. The major intermediates in the intrinsic termination pathway are depicted in schematic form. Three alternative routes to EC disruption by hairpin completion are depicted. The version of hairpin invasion depicted corresponds to the specific conformational change model of Epshtein et al.12 The changes in TL conformation in different intermediates are emphasized by color changes but remain speculative.
Pausing at the intrinsic terminator
In the first step of termination, a transcriptional pause halts nucleotide addition, allowing the terminator hairpin to form while the U-tract RNA is still within the RNA:DNA hybrid. Prior to hairpin formation, the pause is induced principally by the U-tract itself.32 Although the intrinsic-terminator-associated pause has not been extensively dissected, studies of other transcriptional pause signals make it highly likely that the sequences of the exiting RNA, the nucleotides in the active site, and the downstream DNA duplex also contribute to the duration of this pause.39–42 An important unanswered question is whether the intrinsic-terminator-associated pause involves backtracking; backtracking would inhibit formation of the terminator hairpin by protecting greater amounts of RNA within the exit channel of the paused RNAP. A U-tract preceded by GC-rich RNA is generally thought to favor backtracking,28,43 and some researchers argue that all pausing involves backtracking.44,45 However, strong evidence for the existence of nonbacktracked pauses has been reported.42,46,47
Terminator hairpin nucleation
Hairpin nucleation occurs by closure of the hairpin loop by one to several base pairs.38,48 This can be as fast as microseconds, but varies with sequence context; understanding of the kinetics of hairpin formation is still in its infancy. Current understanding suggests that multiple routes are possible, ranging from initial loop formation to initial formation of the first few hairpin base pairs.49 Hairpin formation is likely promoted by the flap and zinc binding domains at the mouth of the RNA exit channel.12 Upon nucleation, the stem likely extends quickly to within 1–2 nt of the upstream end of the hybrid before encountering a significant barrier posed by the lid domain (Fig. 1c).
An important and underappreciated contribution to hairpin nucleation is competition from upstream RNA structures (Fig. 2). Although alternative structures that compete with a terminator hairpin are well known from their roles in transcriptional attenuation mechanisms,50 competing structures likely play a much more general role in limiting the efficiency of termination. Indeed, eliminating upstream RNA structure using mild force in single-molecule experiments or by sequestration with oligonucleotides increases termination efficiency even when alternative structures are not obvious.14 Such competition of upstream RNA for hairpin nucleation likely explains reports that promoter-proximal sequence51,52 or complementary alterations to terminator hairpin structure38,53 can alter intrinsic termination efficiency. Studies of termination should be constructed with care to avoid these potentially confounding effects.
Possible role of the partially formed terminator hairpin in pausing
Once the stem extends to the lid, the configuration of the EC is remarkably similar to that of a hairpin-stabilized, paused EC in which a hairpin in the RNA exit channel that leaves 11–12 unpaired nucleotides in the 3′-proximal RNA is known to prolong pausing (Fig. 3).56 The extent to which the partially formed terminator hairpin contributes to pausing at terminators is unknown. However, it is unlikely to be essential because interactions of the hairpin with the RNAP flap domain, which are required for hairpin stabilization of the his pause, contribute only modestly to termination efficiency at several terminators tested.56 Thus, the flap may assist hairpin nucleation or extension, but does not play an essential role in pausing as it does at the hairpinstabilized his pause.56,57
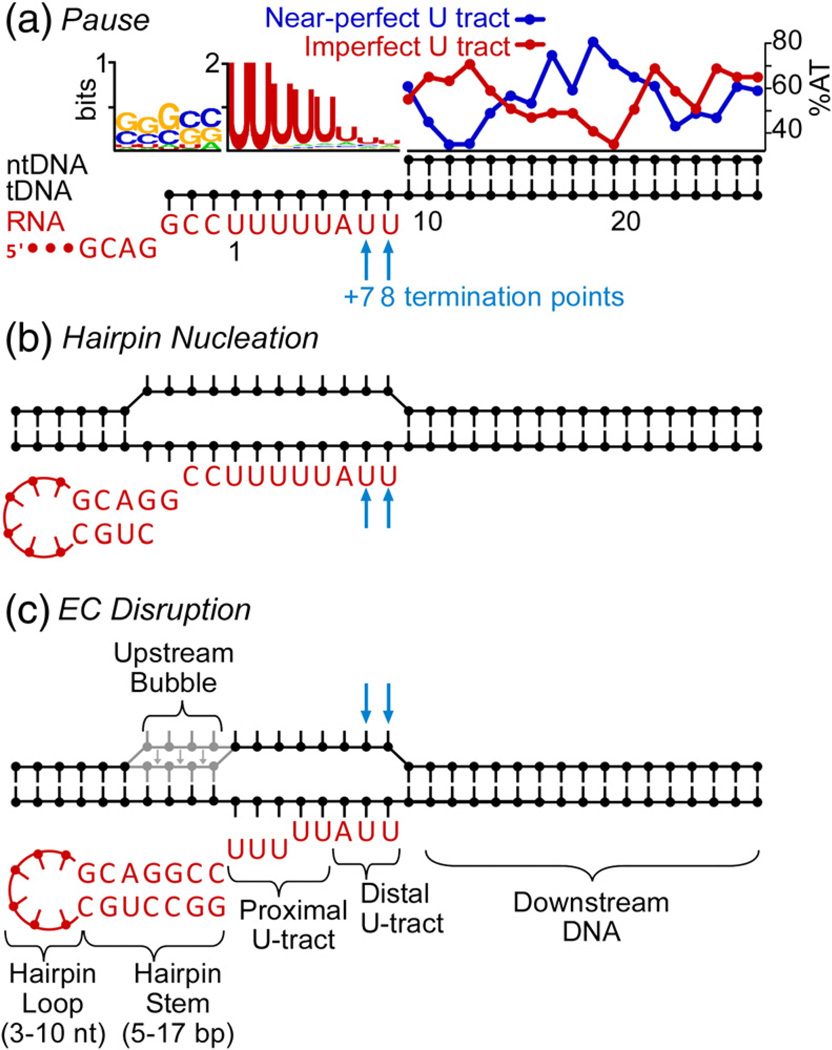
Fig. 3.
Sequence features of intrinsic terminators. Canonical RNA sequences (red) are depicted paired to a DNA scaffold with paired and unpaired nucleotides depicted as filled circles and lines. The sequence of λtR2 is used to depict canonical features in the RNA, except the hairpin loop, which is shown as filled circles and lines. (a) Configuration at the pause step, with portions of the DNA scaffold omitted. The extent of conservation in the RNA 3′ stem and in the U-tract is shown above the DNA as information content using WebLogo (www.weblogo.berkeley.edu).54,55 The %AT for downstream DNA of 50 near-perfect U-tract terminators (blue) and 50 imperfect U-tract terminators (red) selected as described in the text is shown. The variations in %AT at positions 11–13 and 19–21 (numbered relative to the U-tract) are significantly different from a randomized sequence (p≤0.05; t-test). (b) RNA/DNA configuration after hairpin nucleation. (c) RNA/DNA configuration during EC disruption, with canonical terminator features labeled.
EC disruption
In the next step of termination, the terminator hairpin extends to ≤8 nt from the terminated RNA 3′-end. This hairpin extension melts ~3 bp of the RNA:DNA hybrid by extracting the RNA strand from the hybrid; by rearrangements of RNAP involving the lid, the exit channel, and the main cleft; or both.27,32 Hybrid melting disrupts and destabilizes the EC to the point that dissociation becomes favorable; this appears to be the energetically limiting step in termination.14 In principle, dissociation could occur by initial release of RNA followed by bubble collapse and DNA release, initial release of DNA followed by RNA release, or near-simultaneous release of both RNA and DNA. The order of these events, whether they are obligatory or stochastic, and whether they are universal or differ among terminators is unknown.
Commitment to the termination pathway
Two key questions about intrinsic termination are whether the EC becomes irreversibly committed to termination prior to dissociation and which step in the process of termination is rate limiting. For the his terminator and the λtR2 terminator, it has been argued that commitment occurs prior to EC dissociation.31,32 Single-molecule observations revealed that a long-lived state unable to resume elongation forms at the his terminator prior to DNA release;31 this complex retains RNA until DNA release.58 A salt-sensitive “trapped” complex has been isolated and studied at the λtR2 terminator,12,32 although it has been suggested to be a binary RNA– RNAP complex similar to a binary complex that forms after RNAP dissociates from the trpL terminator.33,34
In principle, any step in the termination pathway prior to a committed intermediate can contribute to the efficiency of termination. For instance, the rate of paused EC formation relative to the rate of elongation past sites of potential termination may set an upper limit on termination efficiency. Escape from the paused EC can also occur and reduce termination efficiency. Thus, rather than a simple partition between elongation and dissociation as envisioned by early models of termination,30 the aggregate consequence of several steps in a termination pathway may determine overall efficiency. This may explain the complex ways that solute concentration, supercoiling, and temperature influence termination efficiency,25,59 but it also makes assigning sequence effects on termination complicated.
Sequence Conservation of Intrinsic Terminators
The conservation of intrinsic terminator sequence determinants has important consequences both for the mechanism of termination (specifically whether termination occurs by a single pathway or by alternative pathways) and for the bioinformatic identification of terminators in genome sequences. To illustrate the sequence conservation of intrinsic terminators, we examined E. coli terminators catalogued in RegulonDB†.60 Because even terminators compiled from the literature may not be experimentally verified (see below), we identified a subset of 100 terminators that matched the predictions of the best available prediction algorithm, TransTermHP.9 These “gold-standard” terminators highlight the key sequence features of intrinsic terminators (Fig. 3).
The terminator hairpin stem varies from 5 bp to 17 bp, with an average of ~8 bp, and exhibits strong bias for GC at the five positions nearest the U-tract with a modest preference for G at −3 (−1 is the 3′-most hairpin nucleotide). This is consistent with the importance of the bottom of the stem in supplying the energy that destabilizes the EC in the final step of termination.14,27,32 The preference for G at −3 (−10 relative to termination at U7) may reflect the role of G at −10 in favoring pausing, presumably by formation of a 10-bp hybrid.61,62 The terminator loops vary from 3 nt to 10 nt, with an average of ~4 (70% of the terminators had loops of 3 nt or 4 nt). The prevalence of these so-called tetraloops in intrinsic terminator hairpins likely reflects their ability to stabilize RNA structures.63,64
The U-tract exhibited near-universal presence of at least two Us adjacent to the hairpin, a strong bias for U in the proximal 5-nt segment of the U-tract, and significantly greater sequence diversity in the distal 3-nt segment of the U-tract (Fig. 3a). About half the terminators had a perfect or near-perfect U-tract (at most one A in positions 4–8 of the U-tract), whereas nearly all the remainder contained at least two non- U residues with at least one C or G in the distal U-tract (imperfect U-tract; Fig. 3a). The downstream DNA sequence exhibited marked differences in % AT for the near-perfect versus imperfect U-tract classes of terminators. Imperfect U-tract terminators exhibited high %AT at positions +10–12, whereas near-perfect U-tract terminators exhibited low %AT at the same positions. The two classes of terminators exhibited the opposite sequence bias at positions +18–19. These sequence patterns are consistent with the evidence that downstream DNA can compensate for an imperfect U-tract at the T7 terminator36,65 and suggest that the +10–12 region may affect the ease of duplex melting during hypertranslocation. The +18–19 region corresponds to a contact made by the clamp domain (+10–11 relative to the active site).17
The Mechanism of Intrinsic Termination
Although the basic steps in intrinsic termination are relatively clear (Figs. 2 and 3), the structural changes that destabilize and dissociate the EC and the extent to which these vary at terminators with different sequences are at best partially understood. Although mechanistic models have often been divided into so-called rigid-body models that emphasize changes to the thermodynamic stability of the RNA/DNA scaffold and conformational change models that emphasize conformational changes in RNAP, this is a false dichotomy because changes to the structures of both the scaffold and RNAP must occur during termination. Thus, we will instead consider what is known about these two aspects of intrinsic termination mechanisms: structural changes in the nucleic acid scaffold and conformational changes in RNAP.
Changes in the nucleic acid scaffold during intrinsic termination
Careful examination of the proximal U-tract using cross-linking and chemical probing reveals that the upstream 3 bp in the RNA:DNA hybrid, corresponding to the first three bases of the U-tract, melts upon terminator hairpin extension.27,32 Three models have been proposed to explain this melting: hybrid shearing, hypertranslocation, and hairpin invasion (Fig. 2).12,14,16,27,56,66,67 In hybrid shearing, extension of the hairpin pulls the RNA out the exit channel by transiently breaking and reforming base pairs in the hybrid as the RNA shifts out of register with the DNA strand. In hypertranslocation, extension of the hairpin pulls the RNA out the exit channel but retains the register of the RNA:DNA hybrid by translocation of both RNA and DNA without accompanying nucleotide addition. In hairpin invasion, the hairpin extends into the main cleft of RNAP rather than pulling RNA out the exit channel and causes hybrid melting due to steric constraints in the main cleft.
Several lines of evidence favor the hybrid shearing and hypertranslocation models and suggest that the selection between them is governed by the ease of shearing the U-tract versus the ease of translocating the DNA bubble. A switch between these two events was observed in single-molecule experiments that detect the ability of force that assists or opposes translocation to alter termination efficiency; an imperfect U-tract terminator (t500) exhibited force dependence, whereas as perfect and near-perfect U-tract terminators did not (his and λtR2).14 The imperfect U-tract t500 terminator is also inhibited by blocking translocation with a roadblock or blocking DNA unwinding with a cross-link.16 These results suggest that hybrid shearing occurs when the hybrid is weak but that hypertranslocation becomes favorable when the hybrid is stronger. The high % AT at positions +10–12 of imperfect U-tract terminators (Fig. 3a), which would favor hypertranslocation, suggests a general use of hypertranslocation at imperfect U-tract terminators. This view is consistent with findings that mismatches in the upstream portion of the bubble inhibit termination,68 although reannealing could also occur in a hairpin invasion model.
The key observation favoring the hairpin invasion model is the retention of a 3′-RNA-nt cross-link to the RNAP active site in an inactivated termination intermediate formed at λtR2.12 Although this model would also be consistent with the lack of effect on termination efficiency at λtR2 in DNA-pulling experiments (because no translocation is involved), it is difficult to explain why stabilization of the distal hybrid in an imperfect U-tract terminator leads to a requirement for hypertranslocation if hairpin invasion alone can dissociate an EC. A comparable examination of RNA 3′-end location at terminators found to hypertranslocate and during steps between formation of the termination intermediate and EC dissociation would be instructive.
Conformational changes in RNAP during intrinsic termination
Given that the EC is held together by interactions of flexible domains with RNA and DNA (e.g., the clamp, flap lobe, and protrusion; Figs 1a and 2), at least some conformational changes seem certain to occur during termination regardless of the mechanism that alters the scaffold structure. However, the nature and role of these RNAP conformational changes is the least understood aspect of the mechanism. The clamp domain has been observed in both open and closed conformations,69–71 is postulated to close during promoter binding,72,73 and should favor termination if opened. Even the initial step of transcriptional pausing is proposed to involve some clamp movement; the TL is thought to be trapped in an inactive configuration that is linked to nascent RNA hairpins, the hybrid, and downstream DNA through the bridge helix and movements of the clamp.46,56,74 A recent report on an EC with a more open clamp conformation presumably in response to an exit channel hairpin strongly supports this view.71 The hairpin in this structure matches the configuration of the partially formed terminator hairpin and the his pause hairpin (extends to −12; Figs 1c, 2, and 3b).46,56 The inability to resolve the hairpin and the presence of the transcription inhibitor Ghf1 in the secondary channel of this relatively low-resolution structure (~4.3 Å) limit interpretation of this result, but minimally, it makes it likely that clamp opening participates both in hairpin stabilization of pauses and in intrinsic termination.
Clamp involvement in termination is also supported by the ability of sequence changes in the downstream DNA at the point of clamp contact and a deletion in this clamp region to alter termination efficiency,12 as well as the bias in sequence composition at this point (+19–21 from the U-tract or +9–11 from the active site; Fig. 3a). Clamp changes in the RNA exit channel (e.g., deletion of the zinc binding domain),12 deletion of the flap tip,12,56 and changes in contacts to the RNA:DNA hybrid opposite the clamp67 also have been reported to affect termination efficiency. At least some of these effects occur after pausing, as they can be detected using halted ECs.12 Understanding the extent, timing, and mechanistic importance of clamp opening is a fertile area for future study, for instance, by using FRET and blocking movements using cross-linking.
Recently, a detailed set of conformational changes in combination with hairpin invasion has been proposed as an “allosteric” model of intrinsic termination (although allostery is usually defined as a change in the activity of a catalytic site caused by the binding of a diffusible effector molecule to a distinct allosteric site).12 In this version of the hairpin invasion mechanism, the terminator hairpin is proposed to dissociate the EC, while the RNA 3′-end remains in the active site by sweeping across the main cleft, disrupting EC-stabilizing contacts, and causing TL folding by direct contact to the TL.12 TL folding is proposed to dissociate the EC. Although the energy source for this extensive hairpin motion is unclear, the model makes several testable predictions. First, TL folding or partial folding can be blocked to determine if it is required for termination as it is for nucleotide addition.18 Second, the model predicts a many-angstrom movement of the hairpin loop toward the downstream DNA that should be readily detectable by FRET between probes in the hairpin loop and the downstream DNA. Finally, since the model was generated using the λtR2 terminator sequence that does not require hypertranslocation, it would be instructive to apply the same tests to terminators with imperfect U-tracts for which hypertranslocation is now indicated.
Intrinsic Termination in Other Bacteria
Remarkably, most knowledge about intrinsic termination mechanisms comes from study of a handful of terminators (e.g., λtR2), with only minimal effort to sample known sequence and structural diversity.59,65 Thus, mechanistic study of a wider variety of terminator sequences is highly desirable. This is especially important because canonical intrinsic terminators (Fig. 3) are absent downstream from genes in many bacteria, including Mycobacteria, Helicobacter, Treponema, Synechocystis, Mycoplasma, and Borrelia.2,3,8 In many cases, other types of RNA structures can be identified after genes,8 which led to the suggestion that novel mechanisms of intrinsic termination operate in these bacteria. For two reasons, we wish to caution against assuming that these structures operate as intrinsic terminators, especially in bioinformatic analyses of bacterial genomes. First, even the sets of intrinsic terminators often relied on as being experimentally validated (e.g., in RegulonDB or in d'Aubenton Carafa et al.1) include sequences that, although near sites of in vivoRNA3′-end formation, were identified by visual inspection and lack canonical features (e.g., the E. coli trpR and hupB terminators).60,75,76 True experimental validation of an intrinsic terminator requires demonstrating the following: (1) it causes dissociation of EC during in vitro transcription as detected by release of RNA and DNA from RNAP; (2) it generates terminated RNA 3′-ends before readthrough transcripts appear during synchronized in vitro transcription; (3) it generates the terminated RNA 3′-ends in vivo; and (4) it significantly reduces synthesis of RNA downstream from the site in vivo. Reports of efficient intrinsic termination at noncanonical sequences do not meet all these criteria,77–79 which are necessary because terminator-like structures in RNA may be present after genes for many other reasons. For example, rather than causing intrinsic termination, such structures could stabilize mRNA against 3′ exonucleases, guide processing of RNAs by binding nucleases, mediate RNA–RNA interactions important for regulation, pause or arrest transcription without causing intrinsic termination, facilitate termination by binding unknown termination proteins, or guide DNA uptake in horizontal gene transfer.9 To emphasize this point, we offer two instructive examples. The E. coli trp operon contains an “obvious” intrinsic terminator after trpA and corresponding to the 3′-end of the trp mRNA. Nonetheless, termination is inefficient at this site and mostly occurs further downstream by Rho-dependent termination followed by processing back to the apparent terminator.80 Similarly, the E. coli asnU gene is followed by a GC-rich hairpin and a U-tract that deviates from consensus only by the presence of a single A between the hairpin and the U-tract. Nonetheless, essentially all termination of asnU in vivo is caused by Rho.81
Clearly, much of great interest remains to be learned about transcriptional termination in diverse bacteria. Given the absence of obvious intrinsic terminators in some bacteria, it seems likely that new termination mechanisms and new termination proteins will be uncovered. However, extrapolation from existing knowledge without accompanying biochemical study that distinguishes intrinsic termination from factor-dependent termination and the many other explanations for the appearance of RNA structures and RNA 3′-ends will be more confusing than illuminating. As molecular biology transitions to an era that incorporates genome-scale data sets and computational prediction, experimental verification becomes especially important.
Although the mechanisms of termination by archaeal and eukaryotic RNAPs are also of significant interest, neither intrinsic nor factor-dependent termination mechanisms for archaeal or eukaryotic RNAPs that match the bacterial paradigms have so far been described. Commonalities in the structures of bacterial, archaeal, and eukaryotic transcription complexes suggest that termination mechanisms must overcome similar energetic and structural barriers. Thus, the apparent divergence of mechanisms by which termination occurs in bacteria, archaea, and eukaryotes suggests that their termination pathways have evolved independently.
Introduction to Rho-Dependent Termination
The key distinction between intrinsic termination and Rho-dependent termination is that the latter requires the participation of Rho, a homohexameric ring protein that binds to the nascent RNA transcript and then threads RNA 5′→3′ through the center of the ring as an ATP-powered translocase. Once the nascent RNA passes through the ring, Rho dissociates RNAP from RNA and template DNA. Recent work has increased our overall understanding of the signals and sequence elements required for Rho-dependent termination, clarified the nature of Rho as a translocase/helicase, provided refined models for the termination process, and suggested new possibilities for regulation by factor-dependent termination in vivo.
The Rho Termination Signal and Steps in the Pathway of Rho Termination
Rho binding to RNA
Rho termination is governed by sequences in the nascent RNA and template DNA that act at three steps: (1) Rho binding to RNA and activation of translocase activity, (2) translocation of RNA through Rho, and (3) pausing of the EC at the site of termination (Fig. 4). In the first step, Rho binds to C-rich unstructured RNA, which triggers the RNA-dependent ATPase activity that powers translocation (reviewed elsewhere83). Rho has the highest affinity for synthetic poly(C) RNA, which maximally stimulates ATPase activity.84 Natural Rho binding sites, which are called rut (Rho utilization) sites and lie in RNA upstream from points of termination, are ~80 nt in length with high C content and relatively little secondary structure.85–87 Unpaired C residues within the rut site are important for termination, since blocking rut with complementary oligonucleotides or replacing rut with highly structured RNA greatly reduces termination.88 A consensus rut sequence is not required for termination; DNA encoding CArich RNA or completely synthetic sequences consisting mostly of C and T residues is sufficient to elicit termination at λtR1 89 or an otherwise non-terminator site,90 respectively, by acting as artificial rut sites. The observed depletion of G residues from rut sites81,91 also plays an indirect role in Rho binding by making the RNA less likely to form strong secondary structures with G:C base pairing. Sequence comparisons of known rut sites reveal few common features (other than C-richness),92 making bioinformatic prediction of Rho-dependent terminators problematic. Combining high-resolution maps of Rho termination obtained experimentally81 with genomic sequence data may help to uncover similarities between rut sites that have previously gone unnoticed.
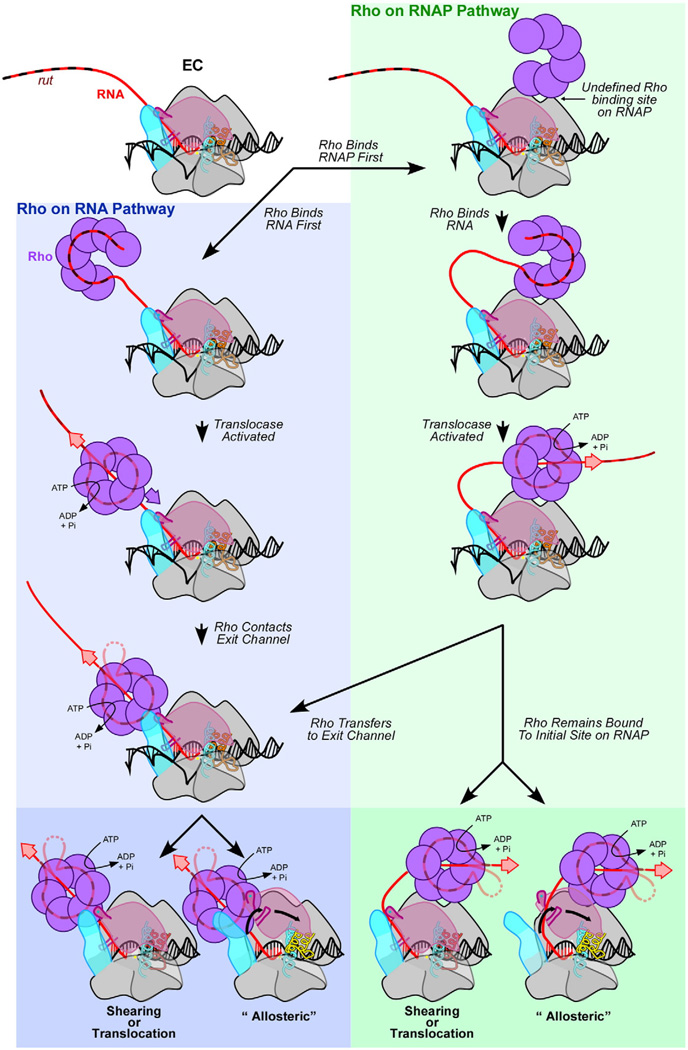
Fig. 4.
Steps prior to Rho termination. The blue panel (left) depicts a model in which Rho binds only to RNA,82 whereas the green panel (right) depicts an alternate model in which Rho binds directly to RNAP.13 rut RNA may remain bound to the Rho-NTD during translocation (tethered tracking model), forming a loop between the primary and secondary RNA-binding sites (shown as partially transparent RNA), or rut RNA may be released during translocation (simple translocation model, shown as opaque RNA). Both RNA extraction and conformational change models are possible in either pathway, as depicted in the darker panels (bottom).
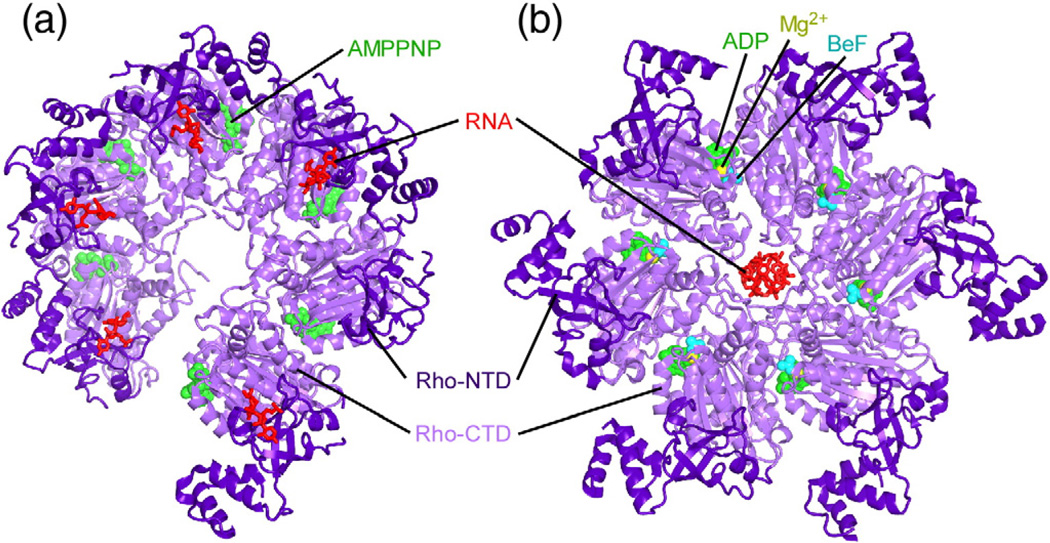
Structural studies have provided atomic-level views of RNA binding and subsequent isomerization by Rho. Both crystallographic and electron microscopy results suggest that Rho initially binds to RNA in an open, “lock-washer” conformation (Fig. 5a)93 and then isomerizes into a closed ring as RNA transfers to the central cavity (Fig. 5b).94 Electron microscopy images depict Rho hexamers in either a closed state or a “notched” state when a short (23 nt) RNA cofactor is present.95 When larger RNAs that exceed the capacity of the primary site (100 nt) are added, the notched population of Rho hexamers converts to the closed state. In crystals of open Rho, RNA is bound to the N-terminal domain (NTD), consistent with previous structural work indicating that the NTD functions as the primary or high-affinity RNA binding site (Fig. 5a).96,97 RNA associates with a cleft in the NTD that is only large enough to fit pyrimidines and exhibits a preferred interaction with C residues.97 Two independent structures revealed closed forms of Rho with RNA bound to the C-terminal ATPase domain, which contains the secondary or low-affinity binding site (not shown; Fig. 5b).94,98 However, the RNA makes different contacts in the two structures, confusing identification of the secondary binding site and understanding of its strict specificity for RNA. The RNA-dependent transition between open and closed states is hypothesized to represent a switch between RNA loading and translocation-competent forms of Rho;98 however, large-scale changes in Rho conformation may also occur during translocation.99
Fig. 5.
Crystal structures of Rho. (a) The open form of Rho (1pvo) bound to AMPPNP (phosphoaminophosphonic acid-adenylate ester, an ATP analogue) and RNA.93 (b) The closed asymmetric form of Rho (3ice) bound to ADP-BeF (adenosine-5′-diphosphate, an ATP analogue; BeF, a phosphate mimic) and RNA.94
RNA translocation through Rho
In the second step of Rho termination, RNA is translocated 5′→3′ through the central cavity of Rho, but it can be impeded by structural blocks, such as RNA hairpins, other RNA-binding proteins, or ribosomes. Thus, the second component of a Rho signal is an unimpeded RNA segment that facilitates translocation. Recent evidence suggests that Rho may be capable of bypassing certain RNA structures by binding to the single-stranded regions around the structure, effectively “stepping around” RNA hairpins.100 Alternatively, the strong translocase activity of Rho, which can displace streptavidin from a biotinylated RNA,101 may directly melt RNA structures and displace some RNA-binding proteins.
Biochemical studies have led to several detailed models for RNA translocation through Rho (reviewed elsewhere99,102), which can be divided into two classes that are relevant to termination. In the tethered tracking model, rut RNA remains bound to the primary site of Rho during translocation,103 whereas in simple translocation models, RNA contacts only the central cavity of Rho during translocation. A structural model of a closed Rho hexamer extrapolated from a dimer crystal structure minus the 65 C-terminal residues (which were unresolved) showed RNA bound to both the primary site and the C-terminal domain (CTD); this model is consistent with the tethered tracking model.98 However, biochemical tests proved inconsistent with the location of RNA in the CTD,104 and a more recent closed Rho structure reveals an asymmetric hexamer with RNA in the central cavity but not bound to the primary site (Fig. 4b).94 Thus, current structural data, while not excluding the tethered tracking model, are more consistent with a simple translocation model. However, active ATP hydrolysis by Rho increases RNA protection from nuclease digestion,105 suggesting that Rho could retain RNA contacts during translocation. Further study is needed to distinguish whether RNA contacts with the primary site are lost during translocation, and if so, how these strong contacts are lost. One complication of some existing studies is reliance on Rho bearing N- or C-terminal His tags, which are known to compromise Rho function in vivo106 and in vitro.107
EC pausing at the site of termination
In the final step of Rho termination, Rho dissociates an EC halted at a pause site. Thus, the third component of Rho termination is a pause sequence that renders the EC susceptible to Rho termination. Several studies of natural and arbitrary sequences establish a correlation between RNAP pausing and the positions of Rho termination.108–112 However, not all RNAP pause sites on the DNA template function as efficient Rho termination sites,108 and some, such as the his pause, inhibit Rho termination.113 The presence of an RNA hairpin in the exit channel at the his pause site may interfere with RNA translocation and termination; also, arrested and highly backtracked ECs are poor substrates for Rho termination.113 Although general determinants of pausing likely apply to pauses at Rho termination sites, additional factors such as distance from the rut site, stability of the EC,30 sequence of the 3′-end of the RNA in the active site of RNAP,13 susceptibility to backtracking, and structure of the nascent RNA113 contribute to the efficiency of termination. This complexity further complicates predicting sites of Rho termination based on sequence.
Kinetic coupling
Rho translocates on RNA being actively synthesized by RNAP. Termination is thought to begin when little or no RNA remains between Rho and RNAP. Thus, competing rates of RNA chain growth and Rho translocation dictate the likelihood of termination. This is referred to as kinetic coupling and is supported by experiments showing that pause-susceptible RNAPs or reduced RNAP elongation rates increase the efficiency of Rho termination.114
Rho Association with ECs
Recent studies suggest that Rho associates with ECs in vivo even when not engaged in termination115 and that Rho may bind directly to RNAP.13 Rho–EC association in vivo is indicated by very similar genome-wide distributions of RNAP and Rho in ChIP-chip experiments.115 Rho–RNAP association in vitro is based on retention of Rho by bead-immobilized ECs with transcripts too short to emerge from the RNA exit channel.13 In addition, wild-type Rho fails to terminate ECs that have been pre-incubated with a termination-defective mutant Rho, implying that mutant Rho is bound to the EC stably enough to prevent binding of active Rho.13
Possible RNAP–Rho association in vivo raises important questions. First, is Rho bound to all ECs? The ChIP-chip assay only reports enrichment, not stoichiometry; thus, it is currently unclear if every EC binds Rho.115 Rho is reported to be ~0.1% of the total protein in E. coli,116 corresponding to ~1400 Rho hexamers per cell. Interestingly, ~1250 of the 13,000 RNAP molecules are in ECs;117 thus, enough Rho appears present to bind every EC. However, if Rho can bind RNAP not in an EC, as current results suggest,13 then not every EC would contain Rho. Better quantitation of the amount of Rho present in cells and more direct tests for EC– Rho association in vivo are needed.
Second, what is the function of Rho–EC association? The local concentration of Rho could be higher, which would allow Rho to engage RNA rapidly when a rut site becomes exposed.13 Binding of RNAP-associated Rho to rut would cause an RNA loop to form between the RNA exit channel and the central cavity of Rho.13 This RNA loop would then be translocated through Rho, possibly altering the trajectory of RNA coming out of the exit channel. When the RNA loop becomes taut, Rho could either remain bound the initial site on RNAP or transfer to the RNA exit channel. Although this could impact the mechanism of termination, it does not change the concept of kinetic coupling: termination requires that the rate at which RNA is pulled through Rho still must exceed the rate at which RNAP extends the nascent RNA chain whether the RNA loops between an RNAP–Rho complex or tethers Rho to RNAP.
Validation of the Rho–RNAP association model requires definition of the binding determinants on Rho and RNAP that, when altered, disrupt Rho– RNAP association and alter Rho termination in vivo and in vitro. Given the extensive history of Rho and RNAP genetics, it is notable that such determinants have not been reported to date.
The Mechanism of Rho Termination
Despite major advances in understanding the structural and biochemical properties of the Rho hexamer, the detailed mechanisms by which Rho dissociates the EC have remained obscure. As for intrinsic termination, changes both in the nucleic acid scaffold and inRNAP conformation are likely to occur during Rho termination. The major classes of the mechanisms are only superficially altered by whether or not Rho is pre-bound to ECs (Fig. 4).
Changes in the nucleic acid scaffold during Rho termination
Rho is a powerful molecular motor capable of applying force through RNA translocation,101 as well as an RNA:DNA helicase;118 these activities are thought to supplant the function of the terminator hairpin in each of the possible classes of termination mechanisms (Fig. 2). In hybrid shearing, RNA is translocated through Rho until the RNA becomes taut, resulting in a pulling force that indirectly disrupts the RNA:DNA hybrid.119 In hypertranslocation, Rho exerts a pushing force that causes RNAP to translocate forward on the DNA template without extension of the RNA chain.15 In the invasion model, the RNA 3′-end remains in the RNAP active site, and the helicase activity of Rho is used to directly unwind the RNA:DNA hybrid.13
For hybrid shearing, Rho must generate sufficient force to shear the RNA:DNA hybrid,119 which can be more stable than the U-tract hybrids found at intrinsic terminators. Although Rho can generate >200 pN of force (based on its ability to displace a streptavidin bead),101 the force required to shear non-U-tract hybrids is unknown because it is greater than 30 pN at which other linkages break in force-clamp experiments.120 Direct, single-molecule measurements of these forces would be invaluable to test the physical plausibility of hybrid shearing by Rho.
The hypertranslocation model is supported by the findings that Rho allows RNAP to elongate by two additional nucleotides against a downstream roadblock and that Rho termination is inhibited when upstream DNA strands in the transcription bubble cannot reanneal.15 However, it remains unknown if Rho can create a hypertranslocated EC in which the RNA 3′-end no longer resides in the active site. Although inhibition of Rho termination by mutations that prevent DNA reannealing in the upstream bubble is consistent with hypertranslocation,15 less is known about the effects of downstream bubble unwinding on termination. The hypertranslocation model predicts that inhibiting downstream unwinding would inhibit both forward translocation by RNAP and termination, as has been observed for some classes of intrinsic terminators. An unresolved question is whether hypertranslocation during Rho termination is dependent on hybrid sequence in a way that parallels the sequence dependence observed in intrinsic termination, making tests of hypertranslocation on templates encoding different Rho-dependent terminators highly desirable.
The invasion model is supported by cross-linking data suggesting that the RNA 3′-end remains in the RNAP active site during termination.13 These crosslinking experiments were performed on inactivated intermediates, which may or may not be on the termination pathway. Interestingly, the position of RNA 3′-end cross-linking shifts slightly in the presence of Rho, suggesting that either the conformation of protein components in the active-site changes or the RNA 3′-end moves independent of a conformational change. The invasion model also posits that Rho uses its helicase activity to directly unwind the upstream end of the RNA:DNAhybrid.13 However, since the hybrid is buried within RNAP, significant conformational changes would need to take place for Rho to have direct access to the hybrid.
Conformational changes in RNAP during Rho termination
Most models for Rho termination are silent about potential conformational changes in RNAP that may coincide with alterations to the nucleic acid scaffold of the EC. However, this does not mean that hybrid shearing or hypertranslocation would occur without RNAP conformational changes. These would be favored by the same changes discussed for intrinsic termination (e.g., clamp opening). Further, bubble collapse, as envisioned when forward translocation is blocked, is physically implausible without conformational changes in RNAP.15
A specific conformational change model has been proposed in which Rho “pushes” against the RNAP lid domain, causing clamp movement and subsequent unfolding of the TL.13 TL unfolding is proposed to cause further clamp opening, loss of nucleic acid contacts, and irreversible inactivation of the EC. This proposal is based on the findings that tagetitoxin, which binds directly to the TL,17 inhibits Rho termination113 and that substitutions in the TL enhance Rho termination.13 Interestingly, in the proposed conformational change model of Rho termination, the unfolded TL destabilizes the EC,13 whereas in the conformational change model of intrinsic termination, TL folding is proposed to destabilize the EC.12 Ultimately, further experiments involving alterations that stabilize, destabilize, or eliminate folding of the TL will be needed to clarify the role of TL conformational changes in Rho termination.
Physiological Functions and Targets of Rho
Rho termination plays a variety of roles in cellular physiology, including formation of transcript 3′- ends,121 prevention of persistent RNA:DNA hybrids (R-loops) that form during transcription,122 and the enforcement of transcription and translation coupling through polarity.123 Recent studies have added new roles for Rho termination: “silencing” transcription of foreign (horizontally transferred) DNA,124 suppression of antisense transcription,81 and elimination of stalled ECs that interfere with DNA replication.125
Targets of Rho
Genome-wide analysis of Rho termination using RNAP ChIP-chip in the presence of the Rho inhibitor bicyclomycin (BCM) revealed ~200 putative Rho termination sites in the E. coli K-12 genome,81 ~10 of which were known prior to the study (reviewed elsewhere92). Rho termination sites were found downstream of stable RNAs, including ~1/3 of all tRNA operons and 7 annotated small RNAs. Rho termination of stable RNAs is unexpected because such RNAs are highly structured. One potential explanation of this result is that Rho could bind to the unstructured “tail” regions of the stable RNA prior to processing.126 However, it is also possible that certain stable RNAs could act as aptamers that bind directly to Rho.81 In vitro mapping of Rho binding sites within stable RNA transcripts should help clarify this issue.
Silencing of foreign DNA
Expression array profiling of BCM-treated cells found a general upregulation of genes within cryptic prophages and other horizontally transferred (i.e., foreign) elements, indicating that Rho silences transcription of these elements.124 RNAP ChIPchip in BCM-treated cells also identified a statistically significant association of Rho-dependent terminators with foreign DNA.81 There are several possible explanations for this association. First, foreign DNA may have a greater number of Rho terminators involved in regulation, such as the timm terminator that suppresses the induction of toxic genes in the rac prophage.124 Second, insertion of foreign DNA into active transcription units may require Rho termination in the foreign segments to compensate for loss of natural terminators; this is hypothesized to occur when phage integrases remove intrinsic terminators from tRNA genes during recombination.81 Third, suboptimal codon usage in foreign DNA could inhibit translation and could expose rut sites in the RNA to which Rho can bind and terminate transcription.124
Suppression of antisense transcription
Antisense transcription may be a general target of Rho termination in vivo. Twenty-four novel antisense transcripts were identified in RNAP ChIP-chip experiments because Rho inhibition with BCM increased transcription in a direction opposite to the annotated gene,81 but this number is thought to be a significant underestimate of the number of Rhoterminated antisense transcripts due to technical limitations. Recently reported deep RNA sequencing of E. coli detected ~1000 antisense RNA sequences.127 Rho termination is better suited to terminate antisense transcription than intrinsic termination because coding requirements on the sense strand make encoding an antisense intrinsic terminator problematic.81 Global, strand-specific RNA quantification should help determine the extent of antisense transcript suppression by Rho.
The Mechanism of Rho-Dependent Polarity
Rho termination of untranslated RNA transcripts causes polarity, which is the decrease in expression of distal genes in an operon caused by premature stop codons or inefficient translation in an upstream gene within the same operon. Although models for polarity have existed for several decades, recent studies reveal added complexity in ways translating ribosomes suppress Rho function. We will describe the two prevailing models for polarity, Rho– ribosome competition for RNA and Rho–ribosome competition for NusG, and suggest how they can be combined into a two-checkpoint model to detect translation failure in mRNAs. We note that these concepts are similar irrespective of whether Rho binds RNAP in vivo.
Rho–ribosome competition for RNA
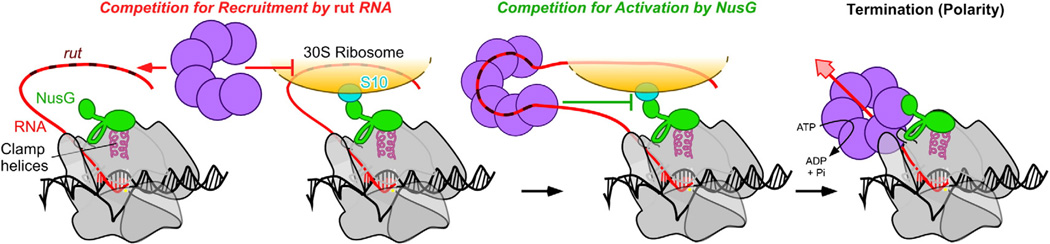
In the RNA competition model, Rho is prevented from associating with nascent RNA in the presence of the ribosome (Fig. 6).128 During active translation, potential Rho binding sites on the RNA (rut sites) are occupied by the ribosome. Since RNA binding by Rho is a prerequisite for Rho termination, physical occlusion of Rho binding sites by ribosomes prevents Rho activation and, thus, termination. Under conditions where translation is optimal, Rho can only bind rut sites 5′ of the ribosome, and Rho termination is suppressed. When translation slows or terminates, Rho can bind rut near RNAP, termination occurs, and polarity results.
Fig. 6.
Two-checkpoint mechanism to suppress Rho termination in protein-coding genes (polarity). Competition can occur for recruitment of rut RNA sequestered in the ribosome and for the NusG-CTD, which binds ribosomal protein S10 or Rho. Because NusG affects Rho dissociation from ECs but not recruitment, the two mechanisms may operate as sequential checks to determine whether an mRNA can be translated.
Rho–ribosome competition for NusG
In the NusG competition model, Rho and the ribosome are alternate targets for the NusG-CTD (Fig. 6). NusG is a small protein (21 kDa in E. coli) that enhances Rho termination. NusG has two conserved domains (NTD and CTD). The NusG-NTD binds to RNAP via interactions with the β′ clamp helices,129 and the NusG-CTD binds to Rho.130,131 Both interactions with RNAP and Rho are needed for termination enhancement by NusG,129 which increases the rate of RNA release by an unknown mechanism.131 Recent NMR studies have found that the NusG-CTD binds to ribosomal protein S10 (also known as NusE).130 Binding of the NusG-CTD to Rho or S10 is mutually exclusive,130 but the affinity of NusG for Rho is much greater than that for S10 (Ksd≈12 nM versus 50 µM, respectively).130,132 The surface of S10 bound by the NusG-CTD is exposed when S10 is in the ribosome, consistent with idea that the EC and the ribosome are physically linked by the NusG–S10 interaction.130 If transcription is actively coupled with translation, the ribosome binds to the NusG-CTD, which blocks the ability of NusG to enhance Rho-dependent termination. However, if the ribosome dissociates due to a premature translation stop codon or if translation is inefficient, Rho has access to the NusG-CTD and can cause polarity by terminating transcription.
Several important questions remain about the NusG-based model of polarity. Due to the large difference in affinity for NusG between S10 and Rho,130 the ribosome must remain close to the EC to win the competition,133 or additional factors present in the translation-coupled EC must prevent Rho association or action. Several ribosomal proteins are known to associate with RNAP (reviewed elsewhere124)134,135 and could serve as potential “coupling factors.” Some Rho-dependent terminators may not require NusG for efficient termination;136 thus, competition for NusG may be relevant only for some Rho termination sites; other sites may be governed only by competition for rut RNA (Fig. 6).
Two-checkpoint model for polarity
A combination of the RNA and NusG competition models may operate as a two-checkpoint mechanism, in which ribosome competition for RNA blocks Rho recruitment, and S10 binding to NusG prevents Rho dissociation of ECs. This two-checkpoint mechanism is consistent with the known biochemical properties of NusG, in that NusG has no effect on the binding of RNA to Rho. Because Rho must first bind RNA to act on the EC, RNA binding would be the first of the two checkpoints. Experiments using mutants of S10 that fail to bind to NusG will be critical in assessing the importance of the NusG–S10 interaction to polarity.
Conclusions
The steps in intrinsic and Rho-dependent termination, the signals that provoke them, and some of the ways they are regulated are now relatively well understood. However, the detailed mechanisms by which changes to the nucleic acid scaffold and to the structure of RNAP destabilize and dissociate the EC remain under study. Strong evidence suggests an energetic trade-off between ease of hybrid shearing and hypertranslocation at different intrinsic terminator sequences that determines which pathway operates, but the rules that govern this switch, the nature of RNAP conformation changes, and the mechanistic importance of these RNAP conformational changes await further study. Rho termination may operate by similar mechanisms, but the rules governing them and the roles of RNAP conformational changes are even less clear. The exciting discovery of the NusG interaction with a ribosomal protein suggests a possible two-checkpoint mechanism by which Rho termination can halt synthesis of untranslatable mRNAs. Finally, elucidation of both intrinsic and factor-dependent termination pathways in diverse bacteria promises to be a fertile area for exciting and important advances in future studies of transcription termination in bacteria. Even after 40 years, a complete understanding of termination mechanisms remains an elusive goal.
Abbreviations used
- RNAP
RNA polymerase
- EC
elongation complex
- FRET
fluorescence resonance energy transfer
- TL
trigger loop
- BCM
bicyclomycin
- NTD
N-terminal domain
- CTD
C-terminal domain.
Footnotes
References
- 1.d'Aubenton Carafa Y, Brody E, Thermes C. Prediction of rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem–loop structures. J. Mol. Biol. 1990;216:835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]
- 2.Washio T, Sasayama J, Tomita M. Analysis of complete genomes suggests that many prokaryotes do not rely on hairpin formation in transcription termination. Nucleic Acids Res. 1998;26:5456–5463. doi: 10.1093/nar/26.23.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ermolaeva MD, Khalak HG, White O, Smith HO, Salzberg SL. Prediction of transcription terminators in bacterial genomes. J. Mol. Biol. 2000;301:27–33. doi: 10.1006/jmbi.2000.3836. [DOI] [PubMed] [Google Scholar]
- 4.Lesnik EA, Sampath R, Levene HB, Henderson TJ, McNeil JA, Ecker DJ. Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res. 2001;29:3583–3594. doi: 10.1093/nar/29.17.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Unniraman S, Prakash R, Nagaraja V. Conserved economics of transcription termination in eubacteria. Nucleic Acids Res. 2002;30:675–684. doi: 10.1093/nar/30.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hosid S, Bolshoy A. New elements of the termination of transcription in prokaryotes. J. Biomol. Struct. Dyn. 2004;22:347–354. doi: 10.1080/07391102.2004.10507006. [DOI] [PubMed] [Google Scholar]
- 7.de Hoon MJ, Makita Y, Nakai K, Miyano S. Prediction of transcriptional terminators in Bacillus subtilis and related species. PLoS Comput. Biol. 2005;1:e25. doi: 10.1371/journal.pcbi.0010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitra A, Angamuthu K, Jayashree HV, Nagaraja V. Occurrence, divergence and evolution of intrinsic terminators across eubacteria. Genomics. 2009;94:110–116. doi: 10.1016/j.ygeno.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Kingsford CL, Ayanbule K, Salzberg SL. Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biol. 2007;8:R22. doi: 10.1186/gb-2007-8-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitra A, Angamuthu K, Nagaraja V. Genome-wide analysis of the intrinsic terminators of transcription across the genus Mycobacterium. Tuberculosis (Edinb) 2008;88:566–575. doi: 10.1016/j.tube.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 11.Mitra A, Kesarwani AK, Pal D, Nagaraja V. WebGeSTer DB—a transcription terminator database. Nucleic Acids Res. 2010;39:D129–D135. doi: 10.1093/nar/gkq971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Epshtein V, Cardinale CJ, Ruckenstein AE, Borukhov S, Nudler E. An allosteric path to transcription termination. Mol. Cell. 2007;28:991–1001. doi: 10.1016/j.molcel.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 13.Epshtein V, Dutta D, Wade J, Nudler E. An allosteric mechanism of Rho-dependent transcription termination. Nature. 2010;463:245–249. doi: 10.1038/nature08669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larson MH, Greenleaf WJ, Landick R, Block SM. Applied force reveals mechanistic and energetic details of transcription termination. Cell. 2008;132:971–982. doi: 10.1016/j.cell.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park JS, Roberts JW. Role of DNA bubble rewinding in enzymatic transcription termination. Proc. Natl Acad. Sci. USA. 2006;103:4870–4875. doi: 10.1073/pnas.0600145103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santangelo TJ, Roberts JW. Forward translocation is the natural pathway of RNA release at an intrinsic terminator. Mol. Cell. 2004;14:117–126. doi: 10.1016/s1097-2765(04)00154-6. [DOI] [PubMed] [Google Scholar]
- 17.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 18.Vassylyev D, Vassylyeva M, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 19.Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 20.Komissarova N, Kashlev M. Functional topography of nascent RNA in elongation intermediates of RNA polymerase. Proc. Natl. Acad. Sci. USA. 1998;95:14699–14704. doi: 10.1073/pnas.95.25.14699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korzheva N, Mustaev A, Kozlov M, Malhotra A, Nikiforov V, Goldfarb A, Darst SA. A structural model of transcription elongation. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- 22.Andrecka J, Treutlein B, Arcusa MA, Muschielok A, Lewis R, Cheung AC, et al. Nano positioning system reveals the course of upstream and nontemplate DNA within the RNA polymerase II elongation complex. Nucleic Acids Res. 2009;37:5803–5809. doi: 10.1093/nar/gkp601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zaychikov E, Denissova L, Heumann H. Translocation of the Escherichia coli transcription complex observed in the registers 11 to 20: “jumping” of RNA polymerase and asymmetric expansion and contraction of the “transcription bubble”. Proc. Natl Acad. Sci. USA. 1995;92:1739–1743. doi: 10.1073/pnas.92.5.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang D, Landick R. Nuclease cleavage of the upstream half of the nontemplate strand DNA in an E. coli transcription elongation complex causes upstream translocation and transcriptional arrest. J. Biol. Chem. 1997;272:5989–5994. doi: 10.1074/jbc.272.9.5989. [DOI] [PubMed] [Google Scholar]
- 25.Wilson KS, von Hippel PH. Stability of Escherichia coli transcription complexes near an intrinsic terminator. J. Mol. Biol. 1994;244:36–51. doi: 10.1006/jmbi.1994.1702. [DOI] [PubMed] [Google Scholar]
- 26.Nudler E, Avetissova E, Markovtsov V, Goldfarb A. Transcription processivity: protein–DNA interactions holding together the elongation complex. Science. 1996;273:211–217. doi: 10.1126/science.273.5272.211. [DOI] [PubMed] [Google Scholar]
- 27.Komissarova N, Becker J, Solter S, Kireeva M, Kashlev M. Shortening of RNA:DNA hybrid in the elongation complex of RNA polymerase is a prerequisite for transcription termination. Mol. Cell. 2002;10:1151–1162. doi: 10.1016/s1097-2765(02)00738-4. [DOI] [PubMed] [Google Scholar]
- 28.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA–DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 29.Korzheva N, Mustaev A, Nudler E, Nikiforov V, Goldfarb A. Mechanistic model of the elongation complex of Escherichia coli RNA polymerase. Cold Spring Harbor Symp. Quant. Biol. 1998;63:337–345. doi: 10.1101/sqb.1998.63.337. [DOI] [PubMed] [Google Scholar]
- 30.von Hippel PH, Yager TD. The elongation–termination decision in transcription. Science. 1992;255:809–812. doi: 10.1126/science.1536005. [DOI] [PubMed] [Google Scholar]
- 31.Yin H, Artsimovitch I, Landick R, Gelles J. Nonequilibrium mechanism of transcription termination from observations of single RNA polymerase molecules. Proc. Natl Acad. Sci. USA. 1999;96:13124–13129. doi: 10.1073/pnas.96.23.13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gusarov I, Nudler E. The mechanism of intrinsic transcription termination. Mol. Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- 33.Berlin V, Yanofsky C. Release of transcript and template during transcription termination at the trp operon attenuator. J. Biol. Chem. 1983;258:1714–1719. [PubMed] [Google Scholar]
- 34.Kashlev M, Komissarova N. Transcription termination: primary intermediates and secondary adducts. J. Biol. Chem. 2002;277:14501–14508. doi: 10.1074/jbc.M200215200. [DOI] [PubMed] [Google Scholar]
- 35.Rosenberg M, Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu. Rev. Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- 36.Telesnitsky A, Chamberlin MJ. Terminator-distal sequences determine the in vitro efficiency of the early terminators of bacteriophages T3 and T7. Biochemistry. 1989;28:5210–5218. doi: 10.1021/bi00438a044. [DOI] [PubMed] [Google Scholar]
- 37.Martinez-Trujillo M, Sanchez-Trujillo A, Ceja V, Avila-Moreno F, Bermudez-Cruz RM, Court D, Montanez C. Sequences required for transcription termination at the intrinsic lambdatI terminator. Can. J. Microbiol. 2010;56:168–177. doi: 10.1139/w09-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson KS, von Hippel PH. Transcription termination at intrinsic terminators: the role of the RNA hairpin. Proc. Natl Acad. Sci. USA. 1995;92:8793–8797. doi: 10.1073/pnas.92.19.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan C, Wang D, Landick R. Spacing from the transcript 3′ end determines whether a nascent RNA hairpin interacts with RNA polymerase to prolong pausing or triggers termination. J. Mol. Biol. 1997;268:54–68. [Google Scholar]
- 40.Chan CL, Landick R. Dissection of the his leader pause site by base substitution reveals a multipartite signal that includes a pause RNA hairpin. J. Mol. Biol. 1993;233:25–42. doi: 10.1006/jmbi.1993.1482. [DOI] [PubMed] [Google Scholar]
- 41.Lee DN, Phung L, Stewart J, Landick R. Transcription pausing by Escherichia coli RNA polymerase is modulated by downstream DNA sequences. J. Biol. Chem. 1990;265:15145–15153. [PubMed] [Google Scholar]
- 42.Kireeva ML, Kashlev M. Mechanism of sequence-specific pausing of bacterial RNA polymerase. Proc. Natl Acad. Sci. USA. 2009;106:8900–8905. doi: 10.1073/pnas.0900407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Komissarova N, Kashlev M. RNA polymerase switches between inactivated and activated states by translocating back and forth along the DNA and the RNA. J. Biol. Chem. 1997;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 44.Mejia YX, Mao H, Forde NR, Bustamante C. Thermal probing of E. coli RNA polymerase off-pathway mechanisms. J. Mol. Biol. 2008;382:628–637. doi: 10.1016/j.jmb.2008.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Depken M, Galburt EA, Grill SW. The origin of short transcriptional pauses. Biophys. J. 2009;96:2189–2193. doi: 10.1016/j.bpj.2008.12.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol. Cell. 2007;27:406–419. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 47.Landick R. Transcriptional pausing without backtracking. Proc. Natl Acad. Sci. USA. 2009;106:8797–8798. doi: 10.1073/pnas.0904373106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woodside MT, Behnke-Parks WM, Larizadeh K, Travers K, Herschlag D, Block SM. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc. Natl Acad. Sci. USA. 2006;103:6190–6195. doi: 10.1073/pnas.0511048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma H, Proctor DJ, Kierzek E, Kierzek R, Bevilacqua PC, Gruebele M. Exploring the energy landscape of a small RNA hairpin. J. Am. Chem. Soc. 2006;128:1523–1530. doi: 10.1021/ja0553856. [DOI] [PubMed] [Google Scholar]
- 50.Henkin TM, Yanofsky C. Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/ antitermination decisions. BioEssays. 2002;24:700–707. doi: 10.1002/bies.10125. [DOI] [PubMed] [Google Scholar]
- 51.Goliger JA, Yang XJ, Guo HC, Roberts JW. Early transcribed sequences affect termination efficiency of Escherichia coli RNA polymerase. J. Mol. Biol. 1989;205:331–341. doi: 10.1016/0022-2836(89)90344-6. [DOI] [PubMed] [Google Scholar]
- 52.Telesnitsky AP, Chamberlin MJ. Sequences linked to prokaryotic promoters can affect the efficiency of downstream termination sites. J. Mol. Biol. 1989;205:315–330. doi: 10.1016/0022-2836(89)90343-4. [DOI] [PubMed] [Google Scholar]
- 53.Cheng SW, Lynch EC, Leason KR, Court DL, Shapiro BA, Friedman DI. Functional importance of sequence in the stem–loop of a transcription terminator. Science. 1992;254:1205–1207. doi: 10.1126/science.1835546. [DOI] [PubMed] [Google Scholar]
- 54.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Toulokhonov I, Landick R. The flap domain is required for pause RNA hairpin inhibition of catalysis by RNA polymerase and can modulate intrinsic termination. Mol. Cell. 2003;12:1125–1136. doi: 10.1016/s1097-2765(03)00439-8. [DOI] [PubMed] [Google Scholar]
- 57.Kuznedelov KD, Komissarova NV, Severinov KV. The role of the bacterial RNA polymerase beta subunit flexible flap domain in transcription termination. Dokl., Biochem. Biophys. 2006;410:263–266. doi: 10.1134/s1607672906050036. [DOI] [PubMed] [Google Scholar]
- 58.Pyun J. Single-Molecule Studies of Active and Static Transcription Complexes. Waltham, MA: Brandeis University; 2005. [Google Scholar]
- 59.Reynolds R, Bermudez-Cruz RM, Chamberlin MJ. Parameters affecting transcription termination by Escherichia coli RNA polymerase. I. Analysis of 13 rho-independent terminators. J. Mol. Biol. 1992;224:31–51. doi: 10.1016/0022-2836(92)90574-4. [DOI] [PubMed] [Google Scholar]
- 60.Gama-Castro S, Salgado H, Peralta-Gil M, Santos-Zavaleta A, Muniz-Rascado L, Solano-Lira H, et al. RegulonDB version 7.0: transcriptional regulation of Escherichia coli K-12 integrated within genetic sensory response units (Gensor Units) Nucleic Acids Res. 2010;39:D98–D105. doi: 10.1093/nar/gkq1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Herbert KM, Greenleaf WJ, Block SM. Single-molecule studies of RNA polymerase: motoring along. Annu. Rev. Biochem. 2008;77:149–176. doi: 10.1146/annurev.biochem.77.073106.100741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kyzer S, Ha KS, Landick R, Palangat M. Direct versus limited-step reconstitution reveals key features of an RNA hairpin-stabilized paused transcription complex. J. Biol. Chem. 2007;282:19020–19028. doi: 10.1074/jbc.M701483200. [DOI] [PubMed] [Google Scholar]
- 63.Antao VP, Lai SY, Tinoco I., Jr A thermodynamic study of unusually stable RNA and DNA hairpins. Nucleic Acids Res. 1991;19:5901–5905. doi: 10.1093/nar/19.21.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Antao VP, Tinoco I., Jr Thermodynamic parameters for loop formation in RNA and DNA hairpin tetraloops. Nucleic Acids Res. 1992;20:819–824. doi: 10.1093/nar/20.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reynolds R, Chamberlin MJ. Parameters affecting transcription termination by Escherichia coli RNA. II. Construction and analysis of hybrid terminators. J. Mol. Biol. 1992;224:53–63. doi: 10.1016/0022-2836(92)90575-5. [DOI] [PubMed] [Google Scholar]
- 66.Macdonald LE, Zhou Y, McAllister WT. Termination and slippage by bacteriophage T7 RNA polymerase. J. Mol. Biol. 1993;232:1030–1047. doi: 10.1006/jmbi.1993.1458. [DOI] [PubMed] [Google Scholar]
- 67.Yarnell WS, Roberts JW. Mechanism of intrinsic transcription termination and antitermination. Science. 1999;284:611–615. doi: 10.1126/science.284.5414.611. [DOI] [PubMed] [Google Scholar]
- 68.Ryder AM, Roberts JW. Role of the nontemplate strand of the elongation bubble in intrinsic transcription termination. J. Mol. Biol. 2003;334:205–213. doi: 10.1016/j.jmb.2003.09.039. [DOI] [PubMed] [Google Scholar]
- 69.Cramer P, Bushnell D, Kornberg R. Structural basis of transcription: RNA polymerase II at 2.8 Å resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 70.Darst SA, Opalka N, Chacon P, Polyakov A, Richter C, Zhang G, Wriggers W. Conformational flexibility of bacterial RNA polymerase. Proc. Natl Acad. Sci. USA. 2002;99:4296–4301. doi: 10.1073/pnas.052054099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tagami S, Sekine S, Kumarevel T, Hino N, Murayama Y, Kamegamori S, et al. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature. 2010;468:978–982. doi: 10.1038/nature09573. [DOI] [PubMed] [Google Scholar]
- 72.Mukhopadhyay J, Das K, Ismail S, Koppstein D, Jang M, Hudson B, et al. The RNA polymerase “switch region” is a target for inhibitors. Cell. 2008;135:295–307. doi: 10.1016/j.cell.2008.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Landick R. RNA polymerase clamps down. Cell. 2001;105:567–570. doi: 10.1016/s0092-8674(01)00381-6. [DOI] [PubMed] [Google Scholar]
- 74.Toulokhonov I, Artsimovitch I, Landick R. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science. 2001;292:730–733. doi: 10.1126/science.1057738. [DOI] [PubMed] [Google Scholar]
- 75.Kohno K, Wada M, Kano Y, Imamoto F. Promoters and autogenous control of the Escherichia coli hupA and hupB genes. J. Mol. Biol. 1990;213:27–36. doi: 10.1016/S0022-2836(05)80119-6. [DOI] [PubMed] [Google Scholar]
- 76.Gunsalus RP, Yanofsky C. Nucleotide sequence and expression of Escherichia coli trpR the structural gene for the trp aporepressor. Proc. Natl Acad. Sci. USA. 1980;77:7117–7121. doi: 10.1073/pnas.77.12.7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Unniraman S, Prakash R, Nagaraja V. Alternate paradigm for intrinsic transcription termination in eubacteria. J. Biol. Chem. 2001;276:41850–41855. doi: 10.1074/jbc.M106252200. [DOI] [PubMed] [Google Scholar]
- 78.Ingham CJ, Hunter IS, Smith MC. Rho-independent terminators without 3′ poly-U tails from the early region of actinophage øC31. Nucleic Acids Res. 1995;23:370–376. doi: 10.1093/nar/23.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abe H, Aiba H. Differential contributions of two elements of rho-independent terminator to transcription termination and mRNA stabilization. Biochimie. 1996;78:1035–1042. doi: 10.1016/s0300-9084(97)86727-2. [DOI] [PubMed] [Google Scholar]
- 80.Wu AM, Christie GE, Platt T. Tandem termination sites in the tryptophan operon of Escherichia coli. Proc. Natl Acad. Sci. USA. 1981;78:2913–2917. doi: 10.1073/pnas.78.5.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Peters JM, Mooney RA, Kuan PF, Rowland JL, Keles S, Landick R. Rho directs widespread termination of intragenic and stable RNA transcription. Proc. Natl Acad. Sci. USA. 2009;106:15406–15411. doi: 10.1073/pnas.0903846106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Banerjee S, Chalissery J, Bandey I, Sen R. Rho-dependent transcription termination: more questions than answers. J. Microbiol. 2006;44:11–22. [PMC free article] [PubMed] [Google Scholar]
- 83.Richardson JP. Loading Rho to terminate transcription. Cell. 2003;114:157–159. doi: 10.1016/s0092-8674(03)00554-3. [DOI] [PubMed] [Google Scholar]
- 84.Lowery-Goldhammer C, Richardson JP. An RNA-dependent nucleoside triphosphate phosphohydrolase (ATPase) associated with rho termination factor. Proc. Natl Acad. Sci. USA. 1974;71:2003–2007. doi: 10.1073/pnas.71.5.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McSwiggen JA, Bear DG, von Hippel PH. Interactions of Escherichia coli transcription termination factor rho with RNA. I. Binding stoichiometries and free energies. J. Mol. Biol. 1988;199:609–622. doi: 10.1016/0022-2836(88)90305-1. [DOI] [PubMed] [Google Scholar]
- 86.Chen CY, Richardson JP. Sequence elements essential for ρ-dependent transcription termination at λtR1. J. Biol. Chem. 1987;262:11292–11299. [PubMed] [Google Scholar]
- 87.Zalatan F, Platt T. Effects of decreased cytosine content on rho interaction with the rho-dependent terminator trp t′ in Escherichia coli. J. Biol. Chem. 1992;267:19082–19088. [PubMed] [Google Scholar]
- 88.Chen CY, Galluppi GR, Richardson JP. Transcription termination at λ tR1 is mediated by interaction of rho with specific single-stranded domains near the 3′ end of cro mRNA. Cell. 1986;46:1023–1028. doi: 10.1016/0092-8674(86)90701-4. [DOI] [PubMed] [Google Scholar]
- 89.Hart CM, Roberts JW. Rho-dependent transcription termination. Characterization of the requirement for cytidine in the nascent transcript. J. Biol. Chem. 1991;266:24140–24148. [PubMed] [Google Scholar]
- 90.Guerin M, Robichon N, Geiselmann J, Rahmouni AR. A simple polypyrimidine repeat acts as an artificial Rho-dependent terminator in vivo and in vitro. Nucleic Acids Res. 1998;26:4895–4900. doi: 10.1093/nar/26.21.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Alifano P, Rivellini F, Limauro D, Bruni CB, Carlomagno MS. A consensus motif common to all Rho-dependent prokaryotic transcription terminators. Cell. 1991;64:553–563. doi: 10.1016/0092-8674(91)90239-u. [DOI] [PubMed] [Google Scholar]
- 92.Ciampi MS. Rho-dependent terminators and transcription termination. Microbiology. 2006;152:2515–2528. doi: 10.1099/mic.0.28982-0. [DOI] [PubMed] [Google Scholar]
- 93.Skordalakes E, Berger JM. Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading. Cell. 2003;114:135–146. doi: 10.1016/s0092-8674(03)00512-9. [DOI] [PubMed] [Google Scholar]
- 94.Thomsen ND, Berger JM. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell. 2009;139:523–534. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gogol EP, Seifried SE, von Hippel PH. Structure and assembly of the Escherichia coli transcription termination factor rho and its interaction with RNA. I. Cryoelectron microscopic studies. J. Mol. Biol. 1991;221:1127–1138. doi: 10.1016/0022-2836(91)90923-t. [DOI] [PubMed] [Google Scholar]
- 96.Briercheck DM, Wood TC, Allison TJ, Richardson JP, Rule GS. The NMR structure of the RNA binding domain of E. coli rho factor suggests possible RNA–protein interactions. Nat. Struct. Biol. 1998;5:393–399. doi: 10.1038/nsb0598-393. [DOI] [PubMed] [Google Scholar]
- 97.Bogden CE, Fass D, Bergman N, Nichols MD, Berger JM. The structural basis for terminator recognition by the Rho transcription termination factor. Mol. Cell. 1999;3:487–493. doi: 10.1016/s1097-2765(00)80476-1. [DOI] [PubMed] [Google Scholar]
- 98.Skordalakes E, Berger JM. Structural insights into RNA-dependent ring closure and ATPase activation by the Rho termination factor. Cell. 2006;127:553–564. doi: 10.1016/j.cell.2006.08.051. [DOI] [PubMed] [Google Scholar]
- 99.Boudvillain M, Nollmann M, Margeat E. Keeping up to speed with the transcription termination factor Rho motor. Transcr. 2010;1:70–75. doi: 10.4161/trns.1.2.12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schwartz A, Walmacq C, Rahmouni AR, Boudvillain M. Noncanonical interactions in the management of RNA structural blocks by the transcription termination rho helicase. Biochemistry. 2007;46:9366–9379. doi: 10.1021/bi700493m. [DOI] [PubMed] [Google Scholar]
- 101.Schwartz A, Margeat E, Rahmouni AR, Boudvillain M. Transcription termination factor rho can displace streptavidin from biotinylated RNA. J. Biol. Chem. 2007;282:31469–31476. doi: 10.1074/jbc.M706935200. [DOI] [PubMed] [Google Scholar]
- 102.Patel SS. Structural biology: steps in the right direction. Nature. 2009;462:581–583. doi: 10.1038/462581a. [DOI] [PubMed] [Google Scholar]
- 103.Steinmetz EJ, Platt T. Evidence supporting a tethered tracking model for helicase activity of Escherichia coli Rho factor. Proc. Natl Acad. Sci. USA. 1994;91:1401–1405. doi: 10.1073/pnas.91.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rabhi M, Gocheva V, Jacquinot F, Lee A, Margeat E, Boudvillain M. Mutagenesis-based evidence for an asymmetric configuration of the ring-shaped transcription termination factor rho. J. Mol. Biol. 2011;405:497–518. doi: 10.1016/j.jmb.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 105.Galluppi GR, Richardson JP. ATP-induced changes in the binding of RNA synthesis termination protein Rho to RNA. J. Mol. Biol. 1980;138:513–539. doi: 10.1016/s0022-2836(80)80016-7. [DOI] [PubMed] [Google Scholar]
- 106.Balasubramanian K, Stitt BL. Evidence for amino acid roles in the chemistry of ATP hydrolysis in Escherichia coli Rho. J. Mol. Biol. 2010;404:587–599. doi: 10.1016/j.jmb.2010.09.039. [DOI] [PubMed] [Google Scholar]
- 107.Miwa Y, Horiguchi T, Shigesada K. Structural and functional dissections of transcription termination factor rho by random mutagenesis. J. Mol. Biol. 1995;254:815–837. doi: 10.1006/jmbi.1995.0658. [DOI] [PubMed] [Google Scholar]
- 108.Kassavetis GA, Chamberlin MJ. Pausing and termination of transcription within the early region of bacteriophage T7 DNA in vitro. J. Biol. Chem. 1981;256:2777–2786. [PubMed] [Google Scholar]
- 109.Morgan WD, Bear DG, von Hippel PH. Rho-dependent termination of transcription. II. Kinetics of mRNA elongation during transcription from the bacteriophage λ PR promoter. J. Biol. Chem. 1983;258:9565–9574. [PubMed] [Google Scholar]
- 110.Lau LF, Roberts JW. ρ-Dependent transcription termination at λtR1 requires upstream sequences. J. Biol. Chem. 1985;260:574–584. [PubMed] [Google Scholar]
- 111.Stewart V, Landick R, Yanofsky C. Rhodependent transcription termination in the trypto-phanase operon leader region of Escherichia coli K-12. J. Bacteriol. 1986;166:217–223. doi: 10.1128/jb.166.1.217-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Galloway JL, Platt T. Signals sufficient for rho-dependent transcription termination at trp t′ span a region centered 60 base pairs upstream of the earliest 3′ end point. J. Biol. Chem. 1988;263:1761–1767. [PubMed] [Google Scholar]
- 113.Dutta D, Chalissery J, Sen R. Transcription termination factor rho prefers catalytically active elongation complexes for releasing RNA. J. Biol. Chem. 2008;283:20243–20251. doi: 10.1074/jbc.M801926200. [DOI] [PubMed] [Google Scholar]
- 114.Jin DJ, Burgess RR, Richardson JP, Gross CA. Termination efficiency at rho-dependent terminators depends on kinetic coupling between RNA polymerase and rho. Proc. Natl Acad. Sci. USA. 1992;89:1453–1457. doi: 10.1073/pnas.89.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mooney RA, Davis SE, Peters JM, Rowland JL, Ansari AZ, Landick R. Regulator trafficking on bacterial transcription units in vivo. Mol. Cell. 2009;33:97–108. doi: 10.1016/j.molcel.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Imai M, Shigesada K. Studies on the altered rho factor in a nitA mutants of Escherichia coli defective in transcription termination. I. Characterization and quantitative determination of rho in cell extracts. J. Mol. Biol. 1978;120:451–466. doi: 10.1016/0022-2836(78)90348-0. [DOI] [PubMed] [Google Scholar]
- 117.Grigorova IL, Phleger NJ, Mutalik VK, Gross CA. Insights into transcriptional regulation and sigma competition from an equilibrium model of RNA polymerase binding to DNA. Proc. Natl Acad. Sci. USA. 2006;103:5332–5337. doi: 10.1073/pnas.0600828103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Brennan CA, Dombroski AJ, Platt T. Transcription termination factor rho is an RNA–DNA helicase. Cell. 1987;48:945–952. doi: 10.1016/0092-8674(87)90703-3. [DOI] [PubMed] [Google Scholar]
- 119.Richardson JP. Rho-dependent termination and ATPases in transcript termination. Biochim. Biophys. Acta. 2002;1577:251–260. doi: 10.1016/s0167-4781(02)00456-6. [DOI] [PubMed] [Google Scholar]
- 120.Dalal RV, Larson MH, Neuman KC, Gelles J, Landick R, Block SM. Pulling on the nascent RNA during transcription does not alter kinetics of elongation or ubiquitous pausing. Mol. Cell. 2006;23:231–239. doi: 10.1016/j.molcel.2006.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Roberts JW. Termination factor for RNA synthesis. Nature. 1969;224:1168–1174. doi: 10.1038/2241168a0. [DOI] [PubMed] [Google Scholar]
- 122.Harinarayanan R, Gowrishankar J. Host factor titration by chromosomal R-loops as a mechanism for runaway plasmid replication in transcription termination-defective mutants of Escherichia coli. J. Mol. Biol. 2003;332:31–46. doi: 10.1016/s0022-2836(03)00753-8. [DOI] [PubMed] [Google Scholar]
- 123.De Crombrugghe B, Adhya S, Gottesman M, Pastan I. Effect of Rho on transcription of bacterial operons. Nat., New Biol. 1973;241:260–264. doi: 10.1038/newbio241260a0. [DOI] [PubMed] [Google Scholar]
- 124.Cardinale CJ, Washburn RS, Tadigotla VR, Brown LM, Gottesman ME, Nudler E. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science. 2008;320:935–938. doi: 10.1126/science.1152763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Washburn RS, Gottesman ME. Transcription termination maintains chromosome integrity. Proc. Natl Acad. Sci. USA. 2010;108:792–797. doi: 10.1073/pnas.1009564108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rossi J, Egan J, Hudson L, Landy A. The tyrT locus: termination and processing of a complex transcript. Cell. 1981;26:305–314. doi: 10.1016/0092-8674(81)90199-9. [DOI] [PubMed] [Google Scholar]
- 127.Dornenburg JE, Devita AM, Palumbo MJ, Wade JT. Widespread antisense transcription in Escherichia coli. MBio. 2010;1:e00024–10. doi: 10.1128/mBio.00024-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Adhya S, Gottesman M. Control of transcription termination. Annu. Rev. Biochem. 1978;47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- 129.Mooney RA, Schweimer K, Rosch P, Gottesman M, Landick R. Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J. Mol. Biol. 2009;391:341–358. doi: 10.1016/j.jmb.2009.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Burmann BM, Schweimer K, Luo X, Wahl MC, Stitt BL, Gottesman ME, Rosch PA. NusE:NusG complex links transcription and translation. Science. 2010;328:501–504. doi: 10.1126/science.1184953. [DOI] [PubMed] [Google Scholar]
- 131.Chalissery J, Muteeb G, Kalarickal NC, Mohan S, Jisha V, Sen R. Interaction surface of the transcription terminator rho required to form a complex with the C-terminal domain of the antiterminator NusG. J. Mol. Biol. 2011;405:49–64. doi: 10.1016/j.jmb.2010.10.044. [DOI] [PubMed] [Google Scholar]
- 132.Pasman Z, von Hippel PH. Regulation of rho-dependent transcription termination by NusG is specific to the Escherichia coli elongation complex. Biochemistry. 2000;39:5573–5585. doi: 10.1021/bi992658z. [DOI] [PubMed] [Google Scholar]
- 133.Proshkin S, Rahmouni AR, Mironov A, Nudler E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010;328:504–508. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Squires CL, Zaporojets D. Proteins shared by the transcription and translation machines. Annu. Rev. Microbiol. 2000;54:775–798. doi: 10.1146/annurev.micro.54.1.775. [DOI] [PubMed] [Google Scholar]
- 135.Torres M, Condon C, Balada JM, Squires C, Squires CL. Ribosomal protein S4 is a transcription factor with properties remarkably similar to NusA, a protein involved in both nonribosomal and ribosomal RNA antitermination. EMBO J. 2001;20:3811–3820. doi: 10.1093/emboj/20.14.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sullivan SL, Gottesman ME. Requirement for E. coli NusG protein in factor-dependent transcription termination. Cell. 1992;68:989–994. doi: 10.1016/0092-8674(92)90041-a. [DOI] [PubMed] [Google Scholar]