Abstract
In Rhodococcus ruber IFP 2001, Rhodococcus zopfii IFP 2005, and Gordonia sp. strain IFP 2009, the cytochrome P450 monooxygenase EthABCD catalyzes hydroxylation of methoxy and ethoxy residues in the fuel oxygenates methyl tert-butyl ether (MTBE), ethyl tert-butyl ether (ETBE), and tert-amyl methyl ether (TAME). The expression of the IS3-type transposase-flanked eth genes is ETBE dependent and controlled by the regulator EthR (C. Malandain et al., FEMS Microbiol. Ecol. 72:289–296, 2010). In contrast, we demonstrated by reverse transcription-quantitative PCR (RT-qPCR) that the betaproteobacterium Aquincola tertiaricarbonis L108, which possesses the ethABCD genes but lacks ethR, constitutively expresses the P450 system at high levels even when growing on nonether substrates, such as glucose. The mutant strain A. tertiaricarbonis L10, which is unable to degrade dialkyl ethers, resulted from a transposition event mediated by a rolling-circle IS91-type element flanking the eth gene cluster in the wild-type strain L108. The constitutive expression of Eth monooxygenase is likely initiated by the housekeeping sigma factor σ70, as indicated by the presence in strain L108 of characteristic −10 and −35 binding sites upstream of ethA which are lacking in strain IFP 2001. This enables efficient degradation of diethyl ether, diisopropyl ether, MTBE, ETBE, TAME, and tert-amyl ethyl ether (TAEE) without any lag phase in strain L108. However, ethers with larger residues, n-hexyl methyl ether, tetrahydrofuran, and alkyl aryl ethers, were not attacked by the Eth system at significant rates in resting-cell experiments, indicating that the residue in the ether molecule which is not hydroxylated also contributes to the determination of substrate specificity.
INTRODUCTION
Synthetic alkyl and aryl ethers are widely used in industry and agriculture as organic solvents, detergents, and pesticides, among other uses. The most prominent examples of important xenobiotic dialkyl ethers are the fuel oxygenates methyl tert-butyl ether (MTBE), ethyl tert-butyl ether (ETBE), tert-amyl methyl ether (TAME), and tert-amyl ethyl ether (TAEE), which since the late 1970s have been added to gasoline for optimizing combustion and thus reducing carbon monoxide emissions (1). The intentional and unintentional release of synthetic ethers both lead to ubiquitous pollution of water resources. Due to the low reactivity of the ether bridge, all of these pollutants generally possess a high degree of resistance to abiotic and biotic attack and tend to persist in the environment (2). In the case of MTBE, its detection at the ppb level or even higher concentrations in innumerable drinking water sources (3) has already led to a ban of its use as a gasoline additive in the United States (4).
In principle, dialkyl and alkyl aryl ethers can be degraded in aerobic microorganisms by monooxygenase-catalyzed hydroxylation to hemiacetals, which can spontaneously dismutate in aqueous solutions to the corresponding alcohols plus aldehydes/ketones (2). However, corresponding monooxygenases are not widespread for all synthetic ethers, as has been demonstrated for MTBE and the other aforementioned fuel oxygenate ethers. Only a couple of enzymes are currently known to attack these chemicals at significant rates. Among them are different AlkB-type and cytochrome P450 systems (5, 6). Interestingly, these monooxygenases differ not only in their substrate specificity (e.g., hydroxylation of only methoxy or methoxy and ethoxy groups of tert-alkyl ethers) but also in their isotope fractionation, and thus a critical evaluation of two-dimensional (D/H and 13C/12C) isotope analysis is needed for assessing degradation activities at contaminated sites (5).
The best-studied fuel oxygenate monooxygenase for MTBE and ETBE hydroxylation is the cytochrome P450 EthABCD system found in several Gram-positive bacterial strains of the genera Rhodococcus, Gordonia, and Mycobacterium (7, 8, 9). The corresponding genes are located in the ethRABCD gene cluster encoding an AraC/XylS-type positive transcriptional regulator (ethR), a ferredoxin reductase (ethA), a cytochrome P450 monooxygenase (ethB), a ferredoxin (ethC), and a protein with an unknown function (ethD). This cytochrome P450 system was classified as the first member of the CYP249A1 family (8, 9). In Rhodococcus ruber IFP 2001 and other strains, expression of the eth genes is induced by ETBE but not by MTBE or TAME (9). In addition, the eth gene cluster can undergo homologous recombination using two identical IS3-type class II transposases located on either side of the gene cluster (Fig. 1), orientated in the same direction and forming a hairpin and releasing the eth genes (7, 8). Surprisingly, a similar Eth system is also present in the fuel oxygenate-degrading betaproteobacterium Aquincola tertiaricarbonis L108 (10, 11). However, unlike the Gram-positive eth-bearing strains, which stoichiometrically accumulate the tert-alcohol metabolites during ether degradation, Aquincola tertiaricarbonis L108 possesses metabolic pathways for complete mineralization. tert-Butyl alcohol (TBA) from MTBE and ETBE is degraded via hydroxylation to 2-methyl-1,2-propanediol by the monooxygenase MdpJK (12, 13) and undergoes further enzymatic oxidation to 2-hydroxyisobutyric acid, which is then activated by coenzyme A (CoA) and isomerized to the common metabolite 3-hydroxybutyryl-CoA by the cobalamin-dependent mutase HcmAB (14, 15). In the same strain, the TAME and TAEE metabolite tert-amyl alcohol (TAA) is desaturated by MdpJK to the hemiterpene 2-methyl-3-buten-2-ol, which is isomerized to prenol, oxidized to 3-methylcrotonyl-CoA, and then degraded via the leucine pathway (16). Consequently, strain L108 is able to grow on all four fuel oxygenate ethers (16, 17).
Fig 1.
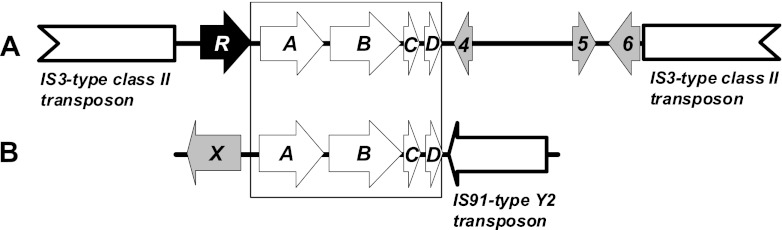
Comparison of the eth gene regions sequenced thus far in R. ruber IFP 2001 (20,138 bp, GenBank/EMBL/DDBJ database number AF333761) (A) and A. tertiaricarbonis L108 (6,909 bp, KC333876) (B), which encode fuel oxygenate ether monooxygenase Eth, consisting of ferredoxin reductase EthA, cytochrome P450 monooxygenase EthB, ferredoxin EthC, the unknown-function protein EthD, and other proteins. The box marks the DNA region showing 99.9% identical nucleotide sequences in the two strains. In contrast, strain IFP 2001 ethABCD genes are flanked by two identical copies of a 5.5-kb IS3-type class II transposon, the ethR gene, and other open reading frames (4 to 6), whereas the four eth genes in strain L108 are flanked by a 957-bp gene fragment (X) encoding a protein showing 99% sequence identity to the C-terminal part of a putative peptidyl-prolyl cis-trans isomerase from the MTBE-degrading strain Methylibium petroleiphilum PM1 (Mpe_A1955) and one copy of an 1.5-kb IS91-type Y2 transposon element.
In this study, we characterized the cluster of eth genes from A. tertiaricarbonis L108 which is lost in the spontaneous deletion mutant strain L10. The latter strain is unable to degrade any of the four fuel oxygenate ethers. Furthermore, we compared the regulation of eth gene expression in A. tertiaricarbonis L108 with the ETBE-dependent induction previously found in R. ruber IFP 2001, Rhodococcus zopfii IFP 2005, and Gordonia sp. strain IFP 2009 (9). In contrast to the latter strains, A. tertiaricarbonis L108, whose eth gene cluster is lacking the regulator gene ethR, shows constitutive expression of the eth genes when incubated on any fuel oxygenate ether or glucose as a growth substrate. This lack of regulation enables strain L108 to degrade MTBE, ETBE, TAME, and TAEE, as well as diethyl and diisopropyl ether, at high rates.
MATERIALS AND METHODS
Chemicals, bacterial strains, and culture conditions.
For a list of the suppliers and the purities of ethers and other chemicals used in this study, see the supplemental material. A. tertiaricarbonis L108 (14, 18) was cultivated at 30°C in liquid mineral salt medium (MSM) supplemented with vitamins, as described previously (19), on MTBE or other fuel oxygenate ethers at concentrations of 0.3 g liter−1. The spontaneous mutant strain A. tertiaricarbonis L10 (14) was grown in MSM on TBA at 0.5 g liter−1 supplemented with vitamins. Growth was monitored by measuring optical density at 700 nm (OD700).
Resting-cell experiments and chemical analyses.
Precultures were harvested for degradation experiments by centrifugation at 13,000 × g at 4°C for 10 min. After washing twice with nitrogen-free MSM, cells were immediately used as inoculum for resting-cell experiments adjusted to an OD700 value of 2.5, corresponding to a concentration of 1.35 g biomass (dry weight) liter−1 (17). The cell suspensions were incubated with 2.5 mM target ether substrate at 30°C in glass serum bottles sealed with butyl rubber stoppers (19). For monitoring degradation, samples were taken as described previously (19) by puncturing the rubber stoppers with syringes equipped with disposable Luer Lock needles. Volatile ethers, alcohols, and other metabolites were quantified by headspace gas chromatography (GC) using flame ionization detection (FID) employing an Optima Delta 3 column (60 m by 0.32 mm by 0.35 μm; Macherey-Nagel, Düren, Germany) either isothermally at 50°C (MTBE, ETBE, diethyl ether, diisopropyl ether, ethanol, acetaldehyde, isopropanol, acetone, tetrahydrofuran, and TBA) or applying a temperature gradient from 100 to 180°C (TAME, TAEE, n-hexyl methyl ether, and TAA) (19). In the case of aryl alkyl ethers (anisole, phenetole, and isopropoxybenzene), the column oven program was set at 120°C for 2 min and then increased to 250°C at 15°C per min and, finally, kept at 250°C for 0.67 min. The shown data represent mean values and standard deviations of results from at least four replicate experiments.
Batch cultivation.
Strain L108 was grown on fuel oxygenate ethers (either MTBE, ETBE, TAME or TAEE) in serum bottles at 30°C as described above. Cultivation on glucose was done in a 2-liter BioStat B-DCU II bioreactor system (Sartorius Stedim Systems GmbH, Melsungen, Germany) at automatically controlled conditions (temperature at 30°C, pH at 7.0, oxygen concentration in culture at 20% of saturation). Cells of strain L108 pregrown on MTBE were harvested by centrifugation as described above, and washed twice with MSM to remove residual MTBE completely. These cells were immediately used as inoculum (20 mg biomass liter−1) for cultivation on 2.5 g liter−1 glucose in 1.5 liters MSM supplemented with vitamins, 500 mg N liter−1 (as ammonium chloride), and 0.5 ml antifoam solution (Carl Roth GmbH, Germany). For PCR, sterile volumes of 1 ml at an OD700 of 0.5 from the growing cultures were harvested by centrifugation and stored at −80°C. Two replicates were taken at each sampling point, serving for DNA and RNA isolation, respectively. The fermentation experiments were performed in triplicate.
Sequencing of genomic DNA.
For sequencing of the eth gene cluster of strain A. tertiaricarbonis L108, highly pure genomic DNA from an MTBE-grown culture was extracted using the MasterPure DNA purification kit (Epicentre) and sequenced by Illumina's HiSeq 2000 technology (GATC Biotech, Konstanz, Germany). A 6,909-bp sequence, including the ethABCD genes, was obtained (Fig. 1). Sequence similarities were evaluated using the BLAST alignment tool (20). Promoter prediction was done using the prokaryotic promoter prediction tool at http://bioinformatics.biol.rug.nl/websoftware/ppp/ (21). The IS91-type transposase was defined by BLAST and the ISfinder software at http://www-is.biotoul.fr (22).
RT-qPCR, qPCR, and PCR.
For reverse transcription-quantitative PCR (RT-qPCR), RNA was isolated from the frozen cell pellets of ether- and glucose-grown cells using the RNeasy mini kit (Qiagen) and transcribed into cDNA using the RevertAid first-strand cDNA synthesis kit (Thermo Scientific) according to the manufacturers' protocols. The cDNA was directly used as the template for RT-qPCR. For DNA isolation, each frozen cell pellet was thawed and then suspended in 30 μl sterile distilled water. The samples were heated up for 1 min at 350 W in a microwave and centrifuged at 4°C for 3 min at 16,000 × g to get the DNA in the supernatant. DNA content and purity were determined via a NanoDrop ND-1000 UV-Vis spectrophotometer (Thermo Scientific) and adjusted to 5 ng μl−1 with sterile distilled water for qPCR. For nonquantitative PCR testing for the IFP 2001 IS3-type tnp genes, DNA was isolated from 1 ml bacterial culture.
Standard PCR with the Taq PCR master mix kit (Qiagen) with 5 ng DNA in 10 μl was performed under the following conditions: 3 min of initial denaturation at 94°C, followed by 30 cycles of 30 s at 94°C, annealing for 30 s at 60°C, and 1 min kb−1 elongation at 72°C, terminated by a final elongation of 5 min at 72°C. For IS3-type tnp PCR, 10% dimethyl sulfoxide (DMSO) (Sigma-Aldrich) was added to the reaction and DNA from strain IFP 2001 was used as positive control. For qPCR of the eth genes, the following protocol was applied: initial denaturation at 98°C for 2 min, followed by 40 cycles of 3 s at 98°C, 5 s at 60°C, 3 s at 82°C plus plate read followed by the melt curve (65°C to 95°C, increment 0.5°C for 5 s), and final plate read. Based on this protocol, ethC resulted in the highest qPCR efficiency and was chosen as the target for qPCR and RT-qPCR experiments. As control genes, we chose mdpJ and hcmA, which encode key enzymes of TBA metabolism in strain L108, i.e., for the TBA monooxygenase (12, 13), and the large subunit of the 2-hydroxyisobutyryl-CoA mutase (15), respectively, and the housekeeping 16S rRNA gene. The qPCR was processed in triplicates with 5 ng μl−1 DNA for each reaction and target gene by using the SsoFast EvaGreen super mix according to the manufacturer's protocol on the CFX96 real-time system and analyzed by using the CFX Manager software (Bio-Rad). All PCR primer pairs of target gene fragments used in this study are listed in Table 1. For RT-qPCR, 1 μl synthesized cDNA was used as the template.
Table 1.
Primer pairs used for standard PCRs and qPCRs
| Primer name | Sequence (5′ → 3′) | Target gene | Length (bp) |
|---|---|---|---|
| iso-for | TCGGTCCGGCATGGGACCTGAT | L108 isomerase upstream of ethA | 1,164 |
| iso-rev | AGGCGCGGAAGGACCCTG | ||
| ethAup-for | TCGGTCCGGCATGGGACCTGAT | eth promoter region | 1,618 |
| ethA-rev1 | ATCGCGCAGCGACATCGCTGCCTGCA | ||
| ethA-for | CAGGGTCCTTCCGCGCCT | ethA | 1,654 |
| ethA-rev | TCAGCGAGCCGTGGCGACCT | ||
| ethB-for | ATGACACTGTCACTGGCCA | ethB | 1,203 |
| ethB-rev | TCACTTCGGGTAGATCCGCA | ||
| ethC-for | ACGGCATCCTCGCCGAGTGC | ethC | 321 |
| ethC-rev | TCAGAACGCGTCGGGGACCT | ||
| ethD-for | ATGTATCAGATCGTGGCCTG | ethD | 312 |
| ethD-rev | CTAGGTCCGGTCGACCTCAT | ||
| tnp-for | ATTCCTGCGGCTACG | L108 IS91-type Y2 tnp | 1,346 |
| tnp-rev | GACACCGTACACATCACA | ||
| mpdJ-2rev_c | CGTCGACGGCAGCCTGCTGG | mpdJ | 380 |
| mpdJ-3for_c | TGTTGTCATCGGTCGGGTGC | ||
| hcmA-c2-for | GACTTCTTCGAGGAGGTCGC | hcmA | 425 |
| hcmA-RPr-2 | GTGCCACCGCGCTTCTCG | ||
| unibac27f | AGAGTTTGATCTGGCTCAG | 16S rRNA gene | 511 |
| univ519r | GTATTACCGCGGCTGCTG | ||
| tnpR-for | TCGTCGCGTGGCAGAGCGCTACCGCGCG | R. ruber IFP 2001 tnpR | 401 |
| tnpR-rev | CGATCACCGCCCTGGCCGACGCGCCGCC | ||
| tnpA-for a | ATGAACACCGTGACGT | R. ruber IFP 2001 tnpA | 810 |
| tnpA-revC | CGCAGTCAACGTCTCCACGC |
Promoter analysis.
The predicted L108 eth promoter was tagged to the promoterless catechol-dioxygenase gene xylE of the broad-host-range promoter-probe vector pCM130 (23). A 1.6-kb fragment upstream from ethA was amplified by standard PCR. By EcoRI digestion and blunting with mung bean nuclease, the promoter region of about 500 bp (PL108eth) was obtained. With the T4-DNA ligation kit, this fragment was ligated into HindIII-linearized, blunt-ended, and, with Antarctic phosphatase, dephosphorylated pCM130. Each step was done according to the respective protocols with enzymes from NEB. The resulting pCM130::PL108eth was transformed into chemically competent Escherichia coli DH5α according to the rubidium chloride (RbCl) method (24) and selected overnight at 37°C on LB agar with 20 μg ml−1 tetracycline. Grown colonies were sprayed with an aqueous solution of 100 mM catechol. Expression of active catechol dioxygenase resulted in instant yellow-green staining of the colonies due to 2-hydroxymuconic semialdehyde formation. Equally processed religated pCM130 served as negative control.
Nucleotide sequence accession number.
The complete sequence of the ethABCD genetic cluster of strain A. tertiaricarbonis L108 obtained in this study has been deposited in the GenBank/EMBL/DDBJ database under the accession number KC333876.
RESULTS
Sequence of the ethABCD gene cluster in strain L108.
In strains L108 and IFP 2001, the ethABCD gene sequence, including the 103-bp noncoding region directly upstream from ethA, is 99.9% identical (Fig. 1), indicating a recent horizontal gene transfer event. However, the other genes found in the neighborhood close to the eth cluster in strain IFP 2001 (8), comprising the regulator gene ethR, the open reading frames 4 to 6, and the identical copies of the IS3-type class II transposon element, are missing in strain L108. In contrast, in the latter case the ethABCD genes are flanked by a single gene fragment encoding a protein showing 99% sequence identity to the C-terminal part of a putative peptidyl-prolyl cis-trans isomerase from the MTBE-degrading strain Methylibium petroleiphilum PM1 (Mpe_A1955) (25, 26) and a putative IS91-type rolling-circle transposase most similar to a sequence found in the Burkholderiales bacterium JOSHI_001 (ZP_09752764, 73% amino acid identity). The absence of IS3-type elements in the genome of strain L108 was verified by PCR targeting of the IFP 2001 tnpA and tnpR genes.
Spontaneous eth deletion mutant strain L10.
As has been previously reported for MTBE and ETBE (14), the mutant strain L10 did not show any growth or degradation activities when incubated with TAME or TAEE. On the other hand, strain L10 readily degraded TBA and TAA at the same rates as the wild-type strain L108 without accumulation of metabolites (data not shown). Standard PCRs designed for detection of the ethABCD genes, the IS91 element, and the peptidyl-prolyl cis-trans isomerase gene fragment in strain L108 (Table 1) revealed that all these genes were undetectable with DNA from strain L10 (see Fig. S1 in the supplemental material), indicating that a larger DNA fragment was lost during the recombination process.
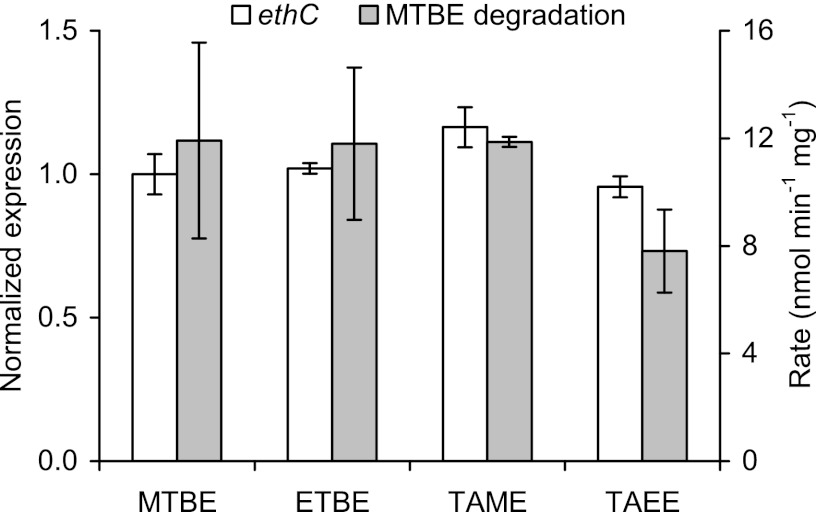
Expression of ethC in strain L108 growing on fuel oxygenate ethers.
For quantifying expression by RT-qPCR, the target gene ethC was chosen as a proxy for the complete eth gene cluster. Cultures of strain L108 exponentially growing on either MTBE, ETBE, TAME, or TAEE as the sole source of energy and carbon were sampled to isolate RNA for RT-qPCR and to test MTBE degradation rates in resting-cell experiments (Fig. 2). In all cases, similar expression levels of the tested target gene ethC were found, indicating that transcription of the eth genes is not particularly induced by ETBE in strain L108, i.e., unlike previously reported results for the eth-bearing Gram-positive strains (9), or by any other fuel oxygenate ether tested. An MTBE degradation rate of about 12 nmol min−1 mg biomass−1 was obtained with resting cells pregrown on either MTBE, ETBE, or TAME. Similar activities have also been measured for MTBE and TAME degradation with strain L108 in a previous study (19). However, only about 70% of the maximal rate was observed with TAEE-grown cells. Most likely, this is not caused by different amounts of the EthABCD monooxygenase but by lower induction of the downstream pathways processing TBA and formaldehyde/formic acid from MTBE. This incomplete degradation could result in an imbalance in reducing equivalents (e.g., NADH) essential for monooxygenase activity. Accordingly, TAEE-grown cells significantly accumulated TBA from MTBE (data not shown).
Fig 2.
RT-qPCR results for the target gene ethC from RNA samples of strain L108 cells exponentially growing on fuel oxygenate ethers as indicated. Normalized expression (ΔΔ quantification cycle [Cq]) with standard deviations determined from results obtained with cDNA of MTBE cultures as the control and expression of 16S rRNA and mpdJ genes as the references. Also shown are MTBE degradation rates in nmol min−1 mg biomass−1 in resting cells derived from the respective ether cultures.
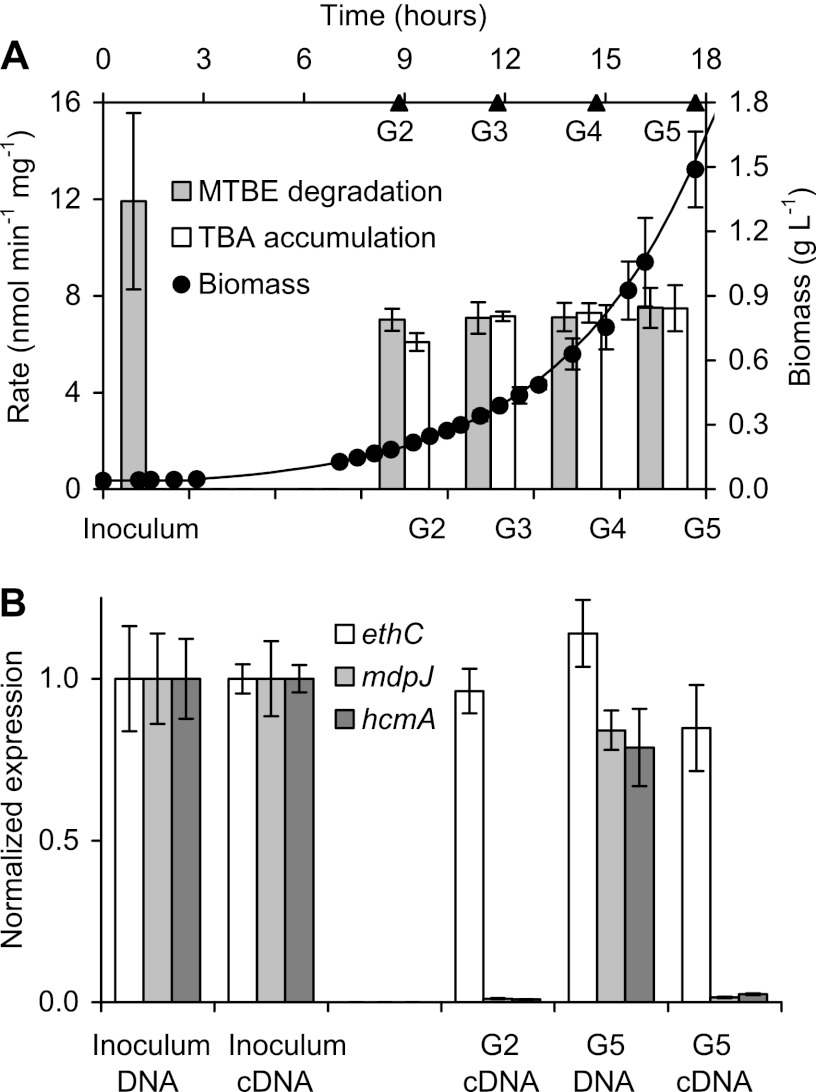
Expression of ethC in strain L108 growing on glucose.
To analyze expression of the EthABCD monooxygenase on a nonether growth substrate, the ethC gene was quantified in glucose-grown cultures. After a lag phase of about 2 h, MTBE-pregrown cells of strain L108 began to grow exponentially on glucose with a generation time of 2.95 h (Fig. 3). RNA samples from the MTBE preculture and from the glucose culture at generations two and five were analyzed. As already shown above for the different ether cultures (Fig. 2), expression of ethC was not affected by glucose as the growth substrate. Even after five generations, expression levels were similar to those obtained for the MTBE preculture (Fig. 3B). In contrast, expression of the other MTBE degradation-related genes mdpJ and hcmA was completely downregulated in glucose-growing cells, whereas these genes were well induced in the MTBE preculture. Hence, this significant repression of TBA and 2-hydroxyisobutyric acid metabolism led to a nearly stoichiometric accumulation of TBA and a reduction of about 40% of Eth monooxygenase activity when MTBE degradation was tested with glucose-grown cells (Fig. 3A), as has also been observed with TAEE-grown cells (Fig. 2). As a control, qPCR with DNA samples proved that all three catabolic genes, ethC, mdpJ, and hcmA, were not subjected to deletion events during the growth of strain L108 on glucose (Fig. 3B).
Fig 3.
MTBE-to-glucose shift experiments. (A) MTBE degradation and TBA accumulation rates in nmol min−1 mg biomass−1 of strain L108 cells pregrown on MTBE (inoculum) and after two to five generations (G2 to G5) of growth on glucose, as indicated. (B) RT-qPCR (cDNA) and qPCR (DNA) results for the target genes ethC, mdpJ, and hcmA from DNA and RNA samples of strain L108 cells pregrown on MTBE (inoculum) and after two and five generations (G2, G5) on glucose. Normalized expression levels (ΔΔCq) were calculated by normalization with control samples (DNA and cDNA from inoculum samples, respectively) and the 16S rRNA gene as reference.
Degradation of other synthetic ethers by strain L108.
For testing degradation capabilities of strain L108, several dialkyl and aryl alkyl ethers not used as fuel oxygenates were tested with cells pregrown on MTBE. In these resting-cell experiments, diethyl and diisopropyl ether were readily degraded, although only at about 30 and 15% of the degradation rate obtained with MTBE, respectively (see Table S1 in the supplemental material). In both cases, accumulation of volatile metabolites was revealed by GC measurements, i.e., ethanol from diethyl ether and acetone from diisopropyl ether. On the other hand, the dialkyl ether n-hexyl methyl ether, the cyclic alkyl ether tetrahydrofuran, and the aryl alkyl ethers anisole, phenetole, and isopropoxybenzene were not degraded at detectable rates by strain L108. Diethyl and diisopropyl ether as well as all other ethers tested were not degraded by the mutant strain L10 pregrown on TBA.
Transcription initiation site upstream of ethA.
Inspection of the eth gene cluster sequences of strains L108 and IFP 2001 revealed that a sigma factor binding site is present upstream of ethA in strain L108 (Fig. 4). Although in the two strains the 103-bp region directly upstream from the ethA translation start is identical, and thus they share a ribosomal binding site, more distant sequences are totally different, and only in strain L108 were the characteristic binding sites at −10 and −35 for a σ70-initiated transcription clearly detected (Fig. 4). Further inspection revealed no promoter regions between ethR and ethA in strain IFP 2001 (data not shown). Integration of the predicted L108 eth promoter region in pCM130 directly upstream of promoterless xylE allowed active expression of the catechol dioxygenase (see Fig. S2 in the supplemental material), establishing this DNA region as a constitutive promoter site.
Fig 4.

Prediction of promoter regions upstream of the ethA gene. A sigma factor binding site that possessed the −10 and −35 regions characteristic for σ70-initiated transcription (score −7.4, E value 2.2) was detected only in strain L108. For inspection, the 468- and 186-bp intergenic spacers upstream of ethA in strains L108 and IFP 2001, respectively, were analyzed using the prokaryotic promoter prediction tool at http://bioinformatics.biol.rug.nl/websoftware/ppp/. Highlighted in bold are the −35 and −10 regions, the +1 transcription start, the ribosomal binding site (RBS), and the translation start of ethA. Boxes indicate identical nucleotide sequences in the two strains.
DISCUSSION
Comparison of the eth gene cluster encoding the cytochrome P450 monooxygenase system EthABCD found in the fuel oxygenate-degrading strains A. tertiaricarbonis L108 and R. ruber IFP 2001 revealed that both gene regions are subjected to transposon-mediated recombination, resulting in eth deletion mutants, e.g., A. tertiaricarbonis L10, that are unable to degrade dialkyl ethers. However, although the high similarity between the ethABCD sequences indicates a recent horizontal gene transfer event, the underlying deletion mechanisms are different (IS3-type transposon element in strain IFP 2001 versus rolling-circle IS91 type in strain L108). In addition, the absence of the regulator EthR, resulting in the constitutive expression of the Eth monooxygenase, enables A. tertiaricarbonis L108 to degrade synthetic dialkyl ethers, including the fuel oxygenates MTBE, ETBE, TAME, and TAEE, at high rates and without any lag phase.
The high sequence similarity (>99%) of the ethABCD genes in all bacterial strains tested thus far indicates that the cluster of eth genes has been recently transferred by horizontal gene transfer. It is likely that the flanking transposon elements were the cause for high mobility. In principle, during the recombination event the excision of the eth genes results in a circular structure, which can then integrate into other genomes (7, 8). Invading transposons are major agents of interspecies gene transfer (27). This is also true for genes related to xenobiotic degradation (28). Interestingly, the eth gene transfer was not restricted to Gram-positive strains of the family Corynebacterineae, but the gene cluster was also transferred to A. tertiaricarbonis L108, belonging to the Betaproteobacteria. However, different transposon systems, the IS3- and IS91-type elements (Fig. 1), have been observed to flank the monooxygenase genes in the two phylogenetically distant groups, indicating a more complex history of the eth gene cluster transfer.
The detection of a gene fragment upstream of ethA in strain L108 which is nearly identical to a sequence found in the betaproteobacterium M. petroleiphilum PM1 (Mpe_A1955) suggests that PM1-related strains and other Gram-negative bacteria might also bear eth genes. Strain PM1, however, does not possess the Eth monooxygenase (29, 30).
Due to the transposon-mediated genetic instability, prolonged cultivation on nonselective growth substrates resulted in the formation of eth deletion mutants, e.g., from R. ruber IFP 2001 (7) and from A. tertiaricarbonis L108 (14). The inability of these mutants to degrade ethers shows that the Eth monooxygenase is responsible for the degradation of all four fuel oxygenates in the wild-type strains. The MTBE-to-glucose shifts revealed, however, that at least in strain L108 the transposon-mediated recombination event does not occur during exponential growth, as the expression and gene copy number of ethC did not differ even after five generations on the nonselective substrate from those of the inoculum. In line with this, it has been demonstrated that transposase activities are typically correlated with stress conditions, such as cold shock, subinhibitory concentrations of antibiotics, or entry into stationary phase, rather than with exponential growth (31, 32, 33).
The Eth monooxygenase system seems to be widespread among Gram-positive bacteria, as it has already been found in several strains of the genera Rhodococcus, Mycobacterium, and Gordonia (7, 34). However, all these bacteria do not possess the tert-alcohol degradation pathways needed for complete mineralization of the fuel oxygenate ethers. On the other hand, strain M. petroleiphilum PM1 can degrade TBA and TAA completely (25, 26) but attacks the ethers by an AlkB-type monooxygenase system (35), which can hydroxylate only MTBE and TAME but not the ethyl ethers (19, 29). Thus far, only the betaproteobacterium A. tertiaricarbonis L108 is known to possess Eth monooxygenase and the complete pathways for the mineralization of tert-alcohols. Interestingly, these degradation capacities combined here in a single strain have been demonstrated in a mixed culture of R. ruber IFP 2001 and strain IFP 2003, another A. tertiaricarbonis strain lacking ether-hydroxylating activity but able to degrade TBA and TAA (18, 36). However, to grow on MTBE this consortium had to be amended with ethanol or isopropanol, obviously because the strain IFP 2001 could not gain energy from the formaldehyde/formic acid released from the ether or from TBA, which was exclusively degraded by the strain IFP 2003 (37).
Previously, Malandain and coworkers (9) demonstrated that the transcriptional regulator EthR was responsible for ETBE-induced expression of the Eth monooxygenase in the Gram-positive strains IFP 2001, IFP 2005, and IFP 2009. Without induction, these strains attacked ETBE at >10-fold reduced rates, and degradation of MTBE and TAME was almost insignificant. This regulation may also explain the results obtained with Rhodococcus sp. strain PEG604, which tested negative on MTBE but likely possesses a CYP249A1 monooxygenase system, as the ethB gene with >99% identity to the sequence found in strain IFP 2001 has been detected in this strain (34). Obviously, the lack of a suitable inducer resulted in insufficient expression of the monooxygenase and, consequently, insignificant degradation of MTBE in strain PEG604. Therefore, the outcome of a combination of genetic analyses and degradation tests performed without consideration of the regulation of gene expression should be interpreted with caution, as it may not reflect the true degradation potential of a certain bacterial strain or enzyme system. Surprisingly, the constitutive expression of the eth genes in strain L108 lacking the positive transcriptional regulator EthR did not result in a 10-fold but only in a 3-fold reduction of the Eth monooxygenase activity compared to the ETBE-induced strain IFP 2001. Obviously, the presence of a σ70-binding site upstream of ethA (Fig. 4) enables relatively high expression rates even without EthR-dependent positive regulation.
In this and also in previous studies (19), it has been found that all fuel oxygenate ethers are degraded at nearly the same rate by strain L108, indicating that the Eth monooxygenase is not specific for one of these compounds. However, ETBE-induced cells of strain IFP 2001 show an 8- to 11-fold decreased degradation activity with MTBE and TAME compared to the high rates observed with the ethyl ether (9). In the latter case, the Eth monooxygenase activity may have been underestimated thus far, as strain IFP 2001 degrades fuel oxygenate ethers only incompletely and significantly converts the methyl ethers only when ethanol or glucose is added as a cosubstrate (5, 38, 39). As a consequence, imbalance in reducing equivalents or oxygen depletion due to additional consumption by the cosubstrates could be a reason for the reduced activity of the Eth monooxygenase in strain IFP 2001.
Besides the fuel oxygenates MTBE, ETBE, TAME, and TAEE, the Eth monooxygenase can also attack other short-chain dialkyl ethers. With strain L108, we have demonstrated the EthABCD-catalyzed degradation of diethyl and diisopropyl ether. In addition, ETBE-induced cells of strain IFP 2001 were already shown to attack n-butyl ethyl ether, di-n-butyl ether, diisopropyl ether, and various short-chain glycol ethers (39). However, synthetic ethers with larger residues, such as n-hexyl methyl ether and the three alkyl aryl ethers tested in this study, anisole, phenetole, and isopropoxybenzene, are not substrates for the Eth monooxygenase. Neither was the cyclic ether tetrahydrofuran. In conclusion, although hydroxylation of methoxy, ethoxy, isopropoxy, and n-butoxy groups has been demonstrated, the second, nonreacting residue in the ether molecule may not exceed the size of tert-amyl as the largest residue that tested positive with the Eth system.
Supplementary Material
ACKNOWLEDGMENTS
We thank C. Dilssner, M. Neytschev, and B. Würz (UFZ) for excellent technical and analytical assistance. We are also indebted to K. Glaser and D. Türkowsky (UFZ) for the introduction to qPCR, T. Weichler (UFZ) for the promoter probe experiment, and T. Unger (UFZ) for providing the vector pCM130.
We are grateful to EMBO (European Molecular Biology Organization) for the financial support of J.P. (ASTF, 262-09).
Footnotes
Published ahead of print 25 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03348-12.
REFERENCES
- 1. Barceló D. 2007. Fuel oxygenates. Springer-Verlag, Berlin, Heidelberg, Germany [Google Scholar]
- 2. White GF, Russell NJ, Tidswell EC. 1996. Bacterial scission of ether bonds. Microbiol. Rev. 60: 216– 232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Moran MJ, Zogorski JS, Squillace PJ. 2005. MTBE and gasoline hydrocarbons in ground water of the United States. Ground Water 43: 615– 627 [DOI] [PubMed] [Google Scholar]
- 4. Weaver JW, Exum LR, Prieto LM. 2010. Gasoline composition regulations affecting LUST sites. EPA 600/R-10/001. Office of Research and Development, U.S. Environmental Protection Agency, Washington, DC: [Google Scholar]
- 5. Rosell M, Gonzales-Olmos R, Rohwerder T, Rusevova K, Georgi A, Kopinke FD, Richnow HH. 2012. Critical evaluation of the 2D-CSIA scheme for distinguishing fuel oxygenate degradation reaction mechanisms. Environ. Sci. Technol. 46: 4757– 4766 [DOI] [PubMed] [Google Scholar]
- 6. Hyman M. 29 October 2012. Biodegradation of gasoline ether oxygenates. Curr. Opin. Biotechnol. [Epub ahead of print.] doi:10.1016/j.copbio.2012.10.005 [DOI] [PubMed] [Google Scholar]
- 7. Béguin P, Chauvaux S, Miras I, François A, Fayolle F, Monot F. 2003. Genes involved in the degradation of ether fuels by bacteria of the Mycobacterium/Rhodococcus group. Oil Gas Sci. Technol. 58: 489– 495 [Google Scholar]
- 8. Chauvaux S, Chevalier F, Le Dantec C, Fayolle F, Miras I, Kunst F, Béguin P. 2001. Cloning of a genetically unstable cytochrome P-450 gene cluster involved in degradation of the pollutant ethyl tert-butyl ether by Rhodococcus ruber. J. Bacteriol. 183: 6551– 6557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Malandain C, Fayolle-Guichard F, Vogel TM. 2010. Cytochrome P450-mediated degradation of fuel oxygenates by environmental isolates. FEMS Microbiol. Ecol. 72: 289– 296 [DOI] [PubMed] [Google Scholar]
- 10. Jechalke S, Rosell M, Martínez-Lavanchy PM, Pérez-Leiva P, Rohwerder T, Vogt C, Richnow HH. 2011. Linking low-level stable isotope fractionation to expression of the cytochrome P450 monooxygenase-encoding ethB gene for elucidation of methyl tert-butyl ether biodegradation in aerated treatment pond systems. Appl. Environ. Microbiol. 77: 1086– 1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Breuer U, Bäjen C, Rohwerder T, Müller RH, Harms H. 2007. MTBE degradation genes of an Ideonella-like bacterium L108, p 103 In Bastiaens L. (ed), Proceedings of the 3rd European conference on MTBE and other fuel oxygenates. VITO, Antwerp, Belgium [Google Scholar]
- 12. Schäfer F, Breuer U, Benndorf D, von Bergen M, Harms H, Müller RH. 2007. Growth of Aquincola tertiaricarbonis L108 on tert-butyl alcohol leads to the induction of a phthalate dioxygenase-related protein and its associated oxidoreductase subunit. Eng. Life Sci. 7: 512– 519 [Google Scholar]
- 13. Schäfer F, Schuster J, Würz B, Härtig C, Harms H, Müller RH, Rohwerder T. 2012. Synthesis of short-chain diols and unsaturated alcohols from secondary alcohol substrates by the Rieske nonheme mononuclear iron oxygenase MdpJ. Appl. Environ. Microbiol. 78: 6280– 6284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rohwerder T, Breuer U, Benndorf D, Lechner U, Müller RH. 2006. The alkyl tert-butyl ether intermediate 2-hydroxyisobutyrate is degraded via a novel cobalamin-dependent mutase pathway. Appl. Environ. Microbiol. 72: 4128– 4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yaneva N, Schuster J, Schäfer F, Lede V, Przybylski D, Paproth T, Harms H, Müller RH, Rohwerder T. 2012. Bacterial acyl-CoA mutase specifically catalyzes coenzyme B12-dependent isomerisation of 2-hydroxyisobutyryl-CoA and (S)-3-hydroxybutyryl-CoA. J. Biol. Chem. 287: 15502– 15511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schuster J, Schäfer F, Hübler N, Brandt A, Rosell M, Härtig C, Harms H, Müller RH, Rohwerder T. 2012. Bacterial degradation of tert-amyl alcohol proceeds via hemiterpene 2-methyl-3-buten-2-ol by employing the tertiary alcohol desaturase function of the Rieske nonheme mononuclear iron oxygenase MdpJ. J. Bacteriol. 194: 972– 981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Müller RH, Rohwerder T, Harms H. 2008. Degradation of fuel oxygenates and their main intermediates by Aquincola tertiaricarbonis L108. Microbiology 154: 1414– 1421 [DOI] [PubMed] [Google Scholar]
- 18. Lechner U, Brodkorb D, Geyer R, Hause G, Härtig C, Auling G, Fayolle-Guichard F, Piveteau P, Müller RH, Rohwerder T. 2007. Aquincola tertiaricarbonis gen. nov., sp. nov., a tertiary butyl moiety-degrading bacterium. Int. J. Syst. Evol. Microbiol. 57: 1295– 1303 [DOI] [PubMed] [Google Scholar]
- 19. Schäfer F, Muzica L, Schuster J, Treuter N, Rosell M, Harms H, Müller RH, Rohwerder T. 2011. Alkene formation from tertiary alkyl ether and alcohol degradation by Aquincola tertiaricarbonis L108 and Methylibium spp. Appl. Environ. Microbiol. 77: 5981– 5987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389– 3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zomer AL, Buist G, Larsen R, Kok J, Kuipers OP. 2007. Time-resolved determination of the CcpA regulon of Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189: 1366– 1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34: D32– D36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marx CJ, Lidstrom ME. 2001. Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology 147: 2065– 2075 [DOI] [PubMed] [Google Scholar]
- 24. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166: 557– 580 [DOI] [PubMed] [Google Scholar]
- 25. Hanson JR, Ackerman CE, Scow KM. 1999. Biodegradation of methyl tert-butyl ether by a bacterial pure culture. Appl. Environ. Microbiol. 65: 4788– 4792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakatsu CH, Hristova K, Hanada S, Meng XY, Hanson J, Scow KM, Kamagata Y. 2006. Methylibium petroleiphilum PM1T gen. nov., sp. nov., a new methyl tert-butyl ether (MTBE) degrading methylotroph of the beta-Proteobacteria. Int. J. Syst. Evol. Microbiol. 56: 983– 989 [DOI] [PubMed] [Google Scholar]
- 27. Wiedenbeck J, Cohan FM. 2011. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol. Rev. 35: 957– 976 [DOI] [PubMed] [Google Scholar]
- 28. Springael D, Top EM. 2004. Horizontal gene transfer and microbial adaptation to xenobiotics: new types of mobile genetic elements and lessons from ecological studies. Trends Microbiol. 12: 53– 58 [DOI] [PubMed] [Google Scholar]
- 29. Hristova KR, Schmidt R, Chakicherla AY, Legler TC, Wu J, Chain PS, Scow KM, Kane SR. 2007. Comparative transcriptome analysis of Methylibium petroleiphilum PM1 exposed to the fuel oxygenates methyl tert-butyl ether and ethanol. Appl. Environ. Microbiol. 73: 7347– 7357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kane SR, Chakicherla AY, Chain PS, Schmidt R, Shin MW, Legler TC, Scow KM, Larimer FW, Lucas SM, Richardson PM, Hristova KR. 2007. Whole-genome analysis of the methyl tert-butyl ether-degrading beta-proteobacterium Methylibium petroleiphilum PM1. J. Bacteriol. 189: 1931– 1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Haniford DB. 2006. Transpososome dynamics and regulation in Tn10 transposition. Crit. Rev. Biochem. Mol. Biol. 41: 407– 424 [DOI] [PubMed] [Google Scholar]
- 32. Pedró L, Banos RC, Aznar S, Madrid C, Balsalobre C, Juárez A. 2011. Antibiotics shaping bacterial genome: deletion of an IS91 flanked virulence determinant upon exposure to subinhibitory antibiotic concentrations. PLoS One 6: e27606 doi:10.1371/journal.pone.0027606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Whitfield CR, Wardle SJ, Haniford DB. 2009. The global bacterial regulator H-NS promotes transpososome formation and transposition in the Tn5 system. Nucleic Acids Res. 37:309– 321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim YH, Engesser KH, Kim SJ. 2007. Physiological, numerical and molecular characterization of alkyl ether-utilizing rhodococci. Environ. Microbiol. 9: 1497– 1510 [DOI] [PubMed] [Google Scholar]
- 35. Schmidt R, Battaglia V, Scow K, Kane S, Hristova KR. 2008. A novel enzyme, MdpA, is involved in MTBE degradation in Methylibium petroleiphilum PM1. Appl. Environ. Microbiol. 74: 6631– 6638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Piveteau P, Fayolle F, Vandecasteele JP, Monot F. 2001. Biodegradation of tert-butyl alcohol and related xenobiotics by a methylotrophic bacterial isolate. Appl. Microbiol. Biotechnol. 55: 369– 373 [DOI] [PubMed] [Google Scholar]
- 37. Piveteau P, Fayolle F, Le Penru Y, Monot F. 2000. Biodegradation of MTBE by cometabolism in laboratory-scale fermentations, p 141–148 In Wickramanyake GB, Gavaskar A, Alleman BC, Magar VS. (ed), Bioremediation and phytoremediation of chlorinated and recalcitrant compounds. Battelle Press, Columbus, OH [Google Scholar]
- 38. Rosell M, Barcelo D, Rohwerder T, Breuer U, Gehre M, Richnow HH. 2007. Variation in 13C/12C and D/H enrichment factors of aerobic bacterial fuel oxygenate degradation. Environ. Sci. Technol. 41: 2036– 2043 [DOI] [PubMed] [Google Scholar]
- 39. Hernandez-Perez G, Fayolle F, Vandecasteele JP. 2001. Biodegradation of ethyl tert-butyl ether (ETBE), methyl tert-butyl ether (MTBE) and tert-amyl methyl ether (TAME) by Gordonia terrae. Appl. Microbiol. Biotechnol. 55: 117– 121 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.