Abstract
Blasticidin S is a peptidyl nucleoside antibiotic produced by Streptomyces griseochromogenes that exhibits strong fungicidal activity. To circumvent an effective DNA uptake barrier system in the native producer and investigate its biosynthesis in vivo, the blasticidin S biosynthetic gene cluster (bls) was engrafted to the chromosome of Streptomyces lividans. However, the resulting mutant, LL2, produced the inactive deaminohydroxyblasticidin S instead of blasticidin S. Subsequently, a blasticidin S deaminase (SLBSD, for S. lividans blasticidin S deaminase) was identified in S. lividans and shown to govern this in vivo conversion. Purified SLBSD was found to be capable of transforming blasticidin S to deaminohydroxyblasticidin S in vitro. It also catalyzed deamination of the cytosine moiety of cytosylglucuronic acid, an intermediate in blasticidin S biosynthesis. Disruption of the SLBSD gene in S. lividans LL2 led to successful production of active blasticidin S in the resultant mutant, S. lividans WJ2. To demonstrate the easy manipulation of the blasticidin S biosynthetic gene cluster, blsE, blsF, and blsL, encoding a predicted radical S-adenosylmethionine (SAM) protein, an unknown protein, and a guanidino methyltransferase, were individually inactivated to access their role in blasticidin S biosynthesis.
INTRODUCTION
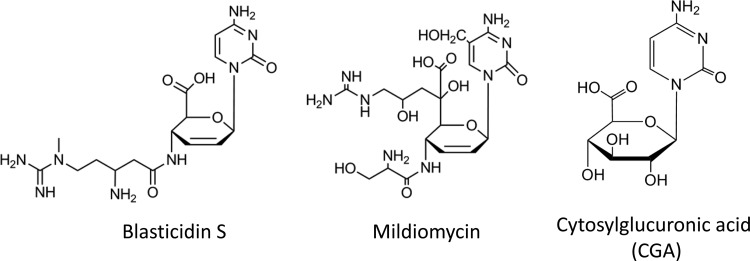
Blasticidin S (Fig. 1) belongs to the peptidyl nucleoside family of antibiotics. This compound, which is produced by Streptomyces griseochromogenes, was first identified in 1958 (1) and was the first antibiotic used to control rice blast in eastern Asia (2). The cytidine core of blasticidin S can bind tightly to the corresponding guanine base at the P site of the 50S subunit of the ribosome through a Watson-Crick base pair (3), which makes it a potent inhibitor of protein synthesis in both prokaryotic and eukaryotic cells (4, 5). There have been three kinds of blasticidin S resistance genes reported to date: BSD from Aspergillus terreus (6), bsr from Bacillus cereus (7), and a blasticidin S acetyltransferase gene from Streptoverticillium sp. (8). Both BSD and bsr encode blasticidin S deaminase (BSD), which can convert blasticidin S to the nontoxic deaminohydroxyblasticidin S, the base moiety of which is converted to uridine, thereby losing the capability of binding to the ribosome. On the other hand, the blasticidin S acetyltransferase gene encodes an acetyltransferase which, along with acetyl-coenzyme A, can block the inhibition of protein synthesis by blasticidin S in a cell-free system (8). Because of these traits, blasticidin S is widely used to select transformed cells that have been engineered to carry a resistance gene in biological research (9).
Fig 1.
Chemical structures of blasticidin S, mildiomycin, and cytosylglucuronic acid.
Puromycin (10), polyoxin (11), nikkomycin (12), and pacidamycin (13) are representative examples of the peptidyl nucleoside family, the biosynthetic pathways of which have been well studied (11, 14). Unlike the above-mentioned peptidyl nucleosides that typically have five-membered rings, the structure of blasticidin S and mildiomycin (Fig. 1) features a glucuronic acid-derived hexose that is coupled to cytosine or hydroxymethyl cytosine. The biosynthetic gene cluster of blasticidin S was first reported in 1998 and heterologously expressed in Streptomyces lividans (15). However, except for some putative intermediates, blasticidin S could not be identified from S. lividans heterologous production. Although the biosynthesis gene cluster for blasticidin S was sequenced and analyzed (16), there were no in vivo studies of the biosynthetic pathway due either to the difficult genetic manipulation system of S. griseochromogenes or to the failure of production of blasticidin S in S. lividans. Hence, the biosynthetic pathway of blasticidin S was proposed largely on the basis of labeled precursor incorporation, biochemical studies, and bioinformatic analysis (17–20), and several steps in the pathway still remain cryptic.
In this study, we identified a blasticidin S deaminase in S. lividans (SLBSD) that is able to transform the cytotoxic blasticidin S to its inactive form, deaminohydroxyblasticidin S. The activity of SLBSD was measured with several nucleosides in vitro. The blasticidin S biosynthetic gene cluster was then engrafted into a S. lividans strain with the SLBSD gene disrupted to generate the mutant WJ2 that produced blasticidin S. Furthermore, to specifically investigate the functions of blsE, blsF, and blsL in blasticidin S biosynthesis in vivo, these genes were individually disrupted, and the fermentation broths of the resultant mutants were analyzed by liquid chromatograph-mass spectrometry (LC-MS).
MATERIALS AND METHODS
Strains and culture conditions.
Key strains and plasmids are listed in Table S1 in the supplemental material. S. lividans 66 and its derivatives were grown at 30°C on soy flour-mannitol (SFM) agar plates for sporulation or in 10.3% TSBY (tryptic soy broth [TSB] supplemented with 10.3% sucrose [wt/vol] and 1% yeast extract [wt/vol]) medium for growth of mycelia. Exconjugants were grown on SFM agar plates containing the appropriate antibiotic (150 μg/ml spectinomycin or 15 μg/ml thiostrepton) or in TSBY (10.3%) medium with the appropriate antibiotic (50 μg/ml spectinomycin or 5 μg/ml thiostrepton). For secondary metabolite analysis, seed cultures were prepared by inoculating a loop of stock culture into a 250-ml baffled flask containing 25 ml of seed medium [per liter, 20 g of glucose, 30 g of soybean flour, 5.0 g of yeast extract, 1.0 g of (NH4)2SO4, 0.5 g of MgSO4 · 7H2O, 3.0 g of CaCO3] and incubated in a rotary shaker for 40 h at 30°C and 200 rpm. Then, 5 ml of this seed culture was transferred into a 500-ml baffled flask containing 50 ml of fermentation medium [per liter, 80 of g glucose, 40 g of soybean flour, 1.0 g of (NH4)2SO4, 0.5 g of MgSO4 · 7H2O, 10.0 g of CaCO3, 0.05 g of FeSO4, 0.4 g of K2HPO4] and incubated another 6 days under the same conditions. For LC-MS analysis of the production of blasticidin S and its intermediates, strains were cultured in TSBY (10.3%) medium in baffled flasks at 30°C and at 220 rpm for 6 days.
General molecular biology methods.
Isolation of total DNA, protoplast preparation, and transformations were performed according to Kieser et al. (21). Manipulations of Escherichia coli strains were conducted according to Sambrook et al. (22). DNA fragments and PCR products were purified from agarose gels (0.8%) using a DNA Gel Extraction Kit (V-Gene Biotechnology Ltd.). The PCR product for SLBSD expression was inserted into pMD18-T vector (TaKaRa, Dalian, China) and verified by sequencing. For the Southern hybridization experiments, all the DNA samples were cleaved with BamHI, separated on an agarose gel (0.8%), and transferred onto Hybond-N+ nylon membranes (Amersham Biosciences). BamHI-digested fosmid 7D11 was labeled with radioactive [α-32P]dCTP (Beijing Furui Bio-Engineering Co., Ltd.) using a Random Priming Kit (Roche) as the probe. The Southern blot hybridization was carried out overnight at 65°C, followed by a high-stringency wash (0.1× SSC [0.15 M NaCl plus 0.015 M sodium citrate], 0.1% [wt/vol] SDS) at the same temperature, and detected with a phosphorimager (Fujifilm). Restriction enzymes, T4 DNA ligase, Taq polymerase, and alkaline phosphatase were purchased from MBI Fermentas. Blasticidin S was purchased from Sigma-Aldrich. Cytosylglucuronic acid was purchased from Shanghai Violetpharm Chemical Technology Co., Ltd. CMP was purchased from Sangon Biotech Co., Ltd. (Shanghai, China).
Bioassay method.
To measure the resistance level of S. lividans and its bsd knockout mutant to blasticidin S, a serially diluted blasticidin S standard was added to an Oxford cup placed on SFM medium plates that were preinoculated with spores of the S. lividans strains. For bioassays, the Streptomyces strains were grown at 30°C for 5 days on SFM plates for the production of secondary metabolite; agar patches were transferred to potato dextrose agar (PDA; 200 g of diced potatoes, 20 g of glucose, and 15 g of agar in 1 liter of tap water) that contained Rhodotorula rubra AS2.166 (see Table S1 in the supplemental material), a yeast that is sensitive to blasticidin S.
LC-MS analysis.
The fermentation broth (25 ml) was harvested and adjusted to pH 5.0 and centrifuged at 12,000 × g for 5 min. The supernatant was applied to Supelclean LC-SCX solid-phase extraction (SPE) columns (bed weight, 500 mg; volume, 3 ml) (Supelco) and washed with 2 ml of water and then with 2 ml of 0.5% NH4OH. The fraction eluting with 3% NH4OH was filtered through a 2-μm-pore-size membrane before injection. The LC-MS analysis was done using an Agilent 1100 LC/MSD (mass-selective detector) with a C18 column (TC-C18; 250 mm by 4.6 mm) (Agilent). The isocratic mobile phase was 10 mM trichloroacetic acid-MeCN, 82:18 (vol/vol), and the flow rate was 0.3 ml/min at room temperature. Elution was monitored with a photodiode array detector at 272 nm, and electrospray ionization (ESI) MS analysis was carried out in the positive mode.
Q-TOF MS analysis.
Quadrupole time of flight (Q-TOF) MS analysis was done using an Agilent 6530 Q-TOF LC-MS with a C18 column (Zorbax StableBond SB-C18; 3.5-μm particle size; 2.1 mm by 150 mm) (Agilent). The isocratic mobile phase was MeCN-water with a gradient from 5% to 100%, and the flow rate was 0.2 ml/min at room temperature. The ion source was ESI, and the analysis was carried out in the positive mode.
Sequencing and bioinformatic analysis.
The genomic library of S. griseochromogenes was constructed in the pCC1FOS vector according to the provided protocol (Epicentre Biotechnologies). DNA sequencing was done at Life Technologies Corporation, Shanghai, China. Open reading frames were predicted using FramePlot, version 3.0 beta (http://watson.nih.go.jp/∼jun/cgi-bin/frameplot-3.0b.pl), and multiple-sequence alignment was performed with BioEdit, version 7.0. Similarity comparisons of nucleotide or amino acid sequences against public databases were done using the BLAST program on the NCBI website (http://ncbi.nlm.nih.gov/blast) (23).
Construction of the recombinant strains.
To integrate the entire blasticidin S gene cluster into the genome of S. lividans, fosmid 7D11, containing the published blasticidin S gene cluster as well as its downstream 8,780-bp sequence, was selected from the genomic library of the blasticidin S native producer S. griseochromogenes. Fosmid 7D11 was then cut by XbaI and SpeI and precipitated with 70% isopropanol and 10% 3 M sodium acetate. The purified digestion products were ligated at the XbaI-SpeI site of pJTU1289 (24) to construct pJTU1780 by rapid screening of the transformants, which contained the complete blasticidin S biosynthesis gene cluster. pJTU1780 was then digested with StuI and ligated with an aadA cassette amplified by the aadA-T primer set, with pIJ779 as the template to replace most of the insertion containing the blasticidin S gene cluster, generating construct pJTU1785. A 4.85-kb dispensable fragment covering the insertion site for the genomic island SLG (accession number EF210454) (34) in the S. lividans HXY16 chromosome was amplified with HXY16 forward and reverse primers and inserted at the EcoRV site of pOJ260 to construct pJTU1528. A 0.25-kb internal region of the 4.85-kb fragment was cut off by StuI, and its flanking 1.6-kb and 3.0-kb fragments were employed as two homologous arms for introduction of foreign DNA sequences. The insertion part of pJTU1785 was excised with EcoRV and ligated with the StuI-cut pJTU1528 to obtain pJTU1786.
To construct the blsE, blsF, and blsL knockout mutants, a 13-kb fragment covering blsE-blsN was excised from pJTU1780 using BglII and ligated with pJTU412 (25) to construct pWJ5. pWJ5 was then modified by module exchanges using ReDirect technology essentially as described previously with the primers Tar-blsE, Tar-blsF, and Tar-blsL (see Table S2 in the supplemental material) (26). The genes blsE, blsF, and blsL were deleted individually by recombinational exchange with an aadA cassette. The unmethylated recombinant plasmids, prepared in E. coli ET12567, were then separately introduced to S. lividans WJ2 by protoplast transformation. The resulting putative double crossover strains were confirmed by PCR with the primers Con-blsE, Con-blsF, and Con-blsL listed in Table S2 in the supplemental material.
Expression and purification of SLBSD.
The SLBSD gene was amplified from the genomic DNA of S. lividans HXY16 by PCR with SLBSD forward and reverse primers (see Table S2 in the supplemental material; NdeI and EcoRI sites are underlined). PCR was carried out with high-fidelity DNA polymerase (KOD-Plus; Toyobo), and products were gel purified, digested with the appropriate enzymes, and cloned into the corresponding restriction sites of the pET28a(+) vector (Novagen). The resulting plasmid, named pWJ4, was used to transform E. coli DH10B for sequencing. For expression studies, the plasmid was introduced to E. coli BL21(DE3)/pLysE (Novagen) and grown overnight at 37°C in liquid LB medium containing chloramphenicol and kanamycin (34 μg/ml and 50 μg/ml, respectively). The seed cultures (10 ml) were used to inoculate 1-liter production cultures of LB medium with the corresponding antibiotics. The cells were grown at 37°C to an optical density at 600 nm (OD600) of 0.6 and then induced with isopropyl-β-d-thiogalactopyranoside (1 mM). The culture was then grown for an additional 5 h at 30°C. After centrifugation, the cells were resuspended in 40 ml of binding buffer (20 mM sodium phosphate, 20 mM imidazole, and 0.5 M NaCl, pH 7.4) and lysed by sonication in an ice bath (10 times for 60 s at 15 W with 60-s pauses). After another round of centrifugation(at 10,000 × g for 1 h), the supernatant was applied to a HisTrap HP column (GE Healthcare) and purified using AKTA fast protein liquid chromatography (FPLC) (GE Healthcare), by eluting with elution buffer (20 mM sodium phosphate, 500 mM imidazole, and 0.5 M NaCl, pH 7.4) in a linear gradient. The purified His-tagged SLBSD was desalted using a HiTrap desalting column (GE Healthcare) and stored in 10 mM Tris-HCl buffer (pH 8.0) with 20% glycerol at −80°C. The purified His-tagged SLBSD was analyzed by 15% SDS-PAGE, and protein concentrations were determined using a Bradford Protein Assay Kit (Bio-Rad).
In vitro assay of the purified SLBSD.
The assays of recombinant SLBSD were carried out at 30°C overnight in a total volume of 100 μl containing Tris-HCl buffer (10 mM, pH 8.0), blasticidin S-mildiomycin-cytosylglucuronic acid (at 0.5 mM, 0.5 mM, and 0.5 mM, respectively), and the corresponding His-tagged SLBSD (10 μM). The reactions were quenched by the addition of chloroform. The products and substrates were analyzed by LC-MS as described above.
Nucleotide sequence accession number.
By chromosomal walking (details not shown), an 8,780-bp DNA fragment immediately downstream of blsN was cloned, sequenced, and deposited under accession number JX244070.
RESULTS
Engraftment of blasticidin S biosynthetic gene cluster to S. lividans.
The blasticidin S producer has a very strong barrier system against introduction of foreign DNA, which made prior genetic engineering of the blasticidin S gene cluster unsuccessful. To bypass this tough genetic manipulation system, attachment of the biosynthetic gene cluster onto the genome of S. lividans has been accomplished to generate a genetically stable heterologous host.
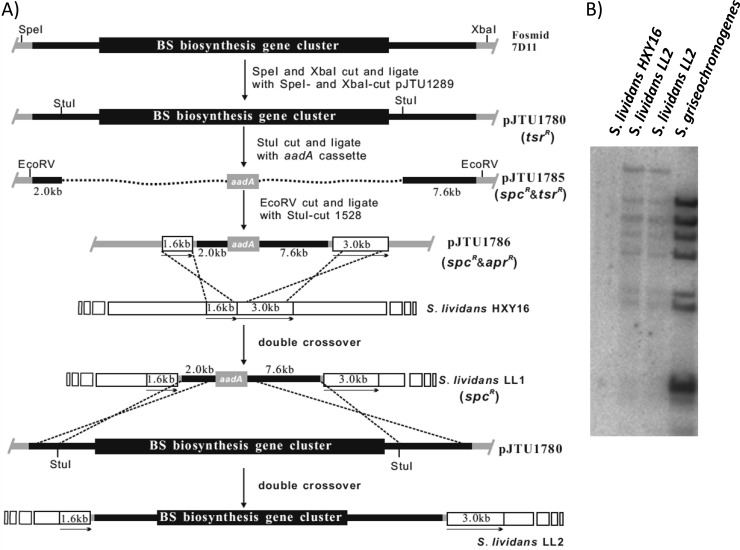
As depicted in Fig. 2A, the complete blasticidin S gene cluster and flanking sequence were excised from 7D11 (see Fig. S1A in the supplemental material) and inserted into pJTU1289 to generate pJTU1780 (see Table S1 and Fig. S1A and B). All internal StuI fragments of the insertion were removed and replaced with a blunt-ended aadA cassette to form pJTU1785 (see Fig. S1C), leaving 2.0-kb and 7.6-kb DNA fragments at the 5′ end and 3′ end of the insertion, respectively. A 4.85-kb dispensable DNA fragment from S. lividans HXY16 on pJTU1528 (see Fig. S1D) was separated into 1.6 kb and 3.0 kb by replacing its internal 0.25-kb StuI fragment with the 2.0 kb-aadA-7.6 kb(EcoRV) fragment from pJTU1785, generating pJTU1786 (see Fig. S1E). The 1.6-kb and 3.0-kb arms were then employed as the target sites for integration of the sandwiched 2.0 kb-aadA-7.6 kb fragment into the S. lividans HXY16 chromosome via double crossover to give mutant S. lividans LL1 (aadA). Finally, pJTU1780 was transferred to S. lividans LL1 to replace the 2.0 kb-addA-7.6 kb fragment with the blasticidin S biosynthetic gene cluster and the flanking sequences. Thus, S. lividans LL2, a mutant strain containing the blasticidin S gene cluster, was constructed and confirmed by Southern blotting (Fig. 2B).
Fig 2.
Construction of the S. lividans-derived strain LL2 that was engrafted with the blasticidin S biosynthetic gene cluster. (A) Schematic representation of the construction of S. lividans LL2. (B) Confirmation of the engraftment of the bls gene cluster to S. lividans HXY16 by Southern blotting. Genomic DNA preparations from S. lividans HXY16 (lane 1), two individual strains of S. lividans LL2 (lanes 2 and 3), and S. griseochromogenes (lane 4) were digested with BamHI, separated by agarose gel electrophoresis, and probed with BamHI-cut pJTU1780. spc, spectinomycin; tsr, thiostrepton; apr, apramycin.
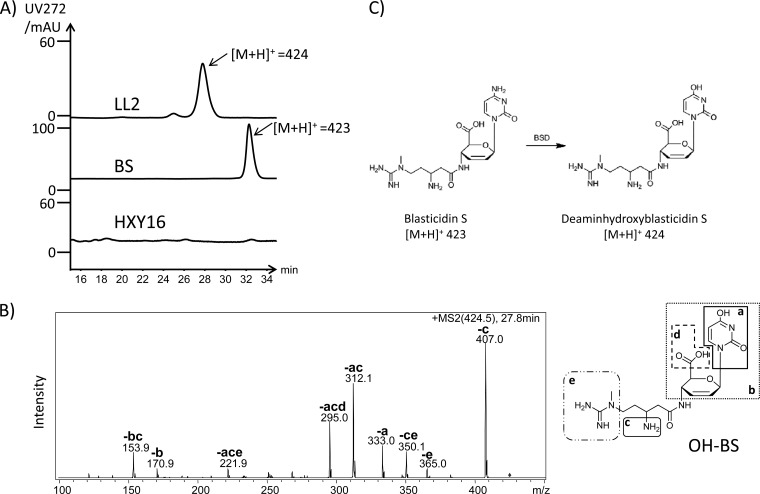
When assayed for antifungal activity, S. lividans LL2 showed a very slight inhibitory effect versus the indicator strain Rhodotorula rubra (see Fig. S2 in the supplemental material). To more accurately measure the blasticidin S production in S. lividans LL2, the fermentation broth was analyzed by LC-MS, but no peak corresponding to a blasticidin S standard was observed in the LC profile. However, a new compound was detected in the fermentation broth with a molecular size one mass unit greater than that of blasticidin S ([M+H]+ of 424) (Fig. 3A). The tandem MS (MS/MS) analysis of this compound identified it as the nontoxic deaminohydroxyblasticidin S (Fig. 3B), which is the known product of blasticidin S deaminase (BSD) (Fig. 3C) (27). To confirm this result, this compound was further purified and analyzed by Q-TOF MS (see Fig. S3 in the supplemental material).
Fig 3.
LC-MS analysis of the metabolite production of S. lividans LL2. (A) High-performance liquid chromatography comparison of the blasticidin S standard to the purified fermentation broth of S. lividans LL2 and S. lividans HXY16. LC-MS analysis revealed the peak presented in the fermentation broth of LL2, having a dominant peak at m/z 424. (B) MS/MS analysis of the m/z 424 parent ion. Each daughter peak (corresponding structures are shown in the inset) labeled could be derived from deaminohydroxyblasticidin S. (C) Blasticidin S was transformed to deaminohydroxyblasticidin S by BSD. BS, blasticidin S; AU, arbitrary units.
Identification of a BSD gene in S. lividans HXY16.
The existence of deaminohydroxyblasticidin S in the fermentation broth of LL2 prompted us to search for a homolog of the Aspergillus terreus BSD in the genome of S. lividans. Hence, we identified a cytidine deaminase (accession number EFD66414) in S. lividans TK24 that showed 51% identity and 67% similarity to the A. terreus BSD. The S. lividans HXY16 strain was also found to contain a BSD homolog, named SLBSD, that is almost identical to the TK24 enzyme except for a single mutation (E112D) (see Fig. S4 in the supplemental material).
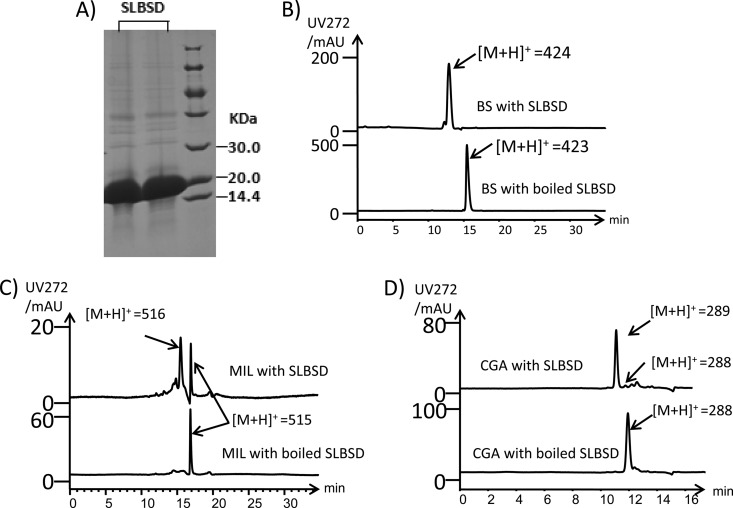
To provide a means to measure the ability of SLBSD to convert blasticidin S to deaminohydroxyblasticidin S, the SLBSD gene was heterologously expressed in E. coli. A purified soluble protein 16.9 kDa in size, consistent with the calculated molecular size of the His-tagged SLBSD, was obtained (Fig. 4A). LC-MS analysis of an overnight reaction incubating SLBSD with blasticidin S showed that blasticidin S ([M+H]+ of 423) was completely transformed to deaminohydroxyblasticidin S ([M+H]+ of 424) (Fig. 4B). Moreover, the substrate specificity of SLBSD was surveyed by examining its activity on mildiomycin, a nucleoside with similar structure to blasticidin S, cytosylglucuronic acid, an intermediate in blasticidin S biosynthesis, and CMP. The deaminase activity was observed with mildiomycin and cytosylglucuronic acid as substrates, but conversion was comparatively low (Fig. 4C and D). However, SLBSD did not catalyze the deamination of CMP (data not shown), suggesting that the hexose moiety may be an essential component for recognition by SLBSD.
Fig 4.
In vitro and in vivo analysis of SLBSD. (A) SDS-PAGE analysis of purified SLBSD. First two lanes, purified SLBSD; right lane, protein marker. The calculated molecular mass of His-tagged SLBSD was 16.9 kDa. (B) LC-MS analysis of products after overnight incubation with SLBSD and boiled SLBSD, with blasticidin S as the substrate. (C) LC-MS analysis of products after overnight incubation with SLBSD and boiled SLBSD, with mildiomycin as the substrate. (D) LC-MS analysis of products after overnight incubation with SLBSD and boiled SLBSD, with cytosylglucuronic acid as the substrate.
Inactivation of the SLBSD gene in S. lividans LL2 leads to the production of blasticidin S.
To stop the conversion of blasticidin S to deaminohydroxyblasticidin S by the SLBSD gene in the blasticidin S-producing S. lividans LL2, the SLBSD gene was disrupted via double crossover. The resulting strain was termed S. lividans WJ2, and the correct genotype was confirmed by PCR (Fig. 5A). LC-MS analysis of the purified fermentation broth of mutant WJ2 grown in production medium revealed a peak with same retention time as the blasticidin S standard. MS analysis of this peak further confirmed the production of blasticidin S in the mutant (Fig. 5B).
Fig 5.
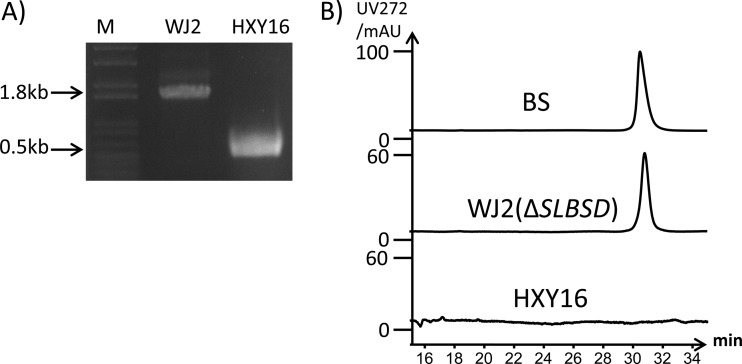
High-performance liquid chromatography analysis of the metabolite production of S. lividans WJ2. (A) Confirmation of the genotype of the SLBSD mutant WJ2 by PCR with primer Con-BKO (see Table S2 in the supplemental material). M, DNA marker; HXY16, PCR product targeting the SLBSD gene from S. lividans HXY16; WJ2, PCR product targeting the SLBSD gene from S. lividans WJ2. (B) High-performance liquid chromatography comparison of the blasticidin S standard to the purified fermentation broth of S. lividans WJ2 and S. lividans HXY16.
To compare the effect of the disruption of the SLBSD gene in S. lividans strains with different genotypes, the SLBSD gene was also inactivated in S. lividans HXY16 via double crossover to generate mutant S. lividans WJ1 (see Fig. S5A in the supplemental material). WJ1, S. lividans HXY16, and WJ2 were then assayed for their resistance levels against blasticidin S at different concentrations (20 μg to 200 μg/ml). The growth of HXY16 was not greatly inhibited by blasticidin S at a concentration of 200 μg/ml, while the SLBSD mutant WJ1 was completely inhibited by blasticidin S at a concentration of 100 μg/ml, as indicated by a clear zone on an agar test plate. Moreover, WJ2, containing the blasticidin S self-resistance gene blsJ, was also not affected by blasticidin S (see Fig. S5B). This observation suggested that the heterologous production of blasticidin S in WJ2 should not significantly affect growth of the host strain.
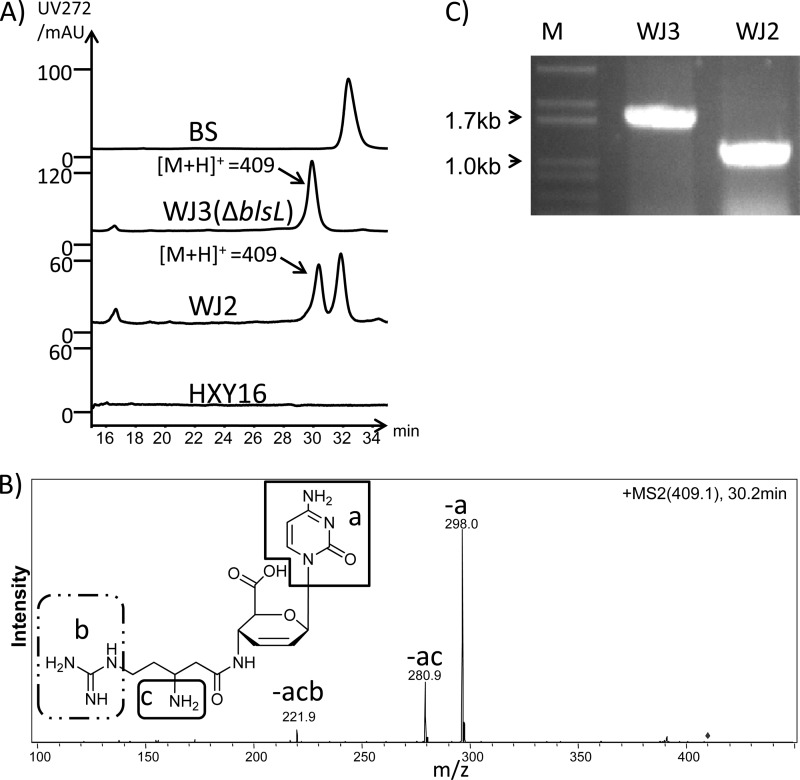
We previously found that the mildiomycin producer, Streptoverticillium rimofaciens ZJU5119, could produce three additional mildiomycin derivatives in the seed/fermentation broth (28). To check the secondary metabolite production by S. lividans WJ2, the strain was grown in the standard seed/fermentation medium, and analysis revealed a new peak exhibiting a dominant ion at [M+H]+ of 409 in the LC profile of the fermentation broth (Fig. 6A). This product was identified as demethylblasticidin S by MS/MS analysis (Fig. 6B) and Q-TOF MS (see Fig. S6 in the supplemental material). However, only blasticidin S was detected when S. griseochromogenes was grown in the same seed/fermentation medium (data not shown).
Fig 6.
Assay of the blasticidin S production of the ΔblsL mutant strain S. lividans WJ3. (A) High-performance liquid chromatography comparison of the blasticidin S standard to the purified fermentation broth of WJ3, WJ2, and HXY16. LC-MS analysis revealed the peak presented in the fermentation broth of LL2 and WJ2, with a dominant peak at m/z 409. (B) MS/MS analysis of the m/z 409 parent ion. Each labeled daughter peak (corresponding structures are shown in the inset) could be derived from demethylblasticidin S. (C) PCR confirmation of the genotype of the ΔblsL mutant WJ3 with primer Con-blsL (see Table S2 in the supplemental material). M, DNA marker; WJ2, PCR product targeting blsL from S. lividans WJ2; WJ3, PCR product targeting blsL from S. lividans WJ3.
Generation of blsE, blsF, and blsL disruption mutants.
To demonstrate the ability to manipulate the blasticidin S gene cluster in a heterologous host, three genes, blsE, blsF, and blsL, were disrupted individually, and the fermentation broths from the resulting mutant strains were analyzed by LC-MS.
Because demethylblasticidin S accumulated in the fermentation broth of WJ2 and was considered an intermediate in the blasticidin S biosynthetic pathway (15), the formation of demethylblasticidin S in WJ2 was investigated. An N-methyltransferase is the predicted product of blsL; thus, this gene was selectively inactivated, and the genotype of the resulting mutant WJ3 was confirmed by PCR (Fig. 6C).
There remain several steps in the biosynthetic pathway to blasticidin S that are unclear. To narrow down the genes involved in blasticidin S assembly, blsF a gene homologous to milL in the mildiomycin gene cluster, which is predicted to encode a protein of unknown function (29), was disrupted. Disruption of blsF (Fig. 7A) did not abolish the production of blasticidin S. On the contrary, the production of blasticidin S was increased in the mutant, S. lividans WJ4 (Fig. 7B). Moreover, in the crude extract of the fermentation broth of WJ4, a known intermediate of the blasticidin S biosynthetic pathway, cytosylglucuronic acid, accumulated (see Fig. S7 in the supplemental material).
Fig 7.
Assay of the blasticidin S production of the ΔblsF and ΔblsE mutant strains, S. lividans WJ4 and S. lividans WJ5, respectively. (A) Confirmation of the ΔblsF mutant WJ4 by PCR with primer Con-blsF (see Table S2 in the supplemental material). M, DNA marker; WJ2, PCR product targeting blsF from S. lividans WJ2; WJ4, PCR product targeting blsF from S. lividans WJ4. (B) High-performance liquid chromatography comparison of the purified fermentation broth of WJ4 to that of WJ2. (C) Confirmation of the ΔblsE mutant WJ5 by PCR with primer Con-blsE (see Table S2). M, DNA marker; WJ2, PCR product targeting blsE from S. lividans WJ2; WJ5, PCR product targeting blsE from S. lividans WJ5. (D) High-performance liquid chromatography comparison of the untreated fermentation broth of WJ5 to that of WJ2 and HXY16.
The accumulation of cytosylglucuronic acid in the WJ4 mutant implied that the enzyme acting on this intermediate was very important in increasing the production of blasticidin S. BLAST analysis showed that BlsE belonged to the radical S-adenosylmethionine (SAM) superfamily, members of which are capable of catalyzing multiple reaction types, such as decarboxylation, methylation, and dehydration (30). In our earlier work, the disruption of milG, a homologue to blsE in the mildiomycin biosynthetic pathway, led to the accumulation of hydroxymethyl cytosylglucuronic acid (29). To analyze the function of blsE in blasticidin S formation, the gene was inactivated, and the resulting mutant, S. lividans WJ5, was confirmed by PCR (Fig. 7C). As expected, production of blasticidin S was abolished in WJ5 (see Fig. S8 in the supplemental material), and the accumulation of cytosylglucuronic acid was observed in the fermentation broth of this mutant (Fig. 7D).
DISCUSSION
As introduction of alien DNA into the chromosome of the blasticidin S native producer S. griseochromogenes was never successful, in vivo functional studies of the role of each gene involved in blasticidin S biosynthesis was hindered. To bypass the strong DNA uptake barrier system of S. griseochromogenes, we set forth to engineer an S. lividans-derived blasticidin S-producing host that was genetically stable and manipulable. However, the resulting S. lividans LL2 mutant that carried the intact blasticidin S gene cluster was found to produce only a blasticidin S derivative, deaminohydroxyblasticidin S; this observation led to the discovery of a blasticidin S deaminase in S. lividans.
BSD was first discovered in Aspergillus terreus in 1975 (31). It was shown to be a zinc-dependent enzyme (27), and the blasticidin S-bound crystal structure of BSD revealed that the position of the zinc ion was very close to C-2 of the cytosine base of blasticidin S (32). This is the site occupied by a hydroxymethyl group in mildiomycin and may thus hinder its coordination to the active zinc ion center. This assumption may in part explain the low in vitro transformation efficiency (Fig. 4C) of mildiomycin to deaminohydroxymildiomycin by SLBSD. Consistent with this assumption, mildiomycin was successfully produced in S. lividans (29), whereas all of the blasticidin S was converted into deaminohydroxyblasticidin S. As for cytosylglucuronic acid, another substrate for SLBSD, this compound was nearly completely transformed into deaminohydroxycytosylglucuronic acid. The detection of cytosylglucuronic acid in the fermentation broth of the earlier heterologous expression experiments (15) might be the result of the high yield of this compound. Finally, the finding that CMP was not recognized by SLBSD might be due to the lack of a carboxyl group on its sugar ring or the negative charge carried by the phosphate group, both of which were essential for the recognition of blasticidin S to BSD as proposed by Kumasaka et al. (32).
The disruption of the SLBSD gene led to the successful heterologous expression of blasticidin S for the first time. This engineered blasticidin S-producing strain provided the necessary means to investigate the blasticidin S biosynthesis pathway in vivo. The disruption of blsF, a homologue of milL that had been confirmed as a gene unnecessary for mildiomycin biosynthesis (29), raised the yields of blasticidin S and other intermediates instead of abolishing their production. This result suggested that blsF might have a negative effect on blasticidin S biosynthesis. Moreover, accumulation of cytosylglucuronic acid and demethylblasticidin S in WJ4 suggested that the production of the cytotoxic blasticidin S repressed or slowed down the metabolic flow toward the final product. However, in the native producer, the coevolution of blasticidin S and a self-resistant mechanism can efficiently counter the cytotoxic activity. A good case for this is that there exist two chromosomal fragments containing putative resistance genes to blasticidin S (15). One fragment contains a transporter-encoding gene, blsJ (16), while the other resistant element is located outside the blasticidin S biosynthetic gene cluster.
The accumulation of cytosylglucuronic acid in the WJ4 mutant may also result from the inefficient expression, or the deficient performance, of some gene(s) or gene products in the heterologous host. Identification of the proteins responsible for directly processing cytosylglucuronic acid to advanced intermediates is therefore very important for fully utilizing the substrates and increasing the yield of blasticidin S in a heterologous expression host. In mildiomycin biosynthesis, disruption of milG, a blsE homolog, led to the accumulation of hydroxymethyl cytosylglucuronic acid (29). This result provided a hint that blsE was a key gene for the accumulation of cytosylglucuronic acid. As expected, disruption of blsE abolished the production of blasticidin S while the accumulation of cytosylglucuronic acid remained.
The wild-type S. griseochromogenes can produce demethylblasticidin S when inoculated in yeast extract-malt extract (YEME) medium (15), and demethylblasticidin S can be transformed to blasticidin S by a cell extract of this strain (33). It is very likely that BlsL alone is in charge of this transformation. In a previous report, the nontoxic leucyl blasticidin S was detected in the heterologous expression strain and a route involving leucyl demethylblasticidin S and leucyl blasticidin S as precursors to the blasticidin S pathway was proposed (16). However, in present study, leucyl blasticidin S was not regularly detected in the fermentation broth. A blasticidin S acetyltransferase was discovered in Streptoverticillium sp. in 1990 (8), and a homolog exists in many Streptomyces strains including S. lividans (accession number ZP_06530511). The formation of leucyl blasticidin S in the original study thus may have resulted from a host resistance or detoxification mechanism rather than from an enzyme in the blasticidin S biosynthetic pathway. Furthermore, the inactivation of blsL led to the production solely of demethylblasticidin S, which suggests that the direct conversion of demethylblasticidin S to blasticidin S is more likely.
In conclusion, blasticidin S is a very important nucleoside antibiotic both in agriculture and in laboratory research. The successful heterologous expression of this antibiotic in S. lividans HXY16 bypassed the difficult genetic manipulation of its native producer and led to the discovery of a blasticidin S deaminase from the S. lividans HXY16. Moreover, the easy genetic manipulation of S. lividans unveiled the function of several genes in the biosynthetic gene cluster. This is very important for the demonstration of the whole biosynthetic pathway of blasticidin S and for increasing its production in both native and heterologous hosts.
Supplementary Material
ACKNOWLEDGMENT
This study was supported in part by grants from the National Natural Science Foundation of China, the Ministry of Science and Technology of China (973 and 863 Programs), the Ministry of Education of China, and the Chen Xing Young Scholars Program of Shanghai Jiao Tong University.
Footnotes
Published ahead of print 1 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03254-12.
REFERENCES
- 1.Takeuchi S, Hirayama K, Ueda K, Sakai H, Yonehara H. 1958. Blasticidin S, a new antibiotic. J. Antibiot. (Tokyo) 11:1–5 [PubMed] [Google Scholar]
- 2.Huang KT, Misato T, Asuyama H. 1964. Selective toxicity of blasticidin S to Piricularia oryzae and Pellicularia sasakii. J. Antibiot. (Tokyo) 17:71–74 [PubMed] [Google Scholar]
- 3.Hansen JL, Moore PB, Steitz TA. 2003. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J. Mol. Biol. 330:1061–1075 [DOI] [PubMed] [Google Scholar]
- 4.Ichikawa Y, Hirata K, Ohbayashi M, Isobe M. 2004. Total synthesis of (+)-blasticidin S. Chemistry 10:3241–3251 [DOI] [PubMed] [Google Scholar]
- 5.Yamaguchi H, Yamamoto C, Tanaka N. 1965. Inhibition of protein synthesis by blasticidin S. I. Studies with cell-free systems from bacterial and mammalian cells. J. Biochem. 57:667–677 [PubMed] [Google Scholar]
- 6.Tamura K, Kimura M, Yamaguchi I. 1995. Blasticidin S deaminase gene (BSD): a new selection marker gene for transformation of Arabidopsis thaliana and Nicotiana tabacum. Biosci. Biotechnol. Biochem. 59:2336–2338 [DOI] [PubMed] [Google Scholar]
- 7.Karreman C. 1998. New positive/negative selectable markers for mammalian cells on the basis of blasticidin deaminase-thymidine kinase fusions. Nucleic Acids Res. 26:2508–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pérez-González JA, Ruiz D, Esteban JA, Jiménez A. 1990. Cloning and characterization of the gene encoding a blasticidin S acetyltransferase from Streptoverticillium sp. Gene 86:129–134 [DOI] [PubMed] [Google Scholar]
- 9.Mamoun CB, Gluzman IY, Goyard S, Beverley SM, Goldberg DE. 1999. A set of independent selectable markers for transfection of the human malaria parasite Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 96:8716–8720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tercero JA, Espinosa JC, Lacalle RA, Jimenez A. 1996. The biosynthetic pathway of the aminonucleoside antibiotic puromycin, as deduced from the molecular analysis of the pur cluster of Streptomyces alboniger. J. Biol. Chem. 271:1579–1590 [DOI] [PubMed] [Google Scholar]
- 11.Chen W, Huang T, He X, Meng Q, You D, Bai L, Li J, Wu M, Li R, Xie Z, Zhou H, Zhou X, Tan H, Deng Z. 2009. Characterization of the polyoxin biosynthetic gene cluster from Streptomyces cacaoi and engineered production of polyoxin H. J. Biol. Chem. 284:10627–10638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bormann C, Huhn W, Zahner H, Rathmann R, Hahn H, Konig WA. 1985. Metabolic products of microorganisms. 228. New nikkomycins produced by mutants of Streptomyces tendae. J. Antibiot. (Tokyo) 38:9–16 [DOI] [PubMed] [Google Scholar]
- 13.Rackham EJ, Gruschow S, Ragab AE, Dickens S, Goss RJ. 2010. Pacidamycin biosynthesis: identification and heterologous expression of the first uridyl peptide antibiotic gene cluster. Chembiochem 11:1700–1709 [DOI] [PubMed] [Google Scholar]
- 14.Bormann C, Mohrle V, Bruntner C. 1996. Cloning and heterologous expression of the entire set of structural genes for nikkomycin synthesis from Streptomyces tendae Tu901 in Streptomyces lividans. J. Bacteriol. 178:1216–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cone MC, Petrich AK, Gould SJ, Zabriskie TM. 1998. Cloning and heterologous expression of blasticidin S biosynthetic genes from Streptomyces griseochromogenes. J. Antibiot. (Tokyo) 51:570–578 [DOI] [PubMed] [Google Scholar]
- 16.Cone MC, Yin X, Grochowski LL, Parker MR, Zabriskie TM. 2003. The blasticidin S biosynthesis gene cluster from Streptomyces griseochromogenes: sequence analysis, organization, and initial characterization. Chembiochem 4:821–828 [DOI] [PubMed] [Google Scholar]
- 17.Grochowski LL, Zabriskie TM. 2006. Characterization of BlsM, a nucleotide hydrolase involved in cytosine production for the biosynthesis of blasticidin S. Chembiochem 7:957–964 [DOI] [PubMed] [Google Scholar]
- 18.Gould SJ, Guo J. 1994. Cytosylglucuronic acid synthase (cytosine: UDP-glucuronosyltransferase) from Streptomyces griseochromogenes, the first prokaryotic UDP-glucuronosyltransferase. J. Bacteriol. 176:1282–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seto H, Furihata K, Yonehara H. 1976. Studies on the biosynthesis of blasticidin S. V. Isolation and structure of pentopyranic acid. J. Antibiot. (Tokyo) 29:595–596 [DOI] [PubMed] [Google Scholar]
- 20.Liu HW, Thorson JS. 1994. Pathways and mechanisms in the biogenesis of novel deoxysugars by bacteria. Annu. Rev. Microbiol. 48:223–256 [DOI] [PubMed] [Google Scholar]
- 21.Kieser T, Bibb MJ, Chater KF, Butter MJ, Hopwood DA. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom [Google Scholar]
- 22.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 23.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He Y, Wang Z, Bai L, Liang J, Zhou X, Deng Z. 2010. Two pHZ1358-derivative vectors for efficient gene knockout in streptomyces. J. Microbiol. Biotechnol. 20:678–682 [DOI] [PubMed] [Google Scholar]
- 25.Sun Y, He X, Liang J, Zhou X, Deng Z. 2009. Analysis of functions in plasmid pHZ1358 influencing its genetic and structural stability in Streptomyces lividans 1326. Appl. Microbiol. Biotechnol. 82:303–310 [DOI] [PubMed] [Google Scholar]
- 26.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 100:1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura M, Sekido S, Isogai Y, Yamaguchi I. 2000. Expression, purification, and characterization of blasticidin S deaminase (BSD) from Aspergillus terreus: the role of catalytic zinc in enzyme structure. J. Biochem. 127:955–963 [DOI] [PubMed] [Google Scholar]
- 28.Li L, He X, Deng Z. 2009. Analysis of mildiomycin and its derivatives from the culture filtrate of Streptoverticillium rimofaciens ZJU5119. J. Shanghai Jiaotong Univ. 43:1–4 [Google Scholar]
- 29.Wu J, Li L, Deng Z, Zabriskie TM, He X. 2012. Analysis of the mildiomycin biosynthesis gene cluster in Streptoverticillum remofaciens ZJU5119 and characterization of MilC, a hydroxymethyl cytosyl-glucuronic acid synthase. Chembiochem 13:1613–1621 [DOI] [PubMed] [Google Scholar]
- 30.Layer G, Heinz DW, Jahn D, Schubert WD. 2004. Structure and function of radical SAM enzymes. Curr. Opin. Chem. Biol. 8:468–476 [DOI] [PubMed] [Google Scholar]
- 31.Yamaguchi I, Shibata H, Seto H, Misato T. 1975. Isolation and purification of blasticidin S deaminase from Aspergillus terreus. J. Antibiot. (Tokyo) 28:7–14 [DOI] [PubMed] [Google Scholar]
- 32.Kumasaka T, Yamamoto M, Furuichi M, Nakasako M, Teh AH, Kimura M, Yamaguchi I, Ueki T. 2007. Crystal structures of blasticidin S deaminase (BSD): implications for dynamic properties of catalytic zinc. J. Biol. Chem. 282:37103–37111 [DOI] [PubMed] [Google Scholar]
- 33.Guo J, Gould S. 1991. Biosynthesis of blasticidin S. Cell-free demonstration of N-methylation as the last step. Bioorg. Med. Chem. Lett. 10:497–500 [Google Scholar]
- 34.He X, Ou HY, Yu Q, Zhou X, Wu J, Liang J, Zhang W, Rajakumar K, Deng Z. 2007. Analysis of a genomic island housing genes for DNA S-modification system in Streptomyces lividans 66 and its counterparts in other distantly related bacteria. Mol. Microbiol. 65:1034–1048 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.