Abstract
During development, a small but significant number of CpG islands (CGIs) become methylated. The timing of developmentally programmed CGI methylation and associated mechanisms of transcriptional regulation during cellular differentiation, however, remain poorly characterized. Here, we used genome-wide DNA methylation microarrays to identify epigenetic changes during human embryonic stem cell (hESC) differentiation. We discovered a group of CGIs associated with developmental genes that gain methylation after hESCs differentiate. Conversely, erasure of methylation was observed at the identified CGIs during subsequent reprogramming to induced pluripotent stem cells (iPSCs), further supporting a functional role for the CGI methylation. Both global gene expression profiling and quantitative reverse transcription-PCR (RT-PCR) validation indicated opposing effects of CGI methylation in transcriptional regulation during differentiation, with promoter CGI methylation repressing and 3′ CGI methylation activating transcription. By studying diverse human tissues and mouse models, we further confirmed that developmentally programmed 3′ CGI methylation confers tissue- and cell-type-specific gene activation in vivo. Importantly, luciferase reporter assays provided evidence that 3′ CGI methylation regulates transcriptional activation via a CTCF-dependent enhancer-blocking mechanism. These findings expand the classic view of mammalian CGI methylation as a mechanism for transcriptional silencing and indicate a functional role for 3′ CGI methylation in developmental gene regulation.
INTRODUCTION
Establishment and maintenance of epigenetic states that govern and stabilize cell fate upon differentiation are crucial for the development of multicellular organisms (1). DNA methylation, which is mitotically heritable, is an important component of mammalian epigenetic gene regulation (2–4). In mammals, DNA methylation occurs predominantly at cytosines preceding guanines (CpG dinucleotides). Although the importance of genomic DNA methylation for normal mammalian development is widely accepted (5–7), it has been proposed that its primary function is to silence transposons and repeats. Hence, the extent to which DNA methylation serves as a general mechanism for regulating gene expression during differentiation remains controversial (8, 9).
Most of the studies aimed at addressing this question have focused on promoters associated with high CpG density, promoter CpG islands (CGIs) (10). It is well established that methylation at promoter CGIs results in self-perpetuating gene silencing either directly, by inhibiting the binding of methylation-sensitive transcriptional activators, or indirectly, by affecting the binding of proteins that orchestrate changes in chromatin conformation (11). Although most promoter CGIs are unmethylated in differentiated mammalian tissues, we and others have shown that methylation occurs at a small but significant number of them and is associated with tissue-specific silencing (12–14). Subsequently, several comprehensive genome-wide studies estimated that 10 to 16% of all CGIs in the human genome are methylated in a tissue-specific fashion, with a significant fraction of these overlapping alternative promoters (15–18). Many important questions regarding the functional significance of tissue-specific CGI methylation, however, remain unanswered. Does de novo CGI methylation occur during early stages of development or during differentiation of adult stem cells? Or, alternatively, is it a secondary consequence of aging and/or environmental exposures? And, while promoter CGIs have been viewed as key epigenetic regulatory elements (19, 20), what is the function of methylation at nonpromoter CGIs?
Recently, several genome-wide studies revealed that gene body methylation is evolutionally conserved and associated with actively transcribed genes (21–25), providing compelling evidence that gene body methylation may be functionally important. In support of this, a genome-wide methylation study in mouse postnatal neural stem cells revealed that Dnmt3a-dependent nonproximal promoter methylation promotes expression of neurogenic genes critical for development (22). One recent study suggested a role of gene body methylation and CTCF in regulating alternative splicing (26). Using CD45 as a model gene system, the authors showed that in several human Burkitt lymphoma cell lines, DNA methylation at the CTCF-binding site regulates the alternative splicing of CD45 exon 5 by local pausing of RNA polymerase II. This mechanistic link between DNA methylation and alternative pre-mRNA splicing was further supported by genome-wide analyses of alternative splicing and CTCF binding in lymphoma cell lines. It remains unclear, however, whether this is a general mechanism. Overall, the mechanisms linking mammalian gene body methylation with transcriptional activation remain largely unknown.
Here, employing differentiation systems of human embryonic stem cells (hESCs), we performed integrated genome-wide analyses to identify epigenetic mechanisms controlling cellular differentiation during early development. In addition to canonical transcriptional repression by methylation at promoter CGIs, we discovered developmentally regulated gene activation by 3′ CGI methylation. Detailed analysis revealed that developmentally programmed methylation at 3′ CGIs confers tissue- and cell-type-specific transcriptional activation. Finally, we provide evidence that CTCF-dependent enhancer-blocking activity at 3′ CGIs serves as a general mechanism to orchestrate transcriptional regulation.
MATERIALS AND METHODS
hESC culture, in vitro differentiation, and reprogramming.
Two hESC lines, H1 (NIH code WA01) and H13 (NIH code WA13), were cultured without feeders under conditioned medium as described previously (27). Random differentiation was induced in these two cell lines as reported previously using differentiation medium containing 20% fetal bovine serum (28, 29). Cells were collected after differentiation at either 21 or 90 days for each cell line. Lineage-specific differentiation to fibroblasts was induced in H1 hESCs as a stable population according to a published protocol (30). Induced pluripotent stem cells (iPSCs) were generated from hESC-derived fibroblasts as previously described using a linked Oct4-Sox2 lentiviral vector (31). For all the experiments including in vitro differentiation and reprogramming, at least two biological replicates were performed.
Human tissue samples.
Normal tissue DNA and RNA samples were purchased from the BioChain Institute (Hayward, CA) and BD Biosciences (San Jose, CA).
DNA methylation microarray.
The methylated CpG island amplification and microarray hybridization (MCAM) procedure was carried out as previously described (12, 32–35). Briefly, 2 μg of genomic DNA was digested with 100 U of methylation-sensitive restriction endonuclease SmaI (New England BioLabs, Ipswich, MA) for 16 h at 20°C (which cuts unmethylated DNA and leaves blunt ends [CCC/GGG]). Subsequently, the DNA was digested with 20 U of SmaI's methylation-insensitive isoschizomer XmaI (New England BioLabs) for 9 h at 37°C (which leaves sticky ends [C/CCGGG]). In total, 500 ng of digested DNA was ligated to 5 nmol of adaptor using T4 DNA ligase (Invitrogen, Grand Island, NY). The adaptors were prepared by incubation of the oligonucleotides RMCA12 (5′-CCGGGCAGAAAG-3′) and RMCA24 (5′-CCACCGCCATCCGAGCCTTTCTGC-3′) at 65°C for 2 min, followed by cooling to room temperature for 60 min. After filling in the overhanging ends of the ligated DNA fragments at 72°C, DNA was amplified under a condition of 95°C for 3 min followed by 25 cycles of 1 min at 95°C and 3 min at 77°C using 100 pmol of RMCA24 primer. MCA products were labeled with Cy5 (red) for differentiated hESCs at either day 21 or day 90 and Cy3 (green) for undifferentiated hESCs using a random primed Klenow polymerase reaction (Invitrogen) at 37°C for 3 h. Labeled samples were then hybridized to a custom-designed Agilent microarray. The 243,000 probes on the custom-designed array cover 92,758 SmaI/XmaI intervals (>80% of human SmaI/XmaI intervals between 60 and 1,500 bp) with an average 2.6 probes/interval. The arrays were washed according to the manufacturer's protocol, scanned on an Agilent scanner, and analyzed using Feature Extraction software (Agilent Technologies, Santa Clara, CA). Array design, reproducibility, and reliability are summarized in Fig. S1 in the supplemental material.
Gene expression microarray.
Total RNA was extracted from hESCs before or after differentiation as described above. Targets for microarray hybridization were generated from the RNA according to manufacturer's instructions (Agilent Technologies). The Agilent whole-human transcriptome array, which contains 41,000 transcripts, was used for gene expression profiling. Hybridization, washing, scanning, and analysis were performed according to the manufacturer's instructions.
DNA methylation microarray analysis.
Based on our earlier studies (12, 34, 35), the DNA methylation microarray analysis was performed at the level of SmaI/XmaI interval; average and median signal intensity, signal ratio, and P value of all probes within each SmaI/XmaI interval were calculated. We first filtered out 8,831 SmaI/XmaI intervals that mapped to multiple genomic locations, and the remaining 83,927 were annotated for (i) chromosome, (ii) chromosomal address of interval start point, (iii) interval length, (iv) overlap with CGI, (v) overlap with repeats, (vi) distance to transcription start site (TSS), and (vii) distance to transcription end site (TES). CGIs were defined as at least 500 bp long with GC contents above 55% and CpG ratios above 0.65 (20). We used the following criteria (12, 34, 35) to identify differentially methylated regions in differentiated hESCs relative to undifferentiated hESCs: (i) median signal ratio of >2 or <0.5, (ii) median upper signal intensity of >1,000, and (iii) median P value log ratio of <0.0001.
Gene expression microarray analysis.
Expression was analyzed by the statistical algorithm in the Agilent two-color microarray-based gene expression analysis using the default parameters. The data from the undifferentiated hESCs were used as a baseline expression for comparison with the differentiated hESCs.
Combined DNA methylation and gene expression analysis.
We integrated the DNA methylation array data with the whole-genome expression array data based on gene annotation. We focused on genes associated with either promoter CGIs or 3′ CGIs that increased methylation at either 21 days or 90 days after induced differentiation. Promoter was defined as 1 kb upstream of the TSS and 300 bp downstream of the TSS and 3′ as 1 kb upstream of the TES and 300 bp downstream of the TES. We were able to obtain gene expression data for 224 genes associated with increased promoter CGI methylation after differentiation (at either 21 or 90 days) and 74 genes associated with increased 3′ CGI methylation after differentiation. To assign equal weights for expression of each gene, we expressed gene expression values in each data set as their respective Z scores calculated as (X − μ)/σ, where X stands for expression value (log10 transformed) of a gene in one sample and μ and σ stand for the mean and standard deviation of that gene among all samples, respectively. To determine the significance of gene expression changes among undifferentiated hESCs, differentiated hESCs at day 21, and differentiated hESCs at day 90, we used analysis of variance to compare gene expression (by Z score) for all genes in four biological replicates per group.
Combined analysis of DNA methylation and histone modifications.
To assess relationships among differentiation-associated DNA methylation changes and genomic regions enriched in bivalent modifications, we downloaded histone modification data from a published genome-wide profile in undifferentiated H1 hESCs (36), which provided H3K4me3 and H3K27me3 status at 93% (32416/34904) of CGI-associated and 92% (51626/55903) of non-CGI-associated regions.
Gene ontology analysis.
Gene ontology enrichment analysis was performed using the GOrilla utility (http://cbl-gorilla.cs.technion.ac.il) (37). We generated a list of 632 genes associated with promoter, intragenic, or 3′ CGIs that showed increased methylation after differentiation. The reference set for the analysis was all genes (n = 9,200) analyzed by the microarray with similar sequence features. The Benjamini-Hochberg procedure (38) was used to control false discovery rate (FDR).
Motif analysis.
Motif analysis was performed as previously reported (12). We generated two sets of sequences, one containing 96 sequences (2-kb window) flanking the center of methylated 3′ CGIs (methylated group) and the other containing 2-kb sequences centered on 1,000 randomly selected 3′ CGIs (reference group). The Fisher exact test was used to identify motifs significantly enriched in the methylated group relative to the reference group.
Combined analysis of DNA methylation and CTCF binding.
To analyze genome-wide associations between DNA methylation and CTCF binding at 3′ CGIs, we downloaded human methylome data comparing DNA methylation profiles of an H1-hESC and a differentiated fibroblast cell (IMR90) from http://neomorph.salk.edu/human_methylome (39). The CTCF-binding signals were downloaded from http://insulatordb.uthsc.edu for IMR90 (40), and from the UCSC genome browser (Hg19) for a skin fibroblast cell line (GEO GSM822281) and a mammary epithelial cell line (HMEC) (41).
Quantitative DNA methylation analyses.
Quantitative bisulfite pyrosequencing for all locus-specific DNA methylation analyses was performed as previously described (42, 43). Primer sequences and PCR conditions for bisulfite pyrosequencing are summarized in Table S1 in the supplemental material. For each assay, setup included positive controls (SssI-treated genomic DNA) and negative controls (whole-genome amplified genomic DNA), mixing experiments to rule out bias, and repeated experiments to assess reproducibility. Annealing temperatures were optimized to overcome PCR bias as previously reported (43). On the basis of methylation at 128 CpG sites measured by bisulfite pyrosequencing as continuous variables, an unsupervised hierarchical clustering was performed (44) using Euclidean distances and an average linkage algorithm. A color-coded cluster image map was generated using the CIMminer (Cluster Image Map program package) (45).
At the human PRR15 gene locus, bisulfite sequencing of multiple cloned PCR products was used to measure methylation quantitatively at 206 CpG sites for a 4.5-kb region (from bp −350 to 4150 relative to TSS). The primer sequences are listed in Table S2 in the supplemental material. For this analysis, we cloned postbisulfite PCR products into the TA vector pCR4-TOPO (Invitrogen), extracted plasmid DNA from 15 to 20 clones with the use of a QIAprep spin miniprep kit (Qiagen, Valencia, CA), and sequenced the DNA at the Sequencing Core Facility at the Baylor College of Medicine. At the mouse Hic1 gene locus, we used multiple bisulfite pyrosequencing as described above to measure methylation quantitatively at 149 CpG sites from bp −673 to 5327 relative to the Hic1a TSS (see Table S3 for primer sequences).
qRT-PCR.
TaqMan quantitative real-time reverse transcription-PCR (qRT-PCR) was carried out in triplicate for human CMYA5, ALOX12, RBM38, PRR15, HIC1, and HOXC5, using probe sets Hs00989056_m1, Hs00911143_g1, Hs00955733_m1, Hs00828414_m1, Hs00948220_m1, and Hs00232747_m1, respectively (Applied Biosystems, Carlsbad, CA). Relative gene expression was calculated by the ratio of the target genes to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Hs02758991_g1) expression on an ABI StepOnePlus detection system. For mouse Hic1, we used probe sets Mm04208063_m1 for Hic1a and Mm04204985_g1for Hic1b and used β-actin (Mm00607939_s1) as a reference.
ChIP and real-time PCR.
Chromatin immunoprecipitation (ChIP) for CTCF was carried out based on a modification of a published method (46). Undifferentiated H1 hESCs (2 × 107) were cross-linked with 1% formaldehyde for 10 min. After washing with cold phosphate-buffered saline (PBS), cell pellets were resuspended in lysis buffer (50 mM Tris-HCl [pH 8.1], 10 mM EDTA, 1% SDS, plus the proteinase inhibitor) and sonicated with a Bioruptor sonicator (Diagenode, Denville, NJ) set at high power and 10 cycles of 30 s on/30 s off. The sonicated chromatin was then diluted and precleaned with 0.5% bovine serum albumin (BSA)-blocked magnetic beads (Invitrogen; 100-02D). At the same time, another two aliquots of blocked magnetic beads were coupled with 20 μl of either anti-CTCF antibody (07-729; Millipore, Billerica, MA) or control IgG (AB-105-C; R&D Systems, Minneapolis, MN). After overnight incubation at 4°C, the coupled beads were washed and mixed with the precleaned chromatin on a rotator overnight at 4°C. Precipitated chromatin was eluted from the beads and reverse cross-linked by heating at 65°C for 4 h. Cellular protein and RNA were removed from the eluate by RNase (100 μg/ml) treatment and proteinase K (200 μg/ml) digestion. DNA fragments were extracted with the Qiagen PCR purification kit. TaqMan real-time PCR was conducted as described above. Primer and probe sets corresponding to regions of interest within the 3′ CGIs of PRR15 and HOXC5, the H19 differentially methylated region (DMR) (as CTCF positive control), and two negative-control regions are summarized in Table S4 in the supplemental material.
Mice, tissue collection, fetal colon dissection, and flow cytometry sorting.
Inbred C57BL/6 and Lgr5-enhanced green fluorescent protein (eGFP)-internal ribosome entry site (IRES)-CreERT2 knock-in mice (Jackson Laboratories, Bar Harbor, ME) were used. Lgr5-eGFP-IRES-CreERT2 knock-in mice were backcrossed to a C57BL/6 background for more than 10 generations. Multiple tissues were collected from C57BL/6 mice at age 34 weeks. To obtain subpopulations of E18.5 mouse colonic cell types, heterozygous Lgr5-eGFP-IRES-CreERT2 knock-in mice were mated with C57BL/6 mice, and the morning of vaginal plug was counted as embryonic day 0.5 (E0.5). At E18.5, fetal colonic tissues were collected under a dissecting microscope. Isolated colons from each litter (∼8 pups) were pooled and washed with cold PBS. The colons were chopped into 1-mm pieces. After rocking in 2 mM EDTA with cold PBS at 4°C overnight, tissue fragments were further incubated with TypLE (Invitrogen) at 37°C for 1 h and neutralized with Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum. After dissociation by pipetting and washing with PBS, cells were passed through a 40-μm cell strainer (BD Biosciences). Isolated single cells were resuspended in cell staining buffer (Biolegend, San Diego, CA) and incubated with phycoerythrin (PE) anti-mouse EpCAM antibody (Biolegend; 118206) on ice for 30 min. Cells were sorted based on enhanced green fluorescent protein (eGFP) and EpCAM expression using a 4-way MoFlo cell sorter (Beckman-Coulter). All applicable institutional and governmental regulations concerning the ethical use of animals were followed during this research. The protocol was approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine.
Immunohistochemistry.
Immunohistochemistry was performed as described previously (47, 48). Tissue slides were dewaxed in xylene, rehydrated in ethanol, and rinsed in PBS. To block endogenous peroxidases, slides were incubated in 3% H2O2 for 30 min at room temperature and then rinsed in PBS. Before primary antibody was applied, slides were incubated in blocking solution, containing 5% sheep serum, 0.2% BSA, and 0.1% Triton X-100 in PBS for 1 h at room temperature. Antibodies used were anti-Hic1 (Abcam; Ab33029; 1:100) and anti-α-smooth muscle actin (anti-α-SMA; A2547; 1:500; Sigma-Aldrich, St. Louis, MO). All antibody staining was performed at 4°C overnight, followed by incubation with antibiotin secondary antibody (Vector Laboratories, Burlingame, CA) diluted 1:1,000. Slides were developed using a DAB kit (Vector Laboratories) and imaged using a DS-Fi1 camera connected to a Nikon E80i stereomicroscope. Images were processed using Nikon imaging software, NIS Elements RA3.2.
Luciferase assays.
The reporter plasmids for promoter, enhancer, and enhancer-blocker assays were constructed using primers described in Table S5 in the supplemental material. For testing fragments, a 920-bp human PRR15 fragment (bp 1766 to 2686 relative to TSS) and a 2,155-bp mouse Hic1 fragment (bp 3073 to 5228 relative to Hic1a promoter) were PCR amplified from genomic DNA. Each fragment was confirmed by sequencing in both directions and subsequently cloned in sense and antisense orientations into the reporter plasmids. To create promoter assay constructs, the testing fragments were inserted into the pGL3-basic vector (Promega, Madison, WI) upstream of the firefly luciferase-encoding region. An endogenous PRR15 promoter (bp −1091 to −1 relative to TSS) was used as a positive control. To generate enhancer assay constructs, a cytomegalovirus (CMV) promoter was inserted into the promoter assay plasmids between the testing fragments and luciferase gene. A CMV enhancer was used as a positive control. The enhancer-blocking reporter plasmids, pIHLIE and pIHLME, containing mouse H19 DMR insulator (H19) and a mutant H19 DMR with only the four CTCF-binding sites substituted (MtH19), respectively, were previously described (49). To construct enhancer-blocking assays, the testing fragments were inserted between the mouse H19 promoter and simian virus 40 (SV40) enhancer by replacing MtH19 in the pIHLME plasmid (49). Plasmid pIHLIE (H19) served as a positive control. Plasmid pIHLME was used as a control for the space effect between promoter and enhancer, and its luciferase activities were used for normalization.
Transfection of cells was performed with equimolar amounts of reporter plasmids by Lipofectamine (Invitrogen) according to the manufacturer's instructions. At 24 h posttransfection, luciferase activity was measured by the dual-luciferase assay kit (Promega) with a GloMax-Multi detection system (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity and presented as the mean and standard deviation of the results from at least three independent experiments. GraphPad Prism 4 software was used to calculate statistical significance based on two-tailed t tests.
RNA interference.
Short hairpin RNA (shRNA) targeting CTCF was designed and prepared as reported previously (50). Briefly, stable knockdown of CTCF was achieved via lentiviral delivery of anti-CTCF shRNA (5′-GGACAGTGTTGACAACTAA-3′) in PLKO.1 vector (Addgene, Cambridge, MA). Scrambled shRNA (Addgene; plasmid 1864) was used as a transfection control. Lentiviral particles were generated and used to transduce HCT116 cells according to Addgene's protocol. After selection with puromycin (5 μg/ml), stable clones were established in 3 to 4 weeks. Relative CTCF mRNA expression level was monitored by TaqMan qRT-PCR (Applied Biosystems; Hs00902008_m1). CTCF protein expression was determined by Western blotting assays using anti-CTCF antibody. Equal protein loading was confirmed by blotting with control antibody against β-actin (Abcam; ab1801).
Microarray data accession numbers.
The data discussed in this publication have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus repository (www.ncbi.nlm.nih.gov/geo) under accession numbers GSE32371 and GPL14617.
RESULTS
CGI methylation is associated with differentiation.
To identify fundamental epigenetic mechanisms that regulate cellular differentiation during early development, we performed genome-wide array-based DNA methylation and gene expression profiling in hESCs at different stages of differentiation. We employed random rather than directed differentiation to gain insights into epigenetic mechanisms important to differentiation in general, rather than those unique to specific lineages. Using stringent criteria to avoid false-positive calls, we identified 3,847 genomic regions that undergo DNA methylation changes upon induced differentiation (see Table S6 in the supplemental material).
Recent genome-wide studies in hESCs suggest that genes involved in early developmental decisions are associated with a bivalent chromatin domain, characterized by trimethylation at both lysine 27 of histone H3 (H3K27me3) and lysine 4 of histone H3 (H3K4me3) (36, 51). We therefore used these published databases to investigate the relationships between differentiation-associated DNA methylation changes and genomic regions marked with both H3K4me3 and H3K27me3 in hESCs. We divided the genomic regions into four categories based on whether methylation was gained or lost during differentiation and whether or not they are associated with a CGI. Interestingly, the bivalent chromatin domain was enriched in hESCs only among CGI-associated regions that gained methylation during subsequent induced differentiation (see Fig. S2A in the supplemental material). A key developmental function for this class of CGIs was further suggested by gene ontology analysis of associated genes, which found significant enrichment for developmental processes, including the multicellular organism process, anatomical structure development, and organ morphogenesis (see Fig. S2B).
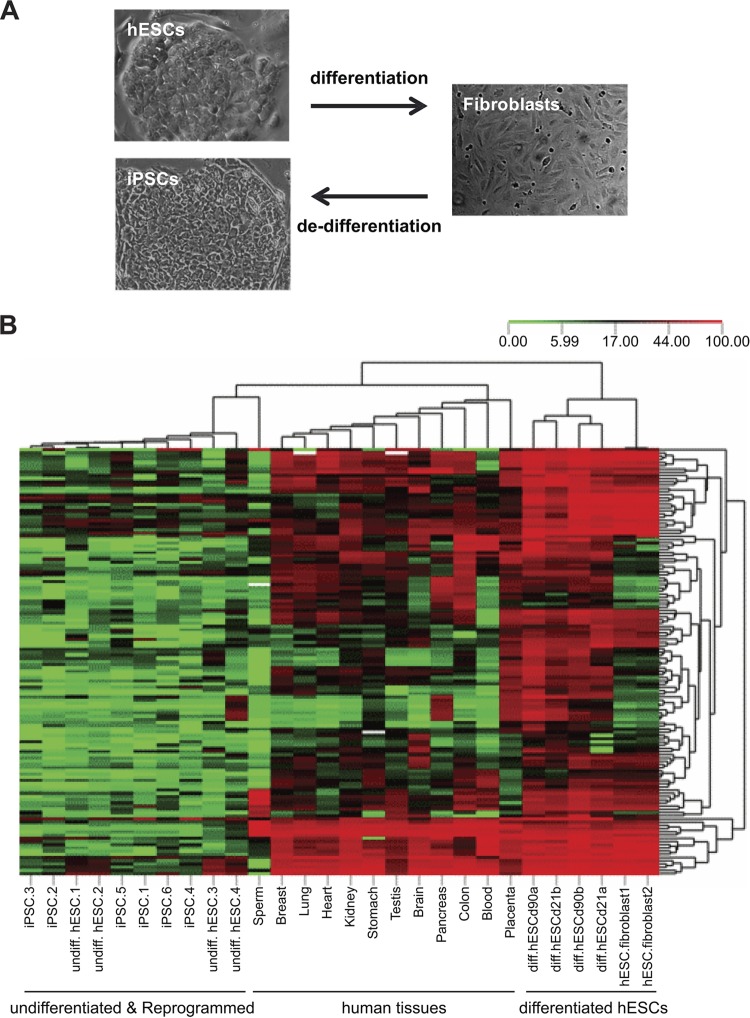
We therefore focused on this methylation-gaining group of CGI-associated genes for further analyses. To validate the array-based results, we performed bisulfite pyrosequencing on over 100 CpG sites in 21 gene-associated CGIs. To determine whether methylation changes identified by in vitro hESC systems recapitulate differentiation in somatic tissues in vivo, we compared methylation profiles in the hESCs before and after differentiation at various time points and in a panel of normal human tissues derived from all three early embryonic germ layers and germ cell and extraembryonic lineages. To further ascertain whether methylation at these CGIs is a developmentally programmed event, we examined methylation in fibroblasts derived from lineage-specific differentiation of hESCs (52), as well as in iPSCs subsequently reprogrammed from these differentiated cells (28) (Fig. 1A). Unsupervised hierarchical clustering based on DNA methylation at all 128 CpG sites revealed a near-perfect correspondence with differentiation state (Fig. 1B). Undifferentiated hESCs clustered together with low methylation, while differentiated hESCs clustered together with remarkably increased methylation at all 128 CpG sites, confirming our microarray results. Further, whereas our methylation microarray approach is nonquantitative, these quantitative data indicate that most (>95%) of these CGIs are unmethylated in undifferentiated hESCs and become de novo methylated upon differentiation. All the normal somatic tissues clustered together in an intermediate zone, consistent with the epigenetic specialization of different cell types compared to randomly differentiated cells, and indicating that DNA methylation at these CGIs is associated with cellular differentiation in vivo. Hence, although the methylation data that we initially generated were based on in vitro differentiation, our ability to validate these associations in various human tissues clearly indicates that they do not simply reflect a cell culture artifact. Fibroblasts differentiated from hESCs clustered with the randomly differentiated cells, exhibiting dense methylation at most CpGs. Most remarkably, methylation at these CGIs was in every case almost completely erased during subsequent reprogramming to iPSCs (Fig. 1B), indicating that erasure of this CGI methylation is associated with dedifferentiation processes. Together, these results provide compelling evidence that DNA methylation at this class of CGIs is associated with both in vitro and in vivo differentiation.
Fig 1.
Unsupervised clustering based on CpG methylation predicts differentiation state. (A) Morphological changes of hESCs after lineage-specific differentiation to fibroblasts and subsequent induced dedifferentiation. (B) Heat map of unsupervised hierarchical clustering analysis based on DNA methylation at 21 CGIs (128 CpG sites) in hESCs at different stages of differentiation and dedifferentiation and among diverse human tissues. Each column represents a sample, and each row represents a CpG site measured by quantitative bisulfite pyrosequencing. Methylation levels range from unmethylated (green) to fully methylated (red).
CGI methylation plays a dual role in transcriptional regulation of developmental genes.
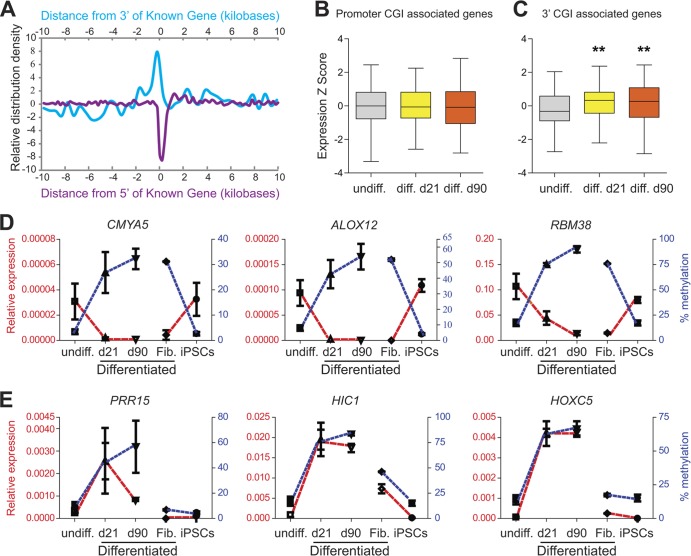
When we compared the genomic localization of these methylation-gaining CGIs with that of all CGIs on the array, we found that they are dramatically underrepresented at promoters (Fig. 2A, purple) but significantly enriched at the 3′ end of known genes (Fig. 2A, blue) (P < 0.0001). To determine whether developmental methylation at these loci is correlated with gene expression, we used human transcriptome microarrays and compared expression levels of genes associated with either promoter or 3′ CGIs. At promoter CGIs, methylation gains during differentiation were not correlated with expression (Fig. 2B). Consistent with a previous study (53), this lack of correlation could be the result of de novo methylation at already transcriptionally silent CGI promoters in undifferentiated stem cells. Alternatively, expression measurements by microarray (especially of low-level expression) are prone to probe and background effects that may confound such correlation. To test this, we performed quantitative measurements of DNA methylation and gene expression at three randomly selected promoter CGI-associated genes (CMYA5, ALOX12, and RBM38) and found excellent inverse correlation between methylation and expression during differentiation of hESCs (Fig. 2D). Moreover, all three promoter CGIs were highly methylated in fibroblasts, and this methylation was lost—and expression was increased—during reprogramming to iPSCs (Fig. 2D).
Fig 2.
Promoter and 3′ CGI methylation exhibit opposite correlations with expression. (A) CGIs that gained methylation during differentiation are underrepresented at 5′ ends (purple) but enriched at the 3′ ends of known genes (blue). (B) Box plot showing normalized gene expression levels (by microarray) for genes associated with increased methylation at promoter CGIs upon induced differentiation. Compared to undifferentiated hESCs, the significance of difference was P = 0.30 at day 21 and P = 0.02 at day 90 (based on expression data of 224 genes in four biological replicates per group). (C) Box plot showing gene expression levels (by microarray) for genes associated with increased methylation at 3′ CGIs upon induced differentiation. Compared to undifferentiated hESCs, the significance of difference was P = 7.35E−08 at day 21 and P = 1.60E−07 at day 90 (based on expression data of 74 genes in four biological replicates per group). (D and E) Quantitative measurements of DNA methylation (blue) and gene expression (red) dynamics at promoter CGI-associated genes (D) and 3′ CGI-associated genes (E) upon induced differentiation and dedifferentiation. In all cases, methylation at promoter CGIs is inversely correlated with expression, but methylation at 3′ CGIs is positively correlated with expression.
The expression microarray analysis showed, surprisingly, that developmental increases in methylation at 3′ CGIs were positively correlated with expression (P = 7.35E−08 at day 21 and P = 1.60E−07 at day 90) (Fig. 2C). We confirmed this association at three 3′ CGI-associated genes (PRR15, HIC1, and HOXC5); during random differentiation, methylation and expression both increased at all 3′ CGIs (Fig. 2E). Further, at the one 3′ CGI that was appreciably methylated in fibroblasts (HIC1), loss of methylation during reprogramming coincided with reduced expression (Fig. 2E).
We expanded the methylation analysis to all CGIs located within the 6 selected genes. Of the 3 genes with increased promoter CGI methylation after differentiation, only RBM38 also contains a 3′ CGI. This CGI was hypermethylated in all samples, regardless of differentiation status (see Fig. S3A in the supplemental material). Each of the 3 selected genes with 3′ CGI methylation after differentiation also contains a promoter CGI. These were essentially unmethylated in all samples (see Fig. S3B). These data indicate that the 5′ and 3′ CGI methylation identified in our screen is uniquely correlated with gene expression changes.
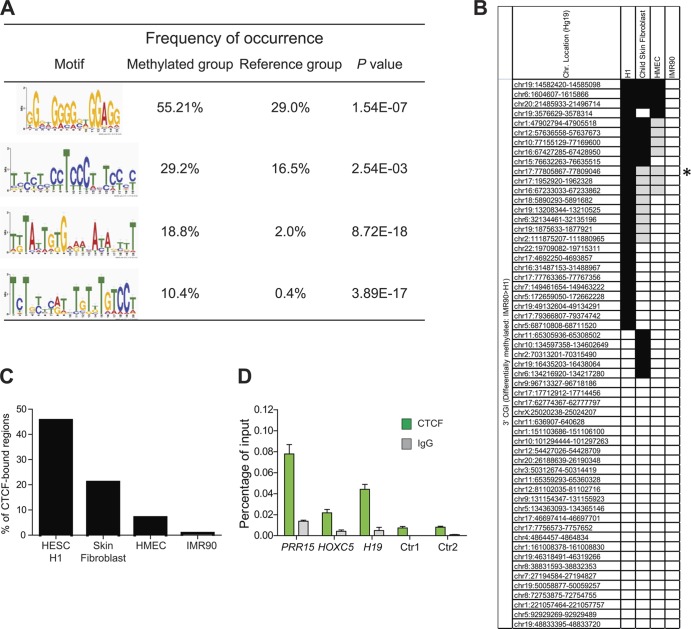
Since CGI methylation has come to be generally viewed as an epigenetic silencing mechanism (10, 54, 55), the identification of developmentally regulated 3′ CGI methylation associated with transcriptional activation was unexpected. To explore the potential underlying mechanism, we searched flanking regions for sequence motifs that may confer shared cis- and/or trans-regulatory mechanisms at these 3′ CGIs. This analysis revealed four sequence motifs significantly enriched relative to reference regions (see Fig. S2 in the supplemental material). The top two motifs were of particular interest because they include multiple “CCCTC” sequences, strongly suggesting the potential for CTCF binding. An evolutionarily conserved transcription factor and key regulator of development (56), CTCF is best known for its DNA methylation-dependent transcriptional regulation at the imprinted IGF2/H19 locus (57). To test the hypothesis that differentiation-associated methylation changes at these regions affect CTCF binding, we exploited published genome-scale DNA methylation (39) and CTCF-ChIP (40, 41) data sets. We identified a set of 3′ CGIs (n = 57) with significantly higher DNA methylation in differentiated IMR90 cells than in undifferentiated H1 hESCs. Whereas a substantial proportion of these (46%) show CTCF binding in the hESCs, the gain in methylation during differentiation is associated with dramatic loss of CTCF binding in the IMR90 cells, as well as in skin fibroblast and mammary epithelial cells (HMEC) (Fig. 3B and C). In addition, we used quantitative ChIP assays in undifferentiated hESCs and confirmed that CTCF binds at the 3′ CGIs of PRR15 and HOXC5 (Fig. 3D).
Fig 3.
Global identification of CTCF binding at 3′ CGIs. (A) Four motifs were significantly enriched in genes with methylated 3′ CGI. The frequency of occurrence was calculated by the number of genes found to match the motif in each group/total number of genes in each group. Fisher's exact test was used to determine if the motifs are significantly enriched in the methylated group (n = 96) compared to the unchanged reference group (n = 2,000). (B) Global analysis of CTCF binding based on published DNA methylation and CTCF-ChIP databases. Each row indicates a 3′ CGI that shows significantly increased methylation in IMR90 compared to undifferentiated H1 hESCs (REF). Their chromosome locations (Hg19 version) are indicated on the left. Each column represents the CTCF-binding signals for H1, skin fibroblast, HMEC, and IMR90, respectively. Black indicates significant CTCF binding, gray indicates significantly reduced binding relative to H1 (i.e., <50%), and white indicates no CTCF binding (the asterisk indicates the 3′ CGI of the HIC1 gene). (C) Summary of CTCF binding based on published databases (40, 41). Significant loss of CTCF binding was observed in differentiated cells relative to undifferentiated hESCs (P = 0.009 for skin fibroblast and 0.0001 for both HMEC and IMR90 [Fisher's exact test]). (D) ChIP validation of CTCF binding at selected 3′ CGIs. Immunoprecipitation was performed with anti-CTCF antibody or control IgG. Immunoprecipitated and 1/10-diluted input DNA samples were used as the templates for TaqMan qPCR. CTCF enrichments at two 3′ CGIs (PRR15 and HOXC5) were analyzed. The H19 imprinting control region was used as a positive control. Two genomic regions predicted to have no CTCF-binding sites (the promoter of the PRR15 gene [Ctr1] and the 3′ non-CGI region of the HOXC5 gene [Ctr2]) were used as negative controls. Error bars are standard deviations (n = 3).
3′ CGI methylation associates with tissue-specific transcriptional activation in vivo.
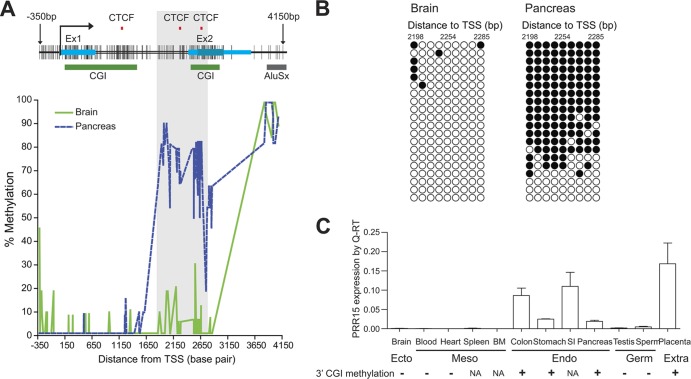
To investigate in greater depth the relationships among CTCF-binding sites, 3′ CGI methylation, and transcriptional regulation during lineage differentiation in vivo, we initially focused on the PRR15 (proline-rich 15) gene. In an animal model, targeted degradation of Prr15 mRNA causes embryonic lethality, indicating a role for PRR15 in early development (58). The human PRR15 gene (which includes two exons) has both a 5′ and a 3′ CGI (Fig. 4A). A CTCF-binding site database (CTCFBSDB) (59) was used to predict CTCF-binding sites around the PRR15 locus. Consistent with the ChIP results, two potential CTCF-binding sites were identified around the 3′ CGI (Fig. 4A). We mapped DNA methylation precisely for 206 CpG sites within a 4.5-kb region encompassing the gene in two normal human tissue types representing two embryonic lineages—brain (ectoderm) and pancreas (endoderm) (Fig. 4A). Whereas the promoter CGI was essentially unmethylated in both tissues, we identified a 920-bp region that was densely methylated in pancreas only. Interestingly, this region (bp 1766 to 2686 relative to TSS) overlaps both the 3′ CGI and its two associated CTCF-binding sites (Fig. 4A). Clonal bisulfite sequencing of this region (Fig. 4B) corroborated the pyrosequencing results and identified both heavily methylated and completely unmethylated molecules within pancreas, suggesting cell-type-specific methylation. More importantly, we found that the strong positive correlation between PRR15 3′ CGI methylation and gene expression observed during in vitro hESC differentiation (Fig. 3E) also extends to multiple tissue lineages in vivo. PRR15 mRNA was detected specifically in endodermal (colon, stomach, small intestine, and pancreas) and extraembryonic (placenta) tissues but not in ectodermal (brain) or mesodermal (blood, heart, spleen, and bone marrow) tissues or in the germ line (sperm and testis) (Fig. 4C). As predicted, in all tissues with DNA available for methylation analysis, we detected 3′ CGI methylation only in PRR15-expressing tissues, supporting the role of 3′ CGI methylation in regulating tissue-specific gene activation.
Fig 4.
Detailed analysis of tissue-specific PRR15 3′ CGI methylation and gene expression. (A) DNA methylation mapping of the PRR15 locus in human brain and pancreas. A diagram of the CpG map is shown at the top. Each vertical line represents a CpG site analyzed for tissue-specific DNA methylation. Red tick marks indicate predicted CTCF-binding sites, blue bars indicate exons, green bars indicate CGIs predicted by the UCSC genome browser, and the gray bar indicates a downstream Alu repeat. A 920-bp region (bp 1766 to 2686 relative to TSS, highlighted in gray) is specifically methylated in pancreas, overlaps both the 3′ CGI and two potential CTCF-binding sites, and was used for further functional characterization. (B) Results of clonal bisulfite sequencing within this region. Each row represents an individual cloned allele. Open circles represent unmethylated CpGs, and filled circles indicate methylation. (C) PRR15 gene expression relative to 3′ CGI methylation in various human tissues. BM, bone marrow; SI, small intestine; NA, DNA sample not available.
Conservation of 3′ CGI methylation and transcriptional activation.
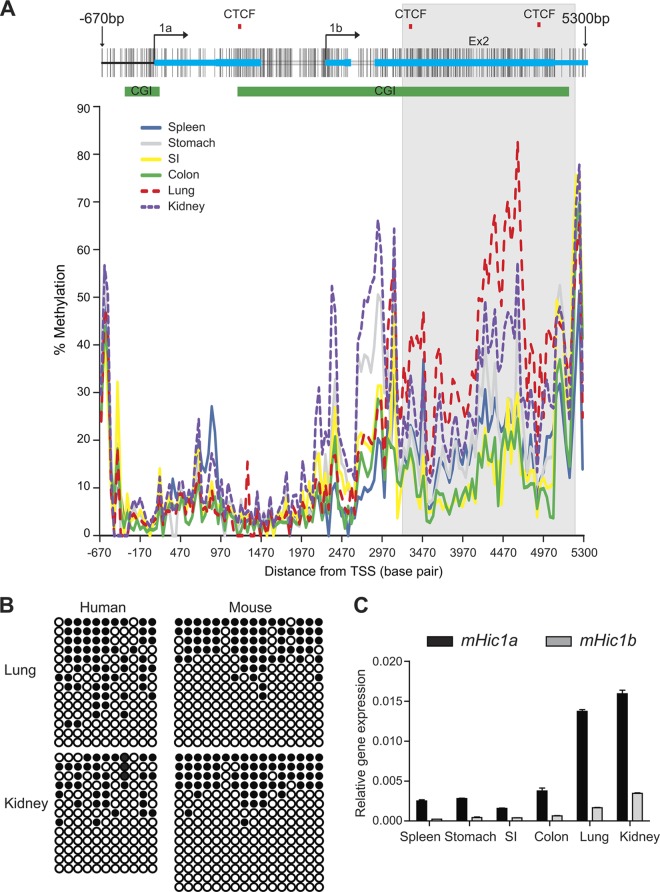
To test whether transcriptional regulation by 3′ CGI methylation extends to other species, we investigated in the mouse an additional gene identified in our human screen, Hic1 (hypermethylated in cancer 1). Hic1 is a well-characterized transcriptional repressor and plays critical roles in embryonic development, tissue morphogenesis, and tumorigenesis (60). Mice deficient in Hic1 die perinatally and exhibit developmental defects in head, face, limbs, and ventral body wall, resembling the Miller-Dieker syndrome in humans (61). Heterozygous loss of Hic1 predisposes mice to tumor development, providing strong evidence that Hic1 is a tumor suppressor gene (62). Remarkably, the exon-intron structure, CGI status, and potential CTCF-binding sites of the Hic1 gene are all conserved in mouse and human, and comparative sequence analysis revealed that there was >90% sequence similarity between the two species (see Fig. S4 in the supplemental material). In both species, Hic1 is transcribed using two alternative promoters (1a and 1b) and spliced onto the same second and last exon (Fig. 5A). The 3′ CGI overlaps promoter 1b and the last two exons. Interestingly, CTCFBSDB predicts three CTCF-binding sites, two of which are located within the 3′ CGI (Fig. 5A, top). The high degree of sequence conservation provides an excellent opportunity to address whether the functional role of 3′ CGI methylation is conserved across species. Indeed, similar patterns of tissue-specific methylation were observed in mouse and human tissues, suggesting functional conservation of 3′ CGI methylation (Fig. 5B). To map the DNA methylation patterns in an approximately 6-kb region at the Hic1 locus, we measured methylation quantitatively for 149 CpG sites in various mouse tissues (Fig. 5A, bottom). Similarly to PRR15, the 5′ CGI was essentially unmethylated in all tissues, and the differentially methylated region was found in the 3′ CGI. To assess the association between methylation and gene expression, we analyzed Hic1 expression separately for the two alternative transcripts. In agreement with previous observations (62, 63), the Hic1a promoter drives the predominant transcript in various tissues (Fig. 5C). Interestingly, expression from both transcripts was positively correlated with 3′ CGI methylation, particularly in the region flanked by two CTCF sites. For instance, relative hypermethylation in lung and kidney (Fig. 5A and B) was associated with strong expression of both transcripts (Fig. 5C). Since it has been previously proposed that gene body methylation regulates differential usage of alternative promoters (15, 16, 26, 64), one might ask whether the 3′ CGI methylation at Hic1a simply acts to repress one of the transcripts, rather than activating transcription per se. The consistency of our results at both alternate transcripts, however, argues against this, suggesting that 3′ CGI methylation regulates tissue-specific expression through a different mechanism.
Fig 5.
Conserved role of 3′ CGI methylation in regulating gene expression. (A) The genomic structure of the mouse Hic1 gene is shown at the top. Alternative promoters at exons 1a and 1b are indicated. The green bars indicate the positions of two evolutionally conserved promoters and 3′ CGIs. The red tick marks indicate predicted CTCF-binding sites. The lower panel is a plot of methylation level versus genomic location for various mouse tissues. A 2-kb region (highlighted in gray) is differentially methylated in multiple tissues, overlaps both the 3′ CGI and two potential CTCF-binding sites, and was used for further functional characterization. Lung and kidney are hypermethylated relative to spleen, stomach, small intestine (SI), and colon tissues. (B) Comparison of tissue-specific methylation in mouse and human by clonal bisulfite sequencing indicates that Hic1 3′ CGI methylation is evolutionarily conserved. Methylation at the core regions of Hic1 3′ CGI was determined in both human (bp 4081 to 4310 relative to 1A promoter) and mouse (bp 4797 to 4947 relative to 1a promoter). Each circle represents a CpG site, and each row represents a single DNA molecule. Closed circles indicate methylation. (C) qRT-PCR shows that increased Hic1 expression from both alternate promoters correlates with 3′ CGI hypermethylation.
3′ CGI methylation mediates cell-type-specific transcriptional activation.
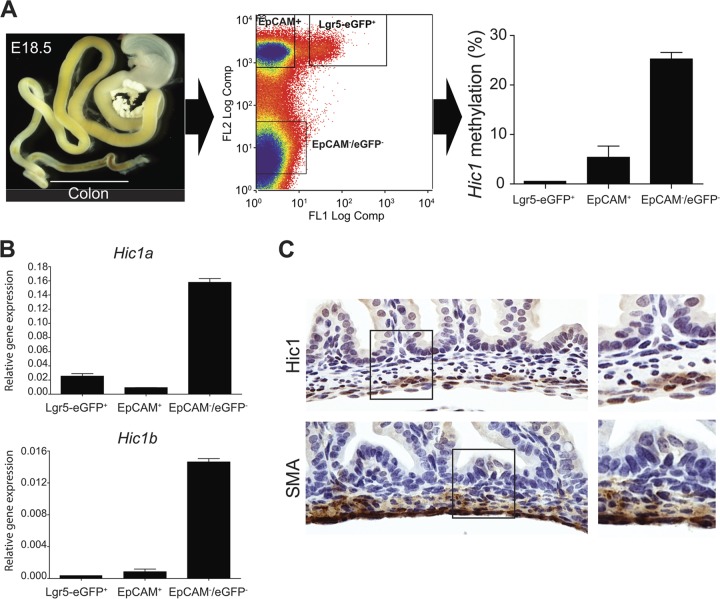
The relatively low levels of both methylation and expression in colon (Fig. 5) suggest that Hic1 3′ CGI methylation might be involved in a minor population of colonic cell types. To test this idea, we used a mouse model to isolate subpopulations of cell types from colonic mucosa. To determine whether 3′ CGI methylation is established during embryonic development, we studied E18.5 mice (cytodifferentiation of undifferentiated endoderm into simple columnar epithelium is apparent by E18.5 [65]). We sorted colonic epithelial stem cells using an Lgr5-eGFP reporter (66) and differentiated epithelial cells using EpCAM (a panepithelial differentiation antigen); mesenchymal cells comprise the remainder (EpCAM−/Lgr5-eGFP−). In E18.5 colon, the Hic1 3′ CGI was methylated specifically in the population of mesenchymal cells (Fig. 6A) and this correlated with increased expression of both transcripts (Fig. 6B). Using immunohistochemistry, we confirmed that Hic1 is exclusively mesenchymal, with particularly robust expression at the outer layer of the muscularis externa (Fig. 6C). Together, these results provide in vivo evidence that 3′ CGI methylation and associated gene activation are established during early development.
Fig 6.
Cell-type-specific 3′ CGI methylation and expression of Hic1 in E18.5 mouse colon. (A) DNA methylation analysis in isolated subpopulation cells from the E18.5 mouse colon. The left panel shows a dissected E18.5 colon. The middle panel shows the separation of colonic epithelial stem cells (Lgr5-eGFP+) and differentiated epithelial cells (EpCAM+) as well as mesenchymal cells (EpCAM−/eGFP−) by flow cytometric cell sorting. In the right panel, Hic1 3′ CGI methylation level among the sorted cells indicates hypermethylation of 3′ CGI specifically in the mesenchymal cells. (B) Expression analysis by qRT-PCR shows that both Hic1 transcripts are expressed specifically in the mesenchymal cells. (C) Immunohistochemistry confirms that Hic1 expression is exclusively mesenchymal (top). α-SMA is used as a marker for mesenchymal cells (bottom). For each staining, there is a magnified view of the boxed area (right panel).
3′ CGI methylation confers transcriptional activation by a CTCF-dependent insulator function.
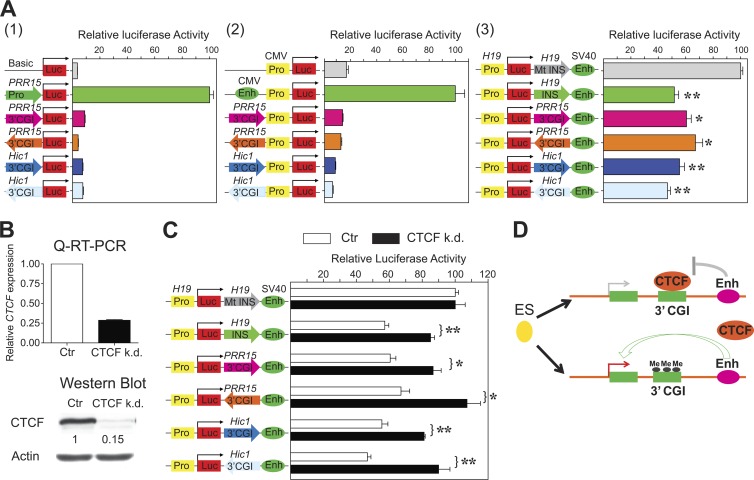
Having identified precisely the tissue- and cell-type-specifically methylated regions for PRR15 (Fig. 4A) and Hic1 (Fig. 5A), we performed detailed functional characterization. We used in vitro luciferase reporter assays to test whether the identified fragments in either sense or antisense orientation exhibit promoter, enhancer, or enhancer-blocking activity. Compared to control constructs, no promoter or enhancer activities were observed for either fragment, in either the sense or the antisense direction (Fig. 7A, panels 1 and 2, respectively). Both fragments, however, did exhibit enhancer-blocking activities, independently of the orientation (Fig. 7A, panel 3). Notably, for both fragments, the enhancer-blocking activities were at levels comparable to that of the H19 insulator (49). We next tested whether insulator function at the identified fragments is, like that at the H19 insulator, regulated by CTCF. We knocked down CTCF by shRNA (Fig. 7B) and measured enhancer-blocking activity using the luciferase reporter constructs. CTCF knockdown abrogated the insulator activities of PRR15 and Hic1 fragments in both orientations, to a degree similar to that at the H19 insulator (Fig. 7C). Collectively, our results indicate that a CTCF-dependent insulator function is involved in transcriptional regulation by 3′ CGI methylation. Furthermore, our results suggest that the ability of CTCF to act as a DNA methylation-sensitive enhancer blocker, well documented at imprinted genes, extends to transcriptional regulation of CGI-associated developmental genes in general (Fig. 7D).
Fig 7.
Functional characterization of tissue-/cell-type-specifically methylated 3′ CGIs reveals CTCF-dependent enhancer-blocking mechanism for transcriptional regulation. (A) Luciferase reporter assays. (1) Promoter assays for the PRR15 and Hic1 3′ CGIs in both sense and antisense orientations. The luciferase activity is shown relative to that obtained with the promoter construct carrying an endogenous PRR15 promoter. (2) Enhancer assays for the PRR15 and Hic1 3′ CGIs in both sense and antisense orientations. The luciferase activity is shown relative to that obtained with the enhancer construct carrying a CMV enhancer. (3) Enhancer-blocking assays for the PRR15 and Hic1 3′ CGIs in both sense and antisense orientations. The luciferase activity is shown relative to that obtained with the enhancer-blocking construct carrying a mutant (Mt) H19 DMR with only the CTCF-binding sites being replaced. *, P < 0.05; **, P < 0.005. (B) RNA interference-mediated CTCF knockdown (k.d.) in HCT116 cells was confirmed by qRT-PCR and Western blot analyses. As a control, cells expressing scrambled shRNA were used. (C) CTCF knockdown in HCT116 cells abrogates enhancer-blocking activity of the PRR15 and Hic1 3′ CGIs in both sense and antisense orientations. Values are given as means and standard deviations of results from more than three independent experiments. (D) Proposed model for how developmentally programmed 3′ CGI methylation regulates tissue-/cell-type-specific transcriptional activation.
DISCUSSION
A long-standing question in developmental epigenetics is whether and to what extent DNA methylation plays a regulatory role in mammalian development. Researchers have taken several approaches to address this question, including comparisons of tissue-specific methylation (13, 15, 18, 67, 68) and measurement of methylation changes at specific stages of mouse development (35, 69–71). We chose to take a different approach, using hESCs as an experimental model to study developmental epigenetics, for the following reasons: (i) developmental changes of DNA methylation in humans cannot be studied directly in vivo and (ii) tissue-specific differences in DNA methylation during the life course do not necessarily reflect developmental processes, because effects of environmental exposures and aging on methylation may be tissue specific (72, 73). On the other hand, we recognize the caveats of our approach: cell culture could induce nonphysiological DNA methylation changes (74, 75), and in vitro differentiation might not accurately recapitulate differentiation in vivo. Our extensive validation studies, including lineage-specific differentiation and dedifferentiation and detailed functional characterization in diverse human tissues and mouse models, however, indicate that our system adequately reflects early embryogenesis and provides an apt model of human developmental epigenetics.
We focused on a group of CGIs that gain methylation upon induced hESC differentiation because of their unique genomic structures, strong association with bivalent histone modifications, and significant enrichment for genes associated with developmental processes. One particularly novel finding of our study is the discovery of dichotomous roles for CGI methylation during development. CGI methylation has been generally viewed as a mechanism of gene silencing. This view has been challenged by recent studies finding that increased gene body methylation correlates with increased transcription genome-wide. Most of these, however, have proposed that the function of intragenic CGI methylation is to silence tissue- and cell-specific alternative promoters, rather than to activate transcription per se. One study (15) estimated that 10% of nonpromoter CGIs are methylated in two somatic tissues, compared with only 3% of promoter CGIs. Using RNA polymerase II occupancy as an annotation for novel transcripts, 20% of nonpromoter CGIs were found to contain alternative promoter activities. Another study of the human brain methylome (16) identified methylation at 34% of intragenic CGIs, approximately 20% of which overlapped alternative promoters. Our results support some aspects of the phenomena described previously, including the strong preference for methylation at nonpromoter CGI methylation. In addition, however, our results highlight the novel finding that a unique class of 3′ CGIs undergoes de novo methylation at early stages of differentiation. Importantly, we find evidence that instead of regulating cryptic alternative promoters, intragenic 3′ CGI methylation controls gene activation through a CTCF-dependent enhancer-blocking mechanism.
In many respects, the regulatory role of 3′ CGIs is reminiscent of chromatin insulator function at imprinting control regions (ICRs). At the H19/Igf2 ICR, for example, paternal-gene-specific methylation of multiple CTCF-binding sites abolishes both CTCF-binding and insulator activity, which allows imprinted Igf2 expression (57). The results of our luciferase reporter assays suggest that the ability of CTCF to act as a DNA methylation-sensitive enhancer blocker, well documented at imprinted genes, may serve as a general developmental mechanism to regulate transcription of 3′ CGI-associated genes. This general model is further supported by our bioinformatic analyses showing enrichment of CTCF-binding sites in the 3′ CGIs that gained methylation during differentiation and genome-wide correlations between increased DNA methylation and decreased CTCF binding in these regions in differentiated cell lines. Our data are complemented by an independent computational analysis (76) which systematically discovered a widespread role for CTCF-based insulation,. It should be pointed out that our enhancer-blocking assay involves heterologous enhancer/promoter sequences and an ectopically expressed plasmid outside its native genomic context. Although many classical insulators were identified using this assay, further experiments are needed to validate the insulator function in vivo and to determine the kinetics of CTCF binding as well as the interactions with higher-order chromatin structure in gene regulation.
The mechanisms involved in establishing developmentally programmed CGI methylation are still unclear. Intriguingly, bivalent histone modifications were found to “premark” these CGIs in undifferentiated hESCs. It is tempting to speculate that local-sequence information (e.g., CpG density) may interact with Trithorax (TrxG) and Polycomb (PcG) complexes that guide the targeting mechanism. Consistent with this conjecture, a previous study (22) suggested that PcG proteins may contribute to the initial recruitment of Dnmt3a in regions outside promoters to facilitate transcription of neurogenic genes. Moreover, recent genome-wide studies (77–79) provide strong evidence supporting a fundamental role of CGI structure in defining the TrxG/PcG chromatin structure in human pluripotent stem cells.
Determining the epigenetic basis of human embryonic stem cell differentiation not only provides new insights into the biology of development and regeneration but also opens new avenues to understand how perturbation of developmental mechanisms may contribute to disease. An attractive hypothesis is that epigenetic variation established during normal development may serve as a substrate for Darwinian selection at the cellular level that underlies aging-associated diseases (80). This is of particular interest for the HIC1 gene, since aberrant promoter CGI hypermethylation is frequently found in many major types of human tumors (81). Hence, our detailed characterization of tissue- and cell-type-specific methylation of the Hic1 3′ CGI may afford new perspectives on the evolution of abnormal DNA methylation in cancer.
In conclusion, our findings provide novel insights into the role of CGI methylation in normal development and cellular differentiation. Transcriptional activation of tissue-specific gene expression by 3′ CGI methylation potentially represents a dramatic expansion of the functional repertoire of DNA methylation in development and disease.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mitsuyoshi Nakao for providing the H19 DMR reporter plasmids and Yi Guo, Wei Zhu, Jinming Shu, Wei Wei, Angelique Nelson, Savannah Cook, and Robert Milczarek for technical assistance. We also thank Adam Gillum for assistance with the figures.
This work was supported by grants from the Sidney Kimmel Foundation to L.S., the USDA (CRIS 6250-5100-050) to L.S. and R.A.W., the NIGMS (P01GM081619-01) to C.W., and the NIDDK (1R01DK081557) to R.A.W. and L.S. and by private funding from the Institute for Stem Cell and Regenerative Medicine to C.W.
Footnotes
Published ahead of print 4 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01124-12.
REFERENCES
- 1. Reik W, Dean W, Walter J. 2001. Epigenetic reprogramming in mammalian development. Science 293: 1089– 1093 [DOI] [PubMed] [Google Scholar]
- 2. Holliday R, Pugh JE. 1975. DNA modification mechanisms and gene activity during development. Science 187: 226– 232 [PubMed] [Google Scholar]
- 3. Riggs AD. 1975. X inactivation, differentiation, and DNA methylation. Cytogenet. Cell Genet. 14: 9– 25 [DOI] [PubMed] [Google Scholar]
- 4. Taylor SM, Jones PA. 1979. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell 17: 771– 779 [DOI] [PubMed] [Google Scholar]
- 5. Bird A. 2007. Perceptions of epigenetics. Nature 447: 396– 398 [DOI] [PubMed] [Google Scholar]
- 6. Li E, Bestor TH, Jaenisch R. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915– 926 [DOI] [PubMed] [Google Scholar]
- 7. Reik W. 2007. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447: 425– 432 [DOI] [PubMed] [Google Scholar]
- 8. Edwards JR, O'Donnell AH, Rollins RA, Peckham HE, Lee C, Milekic MH, Chanrion B, Fu Y, Su T, Hibshoosh H, Gingrich JA, Haghighi F, Nutter R, Bestor TH. 2010. Chromatin and sequence features that define the fine and gross structure of genomic methylation patterns. Genome Res. 20: 972– 980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Illingworth R, Kerr A, DeSousa D, Jorgensen H, Ellis P, Stalker J, Jackson D, Clee C, Plumb R, Rogers J, Humphray S, Cox T, Langford C, Bird A. 2008. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 6: e22 doi:10.1371/journal.pbio.0060022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jones PA. 2012. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13:484– 492 [DOI] [PubMed] [Google Scholar]
- 11. Suzuki MM, Bird A. 2008. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 9: 465– 476 [DOI] [PubMed] [Google Scholar]
- 12. Shen L, Kondo Y, Guo Y, Zhang J, Zhang L, Ahmed S, Shu J, Chen X, Waterland RA, Issa JP. 2007. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 3: 2023– 2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Song F, Smith JF, Kimura MT, Morrow AD, Matsuyama T, Nagase H, Held WA. 2005. Association of tissue-specific differentially methylated regions (TDMs) with differential gene expression. Proc. Natl. Acad. Sci. U. S. A. 102: 3336– 3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. 2007. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 39: 457– 466 [DOI] [PubMed] [Google Scholar]
- 15. Illingworth RS, Gruenewald-Schneider U, Webb S, Kerr AR, James KD, Turner DJ, Smith C, Harrison DJ, Andrews R, Bird AP. 2010. Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet. 6: e1001134 doi:10.1371/journal.pgen.1001134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, Turecki G, Delaney A, Varhol R, Thiessen N, Shchors K, Heine VM, Rowitch DH, Xing X, Fiore C, Schillebeeckx M, Jones SJ, Haussler D, Marra MA, Hirst M, Wang T, Costello JF. 2010. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466:253– 257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rauch TA, Wu X, Zhong X, Riggs AD, Pfeifer GP. 2009. A human B cell methylome at 100-base pair resolution. Proc. Natl. Acad. Sci. U. S. A. 106: 671– 678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Straussman R, Nejman D, Roberts D, Steinfeld I, Blum B, Benvenisty N, Simon I, Yakhini Z, Cedar H. 2009. Developmental programming of CpG island methylation profiles in the human genome. Nat. Struct. Mol. Biol. 16: 564– 571 [DOI] [PubMed] [Google Scholar]
- 19. Illingworth RS, Bird AP. 2009. CpG islands–‘a rough guide'. FEBS Lett. 583: 1713– 1720 [DOI] [PubMed] [Google Scholar]
- 20. Takai D, Jones PA. 2002. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl. Acad. Sci. U. S. A. 99: 3740– 3745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, Ukomadu C, Sadler KC, Pradhan S, Pellegrini M, Jacobsen SE. 2010. Conservation and divergence of methylation patterning in plants and animals. Proc. Natl. Acad. Sci. U. S. A. 107: 8689– 8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE. 2010. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 329: 444– 448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zemach A, McDaniel IE, Silva P, Zilberman D. 2010. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328: 916– 919 [DOI] [PubMed] [Google Scholar]
- 24. Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, Ecker JR. 2006. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell 126: 1189– 1201 [DOI] [PubMed] [Google Scholar]
- 25. Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. 2007. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 39: 61– 69 [DOI] [PubMed] [Google Scholar]
- 26. Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. 2011. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479: 74– 79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ware CB, Wang L, Mecham BH, Shen L, Nelson AM, Bar M, Lamba DA, Dauphin DS, Buckingham B, Askari B, Lim R, Tewari M, Gartler SM, Issa JP, Pavlidis P, Duan Z, Blau CA. 2009. Histone deacetylase inhibition elicits an evolutionarily conserved self-renewal program in embryonic stem cells. Cell Stem Cell 4: 359– 369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stadler B, Ivanovska I, Mehta K, Song S, Nelson A, Tan Y, Mathieu J, Darby GC, Blau CA, Ware C, Peters G, Miller DG, Shen L, Cleary M, Ruohola-Baker H. 2010. Characterization of microRNAs involved in embryonic stem cell states. Stem Cells Dev. 19: 935– 950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ware CB, Nelson AM, Blau CA. 2006. A comparison of NIH-approved human ESC lines. Stem Cells 24: 2677– 2684 [DOI] [PubMed] [Google Scholar]
- 30. Gonzalez R, Loring JF, Snyder EY. 2008. Preparation of autogenic human feeder cells for growth of human embryonic stem cells. Curr. Protoc. Stem Cell Biol. Chapter 1:Unit 1C.5.1–1C.5.15 doi:10.1002/9780470151808.sc01c05s4 [DOI] [PubMed] [Google Scholar]
- 31. Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. 2009. Human induced pluripotent stem cells free of vector and transgene sequences. Science 324: 797– 801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Estecio MR, Yan PS, Ibrahim AE, Tellez CS, Shen L, Huang TH, Issa JP. 2007. High-throughput methylation profiling by MCA coupled to CpG island microarray. Genome Res. 17: 1529– 1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, Yamochi T, Urano T, Furukawa K, Kwabi-Addo B, Gold DL, Sekido Y, Huang TH, Issa JP. 2008. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat. Genet. 40: 741– 750 [DOI] [PubMed] [Google Scholar]
- 34. Waterland RA, Kellermayer R, Laritsky E, Rayco-Solon P, Harris RA, Travisano M, Zhang W, Torskaya MS, Zhang J, Shen L, Manary MJ, Prentice AM. 2010. Season of conception in rural Gambia affects DNA methylation at putative human metastable epialleles. PLoS Genet. 6: e1001252 doi:10.1371/journal.pgen.1001252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Waterland RA, Kellermayer R, Rached MT, Tatevian N, Gomes MV, Zhang J, Zhang L, Chakravarty A, Zhu W, Laritsky E, Zhang W, Wang X, Shen L. 2009. Epigenomic profiling indicates a role for DNA methylation in early postnatal liver development. Hum. Mol. Genet. 18:3026– 3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. 2007. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell 1: 299– 312 [DOI] [PubMed] [Google Scholar]
- 37. Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48 doi:10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hochberg Y, Benjamini Y. 1990. More powerful procedures for multiple significance testing. Stat. Med. 9:811– 818 [DOI] [PubMed] [Google Scholar]
- 39. Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315– 322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128: 1231– 1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43– 49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. 2003. Sensitive and quantitative universal pyrosequencing methylation analysis of CpG sites. Biotechniques 35: 146– 150 [DOI] [PubMed] [Google Scholar]
- 43. Shen L, Guo Y, Chen X, Ahmed S, Issa JP. 2007. Optimizing annealing temperature overcomes bias in bisulfite PCR methylation analysis. Biotechniques 42: 48,50,52. [DOI] [PubMed] [Google Scholar]
- 44. Shen L, Toyota M, Kondo Y, Lin E, Zhang L, Guo Y, Hernandez NS, Chen X, Ahmed S, Konishi K, Hamilton SR, Issa JP. 2007. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc. Natl. Acad. Sci. U. S. A. 104:18654– 18659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Staunton JE, Slonim DK, Coller HA, Tamayo P, Angelo MJ, Park J, Scherf U, Lee JK, Reinhold WO, Weinstein JN, Mesirov JP, Lander ES, Golub TR. 2001. Chemosensitivity prediction by transcriptional profiling. Proc. Natl. Acad. Sci. U. S. A. 98: 10787– 10792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mortazavi A, Leeper Thompson EC, Garcia ST, Myers RM, Wold B. 2006. Comparative genomics modeling of the NRSF/REST repressor network: from single conserved sites to genome-wide repertoire. Genome Res. 16: 1208– 1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen MH, Gao N, Kawakami T, Chuang PT. 2005. Mice deficient in the fused homolog do not exhibit phenotypes indicative of perturbed hedgehog signaling during embryonic development. Mol. Cell. Biol. 25: 7042– 7053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen MH, Li YJ, Kawakami T, Xu SM, Chuang PT. 2004. Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and long-range signaling in vertebrates. Genes Dev. 18: 641– 659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ishihara K, Oshimura M, Nakao M. 2006. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol. Cell 23: 733– 742 [DOI] [PubMed] [Google Scholar]
- 50. Gomes NP, Espinosa JM. 2010. Gene-specific repression of the p53 target gene PUMA via intragenic CTCF-cohesin binding. Genes Dev. 24: 1022– 1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315– 326 [DOI] [PubMed] [Google Scholar]
- 52. Xu C, Jiang J, Sottile V, McWhir J, Lebkowski J, Carpenter MK. 2004. Immortalized fibroblast-like cells derived from human embryonic stem cells support undifferentiated cell growth. Stem Cells 22: 972– 980 [DOI] [PubMed] [Google Scholar]
- 53. Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D. 2008. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell 30: 755– 766 [DOI] [PubMed] [Google Scholar]
- 54. Cedar H, Bergman Y. 30 April 2009. Epigenetic silencing during early lineage commitment. StemBook. Harvard Stem Cell Institute, Cambridge, MA: doi/10.3824/stembook.1.42.1 [PubMed] [Google Scholar]
- 55. Jones PA. 1999. The DNA methylation paradox. Trends Genet. 15: 34– 37 [DOI] [PubMed] [Google Scholar]
- 56. Phillips JE, Corces VG. 2009. CTCF: master weaver of the genome. Cell 137: 1194– 1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bell AC, Felsenfeld G. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405: 482– 485 [DOI] [PubMed] [Google Scholar]
- 58. Purcell SH, Cantlon JD, Wright CD, Henkes LE, Seidel GE, Jr, Anthony RV. 2009. The involvement of proline-rich 15 in early conceptus development in sheep. Biol. Reprod. 81: 1112– 1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bao L, Zhou M, Cui Y. 2008. CTCFBSDB: a CTCF-binding site database for characterization of vertebrate genomic insulators. Nucleic Acids Res. 36: D83– D87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fleuriel C, Touka M, Boulay G, Guerardel C, Rood BR, Leprince D. 2009. HIC1 (Hypermethylated in Cancer 1) epigenetic silencing in tumors. Int. J. Biochem. Cell Biol. 41: 26– 33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Carter MG, Johns MA, Zeng X, Zhou L, Zink MC, Mankowski JL, Donovan DM, Baylin SB. 2000. Mice deficient in the candidate tumor suppressor gene Hic1 exhibit developmental defects of structures affected in the Miller-Dieker syndrome. Hum. Mol. Genet. 9: 413– 419 [DOI] [PubMed] [Google Scholar]
- 62. Chen WY, Zeng X, Carter MG, Morrell CN, Chiu Yen RW, Esteller M, Watkins DN, Herman JG, Mankowski JL, Baylin SB. 2003. Heterozygous disruption of Hic1 predisposes mice to a gender-dependent spectrum of malignant tumors. Nat. Genet. 33: 197– 202 [DOI] [PubMed] [Google Scholar]
- 63. Guerardel C, Deltour S, Pinte S, Monte D, Begue A, Godwin AK, Leprince D. 2001. Identification in the human candidate tumor suppressor gene HIC-1 of a new major alternative TATA-less promoter positively regulated by p53. J. Biol. Chem. 276: 3078– 3089 [DOI] [PubMed] [Google Scholar]
- 64. Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, Wei CL. 2010. Dynamic changes in the human methylome during differentiation. Genome Res. 20: 320– 331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Park YK, Franklin JL, Settle SH, Levy SE, Chung E, Jeyakumar LH, Shyr Y, Washington MK, Whitehead RH, Aronow BJ, Coffey RJ. 2005. Gene expression profile analysis of mouse colon embryonic development. Genesis 41: 1– 12 [DOI] [PubMed] [Google Scholar]
- 66. Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. 2007. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003– 1007 [DOI] [PubMed] [Google Scholar]
- 67. Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, Haefliger C, Horton R, Howe K, Jackson DK, Kunde J, Koenig C, Liddle J, Niblett D, Otto T, Pettett R, Seemann S, Thompson C, West T, Rogers J, Olek A, Berlin K, Beck S. 2006. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet. 38: 1378– 1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, Ji H, Potash JB, Sabunciyan S, Feinberg AP. 2009. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 41: 178– 186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Borgel J, Guibert S, Li Y, Chiba H, Schubeler D, Sasaki H, Forne T, Weber M. 2010. Targets and dynamics of promoter DNA methylation during early mouse development. Nat. Genet. 42: 1093– 1100 [DOI] [PubMed] [Google Scholar]
- 70. Kobayashi H, Sakurai T, Imai M, Takahashi N, Fukuda A, Yayoi O, Sato S, Nakabayashi K, Hata K, Sotomaru Y, Suzuki Y, Kono T. 2012. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet. 8: e1002440 doi:10.1371/journal.pgen.1002440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. 2012. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484:339– 344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kim KM, Shibata D. 2002. Methylation reveals a niche: stem cell succession in human colon crypts. Oncogene 21: 5441– 5449 [DOI] [PubMed] [Google Scholar]
- 73. Maegawa S, Hinkal G, Kim HS, Shen L, Zhang L, Zhang J, Zhang N, Liang S, Donehower LA, Issa JP. 2010. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 20: 332– 340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Jones PA, Wolkowicz MJ, Rideout WM, III, Gonzales FA, Marziasz CM, Coetzee GA, Tapscott SJ. 1990. De novo methylation of the MyoD1 CpG island during the establishment of immortal cell lines. Proc. Natl. Acad. Sci. U. S. A. 87: 6117– 6121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766– 770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Xie X, Mikkelsen TS, Gnirke A, Lindblad-Toh K, Kellis M, Lander ES. 2007. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc. Natl. Acad. Sci. U. S. A. 104: 7145– 7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553– 560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Orlando DA, Guenther MG, Frampton GM, Young RA. 2012. CpG island structure and trithorax/polycomb chromatin domains in human cells. Genomics 100: 320– 326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD, Turner DJ, Illingworth R, Bird A. 2010. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 464: 1082– 1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Issa JP. 2011. Epigenetic variation and cellular Darwinism. Nat. Genet. 43: 724– 726 [DOI] [PubMed] [Google Scholar]
- 81. Wales MM, Biel MA, el Deiry W, Nelkin BD, Issa JP, Cavenee WK, Kuerbitz SJ, Baylin SB. 1995. p53 activates expression of HIC-1, a new candidate tumour suppressor gene on 17p13.3. Nat. Med. 1: 570– 577 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.