Abstract
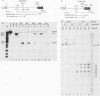
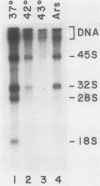
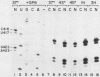
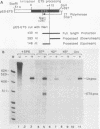
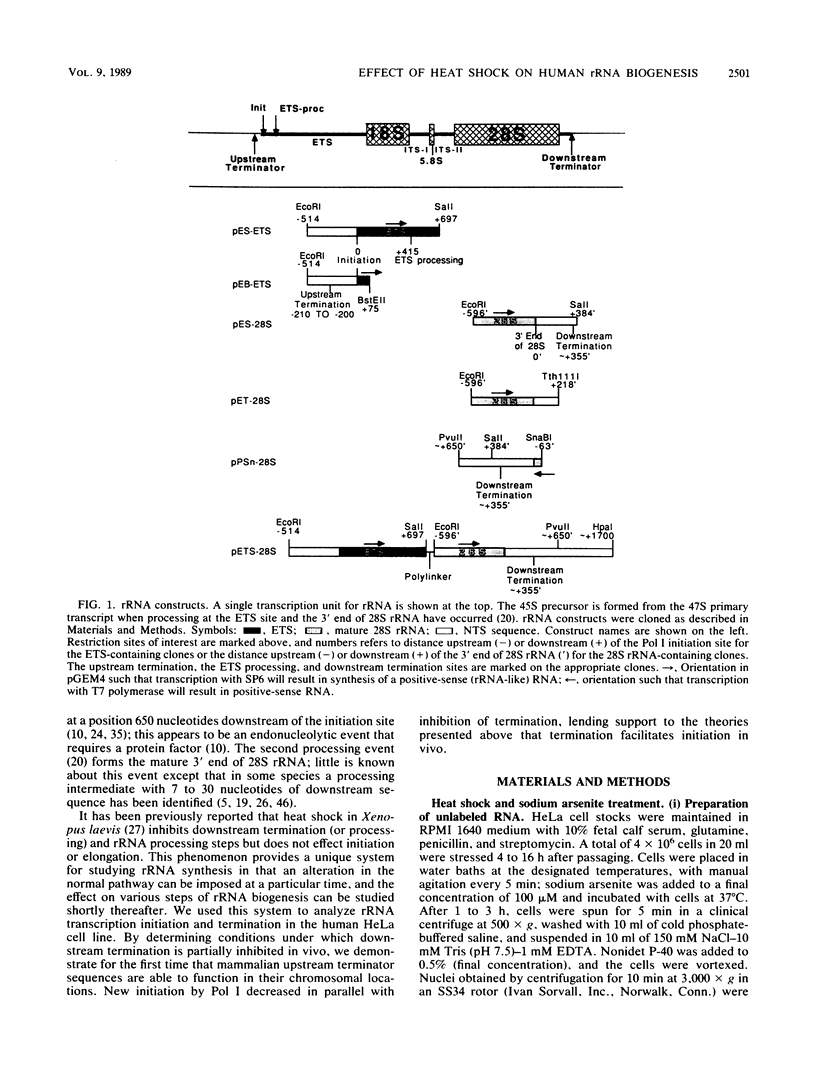
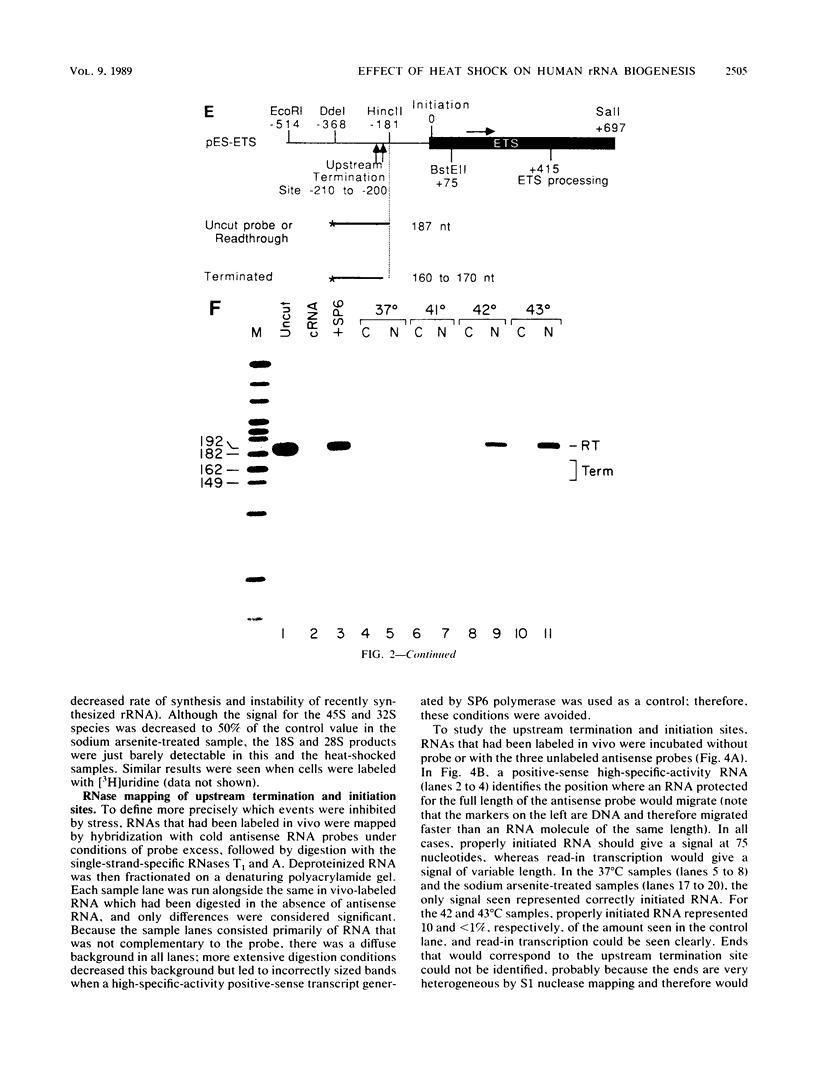
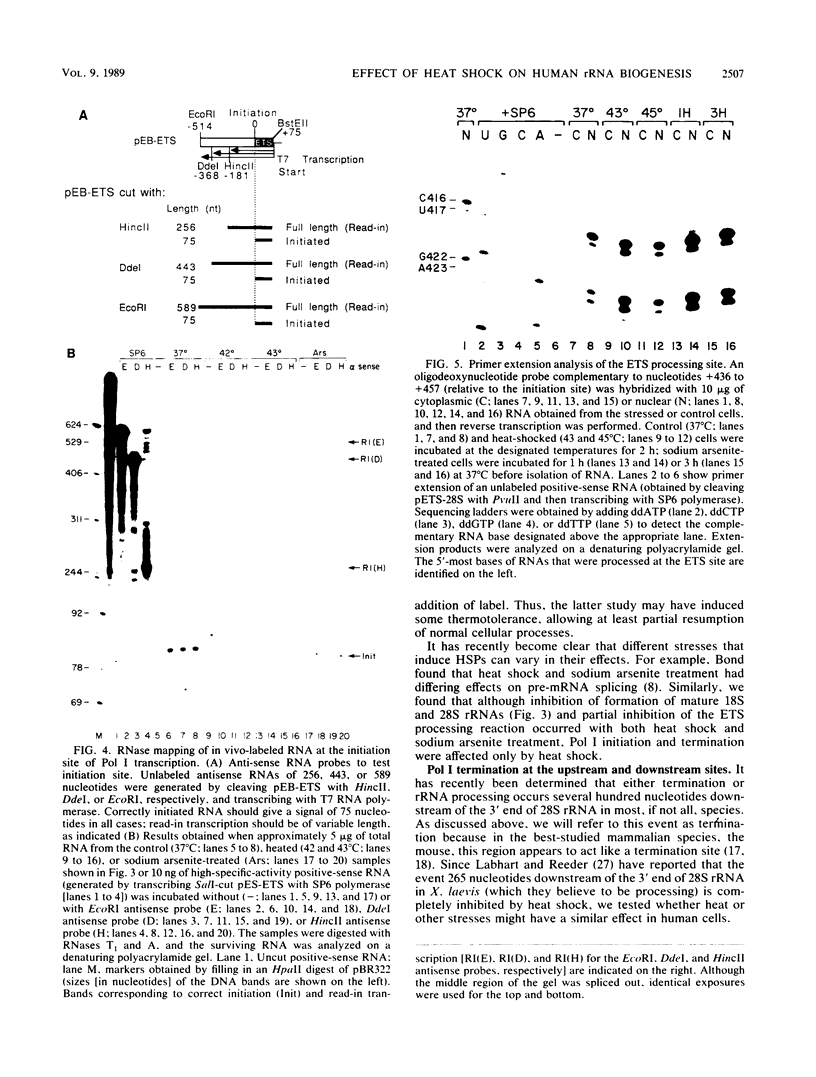
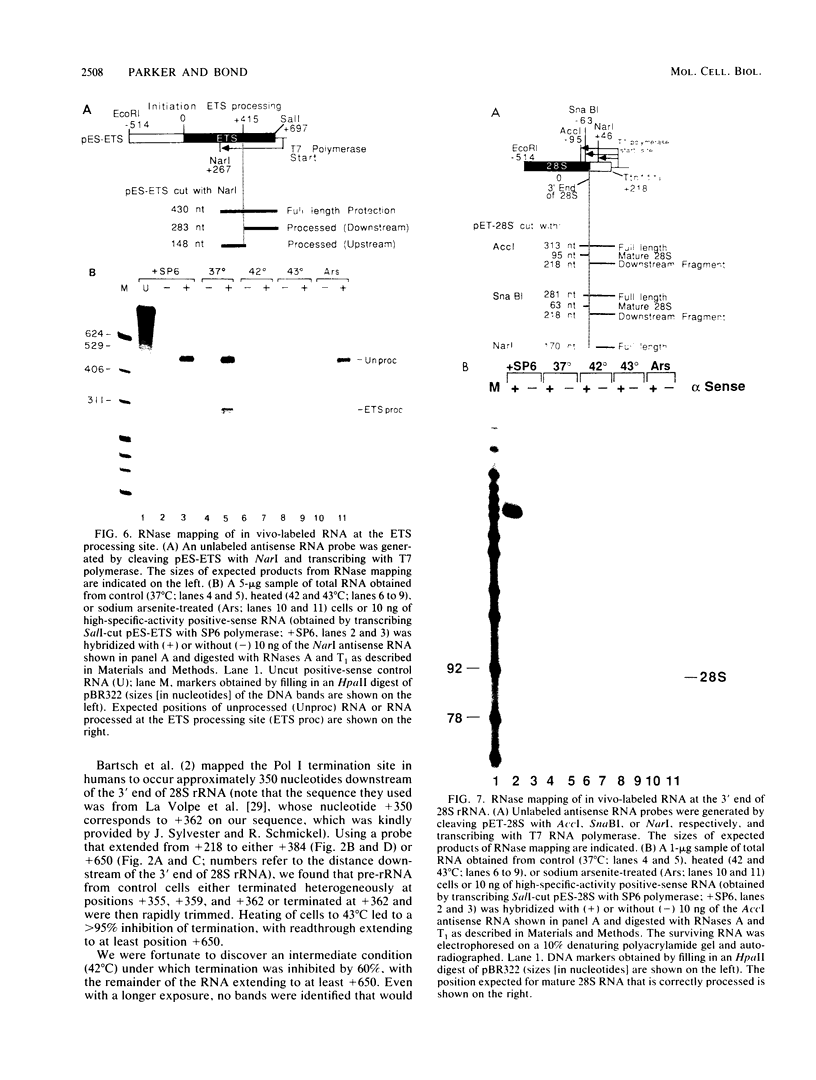
Human rRNA precursor from normal or stressed HeLa cells were studied by S1 nuclease mapping of unlabeled RNA and by antisense RNase mapping of RNA from cells that had been labeled in vivo with [32P]PO4. Heating cells to 43 degrees C decreased the amount of newly synthesized rRNA to less than 5% of the control level and led to greater than 95% inhibition of transcription termination at a region 355 to 362 nucleotides downstream of the 3' end of 28S rRNA, with readthrough continuing into the next transcription unit. Heating of cells to 42 degrees C led to 60% inhibition of termination at this site; 50% of transcripts that extended into the nontranscribed spacer ended in a region 200 to 210 nucleotides upstream of the polymerase I (Pol I) initiation site. This is presumed to be the human upstream transcription termination site because of the absence of RNAs with a 5' end corresponding to this region, the location relative to the Pol I initiation site (which is similar to the location of upstream terminators in other species), and the fact that it is 15 to 25 nucleotides upstream of the sequence GGGTTGACC, which has an 8-of-9 base identity with the sequence 3' of the downstream termination site. Surprisingly, treatment of cells with sodium arsenite, which also leads to the induction of a stress response, did not inhibit termination. Pol I initiation was decreased to the same extent as termination, which lends support to the hypothesis that termination and initiation are coupled. Although termination was almost completely inhibited at 43 degrees C, the majority of the recently synthesized rRNAs were processed to have the correct 3' end of 28S. This finding suggests that 3'-end formation can involve an endonucleolytic cut and is not solely dependent on exonucleolytic trimming of correctly terminated rRNAs.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker S. M., Platt T. Pol I transcription: which comes first, the end or the beginning? Cell. 1986 Dec 26;47(6):839–840. doi: 10.1016/0092-8674(86)90795-6. [DOI] [PubMed] [Google Scholar]
- Bartsch I., Schoneberg C., Grummt I. Evolutionary changes of sequences and factors that direct transcription termination of human and mouse ribsomal genes. Mol Cell Biol. 1987 Jul;7(7):2521–2529. doi: 10.1128/mcb.7.7.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch I., Schoneberg C., Grummt I. Purification and characterization of TTFI, a factor that mediates termination of mouse ribosomal DNA transcription. Mol Cell Biol. 1988 Sep;8(9):3891–3897. doi: 10.1128/mcb.8.9.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman E., Paule M. R. Promoter occlusion during ribosomal RNA transcription. Cell. 1988 Sep 23;54(7):985–992. doi: 10.1016/0092-8674(88)90113-4. [DOI] [PubMed] [Google Scholar]
- Bayev A., Georgiev O. I., Hadjiolov A. A., Nikolaev N., Skryabin K. G., Zakharyev V. M. The structure of the yeast ribosomal RNA genes. 3. Precise mapping of the 18 S and 25 S rRNA genes and structure of the adjacent regions. Nucleic Acids Res. 1981 Feb 25;9(4):789–799. doi: 10.1093/nar/9.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell J., Neilson L., Pellegrini M. Effect of heat shock on ribosome synthesis in Drosophila melanogaster. Mol Cell Biol. 1988 Jan;8(1):91–95. doi: 10.1128/mcb.8.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Bond U. Heat shock but not other stress inducers leads to the disruption of a sub-set of snRNPs and inhibition of in vitro splicing in HeLa cells. EMBO J. 1988 Nov;7(11):3509–3518. doi: 10.1002/j.1460-2075.1988.tb03227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirico W. J., Waters M. G., Blobel G. 70K heat shock related proteins stimulate protein translocation into microsomes. Nature. 1988 Apr 28;332(6167):805–810. doi: 10.1038/332805a0. [DOI] [PubMed] [Google Scholar]
- Craig N., Kass S., Sollner-Webb B. Nucleotide sequence determining the first cleavage site in the processing of mouse precursor rRNA. Proc Natl Acad Sci U S A. 1987 Feb;84(3):629–633. doi: 10.1073/pnas.84.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Winter R. F., Moss T. The ribosomal spacer in Xenopus laevis is transcribed as part of the primary ribosomal RNA. Nucleic Acids Res. 1986 Aug 11;14(15):6041–6051. doi: 10.1093/nar/14.15.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R. J., Koch B. D., Werner-Washburne M., Craig E. A., Schekman R. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature. 1988 Apr 28;332(6167):800–805. doi: 10.1038/332800a0. [DOI] [PubMed] [Google Scholar]
- Ellgaard E. G., Clever U. RNA metabolism during puff induction in Drosophila melanogaster. Chromosoma. 1971;36(1):60–78. doi: 10.1007/BF00326422. [DOI] [PubMed] [Google Scholar]
- Erickson J. M., Rushford C. L., Dorney D. J., Wilson G. N., Schmickel R. D. Structure and variation of human ribosomal DNA: molecular analysis of cloned fragments. Gene. 1981 Dec;16(1-3):1–9. doi: 10.1016/0378-1119(81)90055-x. [DOI] [PubMed] [Google Scholar]
- Financsek I., Mizumoto K., Mishima Y., Muramatsu M. Human ribosomal RNA gene: nucleotide sequence of the transcription initiation region and comparison of three mammalian genes. Proc Natl Acad Sci U S A. 1982 May;79(10):3092–3096. doi: 10.1073/pnas.79.10.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I., Kuhn A., Bartsch I., Rosenbauer H. A transcription terminator located upstream of the mouse rDNA initiation site affects rRNA synthesis. Cell. 1986 Dec 26;47(6):901–911. doi: 10.1016/0092-8674(86)90805-6. [DOI] [PubMed] [Google Scholar]
- Grummt I., Maier U., Ohrlein A., Hassouna N., Bachellerie J. P. Transcription of mouse rDNA terminates downstream of the 3' end of 28S RNA and involves interaction of factors with repeated sequences in the 3' spacer. Cell. 1985 Dec;43(3 Pt 2):801–810. doi: 10.1016/0092-8674(85)90253-3. [DOI] [PubMed] [Google Scholar]
- Grummt I., Rosenbauer H., Niedermeyer I., Maier U., Ohrlein A. A repeated 18 bp sequence motif in the mouse rDNA spacer mediates binding of a nuclear factor and transcription termination. Cell. 1986 Jun 20;45(6):837–846. doi: 10.1016/0092-8674(86)90558-1. [DOI] [PubMed] [Google Scholar]
- Grummt I., Sorbaz H., Hofmann A., Roth E. Spacer sequences downstream of the 28S RNA coding region are part of the mouse rDNA transcription unit. Nucleic Acids Res. 1985 Apr 11;13(7):2293–2304. doi: 10.1093/nar/13.7.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney T., Jr Characterization of mouse 45S ribosomal RNA subspecies suggests that the first processing cleavage occurs 600 +/- 100 nucleotides from the 5' end and the second 500 +/- 100 nucleotides from the 3' end of a 13.9 kb precursor. Nucleic Acids Res. 1985 Jul 11;13(13):4905–4919. doi: 10.1093/nar/13.13.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S., Sollner-Webb B. A transcriptional terminator is a novel element of the promoter of the mouse ribosomal RNA gene. Cell. 1986 Dec 26;47(6):891–900. doi: 10.1016/0092-8674(86)90804-4. [DOI] [PubMed] [Google Scholar]
- Henle K. J., Dethlefsen L. A. Time-temperature relationships for heat-induced killing of mammalian cells. Ann N Y Acad Sci. 1980;335:234–253. doi: 10.1111/j.1749-6632.1980.tb50752.x. [DOI] [PubMed] [Google Scholar]
- Kass S., Craig N., Sollner-Webb B. Primary processing of mammalian rRNA involves two adjacent cleavages and is not species specific. Mol Cell Biol. 1987 Aug;7(8):2891–2898. doi: 10.1128/mcb.7.8.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempers-Veenstra A. E., Oliemans J., Offenberg H., Dekker A. F., Piper P. W., Planta R. J., Klootwijk J. 3'-End formation of transcripts from the yeast rRNA operon. EMBO J. 1986 Oct;5(10):2703–2710. doi: 10.1002/j.1460-2075.1986.tb04554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominami R., Mishima Y., Urano Y., Sakai M., Muramatsu M. Cloning and determination of the transcription termination site of ribosomal RNA gene of the mouse. Nucleic Acids Res. 1982 Mar 25;10(6):1963–1979. doi: 10.1093/nar/10.6.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Volpe A., Simeone A., D'Esposito M., Scotto L., Fidanza V., de Falco A., Boncinelli E. Molecular analysis of the heterogeneity region of the human ribosomal spacer. J Mol Biol. 1985 May 25;183(2):213–223. doi: 10.1016/0022-2836(85)90214-1. [DOI] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Characterization of three sites of RNA 3' end formation in the Xenopus ribosomal gene spacer. Cell. 1986 May 9;45(3):431–443. doi: 10.1016/0092-8674(86)90329-6. [DOI] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Heat shock stabilizes highly unstable transcripts of the Xenopus ribosomal gene spacer. Proc Natl Acad Sci U S A. 1987 Jan;84(1):56–60. doi: 10.1073/pnas.84.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McStay B., Reeder R. H. A termination site for Xenopus RNA polymerase I also acts as an element of an adjacent promoter. Cell. 1986 Dec 26;47(6):913–920. doi: 10.1016/0092-8674(86)90806-8. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesfeld R., Arnheim N. Identification of the in vivo and in vitro origin of transcription in human rDNA. Nucleic Acids Res. 1982 Jul 10;10(13):3933–3949. doi: 10.1093/nar/10.13.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. G., Sollner-Webb B. Transcription of mouse rRNA genes by RNA polymerase I: in vitro and in vivo initiation and processing sites. Cell. 1981 Nov;27(1 Pt 2):165–174. doi: 10.1016/0092-8674(81)90370-6. [DOI] [PubMed] [Google Scholar]
- Mitchelson K., Moss T. The enhancement of ribosomal transcription by the recycling of RNA polymerase I. Nucleic Acids Res. 1987 Nov 25;15(22):9577–9596. doi: 10.1093/nar/15.22.9577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T. A transcriptional function for the repetitive ribosomal spacer in Xenopus laevis. Nature. 1983 Mar 17;302(5905):223–228. doi: 10.1038/302223a0. [DOI] [PubMed] [Google Scholar]
- Parker K. A., Bruzik J. P., Steitz J. A. An in vitro interaction between the human U3 snRNP and 28S rRNA sequences near the alpha-sarcin site. Nucleic Acids Res. 1988 Nov 25;16(22):10493–10509. doi: 10.1093/nar/16.22.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker K. A., Steitz J. A. Structural analysis of the human U3 ribonucleoprotein particle reveal a conserved sequence available for base pairing with pre-rRNA. Mol Cell Biol. 1987 Aug;7(8):2899–2913. doi: 10.1128/mcb.7.8.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham H. R. Hsp70 accelerates the recovery of nucleolar morphology after heat shock. EMBO J. 1984 Dec 20;3(13):3095–3100. doi: 10.1002/j.1460-2075.1984.tb02264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham H. R. Speculations on the functions of the major heat shock and glucose-regulated proteins. Cell. 1986 Sep 26;46(7):959–961. doi: 10.1016/0092-8674(86)90693-8. [DOI] [PubMed] [Google Scholar]
- Sadis S., Hickey E., Weber L. A. Effect of heat shock on RNA metabolism in HeLa cells. J Cell Physiol. 1988 Jun;135(3):377–386. doi: 10.1002/jcp.1041350304. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Tower J. Transcription of cloned eukaryotic ribosomal RNA genes. Annu Rev Biochem. 1986;55:801–830. doi: 10.1146/annurev.bi.55.070186.004101. [DOI] [PubMed] [Google Scholar]
- Tautz D., Dover G. A. Transcription of the tandem array of ribosomal DNA in Drosophila melanogaster does not terminate at any fixed point. EMBO J. 1986 Jun;5(6):1267–1273. doi: 10.1002/j.1460-2075.1986.tb04356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., de Jonge P., Leer R. J., Planta R. J. The transcription termination site of the ribosomal RNA operon in yeast. Nucleic Acids Res. 1980 Nov 25;8(22):5179–5192. doi: 10.1093/nar/8.22.5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warocquier R., Scherrer K. RNA metabolism in mammalian cells at elevated temperature. Eur J Biochem. 1969 Sep;10(2):362–370. doi: 10.1111/j.1432-1033.1969.tb00699.x. [DOI] [PubMed] [Google Scholar]
- Welch W. J., Feramisco J. R. Nuclear and nucleolar localization of the 72,000-dalton heat shock protein in heat-shocked mammalian cells. J Biol Chem. 1984 Apr 10;259(7):4501–4513. [PubMed] [Google Scholar]