Abstract
Cationic antimicrobial peptides are essential components of the innate immune system. As a major family of mammalian antimicrobial peptides, defensins are expressed mainly by mucosal epithelial cells and promyelocytes. Despite the capacity to kill a broad spectrum of bacteria through physical disruption of membranes, most defensins show substantially reduced antibacterial activities in the presence of monovalent and divalent cations, thereby limiting their therapeutic potential, particularly for the treatment of systemic infections. Genome-wide computational screening of the rat genome led to the identification of the gene for a novel α-defensin-related peptide that we termed rattusin. Rattusin shares a highly conserved signal and prosequence with mammalian α-defensins, but instead of the canonical α-defensin six-cysteine motif, rattusin consists of five cysteines with a distinctive spacing pattern. Furthermore, rattusin is preferentially expressed in Paneth cells of the distal small intestine with potent antibacterial activity against a broad range of Gram-negative and Gram-positive bacteria, including antibiotic-resistant strains. The MICs were mostly in the range of 2 to 4 μM, with no appreciable toxicity to mammalian cells at up to 100 μM. In contrast to classical α- and β-defensins, rattusin retained its activity in the presence of physiological concentrations of NaCl and Mg2+, making it an attractive antimicrobial candidate for both topical and systemic applications.
INTRODUCTION
The emergence of antibiotic-resistant pathogens is a major health crisis worldwide (1), and there is an urgent need to develop novel antimicrobial drugs against resistant microbes. As critical components of innate defense mechanisms, cationic antimicrobial peptides are capable of killing a broad spectrum of bacteria, including antibiotic-resistant strains with potential for further exploration as a new class of antimicrobial drugs (2–4). Two major families of antimicrobial peptides, namely, defensins and cathelicidins, exist in vertebrates (5–8). Unlike cathelicidins, which are often devoid of cysteines, defensins are characterized by six cysteines in well-defined spacing patterns forming intramolecular disulfide bonds whose pairings define the three mammalian defensin subfamilies (Fig. 1). Besides the well-described α-, β-, and θ-defensins with six characteristic cysteine residues, two additional subfamilies of α-defensin-related cryptdin-related sequence (CRS) peptides (CRS1C and CRS4C) exist that contain 9 and 11 cysteines, respectively (9). In mammals, α-defensins are produced mainly by promyelocytes and intestinal Paneth cells, whereas β-defensins are widely expressed by diverse mucosal epithelial cells (5–7). In contrast, θ-defensins are uniquely expressed in Old World monkey promyelocytes and accumulate in neutrophils and monocytes, and CRS peptides are exclusive to mice and are abundant in Paneth cell secretory granules (5–7, 9).
Fig 1.
Schematic drawing of the structure of a mammalian defensin precursor. Although all classical α-, β-, and θ-defensins contain six cysteines with different disulfide arrays, three subfamilies of α-defensin-related sequences (defa-rs) with a different number and spacing pattern of cysteines have been found in intestinal Paneth cells of mice (CRS1C and CRS4C) and rats (rattusin). The prosequences of defensins are highly conserved within but not between subfamilies, except for β-defensins, whose prosequences are variable as well. Biologically active, mature sequences of defensins are released from proforms through proteolytic cleavage.
Defensins possess pleotropic functions in host defense. In addition to broad-spectrum antibacterial, antiviral, and antifungal activities, certain α- or β-defensins may be chemotactic for dendritic, mast, monocytic, and T cells (5–7) and may induce maturation of dendritic cells (10). Defensins help in wound healing by inducing vascularization, promoting proliferation of epithelial and fibroblast cells, and augmenting wound closure (5–7, 10). In addition, several human neutrophil α-defensins are capable of neutralizing bacterial toxins (11, 12).
Defensins are encoded by distinct genes and synthesized initially as precursors with conserved signal and propeptide sequences (Fig. 2). Biologically active, mature defensins are generated through specific proteolytic cleavage events within the prosequence. For example, matrix metalloproteinase 7 (MMP7) processes Paneth cell α-defensin precursors into biologically active peptides in mice (13), whereas elastase and proteinase 3 appear to be the convertases for neutrophil α-defensin precursors in humans (14) and an intracellular proteoglycan, serglycin, is involved in the retention of mature peptides in neutrophil granules (15). Human intestinal α-defensin (HD5), on the other hand, is processed by trypsin (16).
Fig 2.

Amino acid sequence alignment of rattusin with representative α-defensins and related peptides in rodents and humans. Dashes were inserted to maximize the alignment. Conserved amino acids are shaded, and mature sequences are underlined. Vertical arrows indicate the known start sites of mature peptides. The length of each mature peptide is also indicated. The signal peptides and prosequences of the peptides are conserved, whereas carboxyl-terminal mature peptides are diversified. Note that the spacing pattern of cysteine residues in rattusin differs from those of all other defensins and defensin-related peptides. Abbreviations: rDefa6, rat α-defensin 6; RatNP4, rat neutrophil peptide α-defensin 4; DEFA5/HD5, human α-defensin 5.
The innate immune role of Paneth cell α-defensins in protecting enteric infections has been well documented. Transgenic mice that express the gene for HD5 are immune to oral Salmonella enterica serovar Typhimurium (S. Typhimurium) infections (17). Also, mice lacking the gene for MMP7 are more susceptible to systemic S. Typhimurium disease and are defective in the clearance of enteric infection because of an inability of Paneth cells to produce mature α-defensins (13). A deficiency in the synthesis of intestinal α-defensins was also recently linked to the pathogenesis of ileal Crohn's disease (18). In addition, Paneth cell α-defensins play a critically important role in regulating the composition of the ileal microbiota (19) with potential implications in health and disease (20).
Although mature defensins are broadly active against both Gram-positive and -negative bacteria, their potential as therapeutics has been hampered by a substantial loss of antibacterial activity in the presence of physiological concentrations of salts (5–7). It is worth noting that although they are essential for chemotaxis (21) and peptide resistance to proteolysis (22), the integrity of conserved disulfide arrays and the spatial structure they impose are not, per se, critical to the microbicidal activity of α- and β-defensins but are required for that of θ-defensins (23).
Through a comprehensive genome-wide computational screening of the genomes of several phylogenetically distant animal species, including primates, rodents, and dogs, we identified a unique α-defensin-related sequence in the rat (24), termed rattusin for Rattus defensin. Rattusin belongs to a new defensin subfamily with a unique cysteine spacing pattern that is distinct from that of all other known peptides (24) (Fig. 1). We now show that the gene for rattusin is abundantly expressed in Paneth cells of the distal small intestine. Desirably, synthetic rattusin possesses potent bactericidal activities that are insensitive to physiological concentrations of NaCl and Mg2+, making it an attractive novel antimicrobial candidate.
MATERIALS AND METHODS
Reverse transcriptase (RT)-PCR.
Different segments of the gastrointestinal tract were collected from healthy 2-month-old Sprague-Dawley rats. Total RNA was extracted with TRIzol (Invitrogen). For each RNA sample, 4 μg was reverse transcribed with random hexamers and SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. The subsequent PCR was carried out as described previously (24). Briefly, 1/40 of the first-strand cDNA was used to amplify rattusin, MMP7, three isoforms of trypsin, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) with gene-specific primers (Table 1). The identities of PCR products were confirmed by agarose gel electrophoresis, followed by gel purification, cloning into T/A cloning vector, and direct DNA sequencing.
Table 1.
Primers sequences used for RT-PCR analysis of rattusin and intestinal proteases
| Gene product | Forward primer | Reverse primer | GenBank accession no. | Product size (bp) |
|---|---|---|---|---|
| Rattusin | GAAGACACTTGTCCTCCTTTCTG | ATGTGGACCTTGATAGCCGATG | AY623756 | 261 |
| MMP7 | AGTGGACAAACTGAGGGAAATG | ACTAAGAACCGAGGCAAGTCTG | NM_012864 | 210 |
| Anionic trypsin | GTTGGAGGATACACCTGCCCG | CTTGATGATCTTGGCAGCATTG | NM_012635 | 213 |
| Mesotrypsin | GGAGGATACACCTGCCAAGAGA | CTTGATGATCTTGGCAGCATTG | NM_001108626 | 210 |
| Cationic trypsin | ATTGATGTCGTTGAGGGTGGT | GAAGCAGTGAAGGGTAGTTCGT | NM_173127 | 244 |
| GAPDH | GTGAAGGTCGGAGTGAACG | GAGATGATGACCCTCTTGGC | NM_017008 | 356 |
In situ hybridization.
A proximal ileal segment was freshly collected from a healthy 2-month-old Sprague-Dawley rat and fixed for 4 h in phosphate-buffered formalin solution. Tissue preparation and in situ hybridization were done largely as described previously (25). Briefly, each paraffin-embedded specimen was cut into 10-μm sections and fixed on poly-l-lysine-treated slides. After dewaxing and rehydration, the slides were treated with 2 μg/ml proteinase K for 30 min at 37°C, followed by prehybridization at 55°C for 1 h in 50% deionized formamide–2.25× SSPE (300 mM NaCl, 20 mM NaH2PO4, 2 mM EDTA, pH 7.4)–10% dextran sulfate,–2.5× Denhardt's solution,–100 μg/ml sheared and denatured herring sperm DNA–100 μg/ml yeast tRNA–5 mM dithiothreitol–40 U/ml RNase inhibitor. Slides were then hybridized in a humidified chamber at 42°C overnight in the same solution containing 1 μg/ml alkaline phosphatase-conjugated, digoxigenin (DIG)-labeled sense or antisense RNA probe for rattusin. After the slides had been washed twice for 5 min each time with 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at room temperature, once for 20 min in 2× SSC, and once for 1 h at 57°C with 0.1× SSC, the DIG Nucleic Acid Detection kit (Roche Applied Science) was used to capture the rattusin mRNA signal in the blocking solution containing 1.25 U/ml of alkaline phosphatase-conjugated anti-DIG Fab fragments. The color was developed in the solution containing 5 mM MgCl2, 0.2 mM 5-bromo-4-chloro-3-indolylphosphate (BCIP), and 0.2 mM nitroblue tetrazolium (NBT) salt overnight at room temperature. After being mounted in VectaMount Mounting Medium (Vector Laboratories), the slides were examined with an Olympus BX51 fluorescence microscope equipped with a DP72 digital camera.
To make sense and antisense rattusin RNA probes, a 126-bp segment located in the less conserved mature region of rattusin mRNA was amplified by RT-PCR from a rat ileal RNA sample with the following primers: forward, GTAAGAAGGACCTTGCAGTG; reverse, CTCATCCTTGGGGTCCATGT. The PCR product was then cloned into the pGEMT-Easy vector (Promega) and sequenced to confirm its identity. The DIG RNA Labeling kit (Roche Applied Science) was used to label sense and antisense RNA probes with DIG-11-UTP by in vitro transcription with SP6 and T7 RNA polymerases, respectively.
Preparation of defensin peptides.
Since MMP7 preferentially cleaves the intestinal defensin sequences prior to leucine in mice (26–28), we reasoned that mature rattusin is likely to start from the first leucine after the prosequence. Therefore, a putatively mature sequence of 31 amino acids (LRVRRTLQCSCRRVCRNTCSCIRLSRSTYAS) was chemically synthesized in the reduced form by standard solid-phase synthesis, purified by reverse-phase high-pressure liquid chromatography (RP-HPLC) to >95% purity, and purchased from Bio-Synthesis Inc. (Lewisville, TX). The mass of the peptide was confirmed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MS) with Voyager DE-PRO (Applied Biosystems, Foster City, CA) housed in the Recombinant DNA/Protein Core Facility at Oklahoma State University.
To oxidize reduced rattusin, a 0.1-mg/ml concentration of the peptide was exposed to O2 gas for 5 min in 50 mM Tris, pH 8.0, and stirred gently for 48 h at room temperature with the cap open as described previously (28). Following air oxidation, refolded rattusin was purified by RP-HPLC on a Vydac C18 column (4.6 by 250 mm; Grace Vydac, Hesperia, CA) and a BioLogic DuoFlow liquid chromatography system (Bio-Rad, Hercules, CA). Buffer A consisted of 5% acetonitrile and 0.18% trifluoroacetic acid (TFA), and buffer B consisted of 90% acetonitrile and 0.15% TFA. The gradient used was 0 to 60% buffer B over 90 min at a flow rate of 1 ml/min. Eluted peptide was lyophilized and stored at −80°C until use. The peptide was reconstituted in 0.01% acetic acid and quantified by measuring UV absorbance at 280 nm on the basis of the extinction coefficients of tyrosine and cysteines present in rattusin (29).
Recombinant mouse Crp4 was produced as an N-terminally six-histidine-tagged fusion protein in Escherichia coli and purified by RP-HPLC as previously described (27, 30). Human intestinal α-defensin 5 (HD5) was chemically synthesized and oxidized as described previously (31).
Bacterial culture and antibacterial assays.
All bacterial strains, including Staphylococcus aureus ATCC 25923, S. aureus ATCC BAA-39, S. aureus ATCC 43300, Listeria monocytogenes ATCC 19115, E. coli O157:H7 ATCC 700728, S. Typhimurium ATCC 14028, S. Typhimurium DT104 ATCC 700408, and Klebsiella pneumoniae ATCC 13883 were purchased from either the American Type Culture Collection (ATCC; Manassas, VA) or MicroBiologics (St. Cloud, MN). Bacteria were grown in tryptic soy broth (TSB) overnight and subcultured for 3 to 4 h at 37°C in a shaking incubator to the mid-log phase. To study the antibacterial spectrum, a modified broth microdilution assay was used (32). Briefly, mid-log-phase bacteria were washed with 25 mM sodium phosphate buffer, pH 7.4, and suspended to 5 × 105 CFU/ml in 25 mM sodium phosphate, pH 7.4, containing 5% TSB with or without 100 mM NaCl. Bacteria (90 μl) were then dispensed into a 96-well tissue culture plate and serial 2-fold dilutions of rattusin or Crp4 (10 μl) were added in duplicate. After overnight incubation at 37°C, the MIC of each peptide was determined as the lowest peptide concentration that gave no visible bacterial growth.
To study the effect of peptide exposure time on bacterial viability, a time-kill assay was used (32). Rattusin, cryptin-4, and HD5 were incubated with 90 μl of 5 × 105 CFU/ml S. aureus ATCC 25923, E. coli ATCC 25922, or E. coli O157:H7 ATCC 700728 in 25 mM sodium phosphate buffer, pH 7.4, containing 1% TSB with or without 100 mM NaCl. Following incubation at 37°C for 10, 30, 60, 120, or 240 min, bacteria were diluted rapidly with ice-cold phosphate-buffered saline and serially plated onto tryptic soy agar plates. Viable bacteria were counted after overnight incubation at 37°C. The effect of Mg2+ on antibacterial activity was studied by incubating rattusin or Crp4 for 4 h with 90 μl of S. aureus (5 × 105 CFU/ml) at 2 μM each or with E. coli O157:H7 at 4 μM each in 25 mM sodium phosphate buffer–1% TSB, pH 7.4, containing 0, 1, 2, or 5 mM MgCl2.
Cytotoxicity assay.
The toxicity of rattusin to human Caucasian colon adenocarcinoma (Caco-2) cells (ATCC, Manassas, VA) was measured with alamarBlue dye (BioSource, Inc., Camarillo, CA) as described previously (33, 34). Briefly, Caco-2 cells were seeded into the wells of a 96-well plate at 5 × 104/ml of Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) and grown overnight in a humidified 5% CO2 incubator. Cells were washed once with DMEM, and then fresh DMEM containing 100 μM rattusin, Crp4, or chicken fowlicidin-1 (32, 33) in the presence or absence of 10% FBS was added. After 18 h of incubation, 10 μl of alamarBlue dye was added and the cells were incubated for another 6 h at 37°C. The plate was read with excitation at 545 nm and emission at 590 nm. The percentage of cell death was calculated as [1 − (Fpeptide − Fbackground)/(Facetic acid − Fbackground)] × 100, where Fpeptide is the fluorescence of cells exposed to 100 μM peptide, Facetic acid is the fluorescence of cells exposed to 0.01% acetic acid only, and Fbackground is the background fluorescence of 10% alamarBlue dye in cell culture medium without cells.
RESULTS
Rattusin, an α-defensin-related peptide, is preferentially expressed in the distal small intestine.
Following a comprehensive screening of the rat genome, we identified the gene for an orphan α-defensin-related protein known as defa-rs1 (24), which we now term rattusin. The gene for rattusin is located within the α-defensin gene cluster in the rat genome, which surprisingly encodes no α-defensin-related CRS peptides seen in mice. Similar to the genes for typical α-defensins, the gene for rattusin is composed of two short exons separated by a 572-bp intron, with the first exon encoding the 5′ untranslated region (UTR) and preprosegment and the second encoding a putatively mature peptide and the 3′ UTR. Although it is conserved in signal and prosequences with rodent α-defensins, the encoded rattusin peptide consists of 5 cysteines in the carboxyl-terminal region, in contrast to the canonical 6-cysteine motif of classical mammalian defensins and the 9 or 11 cysteines of CRS peptides (Fig. 2). Furthermore, the cysteine spacing pattern of rattusin is different from those of classical defensins or CRS peptides (Fig. 2).
Rattusin mRNA is transcribed abundantly in the intestinal tract of the rat (24). To analyze the rattusin expression pattern in greater detail, RT-PCR was performed with RNA samples isolated from different segments of the gastrointestinal tracts of 2-month-old healthy rats. As shown in Fig. 3, rattusin was highly expressed in the distal jejunum and the entire ileum, with peak expression in the proximal ileum. The stomach, duodenum, cecum, or colon showed no evident expression. The result is reminiscent of Crp4 and HD5, two Paneth cell-specific α-defensins that also show a similar expression pattern in the small intestine (35–37).
Fig 3.

mRNA expression patterns of rattusin, MMP7, and three isoforms of trypsin in the rat gastrointestinal tract by RT-PCR. The housekeeping gene for GAPDH was used to normalize template input. Prox., proximal; Mid., middle.
Paneth cell α-defensin precursors are activated by trypsin in humans (16) or by MMP7 in mice (13, 26). To begin identifying the protease that mediates the activation of rattusin, we studied the tissue expression patterns of MMP7 and three major isoforms of trypsin (cationic trypsin, anionic trypsin, and mesotrypsin) (38) in the rat gastrointestinal tract by RT-PCR. MMP7 was preferentially expressed from the distal jejunum to the distal ileum, coinciding with rattusin expression, whereas trypsin isoform mRNAs were more localized to the duodenum (Fig. 3), suggesting that MMP7, a Paneth cell-associated proteinase, is a more likely prorattusin convertase, as is the case in mice (13, 26, 27). In agreement with this, two serine residues, which are preferred MMP7 cleavage sites in rodent enteric α-defensins (26, 27), are conserved in rattusin but not in rat myeloid α-defensin (RatNP4) (Fig. 2). These findings also predicted that rattusin is produced by Paneth cells at the base of small intestinal crypts.
To determine the cell type responsible for the expression of rattusin, in situ hybridization was performed with rat proximal ileal sections and DIG-labeled sense and antisense rattusin RNA probes. The antisense rattusin RNA probe revealed positive brown staining at the base of intestinal crypts, where the control sense strand RNA probe showed no staining (Fig. 4), suggesting that rattusin is produced by Paneth cells, as are the Crp4 and HD5 enteric α-defensins.
Fig 4.

Localization of rattusin mRNA in rat ileal crypts. In situ hybridization was performed with rat ileal sections probed with rattusin-specific, DIG-labeled antisense (A) and sense (B) RNA probes, followed by anti-DIG antibody reaction and color development. Note the positive staining throughout the base of intestinal crypts (indicated by arrows) in panel A but not in panel B.
Rattusin has broad-spectrum, salt-insensitive antibacterial activities.
To evaluate the antibacterial activity of rattusin, we inferred its mature sequence on the basis of the MMP7 cleavage pattern of mouse CRS peptides (28) and synthesized the reduced form of the deduced five-cysteine peptide, refolded the molecule by air oxidation (28), and purified it by RP-HPLC. Because refolding induced changes in the spatial structure and hydrophobicity of rattusin compared to the reduced form, an approximately 5.5-min difference in retention time existed between the two forms (Fig. 5), enabling their separation. Refolded rattusin was assayed for antibacterial activity against several representative Gram-negative and Gram-positive bacteria with a modified broth microdilution assay with or without 100 mM NaCl (32). Rattusin exhibited potent antibacterial activities, with MICs in the range of 2 to 4 μM or 7.3 to 14.6 μg/ml (Table 2). In most cases, rattusin was as potent as Crp4, a highly bactericidal mouse Paneth cell α-defensin (39). Importantly, rattusin also displayed similar antibacterial efficiency against two strains of methicillin-resistant S. aureus (MRSA) and against multidrug-resistant S. Typhimurium DT104 (Table 2). Importantly, the activity of rattusin remained largely unchanged in the presence of NaCl, in sharp contrast to that of Crp4, which was nearly completely abolished by salt (Table 2).
Fig 5.

RP-HPLC profile of reduced and oxidized rattusin. The reduced synthetic peptide was refolded by air oxidation. Refolded rattusin was purified to homogeneity by RP-HPLC. Note that there is an approximately 5.5-min decrease in the retention time of oxidized rattusin due to refolding.
Table 2.
MICs of rattusin and Crp4
| Bacterium | ATCC no. | MIC (μM)a |
|||
|---|---|---|---|---|---|
| Rattusin |
Crp4 |
||||
| Without NaCl | With NaCl | Without NaCl | With NaCl | ||
| Gram negative | |||||
| E. coli O157:H7 | 700728 | 2–4 | 2–4 | 2 | >16 |
| S. Typhimurium | 14028 | 4 | 8 | 8 | >16 |
| S. Typhimurium DT104 | 700408 | 4 | 4–8 | 4–8 | >16 |
| K. pneumoniae | 13883 | 2–4 | 4–8 | 8 | >16 |
| Gram positive | |||||
| S. aureus | 25923 | 4 | 4 | 1–2 | ≥16 |
| L. monocytogenes | 19115 | 4 | 4 | 8 | >16 |
| S. aureus (MRSA) | 43300 | 4 | 4 | 2 | 8 |
| S. aureus (MRSA) | BAA-39 | 4–8 | 4–8 | 1–2 | >16 |
The antibacterial assay was performed in the presence or absence of 100 mM NaCl. The MICs were obtained from two independent experiments.
Disulfide bonds are not necessarily essential for the antibacterial activity of most α- and β-defensins (5–7). To test whether rattusin remains active in the reduced form, different concentrations of reduced and refolded rattusin were incubated with E. coli ATCC 25922 and S. aureus ATCC 25923 for 2 h in 10 mM phosphate buffer, pH 7.4, with or without supplementation with 100 mM NaCl. Bacterial viability was measured by quantitative plating. In the absence of NaCl, reduced rattusin showed a slightly enhanced antibacterial activity against both E. coli and S. aureus compared to that of the refolded form, with the bacteria becoming more sensitive to reduced rattusin, particularly at low concentrations (Fig. 6A and C). However, in the presence of 100 mM NaCl, an obvious reduction of the killing of E. coli by reduced rattusin was observed compared to that by the refolded peptide (Fig. 6B). The reduction of the killing of S. aureus by the reduced peptide became more pronounced (Fig. 6D). However, regardless of the rattusin redox status, bacterial killing efficacy, and possibly MICs, remained largely unchanged with or without NaCl, particularly at high concentrations (Fig. 6). Although NaCl diminished the bacterial sensitivity to reduced rattusin and possibly the kinetics of bacterial killing, its antibacterial potency, as measured by MICs, was not greatly affected. Thus, the peptide conformation established by intramolecular disulfide bonds does not appear to be a critically important determinant of rattusin salt-insensitive antibacterial activity. It is noteworthy that reduced rattusin remained unoxidized in 2 to 4 h under the time-kill assay conditions, as evidenced by the RP-HPLC profile (data not shown). Consistent with data in Table 2, Crp4 had antibacterial potency similar to that of rattusin when assayed in a low-ionic-strength environment but was completely inactivate against both E. coli and S. aureus in 100 mM NaCl (Fig. 6).
Fig 6.
Antibacterial activities of reduced and oxidized forms of rattusin in the presence or absence of 100 mM NaCl. A colony counting assay was conducted following 2 h of incubation of bacteria with each peptide in serial 2-fold dilutions in duplicate. (A) Activity against E. coli ATCC 25922 in the absence of NaCl. (B) Activity against E. coli ATCC 25922 in the presence of NaCl. (C) Activity against S. aureus ATCC 25923 in the absence of NaCl. (D) Activity against S. aureus ATCC 25923 in the presence of NaCl. Data shown are the means ± the standard errors of the means of a representative of two independent experiments.
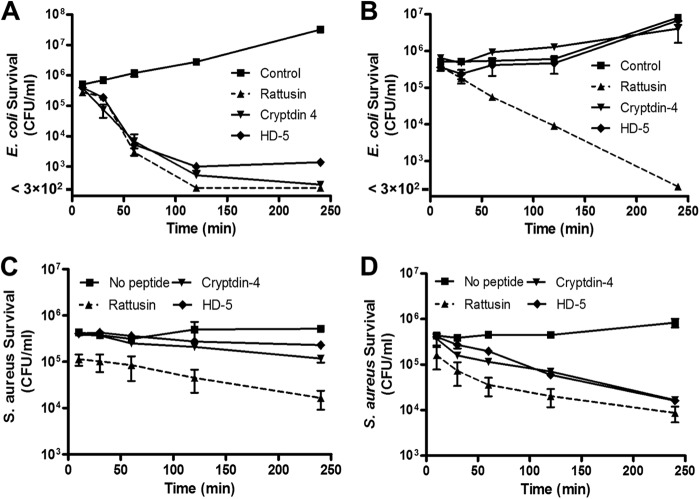
To study the effect of peptide exposure time on bacterial killing, each of the representative species of Gram-negative and Gram-positive bacteria was evaluated with the peptides by the time-kill assay. When E. coli was exposed to 4 μM refolded rattusin, Crp4, and HD5 for 2 h, its survival was reduced by 3 logs in the absence of exogenous NaCl (Fig. 7A), but in 100 mM NaCl, 4 h of exposure to rattusin was required to achieve maximum bacterial killing (Fig. 7B). As before, Crp4 and HD5 were completely inactivated by 100 mM NaCl (Fig. 7B). A similar trend occurred with S. aureus, with survival reduced by 2 logs after 4 h of exposure to 2 μM peptides (Fig. 7C), in which rattusin activity was unaffected by 100 mM NaCl but Crp4 and HD5 were significantly inhibited in the presence of salt (Fig. 7D). To study the effect of salinity further, the antibacterial activity of rattusin was assayed in the presence of increasing concentrations of NaCl, and the peptide maintained its bactericidal effects against both E. coli (Fig. 8A) and S. aureus (Fig. 8B) at NaCl levels of up to 200 mM. Collectively, these results showed that rattusin is a highly bactericidal α-defensin-related peptide with broad-spectrum, salt-insensitive activity.
Fig 7.
Kinetics of killing of E. coli and S. aureus in the presence or absence of 100 mM NaCl by rattusin, Crp4, and HD5. E. coli O157:H7 ATCC 700728 was incubated with 4 μM rattusin, Crp4, or HD5 or an equal volume of 0.01% acetic acid (control) in duplicate with (A) or without 100 mM NaCl (B) for 10, 30, 60, 120, or 240 min. S. aureus ATCC 25923 was incubated with or without each peptide at 2 μM in duplicate in the absence (C) or presence (D) of 100 mM NaCl. Surviving bacteria were plated and counted. Data shown are the means ± the standard errors of the means of two independent experiments.
Fig 8.
Effect of salinity on the antibacterial activity of rattusin. E. coli O157:H7 ATCC 700728 (A, C) and S. aureus ATCC 25923 (B, D) were incubated with 4 and 2 μM rattusin, respectively, with increasing concentrations of NaCl (A, B) or MgCl2 (C, D) for 4 h. Surviving bacteria were plated and counted. Data shown are the means ± the standard errors of the means of two or three independent experiments.
Divalent cations such as Mg2+ often occur at 1 to 2 mM in most biological fluids (40). At low concentrations, divalent cations are known to inhibit the antibacterial activities of cationic peptides (41, 42). To study the effect of Mg2+ on their antibacterial activity, rattusin and Crp4 were incubated with bacteria in the presence of increasing concentrations of MgCl2. There was a dose-dependent loss of the activity of Crp4 against E. coli, leading to complete inactivation at 2 to 5 mM MgCl2 (Fig. 8C). In contrast, rattusin activity was largely unaffected by MgCl2 (Fig. 8C). Against S. aureus, on the other hand, Mg2+ failed to inhibit the activity of either rattusin or Crp4 over the same concentration range (Fig. 8D).
Rattusin displays minimum cytotoxicity.
To examine the cytotoxic effect of rattusin to intestinal epithelial cells, human Caco-2 cells were treated with the peptide at 100 μM for 24 h in the presence or absence of 10% FBS. Fowlicidin-1 (100 μM) was used as a positive reference because it has shown significant cytotoxicity to mammalian cells (32). Similar to Crp4, rattusin lacked cytotoxicity even at 100 μM or 365 μg/ml with or without FBS under conditions in which fowlicidin-1 killed nearly 100% of the Caco-2 cells (Fig. 9) or lysed human erythrocytes (data not shown).
Fig 9.

Absence of cytotoxicity of rattusin and Crp4 to Caco-2 cells. Cells were incubated with 100 μM rattusin, Crp4, or fowlicidin-1 in duplicate for 24 h with or without 10% FBS. Cell viability was measured by an alamarBlue dye-based method. The data shown are representative of two independent experiments.
DISCUSSION
Rattusin is the only α-defensin-related peptide in the rat that contains five cysteine residues with a unique cysteine spacing pattern that differs from that of all known defensins. It will be important to determine its aggregation form and disulfide bonding pattern. Our preliminary data revealed that, upon oxidative refolding, a fraction of synthetic reduced rattusin peptides formed homodimers and even oligomers (data not shown), suggesting that four cysteine residues are likely to form two intramolecular disulfide bonds with the fifth cysteine involved in dimerization with other rattusin molecules. However, the pairing between cysteines appears to be somewhat random, as a mixture of disulfide isomers was observed in refolded rattusin (data not shown). Nevertheless, it is plausible that native mature rattusin is present in covalent dimers or multimers in the rat intestinal tract, given earlier reports on the dimerization of both α- and β-defensins and related peptides consisting of an odd number of cysteines. For example, mouse CRS peptides containing 9 or 11 cysteines form mixtures of covalent homo- and heterodimers in vivo (28), and a mouse β-defensin peptide variant with 5 cysteines also dimerizes (43). Dimeric CRS peptides are 2- to 4-fold more potent than their monomeric forms (28), and several canonical defensins with six cysteines form noncovalent dimers or oligomers upon interaction with bacterial membranes (44). Human intestinal α-defensin HD6, albeit with weak antibacterial activities, promotes mucosal innate defense by assembling into nanonets to entrap and prevent bacteria from attaching to and invading intestinal epithelial cells (45). Oligomerization thus appears to potentiate antibacterial activity. It will be interesting to determine the difference in antibacterial and other functional properties between monomeric and multimeric forms if rattusin indeed oligomerizes, although only monomeric rattusin purified through RP-HPLC was used in all of our functional assays.
Refolded rattusin possesses potent, broad-spectrum antibacterial activities. Furthermore, rattusin maintained potency against both E. coli and S. aureus when the disulfide bonds were reduced, reminiscent of most α- and β-defensins, whose antibacterial activities appear to be independent of their redox status. In fact, human β-defensin 1 exhibits substantially enhanced antibacterial potency when reduced (46), while CRS peptides show altered antibacterial selectivity but not potency following the reduction of disulfide bonds (28). The macrocyclic θ-defensins are exceptions in that their bactericidal activities are nearly abolished when they are reduced (23). Although the integrity of disulfide bonds and spatial structure does not determine the antibacterial activities of most defensins per se, the disulfide arrays are required for other biological functions, such as chemotactic activities (21) and resistance to proteolysis (47). However, it remains unknown whether reduction of the disulfide bonds of rattusin impact its nonbactericidal activities.
In sharp contrast to Crp4, HD5, and most α- and β-defensins (5–7), rattusin maintains its antibacterial activity in the presence of physiological concentrations of NaCl and Mg2+, a prerequisite for potential therapeutics. Although θ-defensins, with a unique cyclic structure, retain their antibacterial activity in the presence of NaCl, reduction of the disulfide bonds diminishes their activity (23). In the case of rattusin, salt insensitivity appears to be independent of structure because the reduced form maintained the antibacterial potency in physiological concentrations of NaCl. Thus, the mechanism of antibacterial action of rattusin is most likely to be different from that of other defensins and defensin-related peptides.
Given the location of their genes within the α-defensin gene cluster (9, 24) and their separate existence in rats and mice, rattusin and CRS peptides must have evolved independently from α-defensins after the divergence of rats and mice 33 million years ago (48). Interestingly, in contrast to rats harboring 12 α-defensins and one α-defensin-related rattusin, mice produce at least 26 functional α-defensins (cryptdins) and 12 α-defensin-related CRS peptides (9, 24), with some even present in a strain-specific manner (49). On the other hand, humans synthesize only six functional α-defensins with a θ-defensin pseudogene and no α-defensin-related sequences (24). Phylogenetic analysis revealed that mammalian α-defensins mostly form species-specific clusters (24), implying that most of these genes may have undergone independent duplications and diversifications within each species after divergence from a common ancestor.
Mammalian α-defensins and their related peptides are antimicrobial components of dense core granules of small intestinal Paneth cells or of circulating neutrophils (5–7). Albeit with direct, broad-spectrum antimicrobial activities, many peptides vary in antimicrobial potency, spectrum, and mechanism (50, 51). For example, HD5 and HD6 coexist in human Paneth cells; however, HD5 is bactericidal whereas HD6 is largely inactive (51). HD5 kills bacteria through physical membrane disruption, while HD6 protects the host by forming nanonets that entrap bacteria and keep them from translocating across the intestinal epithelial barrier (45). Given the differences in the expression patterns and antimicrobial properties of individual α-defensins and defensin-related peptides, it is tempting to speculate that the presence of a distinct repertoire of such peptides in each species may confer a distinct host defense capacity, ensuring optimal host protection from invading pathogens in diverse environmental niches.
The preferential expression of a large number of α-defensins in Paneth cells of the distal small intestine but not the colon may help explain, in part, the fact that the bacterial population in the small intestine is 104- to 106-fold smaller than that in the colon (52). Furthermore, intestinal defensins play a profound role in dictating the makeup of the ileal microflora, as evidenced by obvious alternations in the composition of the microbiota of the small intestine both in mice overexpressing HD5 and in mice deficient in the Paneth cell α-defensin convertase MMP7 (19). Because the reduction of disulfide bonds profoundly affects the biological activities of defensins (21) and some defensins may be naturally present in the reduced form by thioredoxin in the colon (46), we speculate that the repertoire of intestinal defensins may influence the composition of microflora in the large intestine as well.
In summary, rattusin is a rat α-defensin-related peptide and is preferentially expressed in Paneth cells of the distal small intestine. Unlike most other α- and β-defensins that are inactivated by mono- or divalent cations, rattusin stands out as a unique defensin-related peptide, possessing potent, salt-independent bactericidal activities, with essentially no toxicity to mammalian cells. Functionally and structurally distinct from any other intestinal defensins, rattusin may confer on rats unique host defense mechanisms. Furthermore, the desirable antibacterial properties of rattusin may be further exploited for the treatment of cystic fibrosis and Crohn's disease, where impairment of the antibacterial activity or synthesis of defensins proves detrimental to the host (18, 53). Coupled with its low cytotoxicity, rattusin may have potential for use in the treatment of both topical and systemic infections.
ACKNOWLEDGMENTS
This work was supported by USDA grant 2008-35204-04544; Oklahoma Center for the Advancement of Science and Technology grants AR07.2-087, HR07-113, and HR12-051; and Oklahoma Agricultural Experiment Station project H-2811to G.Z. and NIH grants DK044632 and AI059346 to A.J.O.
We thank Yugendar Bomminenni for help with cell culture.
Footnotes
Published ahead of print 4 February 2013
REFERENCES
- 1. Finch RG. 2004. Antibiotic resistance: a view from the prescriber. Nat. Rev. Microbiol. 2:989–994 [DOI] [PubMed] [Google Scholar]
- 2. Zasloff M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389–395 [DOI] [PubMed] [Google Scholar]
- 3. Hancock RE, Sahl HG. 2006. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24:1551–1557 [DOI] [PubMed] [Google Scholar]
- 4. Brogden KA. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3:238–250 [DOI] [PubMed] [Google Scholar]
- 5. Selsted ME, Ouellette AJ. 2005. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 6:551–557 [DOI] [PubMed] [Google Scholar]
- 6. Lehrer RI. 2004. Primate defensins. Nat. Rev. Microbiol. 2:727–738 [DOI] [PubMed] [Google Scholar]
- 7. Ganz T. 2003. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3:710–720 [DOI] [PubMed] [Google Scholar]
- 8. Zanetti M. 2004. Cathelicidins, multifunctional peptides of the innate immunity. J. Leukoc. Biol. 75:39–48 [DOI] [PubMed] [Google Scholar]
- 9. Andersson ML, Karlsson-Sjoberg JM, Putsep KL. 2012. CRS-peptides: unique defense peptides of mouse Paneth cells. Mucosal Immunol. 5:367–376 [DOI] [PubMed] [Google Scholar]
- 10. Yang D, Biragyn A, Hoover DM, Lubkowski J, Oppenheim JJ. 2004. Multiple roles of antimicrobial defensins, cathelicidins, and eosinophil-derived neurotoxin in host defense. Annu. Rev. Immunol. 22:181–215 [DOI] [PubMed] [Google Scholar]
- 11. Kim C, Slavinskaya Z, Merrill AR, Kaufmann SH. 2006. Human alpha-defensins neutralize toxins of the mono-ADP-ribosyltransferase family. Biochem. J. 399:225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim C, Gajendran N, Mittrucker HW, Weiwad M, Song YH, Hurwitz R, Wilmanns M, Fischer G, Kaufmann SH. 2005. Human alpha-defensins neutralize anthrax lethal toxin and protect against its fatal consequences. Proc. Natl. Acad. Sci. U. S. A. 102:4830–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wilson CL, Ouellette AJ, Satchell DP, Ayabe T, Lopez-Boado YS, Stratman JL, Hultgren SJ, Matrisian LM, Parks WC. 1999. Regulation of intestinal alpha-defensin activation by the metalloproteinase matrilysin in innate host defense. Science 286:113–117 [DOI] [PubMed] [Google Scholar]
- 14. Tongaonkar P, Golji AE, Tran P, Ouellette AJ, Selsted ME. 2012. High fidelity processing and activation of the human alpha-defensin HNP1 precursor by neutrophil elastase and proteinase 3. PLoS One 7:e32469 doi:10.1371/journal.pone.0032469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Glenthøj A, Cowland JB, Heegaard NH, Larsen MT, Borregaard N. 2011. Serglycin participates in retention of alpha-defensin in granules during myelopoiesis. Blood 118:4440–4448 [DOI] [PubMed] [Google Scholar]
- 16. Ghosh D, Porter E, Shen B, Lee SK, Wilk D, Drazba J, Yadav SP, Crabb JW, Ganz T, Bevins CL. 2002. Paneth cell trypsin is the processing enzyme for human defensin-5. Nat. Immunol. 3:583–590 [DOI] [PubMed] [Google Scholar]
- 17. Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. 2003. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature 422:522–526 [DOI] [PubMed] [Google Scholar]
- 18. Wehkamp J, Salzman NH, Porter E, Nuding S, Weichenthal M, Petras RE, Shen B, Schaeffeler E, Schwab M, Linzmeier R, Feathers RW, Chu H, Lima H, Jr, Fellermann K, Ganz T, Stange EF, Bevins CL. 2005. Reduced Paneth cell alpha-defensins in ileal Crohn's disease. Proc. Natl. Acad. Sci. U. S. A. 102:18129–18134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, Stoel M, Zhou Y, Sodergren E, Weinstock GM, Bevins CL, Williams CB, Bos NA. 2010. Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 11:76–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tremaroli V, Backhed F. 2012. Functional interactions between the gut microbiota and host metabolism. Nature 489:242–249 [DOI] [PubMed] [Google Scholar]
- 21. Wu Z, Hoover DM, Yang D, Boulegue C, Santamaria F, Oppenheim JJ, Lubkowski J, Lu W. 2003. Engineering disulfide bridges to dissect antimicrobial and chemotactic activities of human beta-defensin 3. Proc. Natl. Acad. Sci. U. S. A. 100:8880–8885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maemoto A, Qu X, Rosengren KJ, Tanabe H, Henschen-Edman A, Craik DJ, Ouellette AJ. 2004. Functional analysis of the alpha-defensin disulfide array in mouse cryptdin-4. J. Biol. Chem. 279:44188–44196 [DOI] [PubMed] [Google Scholar]
- 23. Tang YQ, Yuan J, Osapay G, Osapay K, Tran D, Miller CJ, Ouellette AJ, Selsted ME. 1999. A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated alpha-defensins. Science 286:498–502 [DOI] [PubMed] [Google Scholar]
- 24. Patil A, Hughes AL, Zhang G. 2004. Rapid evolution and diversification of mammalian alpha-defensins as revealed by comparative analysis of rodent and primate genes. Physiol. Genomics 20:1–11 [DOI] [PubMed] [Google Scholar]
- 25. De Block M, Debrouwer D. 1993. RNA-RNA in situ hybridization using digoxigenin-labeled probes: the use of high-molecular-weight polyvinyl alcohol in the alkaline phosphatase indoxyl-nitroblue tetrazolium reaction. Anal. Biochem. 215:86–89 [DOI] [PubMed] [Google Scholar]
- 26. Ayabe T, Satchell DP, Pesendorfer P, Tanabe H, Wilson CL, Hagen SJ, Ouellette AJ. 2002. Activation of Paneth cell alpha-defensins in mouse small intestine. J. Biol. Chem. 277:5219–5228 [DOI] [PubMed] [Google Scholar]
- 27. Shirafuji Y, Tanabe H, Satchell DP, Henschen-Edman A, Wilson CL, Ouellette AJ. 2003. Structural determinants of procryptdin recognition and cleavage by matrix metalloproteinase-7. J. Biol. Chem. 278:7910–7919 [DOI] [PubMed] [Google Scholar]
- 28. Hornef MW, Putsep K, Karlsson J, Refai E, Andersson M. 2004. Increased diversity of intestinal antimicrobial peptides by covalent dimer formation. Nat. Immunol. 5:836–843 [DOI] [PubMed] [Google Scholar]
- 29. Gill SC, von Hippel PH. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319–326 [DOI] [PubMed] [Google Scholar]
- 30. Satchell DP, Sheynis T, Shirafuji Y, Kolusheva S, Ouellette AJ, Jelinek R. 2003. Interactions of mouse Paneth cell alpha-defensins and alpha-defensin precursors with membranes. Prosegment inhibition of peptide association with biomimetic membranes. J. Biol. Chem. 278:13838–13846 [DOI] [PubMed] [Google Scholar]
- 31. Wu Z, Ericksen B, Tucker K, Lubkowski J, Lu W. 2004. Synthesis and characterization of human alpha-defensins 4-6. J. Pept. Res. 64:118–125 [DOI] [PubMed] [Google Scholar]
- 32. Xiao Y, Cai Y, Bommineni YR, Fernando SC, Prakash O, Gilliland SE, Zhang G. 2006. Identification and functional characterization of three chicken cathelicidins with potent antimicrobial activity. J. Biol. Chem. 281:2858–2867 [DOI] [PubMed] [Google Scholar]
- 33. Xiao Y, Dai H, Bommineni YR, Soulages JL, Gong YX, Prakash O, Zhang G. 2006. Structure-activity relationships of fowlicidin-1, a cathelicidin antimicrobial peptide in chicken. FEBS J. 273:2581–2593 [DOI] [PubMed] [Google Scholar]
- 34. Xiao Y, Herrera AI, Bommineni YR, Soulages JL, Prakash O, Zhang G. 2009. The central kink region of fowlicidin-2, an alpha-helical host defense peptide, is critically involved in bacterial killing and endotoxin neutralization. J. Innate Immun. 1:268–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Darmoul D, Ouellette AJ. 1996. Positional specificity of defensin gene expression reveals Paneth cell heterogeneity in mouse small intestine. Am. J. Physiol. 271:G68–G74 [DOI] [PubMed] [Google Scholar]
- 36. Ouellette AJ, Darmoul D, Tran D, Huttner KM, Yuan J, Selsted ME. 1999. Peptide localization and gene structure of cryptdin 4, a differentially expressed mouse Paneth cell alpha-defensin. Infect. Immun. 67:6643–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wehkamp J, Chu H, Shen B, Feathers RW, Kays RJ, Lee SK, Bevins CL. 2006. Paneth cell antimicrobial peptides: topographical distribution and quantification in human gastrointestinal tissues. FEBS Lett. 580:5344–5350 [DOI] [PubMed] [Google Scholar]
- 38. Rinderknecht H, Renner IG, Abramson SB, Carmack C. 1984. Mesotrypsin: a new inhibitor-resistant protease from a zymogen in human pancreatic tissue and fluid. Gastroenterology 86:681–692 [PubMed] [Google Scholar]
- 39. Ouellette AJ. 2004. Defensin-mediated innate immunity in the small intestine. Best Pract. Res. Clin. Gastroenterol. 18:405–419 [DOI] [PubMed] [Google Scholar]
- 40. Bowdish DM, Davidson DJ, Hancock RE. 2005. A re-evaluation of the role of host defence peptides in mammalian immunity. Curr. Protein Pept. Sci. 6:35–51 [DOI] [PubMed] [Google Scholar]
- 41. Friedrich C, Scott MG, Karunaratne N, Yan H, Hancock RE. 1999. Salt-resistant alpha-helical cationic antimicrobial peptides. Antimicrob. Agents Chemother. 43:1542–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bowdish DM, Davidson DJ, Lau YE, Lee K, Scott MG, Hancock RE. 2005. Impact of LL-37 on anti-infective immunity. J. Leukoc. Biol. 77:451–459 [DOI] [PubMed] [Google Scholar]
- 43. Morrison GM, Rolfe M, Kilanowski FM, Cross SH, Dorin JR. 2002. Identification and characterization of a novel murine beta-defensin-related gene. Mamm. Genome 13:445–451 [DOI] [PubMed] [Google Scholar]
- 44. White SH, Wimley WC, Selsted ME. 1995. Structure, function, and membrane integration of defensins. Curr. Opin. Struct. Biol. 5:521–527 [DOI] [PubMed] [Google Scholar]
- 45. Chu H, Pazgier M, Jung G, Nuccio SP, Castillo PA, de Jong MF, Winter MG, Winter SE, Wehkamp J, Shen B, Salzman NH, Underwood MA, Tsolis RM, Young GM, Lu W, Lehrer RI, Baumler AJ, Bevins CL. 2012. Human alpha-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science 337:477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schroeder BO, Wu Z, Nuding S, Groscurth S, Marcinowski M, Beisner J, Buchner J, Schaller M, Stange EF, Wehkamp J. 2011. Reduction of disulphide bonds unmasks potent antimicrobial activity of human beta-defensin 1. Nature 469:419–423 [DOI] [PubMed] [Google Scholar]
- 47. Andersson HS, Figueredo SM, Haugaard-Kedstrom LM, Bengtsson E, Daly NL, Qu X, Craik DJ, Ouellette AJ, Rosengren KJ. 2012. The alpha-defensin salt-bridge induces backbone stability to facilitate folding and confer proteolytic resistance. Amino Acids 43:1471–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nei M, Xu P, Glazko G. 2001. Estimation of divergence times from multiprotein sequences for a few mammalian species and several distantly related organisms. Proc. Natl. Acad. Sci. U. S. A. 98:2497–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shanahan MT, Tanabe H, Ouellette AJ. 2011. Strain-specific polymorphisms in Paneth cell alpha-defensins of C57BL/6 mice and evidence of vestigial myeloid alpha-defensin pseudogenes. Infect. Immun. 79:459–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nuding S, Zabel LT, Enders C, Porter E, Fellermann K, Wehkamp J, Mueller HA, Stange EF. 2009. Antibacterial activity of human defensins on anaerobic intestinal bacterial species: a major role of HBD-3. Microbes Infect. 11:384–393 [DOI] [PubMed] [Google Scholar]
- 51. Ericksen B, Wu Z, Lu W, Lehrer RI. 2005. Antibacterial activity and specificity of the six human alpha-defensins. Antimicrob. Agents Chemother. 49:269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bevins CL. 2006. Paneth cell defensins: key effector molecules of innate immunity. Biochem. Soc. Trans. 34:263–266 [DOI] [PubMed] [Google Scholar]
- 53. Agerberth B, Gudmundsson GH. 2006. Host antimicrobial defence peptides in human disease. Curr. Top. Microbiol. Immunol. 306:67–90 [DOI] [PubMed] [Google Scholar]