Abstract
A carbapenem-resistant clinical isolate of Escherichia coli, which lacked OmpF and OmpC porins, carried a marR mutation and expressed a functional yedS, a normally nontranslated gene. MarR and YedS are described here as having effects on the ability of this strain to resist carbapenems. Additionally, expression of YedS was regulated by the small RNA MicF in a MarA-dependent way. These findings illustrate how broadly bacteria can mutate within a selective clinical setting, in this case, resistance to carbapenems, by altering three porin genes and one regulatory gene.
TEXT
Carbapenems are broad-spectrum β-lactam antibiotics used for the treatment of multidrug-resistant Gram-negative pathogens (1–3). Carbapenem resistance most commonly arises through the acquisition of genes encoding carbapenemases, which hydrolyze carbapenems (3–5). The other chief mechanism of carbapenem resistance in Escherichia coli and other Enterobacteriaceae is decreased bacterial cell permeability due to loss or alteration of the outer membrane porins F and/or C (1, 6–8).
The marRAB operon of E. coli encodes the MarR repressor, the transcriptional regulator MarA, and a putative small protein, MarB (9). MarR represses transcription of marRAB by binding to marO and negatively controlling MarA-dependent expression of other genes in the regulon (10, 11). Upon induction by a variety of compounds (12) or by mutation of marR or marO, the repressor is rendered inactive (10). The resulting overexpression of MarA produces antibiotic resistance by increasing the expression of the major multidrug efflux pump AcrAB-TolC (13, 14) and downregulating the outer membrane protein OmpF via the small RNA (sRNA) MicF (15, 16). In this study, a carbapenem-resistant, non-carbapenemase-producing clinical isolate of E. coli from China (CH4) was investigated to determine the genetic basis for the carbapenem resistance phenotype.
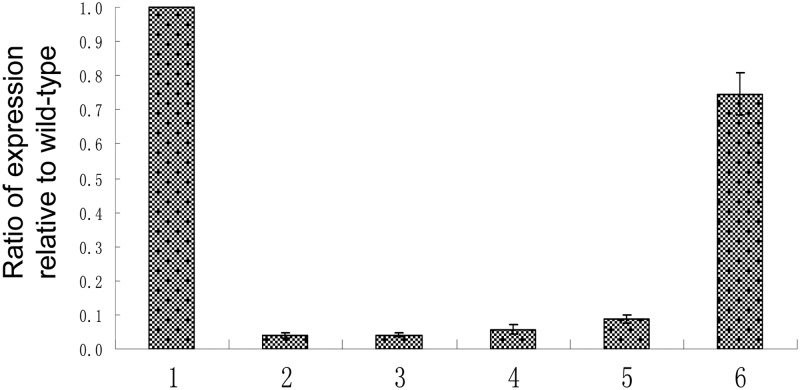
PCR amplification and sequencing using the primers marR-for (5′-ATTAGCGGCCGCATCGGTCAATTCAT) and marR-rev (5′-ATAGGATCCTTACGGCTGCGGATGTA) revealed numerous mutations in the marR open reading frame (ORF) of strain CH4 and other clinical isolates from China (Table 1). We cloned ORFs containing the various marR mutations, using the primers marR-clone-For and marR-clone-Rev (17), into expression vector pET-13a (18), for which expression was controlled by the T7 promoter. Expression of T7 polymerase was induced from plasmid pACT7-Spc (19) via isopropyl-β-d-thiogalactopyranoside (IPTG) in the reporter strain SPC-106, a marO-lacZ fusion that contains a ΔmarR mutation (12). Analysis of LacZ activity (11, 20) showed that the Gly42Arg mutation in the CH4 marR gene did not complement the ΔmarR mutation in this reporter strain (Fig. 1), indicating that this mutation affected the activity of MarR.
Table 1.
Effects of complementation of wild-type MarR and YedSCH4 on carbapenem susceptibility in clinical and laboratory strains of E. coli
| Strain | Mutation(s) in marR | MIC (μg/ml)a |
||
|---|---|---|---|---|
| Imipenem | Meropenem | Ertapenem | ||
| CH4 | Gln42Arg, Gly103Ser, Tyr137His | >32 | 32 | >32 |
| CH4/pACT7/pET13a | >32 | 32 | >32 | |
| CH4/pACT7/pmarRwildtype | 0.25 | 0.38 | 0.25 | |
| CH4/pACT7/pyedSCH4 | 1.5 | 1.5 | 4.0 | |
| CH4/pAC-MarRwt | 0.25 | 0.38 | 0.25 | |
| BL21(DE3)/pET13a | Wild-type marR | 0.047 | 0.023b | 0.047 |
| BL21(DE3)/pyedSCH4 | 0.004 | 0.023b | 0.004 | |
MICs were determined using Etest (bioMérieux). CH4 cells were cultured on LB agar containing kanamycin (800 μg/ml) and spectinomycin (200 μg/ml) when carrying pET13a plasmids and pACT7 plasmids, respectively. BL21(DE3) cells were cultured on agar containing kanamycin (50 μg/ml) when carrying the pET13a plasmid. All cultures were induced with IPTG (0.5 mM).
Meropenem MICs were not affected by overexpression of YedS in BL21(DE3), most likely due to other factors in this strain which affect susceptibility to this carbapenem.
Fig 1.
Reporter gene assay for MarR function. The reporter strain SPC105ΔmarR, which lacks the marR gene, carries a chromosomal PmarII::lacZ transcriptional fusion. Results are expressed as percentages of transcription activity of the control (SPC105ΔmarR bearing pACT7 and pET13a without the insert) and are the means and standard deviations of results from at least three replicated assays. All assays were performed as described previously (20). The origins of the cloned marR genes were as follows: bar 1, none; bar 2, wild-type marR; bar 3, marR gene encoding the Lys62Arg, Gly103Ser, and Tyr137His mutations; bar 4, marR gene encoding the Gly103Ser and Tyr137His mutations; bar 5, marR gene encoding the Ala53Glu, Gly103Ser, and Tyr137His mutations; bar 6, marRCH4 gene encoding the Gln42Arg, Gly103Ser, and Tyr137His mutations.
We then complemented the marR mutation in CH4 by transforming the strain with pET-marRwildtype and pACT7-Spc or pAC-MarRwt (17). MICs were determined. The data showed that the two expression vectors produced similar decreases in resistance in strain CH4 that were not seen with an empty vector control (Table 1). We hypothesized that this effect was due to expression of ompF; however, sequencing showed this gene to be inactivated by a partial deletion mutation. Upon extraction of outer membrane proteins from CH4 derivative strains (listed in Fig. 2), we found an ∼30-kDa protein newly expressed upon addition of wild-type MarR (Fig. 2). This protein was purified and processed for N-terminal sequencing, which revealed the protein to be YedS. The encoding gene, yedS, is a previously described pseudogene which is untranslatable due to a large gap in the ORF in most sequenced strains. Sequence analysis of the yedS gene in strain CH4 showed a complete and translatable gene.
Fig 2.
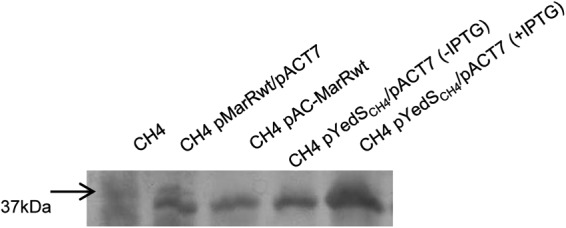
Urea SDS-PAGE analysis of outer membrane proteins. Outer membrane proteins were purified and subjected to gel electrophoresis as described previously (27). The arrow to the left denotes the migration of the 37-kDa molecular weight marker. Each lane was loaded with 5 μg of total outer membrane protein.
Subsequent cloning of yedSCH4 into pET13a via amplification with primers yed-nde-for (5′-GCGCCATATGAAAAGAAAAGTTCTGG) and yed-bam-rev (5′-ATAAGGATCCGAACTGGTAGACGATA) revealed it to be transcribed and translated into a similarly sized outer membrane protein in strain CH4 (Fig. 2). When these plasmid-bearing strains were tested in MIC studies, decreased carbapenem resistance was observed in strains CH4 and BL21(DE3) (Invitrogen) (Table 1), indicating that YedSCH4 is involved in carbapenem resistance.
To investigate the link of yedSCH4 transcription to MarA, we engineered a yedSCH4-lacZ promoter fusion plasmid using primers yedS-lac pro for (5′-GCACCAATTGCCCGGAAAATTCAGAC) and yedS-lac pro rev (5′-AGTCGGATCCTGTATTCCCTTGTGA) and reporter plasmid pRS415 (21). This construct was transformed into lacZ-lacking strains from the Keio collection (22, 23) containing mutations in either marR or marRA (Table 2). When these strains were grown to late log phase at temperatures of 37°C, we found that expression of the yedSCH4 promoter was ∼30% in the marR strain compared to that of its wild-type parent. However, when the marR strain also contained a marA deletion, transcription of yedSCH4-lacZ was equal to that of the parental strain. Suspecting that this relationship was due to the MarA-regulated micF, we transduced (24) a micF::Cm mutation into these strains and observed a restored transcription of yedSCH4-lacZ in all strains (Table 2). Thus, the mar operon controls expression of yedSCH4 via the sRNA micF.
Table 2.
Effects of marR, marA, and micF mutations on the yedSCH4 promotera
| Strain | Relative β-galactosidase value |
|
|---|---|---|
| micF+ | micF::Cm | |
| BW25113 | 1 | 1 |
| CR1000 (ΔmarR) | 0.29 ± 0.13 | 1.01 ± 0.07 |
| CR2000 (ΔmarRA) | 0.96 ± 0.07 | 1.29 ± 0.09 |
Data represent β-galactosidase values relative to that of the wild-type strain BW25113 containing plasmid pRS415-yedSCH4-lacZ, grown under the same conditions, and the standard error of the mean. All strains were grown in LB broth supplemented with ampicillin (100 μg ml−1) to maintain carriage of pRS415-yedSCH4-lacZ.
Our findings implicate the outer membrane protein YedSCH4 in carbapenem sensitivity/resistance. We hypothesize that the maintenance of a functional YedS in strain CH4 is an evolutionary response to the lack of functional OmpF and OmpC. Additionally, the presence of this carbapenem portal presents a selective pressure for this strain to maintain its novel marR mutation, downregulating yedSCH4 expression via MicF and producing resistance to carbapenems. In the absence of carbapenemase, selection may occur for mar mutants which will be resistant to a greater spectrum of antibiotics and potentially have greater virulence (25, 26) than parental strains containing functional marR genes. Our findings suggest how this uniquely selective environment may affect genetic fluidity of the bacterial cell that seeks to survive in response to different insults. The isolate described here has mutated two of its porins, enabled a pseudogene to be expressed, and derepressed the marRAB operon, sufficient to produce a drug-resistant strain. The order in which these mutations occurred is not known; however, the accumulation of so many mutations in a single isolate is a clear display of bacterial adaptation.
Nucleotide sequence accession number.
The sequence of the yedS gene in strain CH4 was deposited in GenBank under accession number JX392406.
ACKNOWLEDGMENT
We thank bioMérieux for providing the Etests used in this work.
Footnotes
Published ahead of print 14 January 2013
REFERENCES
- 1. Chia JH, Siu LK, Su LH, Lin HS, Kuo AJ, Lee MH, Wu TL. 2009. Emergence of carbapenem-resistant Escherichia coli in Taiwan: resistance due to combined CMY-2 production and porin deficiency. J. Chemother. 21:621–626 [DOI] [PubMed] [Google Scholar]
- 2. Hsueh PR, Hoban DJ, Carmeli Y, Chen SY, Desikan S, Alejandria M, Ko WC, Binh TQ. 2011. Consensus review of the epidemiology and appropriate antimicrobial therapy of complicated urinary tract infections in Asia-Pacific region. J. Infect. 63:114–123 [DOI] [PubMed] [Google Scholar]
- 3. Yang Q, Wang H, Sun H, Chen H, Xu Y, Chen M. 2010. Phenotypic and genotypic characterization of Enterobacteriaceae with decreased susceptibility to carbapenems: results from large hospital-based surveillance studies in China. Antimicrob. Agents Chemother. 54:573–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goren MG, Navon-Venezia S, Chmelnitsky I, Carmeli Y. 2010. Carbapenem-resistant KPC-2-producing Escherichia coli in a Tel Aviv Medical Center, 2005 to 2008. Antimicrob. Agents Chemother. 54:2687–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Queenan AM, Bush K. 2007. Carbapenemases: the versatile β-lactamases. Clin. Microbiol. Rev. 20:440–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oteo J, Delgado-Iribarren A, Vega D, Bautista V, Rodriguez MC, Velasco M, Saavedra JM, Perez-Vazquez M, Garcia-Cobos S, Martinez-Martinez L, Campos J. 2008. Emergence of imipenem resistance in clinical Escherichia coli during therapy. Int. J. Antimicrob. Agents 32:534–537 [DOI] [PubMed] [Google Scholar]
- 7. Poirel L, Heritier C, Spicq C, Nordmann P. 2004. In vivo acquisition of high-level resistance to imipenem in Escherichia coli. J. Clin. Microbiol. 42:3831–3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stapleton PD, Shannon KP, French GL. 1999. Carbapenem resistance in Escherichia coli associated with plasmid-determined CMY-4 β-lactamase production and loss of an outer membrane protein. Antimicrob. Agents Chemother. 43:1206–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cohen SP, Hachler H, Levy SB. 1993. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J. Bacteriol. 175:1484–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alekshun MN, Levy SB, Mealy TR, Seaton BA, Head JF. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 A resolution. Nat. Struct. Biol. 8:710–714 [DOI] [PubMed] [Google Scholar]
- 11. Seoane AS, Levy SB. 1995. Characterization of MarR, the repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli. J. Bacteriol. 177:3414–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cohen SP, Levy SB, Foulds J, Rosner JL. 1993. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J. Bacteriol. 175:7856–7862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li XZ, Nikaido H. 2004. Efflux-mediated drug resistance in bacteria. Drugs 64:159–204 [DOI] [PubMed] [Google Scholar]
- 14. Okusu H, Ma D, Nikaido H. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178:306–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cohen SP, McMurry LM, Levy SB. 1988. marA locus causes decreased expression of OmpF porin in multiple-antibiotic-resistant (Mar) mutants of Escherichia coli. J. Bacteriol. 170:5416–5422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chubiz LM, Rao CV. 2011. Role of the mar-sox-rob regulon in regulating outer membrane porin expression. J. Bacteriol. 193:2252–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alekshun MN, Levy SB. 1999. Characterization of MarR superrepressor mutants. J. Bacteriol. 181:3303–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60–89 [DOI] [PubMed] [Google Scholar]
- 19. Maneewannakul K, Maneewannakul S, Ippen-Ihler K. 1992. Sequence alterations affecting F plasmid transfer gene expression: a conjugation system dependent on transcription by the RNA polymerase of phage T7. Mol. Microbiol. 6:2961–2973 [DOI] [PubMed] [Google Scholar]
- 20. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 21. Simons RW, Houman F, Kleckner N. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85–96 [DOI] [PubMed] [Google Scholar]
- 22. Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol 2:2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ruiz C, Levy SB. 2010. Many chromosomal genes modulate MarA-mediated multidrug resistance in Escherichia coli. Antimicrob. Agents Chemother. 54:2125–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nicoloff H, Perreten V, McMurry LM, Levy SB. 2006. Role for tandem duplication and Lon protease in AcrAB-TolC-dependent multiple antibiotic resistance (Mar) in an Escherichia coli mutant without mutations in marRAB or acrRAB. J. Bacteriol. 188:4413–4423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Warner DM, Levy SB. 2012. SoxS increases the expression of the zinc uptake system ZnuACB in an Escherichia coli murine pyelonephritis model. J. Bacteriol. 194:1177–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Casaz P, Garrity-Ryan LK, McKenney D, Jackson C, Levy SB, Tanaka SK, Alekshun MN. 2006. MarA, SoxS and Rob function as virulence factors in an Escherichia coli murine model of ascending pyelonephritis. Microbiology 152:3643–3650 [DOI] [PubMed] [Google Scholar]
- 27. Duval V, Nicoloff H, Levy SB. 2009. Combined inactivation of lon and ycgE decreases multidrug susceptibility by reducing the amount of OmpF porin in Escherichia coli. Antimicrob. Agents Chemother. 53:4944–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]