Abstract
Abnormal activities of histone lysine demethylases (KDMs) and lysine deacetylases (HDACs) are associated with aberrant gene expression in breast cancer development. However, the precise molecular mechanisms underlying the crosstalk between KDMs and HDACs in chromatin remodeling and regulation of gene transcription are still elusive. In this study, we showed that treatment of human breast cancer cells with inhibitors targeting the zinc cofactor dependent class I/II HDAC, but not NAD+ dependent class III HDAC, led to significant increase of H3K4me2 which is a specific substrate of histone lysine-specific demethylase 1 (LSD1) and a key chromatin mark promoting transcriptional activation. We also demonstrated that inhibition of LSD1 activity by a pharmacological inhibitor, pargyline, or siRNA resulted in increased acetylation of H3K9 (AcH3K9). However, siRNA knockdown of LSD2, a homolog of LSD1, failed to alter the level of AcH3K9, suggesting that LSD2 activity may not be functionally connected with HDAC activity. Combined treatment with LSD1 and HDAC inhibitors resulted in enhanced levels of H3K4me2 and AcH3K9, and exhibited synergistic growth inhibition of breast cancer cells. Finally, microarray screening identified a unique subset of genes whose expression was significantly changed by combination treatment with inhibitors of LSD1 and HDAC. Our study suggests that LSD1 intimately interacts with histone deacetylases in human breast cancer cells. Inhibition of histone demethylation and deacetylation exhibits cooperation and synergy in regulating gene expression and growth inhibition, and may represent a promising and novel approach for epigenetic therapy of breast cancer.
Keywords: Histone demethylase, Histone deacetylase, Epigenetics, Breast cancer, Growth inhibition, Gene expression
Introduction
In cancer, histone deacetylases (HDACs) may have abnormally high activity and thus contribute to hypoacetylation and the aberrant silencing of genes [1]. A number of synthetic HDAC inhibitors have been rationally developed. Among the most interesting inhibitors are those that have been designed to target primarily the zinc cofactor at the active site of class I/II HDACs [2, 3]. Some of the HDAC inhibitors have been examined for their ability to alter chromatin structure and re-express aberrantly silenced genes in breast cancer [4–7].
Another important protein that is responsible for abnormal chromatin remodeling and epigenetic silencing of genes is a recently identified histone lysine-specific demethylase 1 (LSD1). LSD1, also known as AOF2 or KDM1A, is the first identified FAD-dependent histone demethylase capable of specifically demethylating mono- and di-methylated lysine 4 of histone H3 (H3K4me1 and H3K4me2) [8, 9]. LSD1 has been typically found in association with HDAC1/2, Co-REST, BHC80, and BRAF35 [8]. Several recent studies have established LSD1 as an important link to the development and progression of cancer and provide a rationale for developing LSD1 inhibitors as a means for therapeutic intervention [10–12]. In our recent studies, we have shown that specific polyamine analogs function as potent inhibitors of LSD1, leading to re-expression of several aberrantly silenced tumor suppressor genes [13–15]. Recently, a second mammalian FAD dependent histone demethylase, LSD2 (also known as AOF1 or KDM1B), has been identified [16–18]. Amino acid sequence analysis shows that LSD1 and LSD2 share 33% overall identity in the amine oxidase domain. However, little is known about the actual biological function of LSD2 in cancer.
Despite the promising results produced by HDAC inhibitors in preclinical studies, lack of specificity may limit the clinical use of these compounds in cancer treatment. One of the major hurdles in predicting the efficacy of HDAC inhibitors is identifying potential selectivity for one or more epigenetic protein complexes and determining the ultimate effect on gene expression. To overcome these obstacles, it is necessary to better explore the epigenetic mechanisms underlying the activity of HDAC inhibitor and seek a more effective and less toxic therapeutic approach by combining HDAC inhibitors with other epi-drugs that can effectively target multiple epigenetic entities. In this study, we define in depth the mechanisms of functional crosstalk between histone demethylase and deacetylase in chromatin remodeling and gene transcription in breast cancer cells.
Methods and materials
Reagents and cell culture conditions
SAHA was purchased from Cayman Chemical (Ann Arbor, MI). TSA and pargyline were obtained from Sigma–Aldrich (St. Louis, MO). MS-275, LBH-589, and PXD-101 were purchased from Selleck Chemicals (Houston, TX). Human breast carcinoma cells MDA-MB-231 were maintained in DMEM medium and MDA-MB-468 cells were maintained in IMEM medium (5% FBS).
Western blots
Whole and nuclear proteins were extracted as previously described [19, 20]. Proteins were fractionated on SDSPAGE gels and transferred onto PVDF membranes. Primary antibodies against H3K4me1, H3K4me2, H3K4me3, H3K9me2, H3K27me2, Acetyl H3, Acetyl H3K9, LSD1, HDAC1, HDAC2, and CoREST were from Millipore. The H3 antibody for normalization was purchased from Abcam (Cambridge, MA).
RNAi
Pre-designed and validated LSD1 siRNA oligonucleotides and non-targeting siRNA (scramble) were purchased from Ambion (Carlsbad, California). Transient transfections were performed using siPORT™ transfection agent (Ambion). After 48 h exposure to a 50 nM LSD1 siRNA, cells were harvested and the lysate was analyzed for LSD1 protein expression. siRNA targeting LSD2 mRNA and a non-targeting scramble oligonucleotides (targeting sequences, LSD2-siRNA: 5-AAGACATTCAAGGAACCGTCT, Scramble: 5-AACTTGCTATGAGAACAAATT) were synthesized using Silencer® siRNAconstruction kit (Ambion). Transient transfection was performed with LipofectAMINE 2000 reagent (Invitrogen). After 24 h of exposure to 70 nM LSD2 siRNA or scramble oligonucleotides, total RNA was extracted using the TRIzol reagent (Invitrogen).
Quantitative real-time RT-PCR
The TaqMan® Gene Expression Assays (Applied Biosystems) were performed to quantify mRNA expression of LSD2, NR4A1, NR4A3, PCDH1, BIK, and RGS16 genes with actin as an internal control.
MTT growth inhibition and drug combination index (CI) analysis
MTT assays were performed using the method as previously described [19]. The median effects (IC50) were determined by CalcuSyn software (Biosoft, Cambridge, UK). The Chou-Talalay median effect/combination index (CI) model was used to determine synergy, additivity, or antagonism of combination treatments [21]. Briefly, cells were treated with each agent individually at its IC50 concentration or fixed fractions of the IC50 concentrations [22]. The agents were also combined in these same dose-fixed ratios to determine the combination index (CI). Synergy was defined as any CI < 1, additivity as CI = 1, and antagonism as any CI > 1.
Microarray analysis of gene expression
Cells were treated with 5 µM SAHA, 2.5 mM pargyline or in combination for 24 h. Total RNA samples from three independent biological replicates (n = 3) were extracted using Qiagen RNeasy kit (Qiagen, Valencia, CA). The array study was performed using Affymetrix GeneChip U133A 2.0 array platform, which contains 20,928 probes representing all functionally characterized genes in the human genome. The data were processed as RMA files (Affymetrix Robust Multi-Array Average) in which the raw intensity data were background corrected, log2 transformed, and then quantile normalized according to Affymetrix recommendations.
Statistical analysis
The Student’s t-test was used to determine the statistical differences between various experimental and control groups. Microarray statistical tests were performed using Significance Analysis of Microarrays software (SAM version 3.09c), which is designed to reduce the risk of Type 1 errors due to multiple testing [23].
Results
Specific HDAC inhibitors increase H3K4 methylation in breast cancer cells
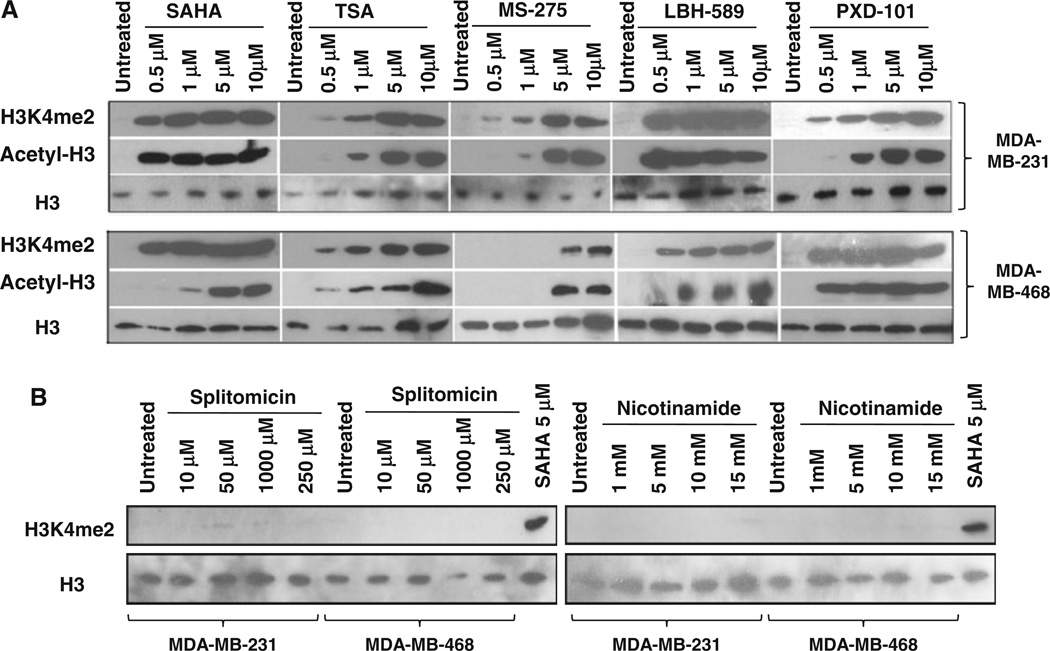
To determine whether histone lysine methylation is functionally linked to activity of histone deacetylase in breast cancer, global nuclear H3K4 methylation was examined after exposure of human breast cancer cells to a variety of HDAC inhibitors. We first tested several clinically relevant inhibitors that have been designed to target primarily the zinc cofactor at the active site of the class I/II HDACs. These inhibitors include hydroxamic acid derivatives SAHA (Vorinostat), TSA (Trichostatin A), LBH589 (Panobinostat), and PXD-101(Belinostat), and a benzamide analog MS-275 (Entinostat). In both MDA-MB-231 and MDA-MB-468 cell lines, exposure to these compounds produced significant global increase of nuclear H3K4me2, which is the specific substrate of LSD1 (Fig. 1a). The enhanced level of histone methylation by HDAC inhibitors parallels the increase of acetylation of histone 3, suggesting that LSD1 is an important epigenetic target of HDAC inhibitors and its activity is intimately associated with HDAC activity. Recently, another class of HDACs, the class III NAD+-dependent sirtuins, has received increasing attention as a potential epigenetic target [24]. To evaluate if inhibition of zinc-independent sirtuins could also affect the LSD1 activity, we applied a pharmacologic approach using splitomicin (SPT) and nicotinamide (NIA), two specific inhibitors against sir2-dependent class III HDACs. Neither of these sirtuin inhibitors altered H3K4me2 level in breast cancer cells (Fig. 1b).
Fig. 1.
Inhibition of LSD1 activity by HDAC inhibitors. a MDA-MB-231 and MDA-MB-468 cells were exposed to indicated HDAC inhibitors for 24 h. b Cells were treated with splitomicin or nicotinamide for 24 h. 30 µg of nuclear protein/lane were analyzed by immunoblots for expression of H3K4me2 or AcH3K9. Nuclear lysate of MDA-MB-231 cells treated with 5 µM SAHA for 24 h was used as positive control for LSD1 inhibition. H3 was used as a loading control
A co-immunoprecipitation study showed that the physical interaction between LSD1 and HDAC1 remained intact under SAHA treatment (Fig. S1). In addition, LSD1 mRNA and protein levels were not altered by increasing concentration of SAHA treatment (Fig. S2). This suggests that inhibition of LSD1 activity by HDAC inhibitors likely occurs through disruption of the functional link between the two enzymes rather than direct interruption of their physical interaction or suppression of the mRNA and protein levels of LSD1.
Inhibition of LSD1 activity leads to enhanced histone acetylation
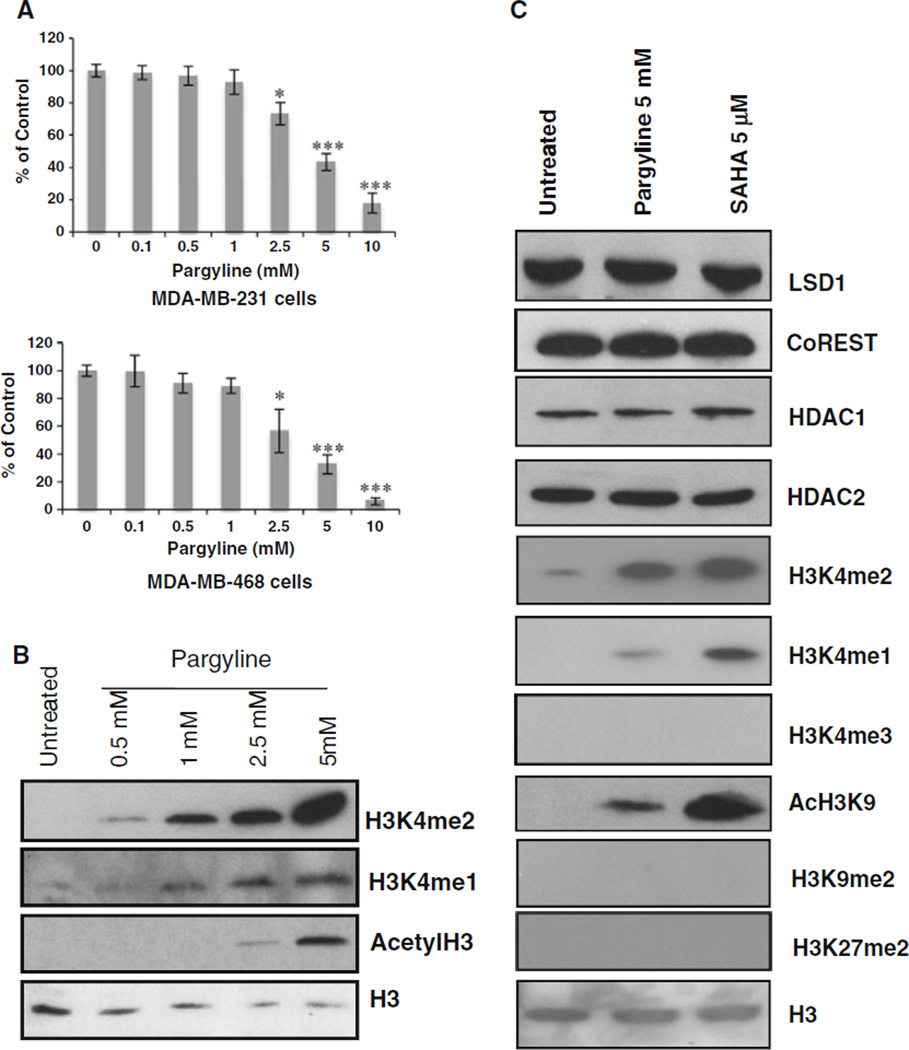
To probe precisely the role of LSD1 in regulation of HDAC activity, MDA-MB-231 and MDA-MB-468 cells were treated with an LSD1 inhibitor, pargyline. MTT study indicates a dose-dependent growth inhibition by pargyline (Fig. 2a). Exposure of MDA-MB-231 cells to pargyline produced significant increase of H3K4me2 and H3K4me1 at concentrations ≥0.5 mM (Fig. 2b). Treatment with pargyline also significantly increased pan acetylation of H3 at concentrations ≥2.5 mM (Fig. 2b).
Fig. 2.
Effects of pargyline on histone marks. a MDA-MB-231 and MDA-MB-468 cells were treated with increasing concentrations of pargyline for 48 h. MTT assays were performed. * P < 0.05, *** P < 0.001, (pargyline vs. control, Student’s t-test). b MDA-MB-231 cells were treated with indicated concentrations of pargyline for 24 h and analyzed by immunoblots for expression of H3K4me2, H3k4me1, and AcH3. c MDA-MB-231 cells were exposed to 5 µM pargyline or 5 mM SAHA for 24 h and analyzed for expression of indicated proteins by immunoblots. H3 was used as a loading control
The exposure of MDA-MB-231 cells to pargyline did not change the expression of LSD1, CoREST, or HDAC1/2, but led to a remarkable increase of H3K4me2 and AcH3K9 (Fig. 2c). Similar results were seen for SAHA treatment (Fig. 2c), suggesting an intimate functional link between LSD1 and HDAC1/2. Neither pargyline nor SAHA increased H3K4me3 or two important repressive marks, H3K9me2 and H3K27me2, suggesting that the inhibition of LSD1 or HDAC activities does not affect the activity of another class of histone demethylase family, the JmjC containing histone demethylases.
LSD1 and LSD2 exhibit distinct effects on HDAC activity
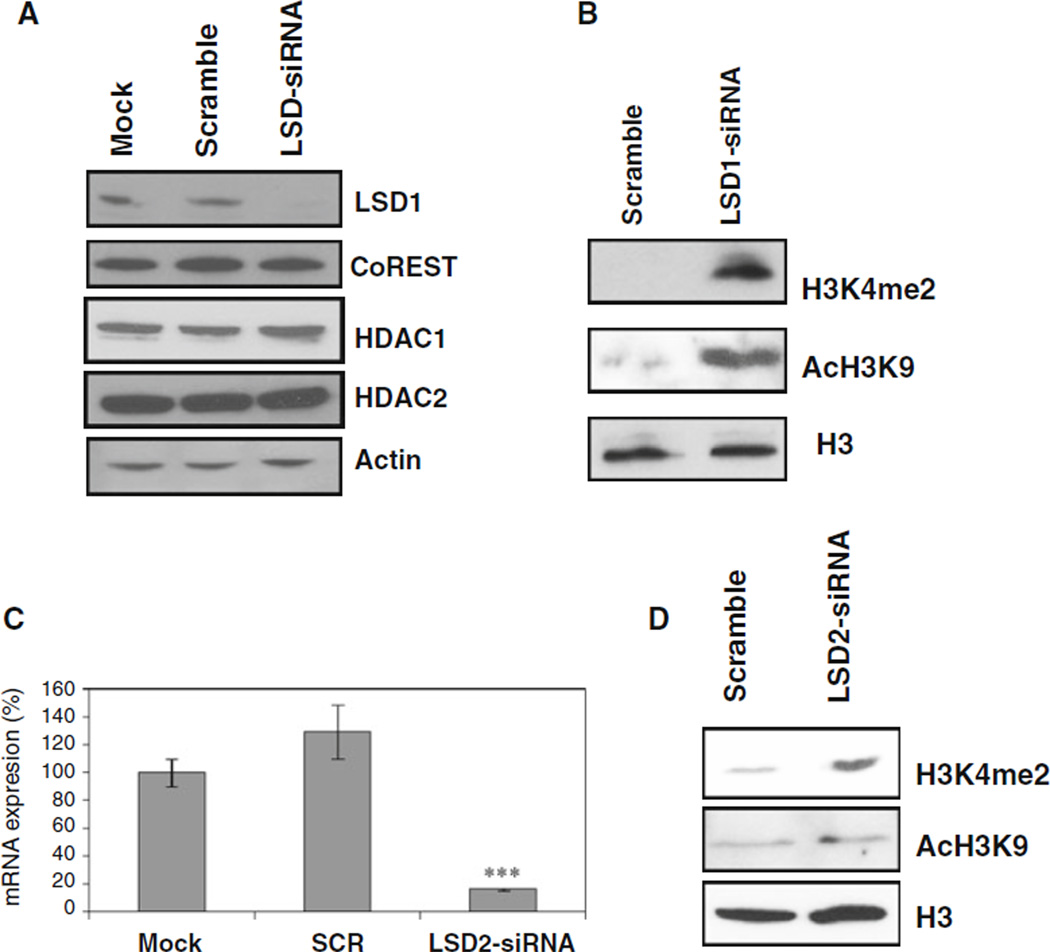
Exposure of MDA-MB-231 cells to LSD1-targeting siRNA resulted in a significant decrease in LSD1 protein without affecting the protein expression of CoREST, HDAC1/2 (Fig. 3a). Similar to pharmacological inhibition, LSD1 siRNA treatment in MDA-MB-231 cells led to significantly increased nuclear levels of H3K4me2 and AcH3K9 (Fig. 3b). This result strengthens the evidence that LSD1 and HDAC activities are indeed functionally associated.
Fig. 3.
Knockdown of LSD1 by siRNA leads to increase of histone acetylation. a MDAMB-231 cells were transfected with mock, scrambled, or LSD1-targeted siRNA oligonucleotides for 48 h and subjected to immunoblotting with indicated antibodies. b After LSD1 siRNA transfection, 30 µg of nuclear protein/lane were analyzed by immunoblots for expression of H3K4me2 and AcH3K9. c After MDA-MB-231 cells were transfected with mock, scrambled, or LSD2 siRNA. mRNA was measured by quantitative real time PCR analysis for LSD2 gene expression. Results represent the mean of three independent experiments, each performed in triplicate ±SD. *** P < 0.001, (siRNA vs. mock or scramble, Student’s t-test). d After LSD2 siRNA transfection, nuclear proteins were analyzed for expression of H3K4me2 and AcH3K9. H3 was used as a loading control
To understand whether LSD2, a new member of the FAD dependent histone demethylase family, in concert with LSD1, interacts with histone deacetylase in human breast cancer cells, we used siRNA approach to knock down LSD2 mRNA expression. Specific LSD2 siRNA oligonucleotides suppressed >90% LSD2 mRNA expression (Fig. 3c) and led to increase of H3K4me2 (Fig. 3d), but failed to alter the global level of AcH3K9 in MDAMB-231 cells (Fig. 3d). These results suggest that LSD2 possesses histone demethylase activity in breast cancer cells, but, unlike LSD1, the activity of LSD2 may not be functionally associated with HDAC activity.
Combined inhibition of histone demethylation and deacetylation exerts synergistic effect on growth inhibition
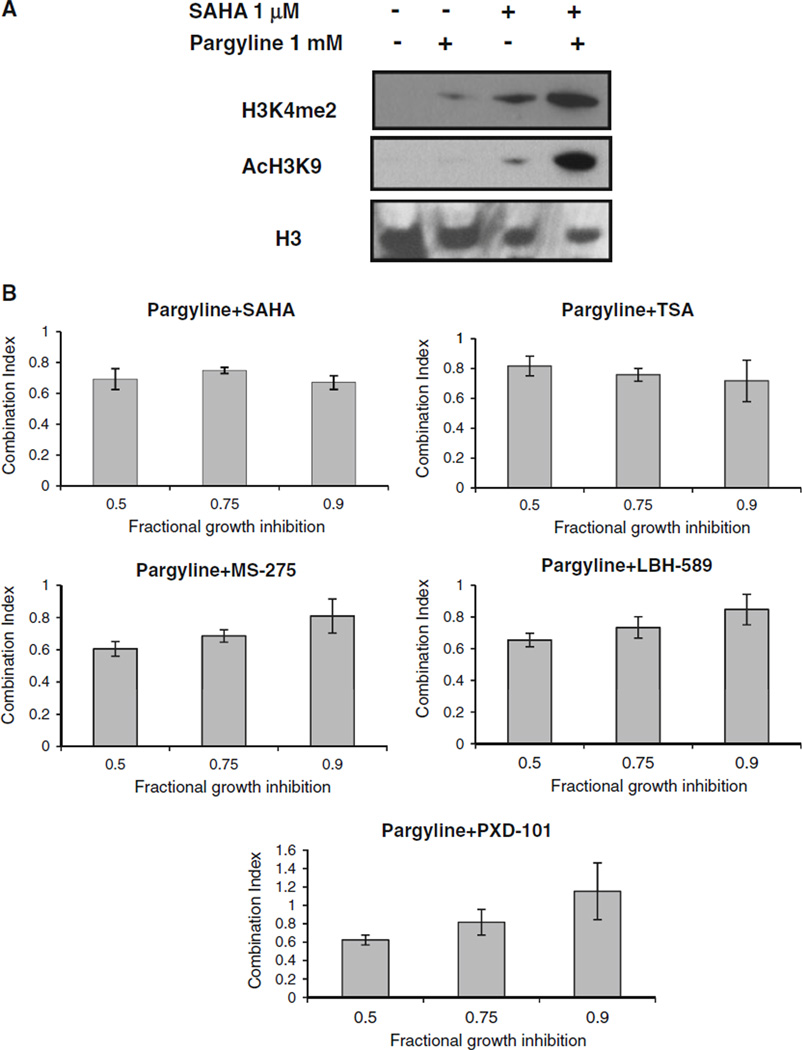
To determine whether combination treatment with LSD1 and HDAC inhibitors could lead to a synergistic effect in chromatin remodeling, we examined the nuclear levels of H3K4me2 and AcH3K9 in MDA-MB-231 cells treated with pargyline or SAHA alone or in combination. Combined treatment resulted in a remarkable increase of H3K4me2 and AcH3K9 (Fig. 4a). To examine whether such chromatin modification also translates to enhanced therapeutic efficacy of the drugs, MDA-MB-231 cells were treated with pargyline and HDAC inhibitors alone or simultaneously for 48 h. The combination index (CI) values were evaluated by using the CalcuSyn program. At very low dose combination (fractional growth inhibition, Fa = 0.9), synergistic growth inhibition (CI < 1) was observed between pargyline and HDAC inhibitors SAHA, TSA, MS-275, and LBH-589 (Fig. 4b). At median or higher dose combination (Fa = 0.5 or 0.75), pargyline exhibited synergy with all the HDAC inhibitors tested (CI < 1) (Fig. 4b). These results indicate that treatment with LSD1 and HDAC inhibitors significantly enhanced growth inhibition when used in combination in human breast cancer cells.
Fig. 4.
LSD1 and HDAC inhibitors exhibit synergistic growth inhibition. a MDA-MB-231 cells were exposed to 1 mM pargyline or 1 µM SAHA alone or in combination for 24 h and were analyzed for expression of H3K4me2 and AcH3K9. H3 was used as a loading control. b Cells were simultaneously treated with pargyline or HDAC inhibitors for 48 h. The combination index (CI) values shown represent the mean ±SD for three independent experiments
Microarray analysis identifies a gene subset related to combination effect with LSD1 and HDAC inhibitors
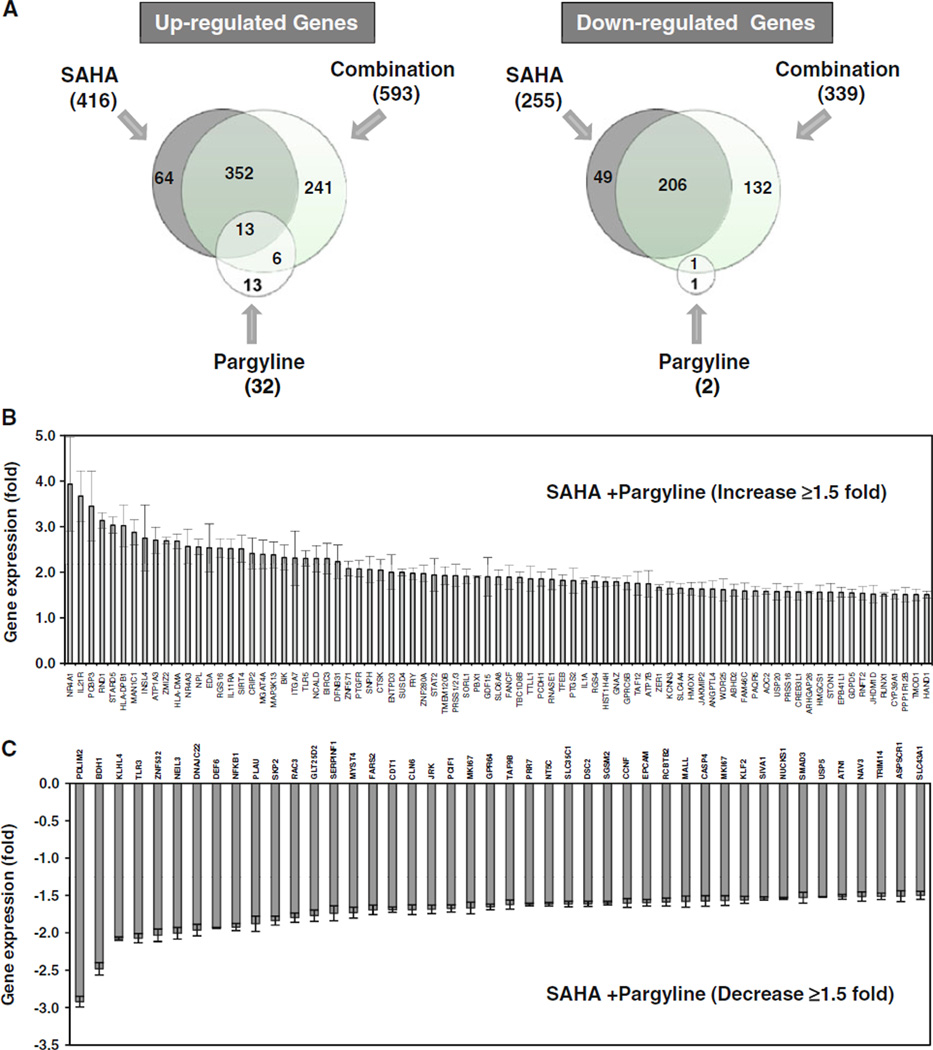
The synergistic effect in growth inhibition by combined treatment with LSD1 and HDAC inhibitors suggests that simultaneous inhibition of both enzymes may have a more profound effect on gene transcription than inhibition of either enzyme function alone. To test this hypothesis, we performed a genome-wide microarray screen of MDA-MB-231 cells to define a comprehensive profile of genes whose expression is altered by combination treatment with LSD1 and HDAC inhibitors. Microarray results showed that expression of a total of 671 genes was changed by 1.5-fold or greater by SAHA alone (416 genes up-regulated and 255 genes down-regulated, P < 0.01) and expression of 34 genes was altered by 1.5-fold or greater following pargyline treatment alone (32 genes up-regulated and 2 genes down-regulated, P < 0.01). Further analysis showed that 932 genes exhibited significant change in their mRNA levels after exposure to both drugs (593 genes up-regulated and 339 genes down-regulated with 1.5-fold or greater, P < 0.01) (Fig. 5a). Among these affected genes, we identified a unique set of 241 genes whose expression was exclusively induced by combination therapy (Fig. 5a), 81 of which displayed a 1.5-fold or greater induction in expression (Fig. 5b). The detailed annotation for these genes is shown in Table 1. The array study also identifies a subset of 132 genes, whose expression was exclusively down-regulated by combination (Fig. 5a). The list of down-regulated genes by SAHA/pargyline with 1.5-fold or greater change (P < 0.01) is shown and annotated in Fig. 5c and Table 2.
Fig. 5.
Genome-wide microarray analysis. a MDA-MB-231 cells were treated with 5 µM SAHA, 2.5 mM pargyline alone or in combination for 24 h. Microarray analysis was performed. Diagrams of up-regulated or down-regulated genes by SAHA, pargyline or combination were shown. b Expression profiles of genes that displayed ≥1.5 fold induction after combined treatment. c Expression profile of genes that displayed ≥1.5 fold reduction after combined treatment. Shown is the mean ±SD for three replicates
Table 1.
Gene expression increased by combined treatment with SAHA and pargyline
| ID | Gene symbol | Gene title | Fold inductiona | SD |
|---|---|---|---|---|
| 202340_x_at | NR4A1 | Nuclear receptor subfamily 4, group A, member 1 | 3.932 | 1.032 |
| 221658_s_at | IL21R | Interleukin 21 receptor | 3.672 | 0.551 |
| 205663_at | PCBP3 | Poly(rC) binding protein 3 | 3.448 | 0.769 |
| 210056_at | RND1 | Rho family GTPase 1 | 3.134 | 0.170 |
| 213820_s_at | STARD5 | StAR-related lipid transfer (START) domain containing 5 | 3.032 | 0.182 |
| 201137_s_at | HLA-DPB1 | Major histocompatibility complex, class II, DP beta 1 | 3.019 | 0.458 |
| 218918_at | MAN1C1 | Mannosidase, alpha, class 1C, member 1 | 2.875 | 0.278 |
| 206549_at | INSL4 | Insulin-like 4 (placenta) | 2.748 | 0.729 |
| 214432_at | ATP1A3 | ATPase, Na+/K+ transporting, alpha 3 polypeptide | 2.701 | 0.287 |
| 221924_at | ZMIZ2 | Zinc finger, MIZ-type containing 2 | 2.692 | 0.075 |
| 217478_s_at | HLA-DMA | Major histocompatibility complex, class II, DM alpha | 2.680 | 0.159 |
| 209959_at | NR4A3 | Nuclear receptor subfamily 4, group A, member 3 | 2.567 | 0.381 |
| 221210_s_at | NPL | N-acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase) | 2.553 | 0.171 |
| 206217_at | EDA | Ectodysplasin A | 2.535 | 0.523 |
| 209324_s_at | RGS16 | Regulator of G-protein signaling 16 | 2.527 | 0.209 |
| 204773_at | IL11RA | Interleukin 11 receptor, alpha | 2.517 | 0.210 |
| 220047_at | SIRT4 | Sirtuin (silent mating type information regulation 2 homolog) 4 (S. cerevisiae) | 2.513 | 0.296 |
| 208978_at | CRIP2 | Cysteine-rich protein 2 | 2.413 | 0.338 |
| 219797_at | MGAT4A | Mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, iso | 2.392 | 0.311 |
| 206249_at | MAP3K13 | Mitogen-activated protein kinase kinase kinase 13 | 2.380 | 0.284 |
| 205780_at | BIK | BCL2-interacting killer (apoptosis-inducing) | 2.321 | 0.276 |
| 216331_at | ITGA7 | Integrin, alpha 7 | 2.311 | 0.595 |
| 210166_at | TLR5 | Toll-like receptor 5 | 2.304 | 0.169 |
| 211685_s_at | NCALD | Neurocalcin delta | 2.300 | 0.287 |
| 210538_s_at | BIRC3 | Baculoviral IAP repeat-containing 3 | 2.300 | 0.335 |
| 221887_s_at | DFNB31 | Deafness, autosomal recessive 31 | 2.231 | 0.365 |
| 206648_at | ZNF571 | Zinc finger protein 571 | 2.080 | 0.158 |
| 207177_at | PTGFR | Prostaglandin F receptor (FP) | 2.074 | 0.180 |
| 205104_at | SNPH | Syntaphilin | 2.060 | 0.289 |
| 202450_s_at | CTSK | Cathepsin K | 2.047 | 0.228 |
| 206191_at | ENTPD3 | Ectonucleoside triphosphate diphosphohydrolase 3 | 1.998 | 0.381 |
| 219389_at | SUSD4 | Sushi domain containing 4 | 1.996 | 0.073 |
| 204072_s_at | FRY | Furry homolog (Drosophila) | 1.973 | 0.127 |
| 216034_at | ZNF280A | Zinc finger protein 280A | 1.973 | 0.176 |
| 205170_at | STAT2 | Signal transducer and activator of transcription 2,113 kDa | 1.941 | 0.354 |
| 219154_at | TMEM120B | Transmembrane protein 120B | 1.928 | 0.192 |
| 216470_x_at | PRSS1/2/3 | Protease, serine, 1 (trypsin 1)///protease, serine, 2 (trypsin 2)///protease | 1.924 | 0.246 |
| 212560_at | SORL1 | Sortilin-related receptor, L(DLR class) A repeats-containing | 1.911 | 0.152 |
| 212148_at | PBX1 | Pre-B-cell leukemia homeobox 1 | 1.902 | 0.014 |
| 221577_x_at | GDF15 | Growth differentiation factor 15///similar to growth differentiation factor 15 | 1.901 | 0.421 |
| 210854_x_at | SLC6A8 | Solute carrier family 6 (neurotransmitter transporter, creatine), member 8 | 1.896 | 0.158 |
| 218689_at | FANCF | Fanconi anemia, complementation group F | 1.892 | 0.261 |
| 219771_at | TBC1D8B | TBC1 domain family, member 8B (with GRAM domain) | 1.887 | 0.121 |
| 205652_s_at | TTLL1 | Tubulin tyrosine ligase-like family, member 1 | 1.857 | 0.273 |
| 203918_at | PCDH1 | Protocadherin 1 | 1.854 | 0.155 |
| 201785_at | RNASE1 | Ribonuclease, RNase A family, 1 (pancreatic) | 1.842 | 0.227 |
| 50221_at | TFEB | Transcription factor EB | 1.824 | 0.119 |
| 204748_at | PTGS2 | Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxyge | 1.817 | 0.272 |
| 210118_s_at | IL1A | Interleukin 1, alpha | 1.809 | 0.062 |
| 204337_at | RGS4 | Regulator of G-protein signaling 4 | 1.797 | 0.130 |
| 208180_s_at | HIST1H4H | Histone cluster 1, H4h | 1.789 | 0.101 |
| 204993_at | GNAZ | Guanine nucleotide binding protein (G protein), alpha z polypeptide | 1.788 | 0.081 |
| 203632_s_at | GPRC5B | G protein-coupled receptor, family C, group 5, member B | 1.768 | 0.152 |
| 209463_s_at | TAF12 | TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa | 1.755 | 0.262 |
| 204624_at | ATP7B | ATPase, Cu++ transporting, beta polypeptide | 1.746 | 0.292 |
| 202452_at | ZER1 | Zer-1 homolog (C. elegans) | 1.665 | 0.061 |
| 205903_s_at | KCNN3 | Potassium intermediate/small conductance calcium-activated channel, subfamily N, | 1.649 | 0.136 |
| 203908_at | SLC4A4 | Solute carrier family 4, sodium bicarbonate cotransporter, member 4 | 1.646 | 0.108 |
| 203665_at | HMOX1 | Heme oxygenase (decycling) 1 | 1.637 | 0.143 |
| 205888_s_at | JAKMIP2 | Janus kinase and microtubule interacting protein 2 | 1.633 | 0.151 |
| 221009_s_at | ANGPTL4 | Angiopoietin-like 4 | 1.632 | 0.181 |
| 219609_at | WDR25 | WD repeat domain 25 | 1.614 | 0.239 |
| 205566_at | ABHD2 | Abhydrolase domain containing 2 | 1.611 | 0.129 |
| 220306_at | FAM46C | Family with sequence similarity 46, member C | 1.589 | 0.175 |
| 220333_at | PAQR5 | Progestin and adipoQ receptor family member V | 1.588 | 0.104 |
| 207064_s_at | AOC2 | Amine oxidase, copper containing 2 (retina-specific) | 1.578 | 0.060 |
| 203965_at | USP20 | Ubiquitin specific peptidase 20 | 1.573 | 0.177 |
| 208165_s_at | PRSS16 | Protease, serine, 16 (thymus) | 1.573 | 0.122 |
| 213059_at | CREB3L1 | cAMP responsive element binding protein 3-like 1 | 1.568 | 0.179 |
| 205069_s_at | ARHGAP26 | Rho GTPase activating protein 26 | 1.568 | 0.019 |
| 205822_s_at | HMGCS1 | 3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) | 1.561 | 0.158 |
| 213413_at | STON1 | Stonin 1 | 1.560 | 0.195 |
| 212339_at | EPB41L1 | Erythrocyte membrane protein band 4.1-like 1 | 1.560 | 0.111 |
| 32502_at | GDPD5 | Glycerophosphodiester phosphodiesterase domain containing 5 | 1.546 | 0.085 |
| 221908_at | RNFT2 | Ring finger protein, transmembrane 2 | 1.537 | 0.149 |
| 221778_at | JHDM1D | Jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae) | 1.521 | 0.192 |
| 210365_at | RUNX1 | Runt-related transcription factor 1 | 1.516 | 0.034 |
| 220432_s_at | CYP39A1 | Cytochrome P450, family 39, subfamily A, polypeptide 1 | 1.516 | 0.097 |
| 201957_at | PPP1R12B | Protein phosphatase 1, regulatory (inhibitor) subunit 12B | 1.508 | 0.153 |
| 203661_s_at | TMOD1 | Tropomodulin 1 | 1.507 | 0.127 |
| 220138_at | HAND1 | Heart and neural crest derivatives expressed 1 | 1.507 | 0.079 |
SD standard deviation
n = 3, gene expression >1.5 fold
Table 2.
Gene expression reduced by combined treatment with SAHA and pargyline
| ID | Gene symbol | Gene title | Fold reductiona | SD |
|---|---|---|---|---|
| 219165_at | PDLIM2 | PDZ and LIM domain 2 (mystique) | 2.921 | 0.072 |
| 211715_s_at | BDH1 | 3-hydroxybutyrate dehydrogenase, type 1 | 2.481 | 0.083 |
| 214591_at | KLHL4 | Kelch-like 4 (Drosophila) | 2.077 | 0.023 |
| 206271_at | TLR3 | Toll-like receptor 3 | 2.071 | 0.061 |
| 220617_s_at | ZNF532 | Zinc finger protein 532 | 2.032 | 0.084 |
| 219502_at | NEIL3 | Nei endonuclease VIII-like 3 (E. coli) | 2.005 | 0.078 |
| 220441_at | DNAJC22 | DnaJ (Hsp40) homolog, subfamily C, member 22 | 1.962 | 0.077 |
| 221293_s_at | DEF6 | Differentially expressed in FDCP 6 homolog (mouse) | 1.934 | 0.007 |
| 209239_at | NFKB1 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | 1.922 | 0.047 |
| 211668_s_at | PLAU | Plasminogen activator, urokinase | 1.879 | 0.102 |
| 203626_s_at | SKP2 | S-phase kinase-associated protein 2 (p45) | 1.836 | 0.059 |
| 206103_at | RAC3 | Ras-related C3 botulinum toxin substrate 3 | 1.796 | 0.062 |
| 209883_at | GLT25D2 | Glycosyltransferase 25 domain containing 2 | 1.769 | 0.075 |
| 202283_at | SERPINF1 | Serpin peptidase inhibitor, clade F (alpha-2 antiplasmin) | 1.738 | 0.101 |
| 212452_x_at | MYST4 | MYST histone acetyltransferase (monocytic leukemia) 4 | 1.729 | 0.072 |
| 204282_s_at | FARS2 | Phenylalanyl-tRNA synthetase 2, mitochondrial | 1.692 | 0.063 |
| 209832_s_at | CDT1 | Chromatin licensing and DNA replication factor 1 | 1.690 | 0.036 |
| 218161_s_at | CLN6 | Ceroid-lipofuscinosis, neuronal 6, late infantile, variant | 1.689 | 0.065 |
| 214692_s_at | JRK | Jerky homolog (mouse) | 1.685 | 0.056 |
| 221762_s_at | PCIF1 | PDX1 C-terminal inhibiting factor 1 | 1.673 | 0.048 |
| 212020_s_at | MKI67 | Antigen identified by monoclonal antibody Ki-67 | 1.667 | 0.078 |
| 206002_at | GPR64 | G protein-coupled receptor 64 | 1.652 | 0.038 |
| 221618_s_at | TAF9B | TAF9B RNA polymerase II, TATA box binding protein (TBP) | 1.621 | 0.062 |
| 219742_at | PRR7 | Proline rich 7 (synaptic) | 1.621 | 0.017 |
| 219214_s_at | NT5C | 5′,3′-nucleotidase, cytosolic | 1.617 | 0.024 |
| 218485_s_at | SLC35C1 | Solute carrier family 35, member C1 | 1.614 | 0.036 |
| 204750_s_at | DSC2 | Desmocollin 2 | 1.613 | 0.035 |
| 212319_at | SGSM2 | Small G protein signaling modulator 2 | 1.602 | 0.024 |
| 204827_s_at | CCNF | Cyclin F | 1.601 | 0.059 |
| 201839_s_at | EPCAM | Epithelial cell adhesion molecule | 1.597 | 0.044 |
| 204759_at | RCBTB2 | Regulator of chromosome condensation and BTB domain containing prot | 1.587 | 0.053 |
| 209373_at | MALL | Mal, T-cell differentiation protein-like | 1.582 | 0.075 |
| 209310_s_at | CASP4 | Caspase 4, apoptosis-related cysteine peptidase | 1.573 | 0.070 |
| 212023_s_at | MKI67 | Antigen identified by monoclonal antibody Ki-67 | 1.569 | 0.066 |
| 219371_s_at | KLF2 | Kruppel-like factor 2 (lung) | 1.559 | 0.047 |
| 222030_at | SIVA1 | SIVA1, apoptosis-inducing factor | 1.539 | 0.023 |
| 222027_at | NUCKS1 | Nuclear casein kinase and cyclin-dependent kinase substrate 1 | 1.538 | 0.013 |
| 205397_x_at | SMAD3 | SMAD family member 3 | 1.529 | 0.074 |
| 206031_s_at | USP5 | Ubiquitin specific peptidase 5 (isopeptidase T) | 1.519 | 0.005 |
| 40489_at | ATN1 | Atrophin 1 | 1.518 | 0.031 |
| 204823_at | NAV3 | Neuron navigator 3 | 1.515 | 0.063 |
| 210846_x_at | TRIM14 | Tripartite motif-containing 14 | 1.511 | 0.044 |
| 218908_at | ASPSCR1 | Alveolar soft part sarcoma chromosome region, candidate 1 | 1.510 | 0.072 |
SD standard deviation
n = 3, gene expression <1.5 fold
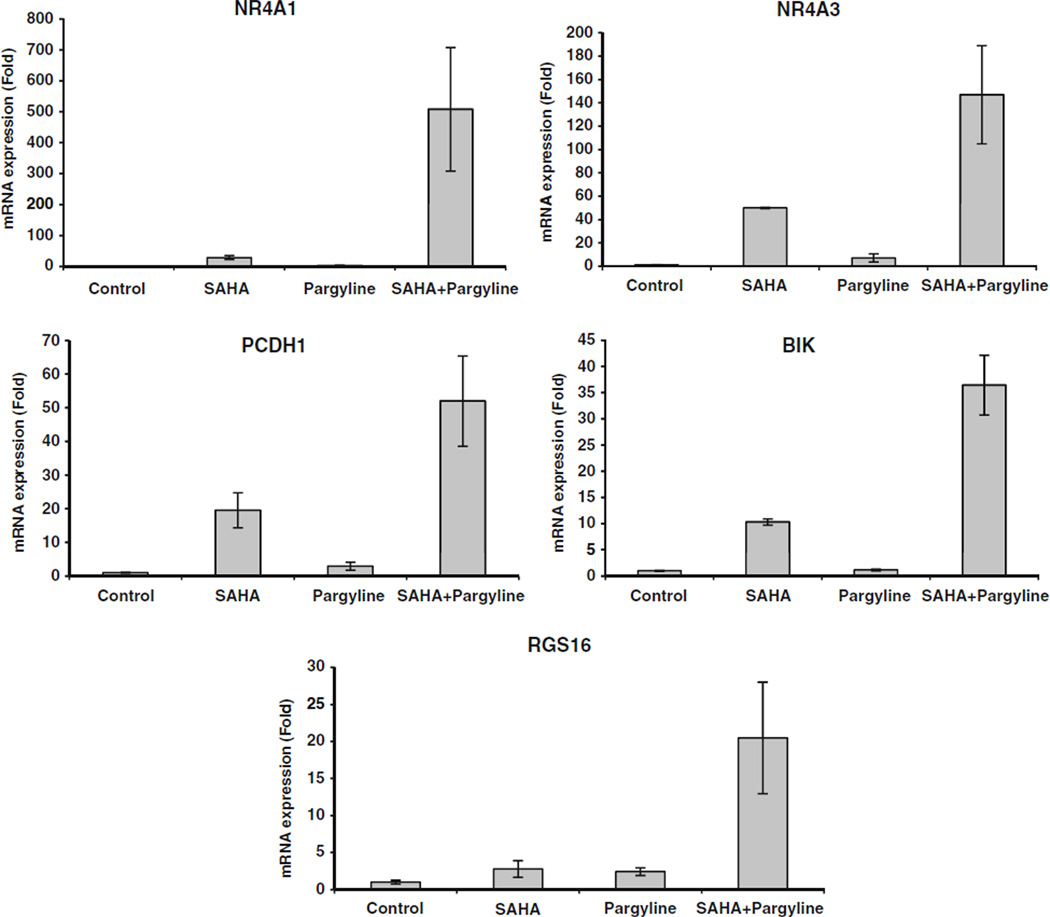
To validate the microarray results, five genes whose expression levels were uniquely induced by combination treatment were selected for evaluation by qPCR because of their potential to play an important role in breast tumorigenesis and therapeutic response. These genes include nuclear receptor subfamily 4, group A, member 1 and 3 (NR4A1, NR4A3), proto-cadherin 1 (PCDH1), BCL2-interacting killer (BIK),and regulator of G-protein signaling 16 (RGS16). The same set of mRNAs used for microarray analysis was reverse transcribed and the qPCR results were consistent with microarray results that the combination resulted in a striking synergistic increase in expression of all the selected genes (Fig. 6).
Fig. 6.
Validation of gene expression induced by combination of SAHA and pargyline. mRNA was measured by quantitative real time PCR for indicated gene expression
We also used DAVID Bioinformatics Resources 6.7 (NIAID/NIH) to categorize the genes into biological groups based on functional similarity. Identified genes have roles in a wide range of cellular functions including cell proliferation and death, cell signaling, transcription regulation, cellular movement, metabolic processes, etc. (Supplementary Table S1).
Discussion
LSD1 has been proposed to demethylate its histone substrate that requires the intimate collaboration between LSD1 and HDAC1/2 [9, 25, 26]. Our current study demonstrated that treatment with zinc dependent class I/II HDAC inhibitors remarkably diminished the activity of LSD1 in breast cancer cells, suggesting that LSD1 is an important downstream target of specific HDAC inhibitors in breast cancer. This is in contrast to a lack of measurable increase of H3K4me2 after treatment with NAD+ dependent class III HDAC inhibitors, splitomicin, and nicotinamide, suggesting the specific interplay between LSD1 and class I/II HDACs, but not class III HDACs. We also demonstrated that either pharmacological inhibition or knockdown of LSD1 expression by siRNA enhanced the acetylation of H3K9, a critical mark for gene activation. These observations indicate that histone demethylation is an important component of the activity of HDAC inhibitors in breast cancer cells.
We showed that LSD2 possesses the activity to demethylate H3K4 in breast cancer cells. This clearly suggests the existence of a more sophisticated FAD-dependent histone demethylase family whose members play a role in chromatin remodeling and transcription regulation in breast cancer. Knockdown of LSD2 did not lead to enhanced lysine acetylation, indicating that, despite structural and catalytic similarities, LSD2 is likely to be part of chromatin-remodeling complexes different from those involving LSD1 and HDACs.
Previous studies have shown the synergistic effects of the combination of DNA methyltransferase and HDAC inhibitors in re-expressing epigenetically silenced genes in cancer cells and leading to clinical responses in patients with leukemia [27, 28]. Our study provides evidence that the HDAC inhibitor and LSD1 inhibitor cooperate to increase both histone methylation and acetylation marks in breast cancer cells, and lead to significant synergy in growth inhibition when used in combination. Through microarray screening, we identified a unique subset of genes whose expression was significantly up-regulated by combined inhibition of LSD1 and HDACs. The enhanced transcriptional activation of these genes appears to lie in the collaboration between the activity of histone methylation and acetylation which are two critical components of transcriptional repressor complexes. Importantly, the genes identified are extensively involved in regulation of cell proliferation, cell signaling, transcription regulation, cellular movement, nucleosomal assembly, and metabolic processes in breast cancer. For example, NR4A1 and NR4A3 act as critical nuclear transcription factors, translocations of which from the nucleus to mitochondria induce apoptosis and reduce migration in breast cancer cells [29, 30]. The integral membrane protein PCDH1 mediates cell– cell adhesion. The epigenetic silencing of PCDH1 has been reported to be associated with cancer-specific differentially methylated regions (DMRs) in breast cancer [31]. RGS16 plays a significant role in G protein signaling and pathways and the activity of RGS16 may inhibit breast cancer cell growth by mitigating phosphatidylinositol 3-kinase signaling [32]. BIK is known to interact with BCL2 in order to enhance programed cell death. Enhanced level of BIK has been reported in apoptosis-inducing reagent treated breast cancer cells [33, 34]. The findings from the array study have added new candidate genes to the existing panel of markers to assess the epigenetic and biological consequences of targeting histone demethylation and deacetylation in breast cancer. The protein expression and activity of the genes modulated by LSD1 and HDAC inhibitor induced chromatin remodeling and growth inhibition should be investigated further. It is anticipated that such studies will identify novel therapeutic targets whose expression may be manipulated to specifically hinder breast cancer cells.
Supplementary Material
Acknowledgments
This work was funded in part by NIH grant CA88843 and the Breast Cancer Research Foundation.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10549-011-1480-8) contains supplementary material, which is available to authorized users.
Contributor Information
Yi Huang, Email: yih26@pitt.edu, University of Pittsburgh Cancer Institute, Pittsburgh, PA, USA; Department of Pharmacology & Chemical Biology, University of Pittsburgh, Pittsburgh, PA, USA.
Shauna N. Vasilatos, University of Pittsburgh Cancer Institute, Pittsburgh, PA, USA
Lamia Boric, University of Pittsburgh Cancer Institute, Pittsburgh, PA, USA.
Patrick G. Shaw, University of Pittsburgh Cancer Institute, Pittsburgh, PA, USA Johns Hopkins University Bloomberg School of Public Health, Baltimore, MD, USA.
Nancy E. Davidson, University of Pittsburgh Cancer Institute, Pittsburgh, PA, USA Department of Pharmacology & Chemical Biology, University of Pittsburgh, Pittsburgh, PA, USA.
References
- 1.Stearns V, Zhou Q, Davidson NE. Epigenetic regulation as a new target for breast cancer therapy. Cancer Invest. 2007;25(8):659–665. doi: 10.1080/07357900701719234. [DOI] [PubMed] [Google Scholar]
- 2.Marks PA, Richon VM, Miller T, Kelly WK. Histone deacetylase inhibitors. Adv Cancer Res. 2004;91:137–168. doi: 10.1016/S0065-230X(04)91004-4. [DOI] [PubMed] [Google Scholar]
- 3.Ficner R. Novel structural insights into class I and II histone deacetylases. Curr Top Med Chem. 2009;9(3):235–240. doi: 10.2174/156802609788085304. [DOI] [PubMed] [Google Scholar]
- 4.Keen JC, Yan L, Mack KM, Pettit C, Smith D, Sharma D, Davidson NE. A novel histone deacetylase inhibitor, scriptaid, enhances expression of functional estrogen receptor alpha (ER) in ER negative human breast cancer cells in combination with 5-aza 2′-deoxycytidine. Breast Cancer Res Treat. 2003;81(3):177–186. doi: 10.1023/A:1026146524737. [DOI] [PubMed] [Google Scholar]
- 5.Zhou Q, Atadja P, Davidson NE. Histone deacetylase inhibitor LBH589 reactivates silenced estrogen receptor alpha (ER) gene expression without loss of DNA hypermethylation. Cancer Biol Ther. 2007;6(1):64–69. doi: 10.4161/cbt.6.1.3549. [DOI] [PubMed] [Google Scholar]
- 6.Yang X, Ferguson AT, Nass SJ, Phillips DL, Butash KA, Wang SM, Herman JG, Davidson NE. Transcriptional activation of estrogen receptor alpha in human breast cancer cells by histone deacetylase inhibition. Cancer Res. 2000;60(24):6890–6894. [PubMed] [Google Scholar]
- 7.Sharma D, Saxena NK, Davidson NE, Vertino PM. Restoration of tamoxifen sensitivity in estrogen receptor-negative breast cancer cells: tamoxifen-bound reactivated ER recruits distinctive corepressor complexes. Cancer Res. 2006;66(12):6370–6378. doi: 10.1158/0008-5472.CAN-06-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 9.Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437(7057):432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 10.Kahl P, Gullotti L, Heukamp LC, Wolf S, Friedrichs N, Vorreuther R, Solleder G, Bastian PJ, Ellinger J, Metzger E, et al. Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer Res. 2006;66(23):11341–11347. doi: 10.1158/0008-5472.CAN-06-1570. [DOI] [PubMed] [Google Scholar]
- 11.Scoumanne A, Chen X. The lysine-specific demethylase 1 is required for cell proliferation in both p53-dependent and - independent manners. J Biol Chem. 2007;282(21):15471–15475. doi: 10.1074/jbc.M701023200. [DOI] [PubMed] [Google Scholar]
- 12.Bradley C, van der Meer R, Roodi N, Yan H, Chandrasekharan MB, Sun ZW, Mernaugh RL, Parl FF. Carcinogen-induced histone alteration in normal human mammary epithelial cells. Carcinogenesis. 2007;28(10):2184–2192. doi: 10.1093/carcin/bgm100. [DOI] [PubMed] [Google Scholar]
- 13.Huang Y, Greene E, Murray Stewart T, Goodwin AC, Baylin SB, Woster PM, Casero RA., Jr Inhibition of lysine-specific demethylase 1 by polyamine analogues results in reexpression of aberrantly silenced genes. Proc Natl Acad Sci USA. 2007;104(19):8023–8028. doi: 10.1073/pnas.0700720104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Y, Stewart TM, Wu Y, Baylin SB, Marton LJ, Perkins B, Jones RJ, Woster PM, Casero RA., Jr Novel oligoamine analogues inhibit lysine-specific demethylase 1 and induce reexpression of epigenetically silenced genes. Clin Cancer Res. 2009;15(23):7217–7228. doi: 10.1158/1078-0432.CCR-09-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Y, Marton LJ, Woster PM, Casero RA. Polyamine analogues targeting epigenetic gene regulation. Essays Biochem. 2009;46:95–110. doi: 10.1042/bse0460007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karytinos A, Forneris F, Profumo A, Ciossani G, Battaglioli E, Binda C, Mattevi A. A novel mammalian flavin-dependent histone demethylase. J Biol Chem. 2009;284(26):17775–17782. doi: 10.1074/jbc.M109.003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, Xu G, Li E, Chen T. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature. 2009;461(7262):415–418. doi: 10.1038/nature08315. [DOI] [PubMed] [Google Scholar]
- 18.Yang Z, Jiang J, Stewart DM, Qi S, Yamane K, Li J, Zhang Y, Wong J. AOF1 is a histone H3K4 demethylase possessing demethylase activity-independent repression function. Cell Res. 2010;20(3):276–287. doi: 10.1038/cr.2010.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Y, Hager ER, Phillips DL, Dunn VR, Hacker A, Frydman B, Kink JA, Valasinas AL, Reddy VK, Marton LJ, et al. A novel polyamine analog inhibits growth and induces apoptosis in human breast cancer cells. Clin Cancer Res. 2003;9(7):2769–2777. [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Y, Keen JC, Hager E, Smith R, Hacker A, Frydman B, Valasinas AL, Reddy VK, Marton LJ, Casero RA, Jr, et al. Regulation of polyamine analogue cytotoxicity by c-Jun in human MDA-MB-435 cancer cells. Mol Cancer Res. 2004;2(2):81–88. [PubMed] [Google Scholar]
- 21.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 22.Hahm HA, Dunn VR, Butash KA, Deveraux WL, Woster PM, Casero RA, Jr, Davidson NE. Combination of standard cytotoxic agents with polyamine analogues in the treatment of breast cancer cell lines. Clin Cancer Res. 2001;7(2):391–399. [PubMed] [Google Scholar]
- 23.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 25.Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19(6):857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 26.Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X, Gozani O, Cheng X, Shi Y. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448(7154):718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21(1):103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 28.Gore SD, Baylin S, Sugar E, Carraway H, Miller CB, Carducci M, Grever M, Galm O, Dauses T, Karp JE, et al. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res. 2006;66(12):6361–6369. doi: 10.1158/0008-5472.CAN-06-0080. [DOI] [PubMed] [Google Scholar]
- 29.Alexopoulou AN, Leao M, Caballero OL, Da Silva L, Reid L, Lakhani SR, Simpson AJ, Marshall JF, Neville AM, Jat PS. Dissecting the transcriptional networks underlying breast cancer: NR4A1 reduces the migration of normal and breast cancer cell lines. Breast Cancer Res. 2010;12(4):R51. doi: 10.1186/bcr2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu Q, Dawson MI, Zheng Y, Hobbs PD, Agadir A, Jong L, Li Y, Liu R, Lin B, Zhang XK. Inhibition of trans-retinoic acid-resistant human breast cancer cell growth by retinoid X receptor-selective retinoids. Mol Cell Biol. 1997;17(11):6598–6608. doi: 10.1128/mcb.17.11.6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novak P, Jensen T, Oshiro MM, Watts GS, Kim CJ, Futscher BW. Agglomerative epigenetic aberrations are a common event in human breast cancer. Cancer Res. 2008;68(20):8616–8625. doi: 10.1158/0008-5472.CAN-08-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang G, Bansal G, Xie Z, Druey KM. RGS16 inhibits breast cancer cell growth by mitigating phosphatidylinositol 3-kinase signaling. J Biol Chem. 2009;284(32):21719–21727. doi: 10.1074/jbc.M109.028407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rao R, Nalluri S, Kolhe R, Yang Y, Fiskus W, Chen J, Ha K, Buckley KM, Balusu R, Coothankandaswamy V, et al. Treatment with panobinostat induces glucose-regulated protein-78 acetylation and endoplasmic reticulum stress in breast cancer cells. Mol Cancer Ther. 2010;9(4):942–952. doi: 10.1158/1535-7163.MCT-09-0988. [DOI] [PubMed] [Google Scholar]
- 34.Ho TF, Ma CJ, Lu CH, Tsai YT, Wei YH, Chang JS, Lai JK, Cheuh PJ, Yeh CT, Tang PC, et al. Undecylprodigiosin selectively induces apoptosis in human breast carcinoma cells independent of p53. Toxicol Appl Pharmacol. 2007;225(3):318–328. doi: 10.1016/j.taap.2007.08.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.