Abstract
Genetic polymorphism in MECP2/IRAK1 on chromosome Xq28 is a confirmed and replicated susceptibility locus for lupus. High linkage disequilibrium in this locus suggests that both MECP2 and IRAK1 are candidate genes for the disease. DNA methylation changes in lupus T cells play a central role in the pathogenesis of lupus, and MeCp-2 (encoded by MECP2) is a master regulator of gene expression and is also known to recruit DNA methyltransferase 1 (DNMT1) during DNA synthesis. Using human T cells from normal individuals with either the lupus risk or the lupus protective haplotype in MECP2/IRAK1, we demonstrate that polymorphism in this locus increases MECP2 isoform 2 mRNA expression in stimulated but not unstimulated T cells. By assessing DNA methylation levels across over 485,000 methylation sites across the entire genome, we further demonstrate that the lupus risk variant in this locus is associated with significant DNA methylation changes, including in the HLA-DR and HLA-DQ loci, as well as interferon-related genes such as IFI6, IRF6, and BST2. Further, using a human MECP2 transgenic mouse, we show that overexpression of MECP2 alters gene expression in stimulated T cells. This includes overexpression of Eif2c2 that regulates the expression of multiple microRNAs (such as miR-21), and the histone demethylase Jhdm1d. In addition, we show that MECP2 transgenic mice develop antinuclear antibodies. Our data suggest that the lupus associated variant in the MECP2/IRAK1 locus has the potential to affect all 3 epigenetic mechanisms: DNA methylation, microRNA expression, and histone modification. Importantly, these data support the notion that variants within the MECP2 gene can alter DNA methylation in other genetic loci including the HLA and interferon-regulated genes, thereby providing evidence for genetic-epigenetic interaction in lupus.
Keywords: MECP2, IRAK1, lupus, epigenetics, polymorphism, DNA methylation, T cells, transgenic mouse
1. Introduction
The etiology of lupus is incompletely understood. However, both genetic and environmental factors are implicated in disease pathogenesis. Indeed, the current paradigm suggests that lupus develops in genetically susceptible individuals upon the exposure to disease-triggering environmental factors. While the genetic etiology of lupus has been supported by a number of confirmed genetic susceptibility loci for the disease, identification of environmental triggers for lupus has been more challenging [1]. Nonetheless, a number of environmental triggers implicated is disease pathogenesis, such as UV light exposure, procainamide, and hydralazine, have been shown to induce autoimmunity by altering T cell DNA methylation [2]. Indeed, abnormal DNA methylation in CD4+ T cells plays an important role in the pathogenesis of lupus [2–7], and DNA demethylation in CD4+ T cells is sufficient to cause lupus in mice [8]. Epigenetic regulation plays an important role in CD4+ T cell responses including differentiation into Th17 cells and regulatory T cells that are implicated in autoimmunity [9]. Importantly, a recent report in identical twins discordant for lupus identified significant DNA methylation changes between the lupus-affected and unaffected twin pairs, emphasizing the role for environmental exposures on inducing pathogenic epigenetic changes [6].
MeCp-2 (methyl-CpG-binding protein 2) is a transcriptional regulator intimately involved in regulating the expression of methylation sensitive genes. The binding of MeCp-2 to methylated CpG sites in gene promoter sequences recruits histone deacetylase complexes, leading to histone tail deacetylation, transcriptionally-repressive chromatin conformation, and silencing of gene expression [10]. MeCp-2 is also known to recruit DNA methyltransferase 1 (DNMT1) during DNA synthesis [11]. Furthermore, recent evidence suggests that MeCp-2 can act as a transcriptional activator via recruiting the transcription factor CREB1 [12], and that MeCp-2 also has a role in regulating RNA splicing [13].
We have previously established and confirmed the genetic association between SNPs located within the gene encoding for MeCp-2 (MECP2) and lupus [14, 15]. Genetic association between lupus and a neighboring gene, IRAK1, has been also established [16, 17]. The two genes, MECP2 and IRAK1 are in high linkage equilibrium, suggesting that lupus-associated variants in this locus might have functional consequences on either of the two genes or possibly both MECP2 and IRAK1 [14, 18].
Herein, we examine the functional effects of the MECP2/IRAK1 lupus-associated polymorphism and demonstrate increased expression of an MECP2 transcript variant in stimulated T cells from normal healthy individuals carrying the lupus risk haplotype in this locus. We further determine the effects of this lupus-associated haplotype on genome-wide DNA methylation in stimulated human T cells, and study the effect of increased MECP2 expression in T cells using a human MECP2 transgenic mouse.
2. Methods
2.1 Human T cell culture and activation
Primary T cells from healthy female European-American donors homozygous for the lupus risk or protective MECP2/IRAK1 haplotypes were studied. These haplotypes were tagged by the SNP rs17435 in this locus. For each sample, T cells were isolated from 1×107 fresh frozen peripheral blood mononuclear cells (PBMCs) obtained from the Oklahoma Immune Cohort collection through the Oklahoma Rheumatic Disease Resources Cores Center biorepository, by negative selection using the Human Pan T Cell Isolation Kit II (Miltenyi Biotec, Cambridge, MA). Briefly, PBMCs were thawed and rested overnight in complete T cell media (RPMI, 10% FBS, 1% penicillin-streptomycin solution and 50uM β-mercaptoethanol). The primary T cells were then isolated and were immediately re-plated with and without stimulation. For T cell stimulation, T cells were re-plated onto anti-CD3-coated (10ug/ml) 12-well plates with complete T cell media containing anti-CD28 (2.5ug/ml) and IL-2 (25U/ml) to begin activation. Cells were incubated at 37°C for time points 6, 12, and 24 hours, after which cells were collected and RNA or DNA isolated. RNA extraction was performed using a combination of TRIzol (Invitrogen, Carlsbad, CA) and RNeasy kits (Qiagen, Valencia, CA), in a hybrid protocol as previously described [15]. DNA was isolated by the DNeasy Kit (Qiagen, Valencia, CA) for subsequent use in the DNA methylation studies.
2.2 Real-Time PCR
Transcript levels of MECP2A (isoform 1), MECP2B (isoform 2), and DNMT1 were measured by real-time PCR using the iScript One Step RT-PCR Kit with SYBR green (Bio-Rad, Hercules, CA) and the Rotor-Gene 3000 Thermocycler (Corbett Research, Australia). Samples were quantified with a NanoDrop 1000 Spectrophotometer (Thermo Scientific, Wilmington, DE) after removing any contaminating genomic DNA using the Turbo DNA-Free Kit (Ambion, Austin, TX) according to manufacturer’s directions. All samples were normalized to the internal control housekeeping gene, ACTB (β-actin), in 20ul reaction volumes with 25ng RNA in each reaction. The PCR protocol was as follows: The following primers were used at 300nM concentrations: MECP2A (isoform 1) forward: 5′-CTGGGATGTTAGGGCTCAGGGA-3′, reverse: 5′-AGAGTGGTGGGCTGATGGCT-3′; MECP2B (isoform 2) forward: 5′-AGGCGAGGAGGAGAGACTGGAA-3′, reverse: 5′-AGAGTGGTGGGCTGATGGCT-3′; DNMT1 forward: 5′-CGACTACATCAAAGGCAGCAACCTG-3′, reverse: 5′-TGGAGTGGACTTGTGGGTGTTCTC-3′; and ACTB forward: 5′-GCACCACACCTTCTACAATGAGC-3′, reverse: 5′-GGATAGCACAGCCTGGATAGCAAC-3′. All of the primers were purchased from Integrated DNA Technologies (Coralville, IA). The PCR conditions used were as follows: 10 minutes at 50°C, 5 minutes at 95°C, and 45 cycles of 95°C for 10 seconds followed by 55°C for 30 seconds. Relative transcript levels were quantified using the 2− (delta delta CT) method [19].
2.3 DNA methylation analysis
Genome-wide DNA methylation was quantified in T cells from healthy individuals with the MECP2/IRAK1 risk and protective haplotypes 24 hours post stimulation using the Illumina Infinium HumanMethylation450 BeadChip platform (San Diego, CA), which allows for the assessment of the methylation status of over 485,000 methylation sites across the entire genome. This platform covers 99% of RefSeq genes, with an average of 17 CpG sites per gene distributed across the promoter, 5′-UTR, first exon, gene body, and 3′-UTR. Validation of the methylation array data was performed using bisulfite sequencing in known hypermethylated and hypomethylated loci as previously described [20].
2.4 MECP2 transgenic mice
Female mice transgenic for human MECP2 (FVB-Tg(MECP2)1Hzo/J), hereafter referred to as MECP2-Tg, and age- and sex-matched wild-type mice (FVB/NJ) were obtained from The Jackson Laboratories (Bar Harbor, Maine). At 8 weeks of age the mice were sacrificed and splenic CD4+ T cells were isolated using indirect labeling and magnetic bead separation (Miltenyi Biotec, Cambridge, MA). Serum samples were also collected at the same time. RNA was isolated from 5×106 purified CD4+ T cells immediately after cell separation. For stimulated CD4+ T cells, another 5×106 cells were stimulated over night with PMA (50ng/ml) and ionomycin (750ng/ml) and then RNA extracted. RNA extraction was performed using a combination of TRIzol (Invitrogen, Carlsbad, CA) and RNeasy kits (Qiagen, Valencia, CA) as previously described [15]. RNA concentration was determined with a NanoDrop 1000 Spectrophotometer (Thermo Scientific, Wilmington, DE) and then qualitatively assessed for degradation with 28:18S ribosomal RNA, using a capillary gel electrophoresis system (Agilent 2100 Bioanalyzer, Agilent, Wilmington, DE). Gene expression profiling was performed in stimulated and unstimulated CD4+ T cells from MECP2-Tg mice and wild-type controls using the MouseWG-6 v2 BeadChip arrays (Illumina).
Sera obtained from MECP2-Tg and wild-type mice at 8 weeks of age were tested for the presence of antinuclear antibody (ANA) and anti-dsDNA antibody using quantitative ELISA assays (Alpha Diagnostic, San Antonio, TX).
2.5 Statistical analysis
Microarray expression analysis was performed and corrected for multiple testing using Significance Analysis of Microarrays (SAM) [21]. Gene ontology, network, and pathway analysis was performed using Ingenuity Pathway Analysis (Redwood City, CA). A Student’s t-test was used in comparisons of continuous variables. DNA methylation analysis was performed using GenomeStudio (Illumina). Differentially methylated loci were defined as CpG loci with an average difference in DNA methylation level of at least 1.2-fold, and differential methylation score of ≥ |30|after adjusting for multiple testing using the Benjamini and Hochberg false discovery rate. Differential methylation score is defined as 10*sgn(βRisk-βProtective)*log10p. β is the average level of DNA methylation on each CpG site and ranges from 0 to 1 (0, completely demethylated; 1, completely methylated).
3. Results
3.1 Effect of the MECP2/IRAK1 lupus-associated haplotype on MECP2 and IRAK1 expression in human T cells
Since a DNA methylation defect in T cells has been previously shown to play an important role in the pathogenesis of lupus, we tested the effect of the lupus-associated MECP2/IRAK1 haplotype on MECP2 mRNA expression in primary T cells isolated from healthy donors. The expression levels of the two known MECP2 transcript variants (MECP2A and MECP2B) were measured in unstimulated and stimulated T cells from individuals homozygous for the risk haplotype (n= 14) and the non-risk haplotype (n= 20). Twenty four hours after stimulation with anti-CD3/anti-CD28 antibodies, the expression of MECP2B was significantly higher in T cells from individuals carrying the lupus-associated MECP2/IRAK1 risk haplotype (P=0.04). There was no difference in MECP2A expression in stimulated T cells, and no difference in the expression of either MECP2 transcript variants in unstimulated T cells (Figure 1).
Figure 1.
Relative mRNA expression of MECP2A (A) and MECP2B (B) in stimulated T cells (A and B, respectively), and unstimulated T cells (C and D, respectively) from healthy individuals with the lupus risk and the non-risk haplotype in the MECP2/IRAK1 locus. Expression is depicted relative to β-actin. NS, not significant.
Next, we examined if the MECP2/IRAK1 haplotype also affects IRAK1 expression in T cells similar to MECP2. We did not observe any difference in IRAK1 mRNA expression as influenced by the presence of the MECP2/IRAK1 lupus-associated haplotype in T cells (Unstimulated, risk= 1.56±0.23, non-risk= 1.36±0.16, P= 0.46; stimulated, risk= 0.87±0.14, non-risk= 0.60±0.093, P= 0.11).
3.2 Effect of the MECP2/IRAK1 lupus-associated haplotype on DNMT1 expression in human T cells
MeCp-2 (encoded by MECP2) represses gene expression primarily by recruiting the histone deacetylase complex to methylated DNA sequences. In addition, MeCp-2 recruits DNMT1 during DNA replication to ensure efficient replication of DNA methylation patterns during DNA synthesis. Further, the expression of Mecp2 has been previously shown to be increased in Dnmt1 knockout mice [22]. Therefore, we tested the expression of DNMT1 in the same normal T cells from individuals with the lupus risk and non-risk MECP2/IRAK1 haplotype but observed no differences in the mRNA levels between the two groups with or without T cell stimulation (P= 0.48 and 0.55, respectively).
3.3 Effect of the MECP2/IRAK1 lupus-associated haplotype on Genome-wide DNA methylation in human T cells
Since the expression of at least one MECP2 transcript variant was increased in stimulated T cells (24 hrs post stimulation) from individuals with the lupus-associated MECP2/IRAK1 haplotype, and MeCp-2 recruits DNMT1 outside of the replication fork, we tested the hypothesis that the lupus-risk haplotype could influence DNA methylation patterns. To do this, we examined global DNA methylation by quantifying the methylation status of over 485,000 methylation sites simultaneously in T cells 24 hours post stimulation from normal individuals with the risk and the non-risk MECP2/IRAK1 haplotype (n=6) using the Infinium HumanMethylation450 BeadChip arrays. Array validation was performed using the same samples by bisulfite sequencing in known hypermethylated and hypomethylated loci as described previously [20], and showed a correlation r2 value of 0.83 between the array data and bisulfite sequencing data.
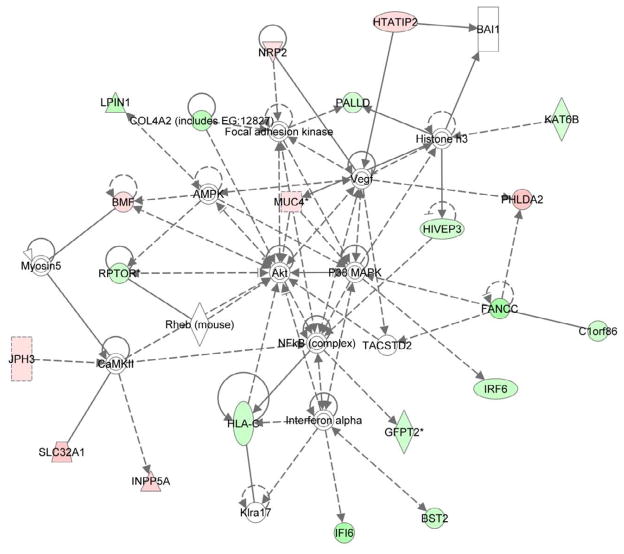
We did not observe a difference between the average global DNA methylation between the risk and protective haplotypes (risk= 0.5562± 0.0021, protective= 0.5576± 0.0022 [mean±SEM], P= 0.64). However, we identified 133 hypermethylated and 112 hypomethylated CpG sites in individuals with the risk compared to the protective MECP2/IRAK1 haplotype (fold difference ≥1.20). Interestingly, the most significant canonical pathways identified in these differentially methylated loci was the protein ubiquitination pathway (P= 9.16×10−3) and the antigen presentation pathway (P= 1.44×10−2). The MECP2/IRAK1 risk haplotype is associated with significant methylation changes in the HLA, and in particular HLA class II (Table 1). The BST2 gene, an interferon-inducible membrane-bound protein that helps restrict the release of retroviral particles, is hypomethylated in individuals with the risk MECP2/IRAK1 haplotype. Other interferon-related genes that are hypomethylated include IFI6 and IRF6. Indeed, network analysis of differentially methylated genes highlight interferon-alpha and NFkB (Figure 2). The complete list of differentially methylated CpG sites between stimulated T cells with and without the MECP2/IRAK1 lupus-associated haplotype are shown in Supplementary Table 1.
Table 1.
CpG sites in the HLA class II loci with differential DNA methylation in T cells between individuals with the lupus-risk and the protective haplotype in MECP2/IRAK
| Target CG ID | Location on Chr. 6 | Gene | Methylation (β) | Ratio | Differential methylation score | |
|---|---|---|---|---|---|---|
| Risk | Protective | |||||
| cg04418355 | 32729174 | HLA-DQB2 | 0.73 | 0.60 | 1.20 | 31.86 |
| cg00103771 | 32525805 | HLA-DRB6 | 0.80 | 0.42 | 1.93 | 340.34 |
| cg06559318 | 32526260 | HLA-DRB6 | 0.70 | 0.36 | 1.93 | 340.34 |
| cg11888470 | 32526021 | HLA-DRB6 | 0.83 | 0.53 | 1.55 | 340.34 |
| cg24638099 | 32526027 | HLA-DRB6 | 0.70 | 0.43 | 1.62 | 340.34 |
Differential methylation score = 10*sgn(βRisk-βProtective)*log10p, where β is the average level of DNA methylation on each CpG site and ranges from 0 to 1 (0, completely demethylated; 1, completely methylated).
Figure 2.
Network analysis of differentially methylated genes in stimulated T cells from normal healthy individuals with the lupus-risk compared to the protective MECP2/IRAK1 haplotype. Green and red colors represent hypomethylated and hypermethylated genes in the risk haplotype, respectively. Colorless genes and molecules are not differentially methylated but brought into the network by the analysis software to denote relationships with differentially methylated genes.
3.4 The effect of MECP2 overexpression in stimulated and unstimulated T cells and autoantibody production in a human MECP2 transgenic mouse
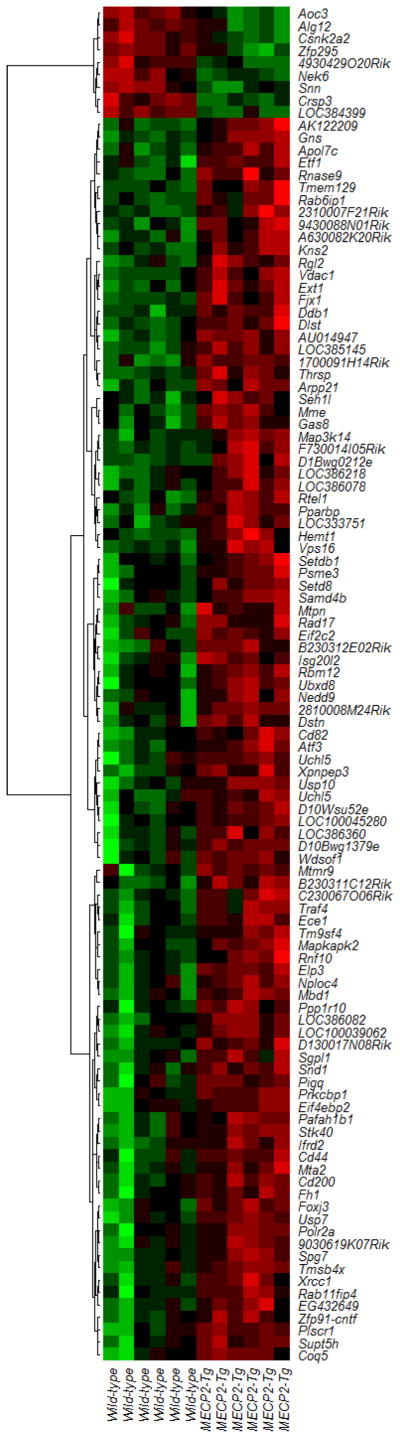
Since MeCp-2 is a transcriptional regulator, and the expression of MECP2 is higher in stimulated T cells from individuals with the lupus-associated risk variants, we studied the effect of MECP2 overexpression using a mouse transgenic for human MECP2 (MECP2-Tg). This mouse includes the entire human MECP2 gene and expresses MeCp-2 at approximately 2-fold wild-type levels [23]. Stimulated and unstimulated CD4+ T cells from the spleen were isolated and gene expression profiles determined using expression microarrays. In unstimulated T cells, we detected 4 overexpressed and 18 underexpressed transcripts in MECP2-Tg mice compared to wild-type controls. However, upon stimulation, we detected 100 significantly overexpressed and 9 underexpressed genes (Figure 3). Network analysis indicated the differentially expressed genes in these stimulated T cells were statistically overrepresented in several networks including “Gene Expression, Post-Translational Modification, Cellular Assembly and Organization”, “Protein Synthesis, Antigen Presentation, Cell-To-Cell Signaling and Interaction”, “Cell Cycle, Cellular Movement, Cell Death”, “Cell Death, Cell Morphology, Cellular Development”, and “Connective Tissue Disorders, Inflammatory Disease, Inflammatory Response”. Interestingly, Eif2c2 (eukaryotic translation initiation factor 2C,2) is among the upregulated genes in MECP2-Tg mice. This gene regulates the expression of a number of microRNAs, including miR-21 which is known to be overexpressed in lupus CD4+ T cells [24]. Several other translation associated factor genes were overexpressed in the human MECP2-Tg mouse including Eif4ebp2 (1.76-fold). Other overexpressed genes in stimulated T cells from the MECP2 transgenic include the histone demethylase Jhdm1d (also known as A630082K20Rik) (1.69-fold), and Cd44 (1.18-fold), a recently reported susceptibility gene for lupus [25]. The expression of Faf2 (Fas associated factor family member 2) is also increased in MECP2-Tg mice (1.38-fold). FAF2 is upregulated in T cells from patients with atopic dermatitis and is suspected to play a role in modulating T cell apoptosis [26]. Canonical pathway analysis highlighted several overlapping pathways including IL-6 signaling (P= 1.62×10−2), IL-17 signaling (P=4.3×10−2), PPARα/RXRα activation (P= 4.63×10−2), and CD40 signaling (P=3.38×10−2).
Figure 3.
A heat map depicting differentially expressed genes in stimulated CD4+ T cells from MECP2-Tg mice compared to controls (n=6). Red and green colors represent upregulated and downregulated transcripts, respectively.
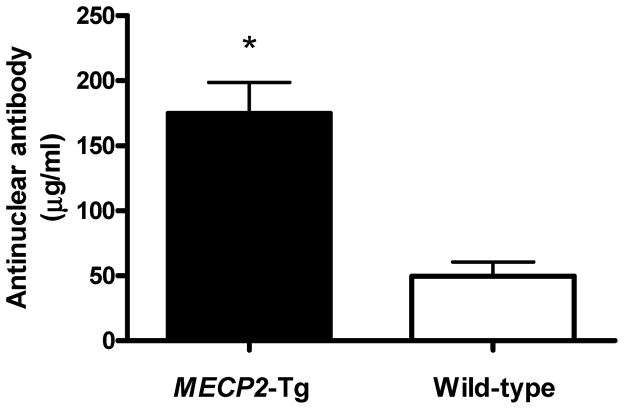
Because the expression of MECP2 is increased in T cells with the risk MECP2/IRAK1 polymorphism, and MECP2-Tg mice differentially express genes involved in inflammatory pathways, we hypothesized that overexpression of MECP2 might result in autoantibody production. Indeed, we detected elevated antinuclear antibody production in the sera obtained from the MECP2-Tg mouse compared to wild-type controls (Figure 4). However, no anti-dsDNA production was detected by ELISA (data not shown).
Figure 4.
Anti-nuclear antibody levels in the serum of mice transgenic for human MECP2 compared to wild-type age- and sex-matched controls (174.7 ± 23.92 versus 49.59 ± 10.87 ug/ml (mean±SD), n=6, P= 0.0008)
4. Discussion
The genetic locus containing MECP2 is a confirmed susceptibility locus for systemic lupus erythematosus, though the causal variant(s) in this locus has not yet been identified. Because T cell DNA methylation defects play an important role in the pathogenesis of lupus, and MeCp-2 is a key regulator for the expression of methylation sensitive genes, we performed studies to characterize the effect of the lupus-associated MECP2/IRAK1 haplotype using human T cells homozygous for the risk and protective haplotypes in this locus. We demonstrated increased expression of one MECP2 transcript variant in stimulated but not unstimulated T cells from normal healthy individuals carrying the lupus-associated haplotype.
Next we performed a genome-wide DNA methylation study in a subset of the same stimulated human T cells to determine if the lupus-associated MECP2/IRAK1 haplotype and the observed difference in MECP2B mRNA expression were associated with DNA methylation changes. Indeed, these data suggest significant wide-spread locus-specific DNA methylation changes, though global DNA methylation levels were not different. We detected significant DNA methylation differences in the HLA region and in particular involving CpG sites in the HLA-class II loci including HLA-DR, and HLA-DQ (Table 1). These data suggest that the lupus-associated MECP2/IRAK1 haplotype is associated with DNA methylation changes which, in turn, might influence other disease-relevant pathways and other susceptibility loci outside of the MECP2/IRAK1 locus. Indeed, HLA-DR contains one of the most robustly associated genetic susceptibility loci for lupus, and both HLA-DR and HLA-DQ are known susceptibility loci for primary Sjögren’s syndrome [27]. We have previously reported the genetic association between MECP2 and primary Sjögren’s syndrome [28]. Therefore, the genetic-epigenetic interaction suggested by our findings can extend beyond lupus and might also play a role in the pathogenesis or genetic susceptibility of other autoimmune diseases. Whether there is also a genetic epistasis effect between the lupus susceptibility loci in the HLA region and MECP2 remains to be determined.
Our network analysis of differentially methylated genes associated with the MECP2/IRAK1 haplotype highlights NFkB and interferon alpha (Figure 2). Several interferon-regulated genes such as BST2, IFI6, and IRF6 were hypomethylated in the presence of the lupus-associated MECP2 variants. We have previously shown that BST2 is hypomethylated in CD4+ T cells from lupus patients compared to controls [20]. This interferon-regulated gene (also known as Tetherin) restricts the release of viral particles, such as HIV-1, by tethering them to the cell surface [29]. We previously suggested that hypomethylation and overexpression of BST2 in lupus CD4+ T cells might provide a plausible mechanistic explanation for the reported lower HIV-1 infection in lupus patients [30]. Indeed, our data suggest that the MECP2 variants associated with increased risk for lupus might be at the same time protective against retroviral infections such as HIV-1, as indicated by hypomethylation of BST2. This phenomenon has been previously reported with other genetic variants, with the classical example being the sickle cell mutation that is protective against malaria [31].
To further examine the extent to which MECP2 upregulation affects T cell function, we used a transgenic mouse that expresses human MECP2. Gene expression profiles in stimulated CD4+ T cells identified significant gene expression differences in the MECP2-Tg mouse compared to wild-type controls. Importantly, these studies found overexpression of Eif2c2 in MECP2-Tg mice. Eif2c2 regulates the expression of miR-21, which has been reported to be overexpressed in lupus CD4+ T cells, and resulting in reduced DNMT1 expression, as well as hypomethylation and overexpression of methylation sensitive genes in CD4+ T cells [24]. Further, the histone demethylase Jhdm1d is ~ 2 times overexpressed in stimulated T cells from MECP2-Tg mice compared to controls, suggesting a possible direct effect of increased MECP2 expression on histone tail modification in T cells.
Our studies suggest that the disease-associated variants in MECP2 could influence all three mechanisms of epigenetic regulation; DNA methylation, histone modification, and microRNA expression, which adds to the functional pathogenic complexity of the lupus-associated variants in this locus.
5. Conclusions
sMECP2 was previously implicated as a susceptibility gene for lupus, yet how it might affect disease etiology was not known. In this study, we identified specific functional consequences of the MECP2 polymorphism that extend beyond the genetic locus containing MECP2 and IRAK1. We found that the risk variant of MECP2 correlates with higher relative expression of isoform 2 (MECP2B) in stimulated T cells. We show evidence for genetic-epigenetic interaction associated with the lupus-associated MECP2 variants, suggested by differential DNA methylation in multiple genetic loci including class II HLA genes. Future studies of disease-associated polymorphism in MECP2 are warranted, both to determine the functional effects in other cell types and to identify the causal variants.
Supplementary Material
Research Highlights.
The lupus variants in MECP2/IRAK1 increase MECP2 expression in stimulated T cells
These variants alter DNA methylation in the HLA and interferon-related genes
MeCP2 overexpression in mice alters gene expression in stimulated T cells.
MeCP2 overexpression is associated with antinuclear antibody production in mice
Acknowledgments
This work was supported by a grant from the Lupus Research Institute, and NIH grants P30 AR053483, P30 RR031152 and P20 GM103456
Footnotes
Financial conflict of interest: None of the authors has any financial conflict of interest to report.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Miller FW, Alfredsson L, Costenbader KH, Kamen DL, Nelson LM, Norris JM, et al. Epidemiology of environmental exposures and human autoimmune diseases: Findings from a National Institute of Environmental Health Sciences Expert Panel Workshop. J Autoimmun. 2012;39:259–71. doi: 10.1016/j.jaut.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jeffries MA, Sawalha AH. Epigenetics in systemic lupus erythematosus: leading the way for specific therapeutic agents. Int J Clin Rheumtol. 2011;6:423–39. doi: 10.2217/ijr.11.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sawalha AH, Jeffries M. Defective DNA methylation and CD70 overexpression in CD4+ T cells in MRL/lpr lupus-prone mice. Eur J Immunol. 2007;37:1407–13. doi: 10.1002/eji.200636872. [DOI] [PubMed] [Google Scholar]
- 4.Sawalha AH, Jeffries M, Webb R, Lu Q, Gorelik G, Ray D, et al. Defective T-cell ERK signaling induces interferon-regulated gene expression and overexpression of methylation-sensitive genes similar to lupus patients. Genes Immun. 2008;9:368–78. doi: 10.1038/gene.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yung RL, Quddus J, Chrisp CE, Johnson KJ, Richardson BC. Mechanism of drug-induced lupus. I. Cloned Th2 cells modified with DNA methylation inhibitors in vitro cause autoimmunity in vivo. J Immunol. 1995;154:3025–35. [PubMed] [Google Scholar]
- 6.Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20:170–9. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hughes T, Sawalha AH. The role of epigenetic variation in the pathogenesis of systemic lupus erythematosus. Arthritis Res Ther. 2011;13:245. doi: 10.1186/ar3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richardson B, Ray D, Yung R. Murine models of lupus induced by hypomethylated T cells. Methods Mol Med. 2004;102:285–94. doi: 10.1385/1-59259-805-6:285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ngalamika O, Zhang Y, Yin H, Zhao M, Gershwin ME, Lu Q. Epigenetics, autoimmunity and hematologic malignancies: A comprehensive review. J Autoimmun. 2012;39:451–65. doi: 10.1016/j.jaut.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–91. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 11.Kimura H, Shiota K. Methyl-CpG-binding protein, MeCP2, is a target molecule for maintenance DNA methyltransferase, Dnmt1. J Biol Chem. 2003;278:4806–12. doi: 10.1074/jbc.M209923200. [DOI] [PubMed] [Google Scholar]
- 12.Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–9. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young JI, Hong EP, Castle JC, Crespo-Barreto J, Bowman AB, Rose MF, et al. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc Natl Acad Sci U S A. 2005;102:17551–8. doi: 10.1073/pnas.0507856102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawalha AH, Webb R, Han S, Kelly JA, Kaufman KM, Kimberly RP, et al. Common variants within MECP2 confer risk of systemic lupus erythematosus. PloS One. 2008;3:e1727. doi: 10.1371/journal.pone.0001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Webb R, Wren JD, Jeffries M, Kelly JA, Kaufman KM, Tang Y, et al. Variants within MECP2, a key transcription regulator, are associated with increased susceptibility to lupus and differential gene expression in patients with systemic lupus erythematosus. Arthritis Rheum. 2009;60:1076–84. doi: 10.1002/art.24360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacob CO, Zhu J, Armstrong DL, Yan M, Han J, Zhou XJ, et al. Identification of IRAK1 as a risk gene with critical role in the pathogenesis of systemic lupus erythematosus. Proc Natl Acad Sci U S A. 2009;106:6256–61. doi: 10.1073/pnas.0901181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacob CO, Reiff A, Armstrong DL, Myones BL, Silverman E, Klein-Gitelman M, et al. Identification of novel susceptibility genes in childhood-onset systemic lupus erythematosus using a uniquely designed candidate gene pathway platform. Arthritis Rheum. 2007;56:4164–73. doi: 10.1002/art.23060. [DOI] [PubMed] [Google Scholar]
- 18.Sawalha AH. Xq28 and lupus: IRAK1 or MECP2? Proc Natl Acad Sci U S A. 2009;106:E62. doi: 10.1073/pnas.0904068106. author reply E3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Jeffries MA, Dozmorov M, Tang Y, Merrill JT, Wren JD, Sawalha AH. Genome-wide DNA methylation patterns in CD4+ T cells from patients with systemic lupus erythematosus. Epigenetics. 2011;6:593–601. doi: 10.4161/epi.6.5.15374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ray D, Wu A, Wilkinson JE, Murphy HS, Lu Q, Kluve-Beckerman B, et al. Aging in heterozygous Dnmt1-deficient mice: effects on survival, the DNA methylation genes, and the development of amyloidosis. J Gerontol A Biol Sci Med Sci. 2006;61:115–24. doi: 10.1093/gerona/61.2.115. [DOI] [PubMed] [Google Scholar]
- 23.Collins AL, Levenson JM, Vilaythong AP, Richman R, Armstrong DL, Noebels JL, et al. Mild overexpression of MeCP2 causes a progressive neurological disorder in mice. Hum Mol Genet. 2004;13:2679–89. doi: 10.1093/hmg/ddh282. [DOI] [PubMed] [Google Scholar]
- 24.Pan W, Zhu S, Yuan M, Cui H, Wang L, Luo X, et al. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J Immunol. 2010;184:6773–81. doi: 10.4049/jimmunol.0904060. [DOI] [PubMed] [Google Scholar]
- 25.Lessard CJ, Adrianto I, Kelly JA, Kaufman KM, Grundahl KM, Adler A, et al. Identification of a systemic lupus erythematosus susceptibility locus at 11p13 between PDHX and CD44 in a multiethnic study. Am J Hum Genet. 2011;88:83–91. doi: 10.1016/j.ajhg.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imai Y, Nakada A, Hashida R, Sugita Y, Tanaka T, Tsujimoto G, et al. Cloning and characterization of the highly expressed ETEA gene from blood cells of atopic dermatitis patients. Biochem Biophys Res Commun. 2002;297:1282–90. doi: 10.1016/s0006-291x(02)02380-x. [DOI] [PubMed] [Google Scholar]
- 27.Sawalha AH, Potts R, Schmid WR, Scofield RH, Harley JB. The genetics of primary Sjogren’s syndrome. Curr Rheumatol Rep. 2003;5:324–32. doi: 10.1007/s11926-003-0012-x. [DOI] [PubMed] [Google Scholar]
- 28.Cobb BL, Fei Y, Jonsson R, Bolstad AI, Brun JG, Rischmueller M, et al. Genetic association between methyl-CpG binding protein 2 (MECP2) and primary Sjogren’s syndrome. Ann Rheum Dis. 2010;69:1731–2. doi: 10.1136/ard.2009.122903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuhl BD, Sloan RD, Donahue DA, Bar-Magen T, Liang C, Wainberg MA. Tetherin restricts direct cell-to-cell infection of HIV-1. Retrovirology. 2010;7:115. doi: 10.1186/1742-4690-7-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burton J, Vera JH, Kapembwa M. HIV and systemic lupus erythematosus: the clinical and diagnostic dilemmas of having dual diagnosis. Int J STD AIDS. 2010;21:845–6. doi: 10.1258/ijsa.2010.010062. [DOI] [PubMed] [Google Scholar]
- 31.Rosenthal PJ. Lessons from sickle cell disease in the treatment and control of malaria. N Engl J Med. 2011;364:2549–51. doi: 10.1056/NEJMcibr1105118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.