Abstract
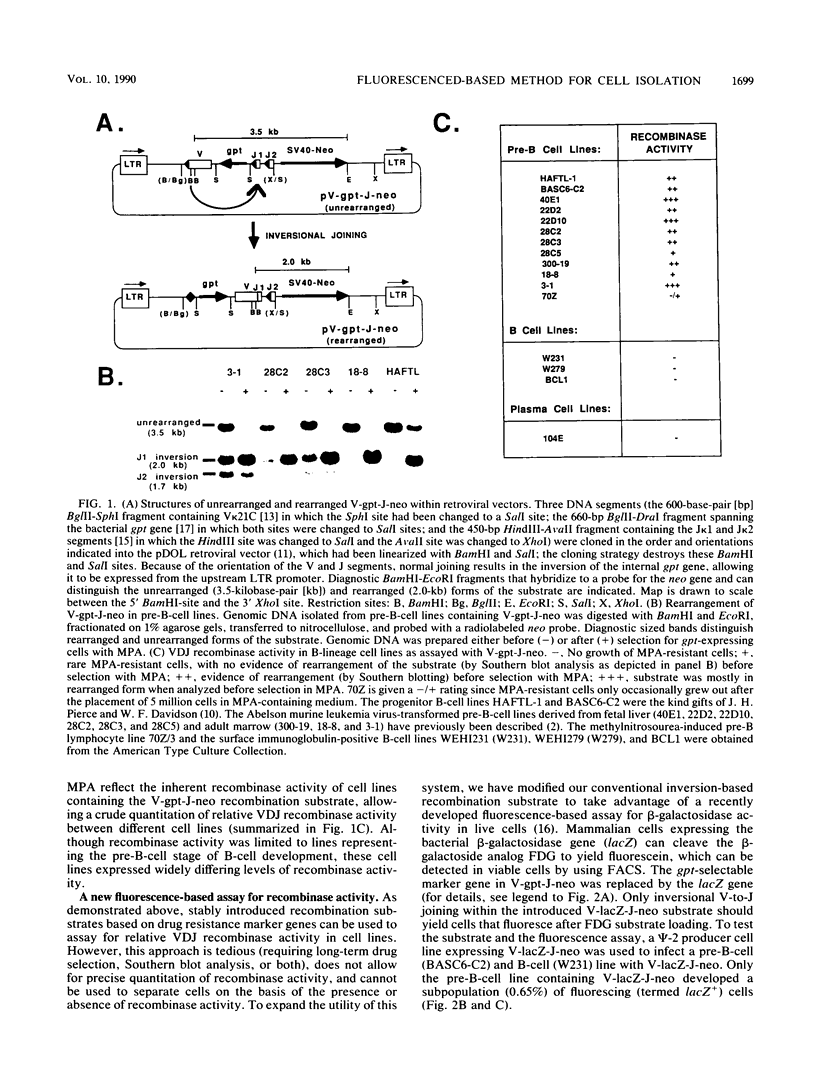
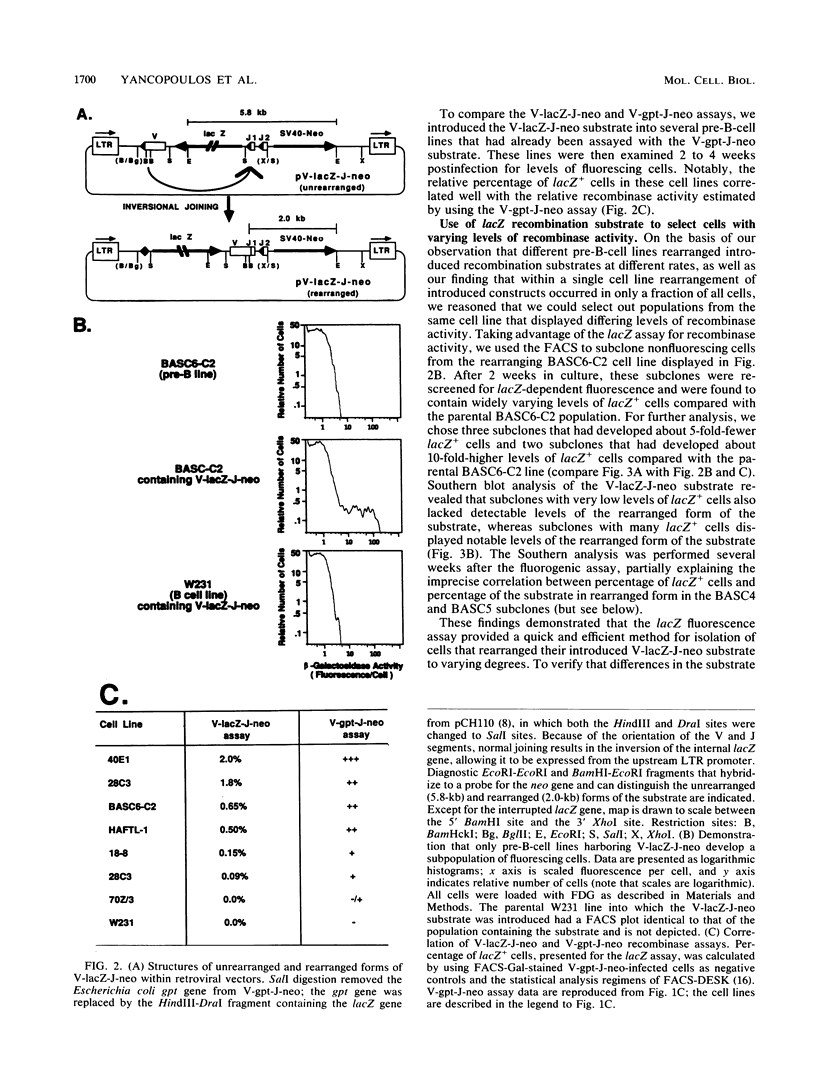
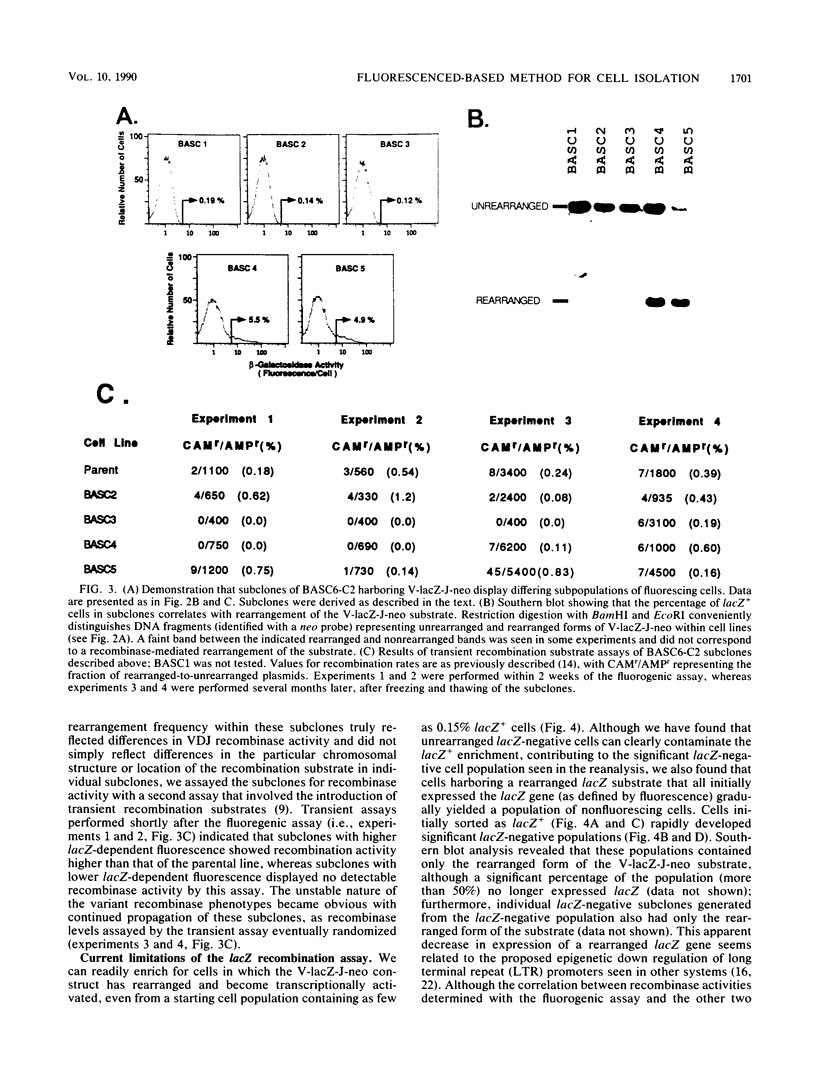
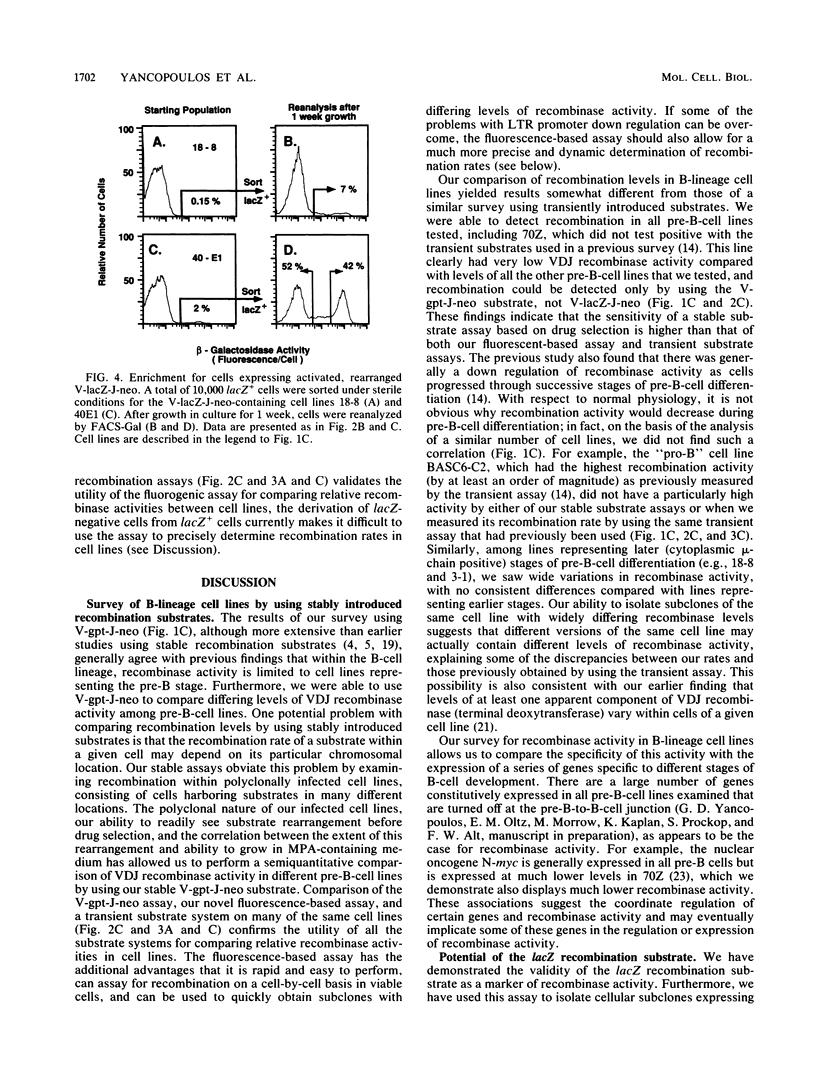
We describe two retroviral vector-based recombination substrate systems designed to assay for lymphoid VDJ recombinase activity in cultured cells. Both substrates incorporate a constitutive dominant marker gene (the simian virus promoter-driven neo gene) to allow selection of cells that stably integrate the substrate. Both substrates also include a second marker gene that becomes transcriptionally active only when inverted by a site-specific recombination event between flanking immunoglobulin variable-region gene segments. The first vector, similar in structure to previous retrovirus-based recombination substrates, utilizes the bacterial guanine-xanthine phosphoribosyltransferase gene (gpt) as its activatable marker; detection of inversion (VDJ recombinase activity) involves drug selection and Southern blotting analyses. We have used this vector to make a more extensive and quantitative survey of VDJ recombinase activity in B-lineage cell lines than has previously been performed with stable substrates, and we have compared our results with those of other studies that use transient recombination substrates. In the second vector, the activatable gene is the bacterial beta-galactosidase gene (lacZ). Detection for inversional activation of this gene is achieved by a fluorogenic assay, termed FACS-Gal, that detects beta-galactosidase activity in viable cells. The latter assay has the unique advantage of rapidly detecting cells that undergo recombination and also allows viable sorting of cells on the basis of the presence or absence of VDJ recombinase activity. We have used the lacZ vector to rapidly quantitate VDJ recombinase activity in B-lineage cell lines and compared the results with those obtained with the gpt vector.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alt F. W., Baltimore D. Joining of immunoglobulin heavy chain gene segments: implications from a chromosome with evidence of three D-JH fusions. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4118–4122. doi: 10.1073/pnas.79.13.4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt F. W., Yancopoulos G. D., Blackwell T. K., Wood C., Thomas E., Boss M., Coffman R., Rosenberg N., Tonegawa S., Baltimore D. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 1984 Jun;3(6):1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell T. K., Alt F. W. Site-specific recombination between immunoglobulin D and JH segments that were introduced into the genome of a murine pre-B cell line. Cell. 1984 May;37(1):105–112. doi: 10.1016/0092-8674(84)90305-2. [DOI] [PubMed] [Google Scholar]
- Blackwell T. K., Moore M. W., Yancopoulos G. D., Suh H., Lutzker S., Selsing E., Alt F. W. Recombination between immunoglobulin variable region gene segments is enhanced by transcription. Nature. 1986 Dec 11;324(6097):585–589. doi: 10.1038/324585a0. [DOI] [PubMed] [Google Scholar]
- Desiderio S. V., Wolff K. R. Rearrangement of exogenous immunoglobulin VH and DJH gene segments after retroviral transduction into immature lymphoid cell lines. J Exp Med. 1988 Feb 1;167(2):372–388. doi: 10.1084/jem.167.2.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dildrop R., Ma A., Zimmerman K., Hsu E., Tesfaye A., DePinho R., Alt F. W. IgH enhancer-mediated deregulation of N-myc gene expression in transgenic mice: generation of lymphoid neoplasias that lack c-myc expression. EMBO J. 1989 Apr;8(4):1121–1128. doi: 10.1002/j.1460-2075.1989.tb03482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early P., Huang H., Davis M., Calame K., Hood L. An immunoglobulin heavy chain variable region gene is generated from three segments of DNA: VH, D and JH. Cell. 1980 Apr;19(4):981–992. doi: 10.1016/0092-8674(80)90089-6. [DOI] [PubMed] [Google Scholar]
- Hall C. V., Jacob P. E., Ringold G. M., Lee F. Expression and regulation of Escherichia coli lacZ gene fusions in mammalian cells. J Mol Appl Genet. 1983;2(1):101–109. [PubMed] [Google Scholar]
- Hesse J. E., Lieber M. R., Gellert M., Mizuuchi K. Extrachromosomal DNA substrates in pre-B cells undergo inversion or deletion at immunoglobulin V-(D)-J joining signals. Cell. 1987 Jun 19;49(6):775–783. doi: 10.1016/0092-8674(87)90615-5. [DOI] [PubMed] [Google Scholar]
- Holmes K. L., Pierce J. H., Davidson W. F., Morse H. C., 3rd Murine hematopoietic cells with pre-B or pre-B/myeloid characteristics are generated by in vitro transformation with retroviruses containing fes, ras, abl, and src oncogenes. J Exp Med. 1986 Aug 1;164(2):443–457. doi: 10.1084/jem.164.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korman A. J., Frantz J. D., Strominger J. L., Mulligan R. C. Expression of human class II major histocompatibility complex antigens using retrovirus vectors. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2150–2154. doi: 10.1073/pnas.84.8.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg M., Siu G., Hood L. E., Shastri N. The molecular genetics of the T-cell antigen receptor and T-cell antigen recognition. Annu Rev Immunol. 1986;4:529–591. doi: 10.1146/annurev.iy.04.040186.002525. [DOI] [PubMed] [Google Scholar]
- Lewis S., Gifford A., Baltimore D. Joining of V kappa to J kappa gene segments in a retroviral vector introduced into lymphoid cells. 1984 Mar 29-Apr 4Nature. 308(5958):425–428. doi: 10.1038/308425a0. [DOI] [PubMed] [Google Scholar]
- Lieber M. R., Hesse J. E., Mizuuchi K., Gellert M. Developmental stage specificity of the lymphoid V(D)J recombination activity. Genes Dev. 1987 Oct;1(8):751–761. doi: 10.1101/gad.1.8.751. [DOI] [PubMed] [Google Scholar]
- Max E. E., Maizel J. V., Jr, Leder P. The nucleotide sequence of a 5.5-kilobase DNA segment containing the mouse kappa immunoglobulin J and C region genes. J Biol Chem. 1981 May 25;256(10):5116–5120. [PubMed] [Google Scholar]
- Nolan G. P., Fiering S., Nicolas J. F., Herzenberg L. A. Fluorescence-activated cell analysis and sorting of viable mammalian cells based on beta-D-galactosidase activity after transduction of Escherichia coli lacZ. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2603–2607. doi: 10.1073/pnas.85.8.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt D., Subramani S. Nucleotide sequence of the Escherichia coli xanthine-guanine phosphoribosyl transferase gene. Nucleic Acids Res. 1983 Dec 20;11(24):8817–8823. doi: 10.1093/nar/11.24.8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano H., Maki R., Kurosawa Y., Roeder W., Tonegawa S. Two types of somatic recombination are necessary for the generation of complete immunoglobulin heavy-chain genes. Nature. 1980 Aug 14;286(5774):676–683. doi: 10.1038/286676a0. [DOI] [PubMed] [Google Scholar]
- Schatz D. G., Baltimore D. Stable expression of immunoglobulin gene V(D)J recombinase activity by gene transfer into 3T3 fibroblasts. Cell. 1988 Apr 8;53(1):107–115. doi: 10.1016/0092-8674(88)90492-8. [DOI] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983 Apr 14;302(5909):575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Xu L., Yee J. K., Wolff J. A., Friedmann T. Factors affecting long-term stability of Moloney murine leukemia virus-based vectors. Virology. 1989 Aug;171(2):331–341. doi: 10.1016/0042-6822(89)90600-4. [DOI] [PubMed] [Google Scholar]
- Yancopoulos G. D., Blackwell T. K., Suh H., Hood L., Alt F. W. Introduced T cell receptor variable region gene segments recombine in pre-B cells: evidence that B and T cells use a common recombinase. Cell. 1986 Jan 31;44(2):251–259. doi: 10.1016/0092-8674(86)90759-2. [DOI] [PubMed] [Google Scholar]
- Zimmerman K. A., Yancopoulos G. D., Collum R. G., Smith R. K., Kohl N. E., Denis K. A., Nau M. M., Witte O. N., Toran-Allerand D., Gee C. E. Differential expression of myc family genes during murine development. 1986 Feb 27-Mar 5Nature. 319(6056):780–783. doi: 10.1038/319780a0. [DOI] [PubMed] [Google Scholar]




