Abstract
Receptor tyrosine kinases (RTKs) exist in equilibrium between tyrosyl-phosphorylated and dephosphorylated states. Despite a detailed understanding of how RTKs become tyrosyl phosphorylated, much less is known about RTK tyrosyl dephosphorylation. Receptor protein tyrosine phosphatases (RPTPs) can play essential roles in the dephosphorylation of RTKs. However, a complete understanding of the involvement of the RPTP subfamily in RTK tyrosyl dephosphorylation has not been established. In this study, we have employed a small interfering RNA (siRNA) screen to identify RPTPs in the human genome that serve as RTK phosphatases. We observed that each RPTP induced a unique fingerprint of tyrosyl phosphorylation among 42 RTKs. We identified EphA2 as a novel LAR substrate. LAR dephosphorylated EphA2 at phosphotyrosyl 930, uncoupling Nck1 from EphA2 and thereby attenuating EphA2-mediated cell migration. These results demonstrate that each RPTP exerts a unique regulatory fingerprint of RTK tyrosyl dephosphorylation and suggest a complex signaling interplay between RTKs and RPTPs. Furthermore, we observed that LAR modulates cell migration through EphA2 site-specific dephosphorylation.
INTRODUCTION
Over the last decade, the field of protein tyrosine phosphatases (PTPs) has matured to a level where it is now fully appreciated that these enzymes play critical roles in the regulation of numerous physiological processes (1, 2). Moreover, recent advances point to the PTP family of enzymes as targets for the cause of several human diseases (1–8). Therefore, understanding the mechanisms of PTP signaling will yield important insight into the molecular pathogenesis of human disease. Although substantial progress has been made toward understanding the mechanisms of PTP signaling, the identification of substrates for the PTPs and understanding how PTP substrates signal when dephosphorylated continue to be active, yet challenging, areas of investigation.
The PTP superfamily consists of 107 PTPs in the human genome (1, 2, 9). The classical PTPs dephosphorylate tyrosyl-phosphorylated proteins and are divided into receptor-like PTPs (RPTPs) and nonreceptor PTPs (2, 10). Although substantial progress toward understanding the function of many of the nonreceptor PTPs has been made (1, 2), much less is known about RPTPs. There are 21 RPTPs in the human genome (9). RPTPs possess a single transmembrane domain, variable extracellular domains, and an intracellular portion containing two PTP domains (in some cases a single PTP domain) (2, 11). Although there are exceptions, ligand binding to RPTPs typically results in the inactivation of RPTPs by inducing dimerization (12–16). In the dimeric state, reciprocal inhibition of the catalytically competent D1 PTP domain occurs whereby the “wedge motif” of one D1 domain occludes the active site of the opposing D1 domain in the dimer (2, 15). The D2 domain is typically inactive, although it provides important regulatory features of RPTP function, such as stabilizing substrate interactions and facilitating RPTP dimerization (13, 17–20).
Unlike their RPTP counterparts, receptor tyrosine kinases (RTKs) are regulated in a distinct manner. The activation of RTKs stems from ligand-induced dimerization that results in the transphosphorylation of tyrosine residues (21). RTKs are coordinately inactivated through several mechanisms, and these can include receptor downregulation and internalization as well as direct dephosphorylation by PTPs. RTKs have been shown to be dephosphorylated by both nonreceptor PTPs and RPTPs. The most notable example of a nonreceptor PTP dephosphorylating an RTK is that of PTP-1B, which directly dephosphorylates the insulin receptor (22). RPTPs have also been shown to dephosphorylate RTKs; for example, PTPRF (leukocyte common antigen related [LAR]) dephosphorylates the insulin receptor (23, 24) and PTPRJ (density-expressed phosphatase [DEP-1]) dephosphorylates vascular endothelial growth factor receptor 2 (VEGFR2) (25). These observations indicate that RTKs are direct targets for both nonreceptor PTPs and RPTPs. However, of the 58 RTKs in the human genome, the majority have no known counteracting PTP with which to account for their dephosphorylation. The identification of the PTPs involved in RTK dephosphorylation and inactivation will provide important insight into the mechanisms dictating RTK signaling.
Although both nonreceptor PTPs and RPTPs are capable of dephosphorylating RTKs, we speculated that because of the relative location in the plasma membrane, RPTPs might play a major role in RTK dephosphorylation. Moreover, the identity of RPTP substrates still remains poorly defined. Therefore, to gain insight into the actions of RPTPs, and to acquire a broader understanding of how RTKs are dephosphorylated, we developed a small interfering RNA (siRNA) screen to identify RPTPs that can function as RTK phosphatases. The siRNA screen revealed that each RPTP, when knocked down by siRNA, exerted a unique pattern of both hyper- and hypo-tyrosyl-phosphorylated RTKs in proliferating MCF10A human breast epithelial cells. These results suggested that RPTPs exhibit unique selectivity toward specific subsets of RTKs. We validated the screen by testing one of the hits that showed that EphA2 was a potential LAR substrate. We subsequently demonstrated that EphA2 is a LAR substrate and that LAR specifically dephosphorylates phosphotyrosyl 930 on EphA2 to control its association with Nck1 and, subsequently, cell migration. The broader implications of this study reveal a complex interplay between RTKs and RPTPs in the control of cell signaling.
MATERIALS AND METHODS
Cell culture.
MCF10A human breast epithelial cells were cultured in Dulbecco's modified Eagle's medium (DMEM)–F-12 containing 5% horse serum (Invitrogen), 20 ng/ml of epidermal growth factor (EGF) (Invitrogen), 10 μg/ml of insulin (Sigma), 0.5 μg/ml of hydrocortisone (Sigma), and 100 ng/ml of cholera toxin (Sigma) as described previously (26). COS7 monkey kidney cells and human embryonic kidney 293 cells were cultured in DMEM containing 10% fetal calf serum with antibiotics. For ephrinA1 stimulation, MCF10A cells were starved for 4 h in serum-free DMEM–F-12 medium and COS7 and 293 cells were starved overnight in serum-free DMEM and then were stimulated with 0.1 to 1.0 μg/ml ephrinA1 for the times indicated below.
Receptor protein tyrosine phosphatase-receptor protein tyrosine kinase screening strategy.
MCF10A cells at approximately 70% confluence in 150-mm culture dishes were transfected with at least two individual siRNAs against each human RPTP. Greater than 70% knockdown efficiency of each RPTP was confirmed between 48 and 96 h posttransfection by reverse transcription-PCR (RT-PCR) (Ambion) (see Tables S1 to S3 in the supplemental material). To limit nonspecific activation of MCF10A cells by EGF and insulin, cells were incubated in growth medium without EGF and insulin for 16 h prior to being lysed. RTK arrays against MCF10A cells that were knocked down with an RPTP siRNA were performed according to the manufacturer's instructions using the Proteome Profile antibody array for human phospho-RTKs (R&D Systems; ARY001). This array kit contains the nitrocellulose membrane array, which is spotted with duplicates of antibodies to the following RTKs: Axl, Dtk, EGF receptor (EGFR), EphA1, EphA2, EphA3, EphA4, EphA6, EphA7, EphB1, EphB2, EphB4, EphB6, ErbB2, ErbB3, ErbB4, fibroblast growth factor receptor 1 (FGFR1), FGFR2 α, FGFR3, FGFR4, Flt-3/Flk-2, hepatocyte growth factor receptor c-MET, insulin-like growth factor 1 (IGF-1) receptor, insulin receptor/CD220, M-CSFR, Mer, MSPR/Ron, MuSK, platelet-derived growth factor receptor α (PDGFRα), PDGFRβ, c-Ret, ROR1, ROR2, SCFR/c-kit, Tie-1, Tie-2, TrkA, TrkB, TrkC, VEGFR1/Flt-1, VEGFR2/KDR, and VEGFR3/Flt-4. Eight positive controls on the array were proprietary to the manufacturer, while the 10 negative controls consisted of various nonspecific IgGs. Both positive and negative controls were used for purposes of normalization. The assay kit provided the following proprietary buffers: array buffer 1, array buffer 2, wash buffer, and antiphosphotyrosine-horseradish peroxidase (antiphosphotyrosine-HRP) antibody. All steps were performed at room temperature. The array was blocked with 2 ml of array buffer 1 for 1 h. After aspiration of array buffer 1, the array was incubated with diluted lysates (500 μg) in an approximate volume of 1.5 ml with array buffer 1 for 2 h. The array was washed with wash buffer three times for 10 min. To detect changes in tyrosyl-phosphorylated RTKs, the array membrane was incubated with 1.5 ml of diluted HRP-conjugated phosphotyrosine antibodies (1: 5,000 dilution) with array buffer 2 on a rocking shaker for 2 h. Finally, the array was washed with wash buffer three times for 10 min and visualized using enhanced chemiluminescence (Amersham Biosciences).
Quantitative assessment of RPTP-RTK screen.
For quantitative assessment of the RTK-RPTP screen, the array was densitometrically scanned, and the relative values of phosphotyrosyl RTK intensities derived from duplicate phospho-RTK spots following knockdown by an individual RPTP were calculated as follows. The average intensity of duplicated RTK spots was measured and subtracted by the average intensity of 10 negative-control spots; this value was normalized to the average intensity of 8 positive-control spots, which was subtracted by the average of 10 negative-control spots. The relative change in each phosphotyrosyl RTK intensity for an individual RPTP-targeting siRNA was calculated by subtraction from the corresponding RTK intensity for nontargeting siRNA. Positive values represent hyper-tyrosyl-phosphorylated RTKs and negative values hypo-tyrosyl-phosphorylated RTKs. The phosphorylation levels of the RTKs were normalized across the 12 RPTPs to yield Z-scores that were plotted on a heat map using the g-plot package in R (version 2.13).
Plasmids and mutagenesis.
Human wild-type EphA2 in pTargetT was a kind gift of Paola Chiarugi (University of L'Aquila, L'Aquila, Italy); EphA2 mutants (tyrosine to phenylalanine) were generated using a QuikChange XL site-directed mutagenesis kit according to the manufacturer's instructions (Stratagene). A variety of LAR constructs were kindly provided by Ruey-Hwa Chen (National Taiwan University, Taipei, Taiwan). Glutathione S-transferase (GST) fusion proteins of LAR PTP domain (PTP-WT) and a PTP domain containing a mutation at cysteine 1548 to serine (PTP-CS) were generated by PCR amplifying amino acids 1351 to 1644 of FLAG-LAR-WT and FLAG-LAR-CS plasmids using a forward primer containing a BamHI site (5′-CGCGGATCCAAGTTCTCCCAGGAG-3′) and a reverse primer containing a BamHI site (5′-CGCGGATCCGCCGAGGTACTCCAG-3′). The LAR PTP domain PCR products were subcloned into pGEX-2TK. All constructs were verified by DNA sequencing at the W. M. Keck Foundation at Yale University.
Cell lysis, immunoblotting, and immunoprecipitation.
Cells were lysed on ice in lysis buffer containing 150 mM NaCl, 50 mM Tris-HCl (pH 7.8), 1 mM EDTA, 1% Nonidet P-40, 1 mM Na3VO4, 10 mM NaF, 1 mM dithiothreitol (DTT), 1 mM benzamidine, 1 mM PMSF, 1 μg/ml of pepstatin A, 5 μg/ml of aprotinin, and 5 μg/ml of leupeptin for 30 min and clarified by centrifugation at 20,800 × g at 4°C for 20 min. Protein concentration was determined using bicinchoninic acid (BCA) reagent according to the manufacturer's instructions (Pierce). For immunoprecipitations, 200 to 500 μg of cell lysate was incubated with 1 μg of rabbit EphA2 polyclonal antibody (Santa Cruz Biotechnology), 1 μg of mouse EphA2 monoclonal antibody (Millipore), 2 μg of mouse FLAG monoclonal antibody (Sigma), or 1 μg of rabbit Nck1 antibody (Millipore) overnight at 4°C. Immune complexes were collected on either protein A- or G-Sepharose (GE Healthcare). Immune complexes were washed three times with lysis buffer and once with STE buffer (100 mM NaCl, 10 mM Tris-HCl [pH 8.0], 1 mM EDTA), and Sepharose beads were heated to 95°C in sample buffer (62.5 mM Tris [pH 6.8], 4% glycerol, 2% SDS, 5% β-mercaptoethanol, and 0.02% bromphenol blue) for 5 min.
For immunoblotting, lysates or immune complexes were resolved by SDS-PAGE and transferred onto Immobilon-P membranes (Millipore). Membranes were first blocked with 5% nonfat dry milk or 5% bovine serum albumin (BSA) in Tris-buffered saline–Tween 20 (TBS-T) for 1 h at room temperature or overnight at 4°C. Primary antibodies were diluted in 5% BSA in TBS-T. Primary antibodies were diluted as follows: mouse monoclonal EphA2 antibody (Millipore), 1:2,000; rabbit anti-EphA2 antibody (Santa Cruz), 1:4,000; mouse antiphosphotyrosine (4G10) serum, 1:20; rabbit polyclonal extracellular signal-regulated kinase 1/2 (ERK1/2) (Santa Cruz Biotechnology), 1:4,000; monoclonal LAR antibody (BD Bioscience), 1:500; mouse FLAG antibody (Sigma), 1:1,000; mouse Nck1 antibody (BD Bioscience), 1:2,000; and rabbit Nck1 antibody (Millipore), 1:2,000. Primary antibodies were used either at room temperature for 2 h or at 4°C overnight. After primary antibody incubations, membranes were washed in TBS-T and incubated in secondary horseradish peroxidase-linked donkey anti-rabbit immunoglobulin antibody or horseradish peroxidase-linked sheep anti-mouse immunoglobulin antibody (GE Healthcare) for 1 h at room temperature at 1:5,000. After secondary antibody incubation, membranes were washed in TBS-T and visualized by using enhanced chemiluminescence reagents (Amersham Biosciences).
For the generation of phospho-specific antibodies to pY930 on human EphA2 receptor, a phosphotyrosine-containing peptide surrounding Y930 (T-E-H-F-M-A-A-G-pY-T-A-I-E-K-V-V) was synthesized and used to immunize rabbits (Proteintech). The resultant rabbit anti-phosphotyrosine 930 EphA2 antiserum (pY930 EphA2) was affinity purified. For immunoblotting, membranes were blocked in BSA and incubated overnight with pY930 EphA2 at a dilution of 1:2,000. Membranes were washed several times with TBS-T and proteins visualized using horseradish peroxidase-linked donkey anti-rabbit immunoglobulin antibody at a dilution of 1:5,000, followed by enhanced chemiluminescence.
Cell migration assay.
Migration assays were performed using a Transwell chamber (Costar) by following the manufacturer's protocol as previously described (27, 28). The Transwell insert membrane containing 8-μm pores was coated with 10 μg/ml of human fibronectin (FN) for 2 h at 25°C. The cells (1 × 105 cells per insert in 250 μl of serum-free medium) were placed in the upper chamber, and growth medium was placed in the lower chamber. After 22 to 24 h of incubation at 37°C, the insert membrane was fixed with 99% methanol (Fisher Scientific) for 10 min, rinsed with phosphate-buffered saline (PBS), stained with 0.2% crystal violet (Fisher Scientific) for 10 min, and washed twice with water. Nonmigrated cells on the upper surface of the membrane were then wiped off with a cotton swab and washed with distilled water twice. Images of migrated cells on the bottom of the membrane were collected on a Zeiss Axiovert S100 microscope (at ×20 and ×40 magnifications) using Axiovision software (Zeiss).
Immunofluorescence and image analysis.
Cells were plated on glass coverslips coated with human FN (20 μg/ml at 25°C for 2 h) in 12-well tissue culture plates and incubated at 37°C for 14 h in growth medium. As described previously (29), cells were fixed with 4% paraformaldehyde for 10 min, permeabilized with 0.2% Triton X-100 for 5 min, and incubated with primary antibody for 45 min and secondary antibodies for 30 min. Images were collected on a Nikon Eclipse Ti2000 epifluorescence microscope running NIS Element software (Nikon) using an oil immersion (60×) lens. For image analysis, an ImageJ plugin, colocalization color map was used for automated quantification and visualization of colocalized fluorescent signals and for calculating the correlation index indicating the fraction of positively correlated pixels in the image (30).
Vanadate competition assay.
Glutathione S-transferase (GST) fusion proteins were expressed in Escherichia coli and purified using the anionic detergent Sarkosyl (N-laurylsarcosine, sodium salt) as described previously (31). For vanadate competition experiments, as described previously (32), 10 μg of GST-PTP-CS fusion proteins was preincubated with 10 mM Na3VO4 for 10 min at 4°C and washed with PBS. GST proteins were resuspended with lysates containing 5 mM iodoacetic acid instead of 1 mM Na3VO4 in lysis buffer, incubated for 3 h, and washed three times with lysis buffer without iodoacetic acid and then with ST buffer (150 mM NaCl and 50 mM Tris-HCl).
RNA isolation and RT-PCR.
RNA was isolated from 60-mm tissue culture dishes when cells were 85 to 95% confluent using TRIzol reagent (Invitrogen) by following the manufacturer's protocol. Total RNA was treated with DNase, and the amount of RNA was determined. RNA was reverse transcribed with Applied Biosystems reverse transcription reagents, and RT-PCR was performed with individual RPTP-specific primers that amplify genomic DNA as a positive control. The sequences of RPTP-specific primers are shown in Table S2 in the supplemental material.
Statistical analysis.
Statistical analyses were performed using Prism software (GraphPad Software). Statistical analyses in three or more groups were performed using a one-way analysis of variance (ANOVA) followed by the Bonferroni's t test for multiple comparisons. Statistical differences between two groups were analyzed using unpaired two-tailed t test.
RESULTS
Screening and identification of functional RPTP-RTK substrate relationships.
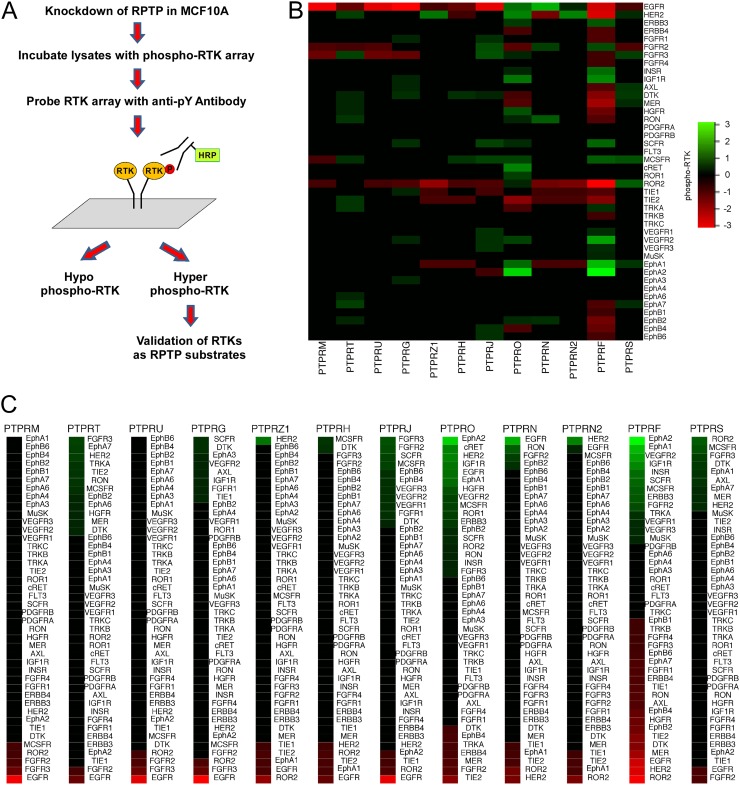
We speculated that RPTPs could serve as essential phosphatases for the dephosphorylation of RTKs because of their relative juxtaposition in the plasma membrane to RTKs. To evaluate the extent to which such RPTP-RTK relationships exist, we developed a strategy in which each of the RPTPs in the human genome was knocked down by siRNA and evaluated for its ability to induce changes in the basal levels of tyrosyl phosphorylation in RTKs expressed in proliferating MCF10A epithelial cells. siRNA RPTP-induced RTK tyrosyl phosphorylation changes were assessed against a RTK tyrosyl phosphorylation array representing 42 of the 58 RTKs in the human genome (Fig. 1A). RTKs that were hyper-tyrosyl phosphorylated following RPTP knockdown represented putative RPTP substrates, whereas those RTKs that were hypo-tyrosyl phosphorylated represented indirect RPTP effects (Fig. 1A).
Fig 1.
siRNA RPTP-RTK tyrosyl phosphorylation screen. (A) Schematic representation of the siRNA RPTP-RTK screen. (B) Heat map of the quantitation of hyper-tyrosyl-phosphorylated (green) and hypo-tyrosyl-phosphorylated (red) RTKs for the indicated siRNA RPTP. (C) Ranked RTKs based on the relative level of hyper-tyrosyl (green) and hypo-tyrosyl (red) phosphorylation in response to knockdown of the indicated RPTP.
Sixteen of the 21 RPTPs in the human genome (9) were confirmed to be expressed in MCF10A breast epithelial cells by PCR (see Tables S1 and S2 in the supplemental material). After siRNA optimization, 12 of the 16 RPTPs could be efficiently knocked down by more than 70% (see Tables S1 and S3 in the supplemental material). In order to detect a wide range of RTKs that may be subject to both hypo- and hyper-tyrosyl phosphorylation upon RPTP knockdown, MCF10A cells were cultured under proliferating conditions in the presence of serum-containing medium without EGF and insulin. Each of the 12 expressed RPTPs were individually knocked down in proliferating MCF10A cells, lysates were prepared and incubated with the Proteome Profiler antibody array. As a control, MCF10A cells were treated with nontargeting (NT) siRNA and these lysates were incubated with the Proteome Profiler antibody array. Following incubation with lysates prepared from RPTP siRNA-treated and NT-treated MCF10A cells, the arrays were incubated with antiphosphotyrosine antibodies (Fig. 1A). The relative value of phosphotyrosine intensity for each RTK was quantitated densitometrically and normalized against the appropriate positive and negative controls. The relative change in the levels of phosphotyrosyl RTK content between NT and RPTP siRNA knockdown for each RPTP was the value obtained when the NT siRNA-treated RTK phosphotyrosyl intensity was subtracted from the RPTP siRNA-treated RTK phosphotyrosyl intensity.
A global analysis of the pattern of RTK tyrosyl phosphorylation when each of the expressed RPTPs was knocked down revealed a distinct fingerprint of RTKs that were either hyper- or hypo-tyrosyl phosphorylated (Fig. 1B and C). PTPRF and PTPRO showed the most changes among the RPTPs, inducing RTK hyper- and hypo-tyrosyl phosphorylation in a broad proportion of the RTKome (Fig. 1B). In contrast, RPTPs such as PTPRU and PTPRM failed to induce RTK hyper-tyrosyl phosphorylation throughout the RTKome (Fig. 1B). Other RPTPs such as PTPRJ, PTPRN, PTPRN2, PTPRZ1, and PTPRS exhibited a more selective complement of hyper- and hypo-tyrosyl-phosphorylated RTKs (Fig. 1B). We performed a rank order analysis for each RPTP against the RTKome. EphA2 was identified as the highest-ranked hit for both PTPRF and PTPRO (Fig. 1C). Interestingly, EphA2 has been shown to be a substrate for PTPRO (33), but it has not been demonstrated to be a PTPRF substrate. Several RTKs that were hyper-tyrosyl phosphorylated had been previously identified to be RPTP substrates. For example, the screen identified the VEGFR2 to be hyper-tyrosyl phosphorylated when PTPRJ was knocked down (Fig. 1B and C), consistent with the report that the VEGFR2 serves as a PTPRJ substrate (25). PTPRF has been demonstrated to dephosphorylate the IGF-1 and insulin receptors (34, 35), both of which were hypertyrosyl phosphorylated in the RPTP-RTK screen (Fig. 1B and C). Taken together, the siRNA RPTP knockdown combined with phosphotyrosyl RTKome analysis revealed that RPTPs target multiple RTKs. Moreover, in addition to identifying known RPTP-RTK relationships, the screen revealed several putative RPTPs that may serve as novel RTK phosphatases.
LAR regulates EphA2 tyrosyl phosphorylation.
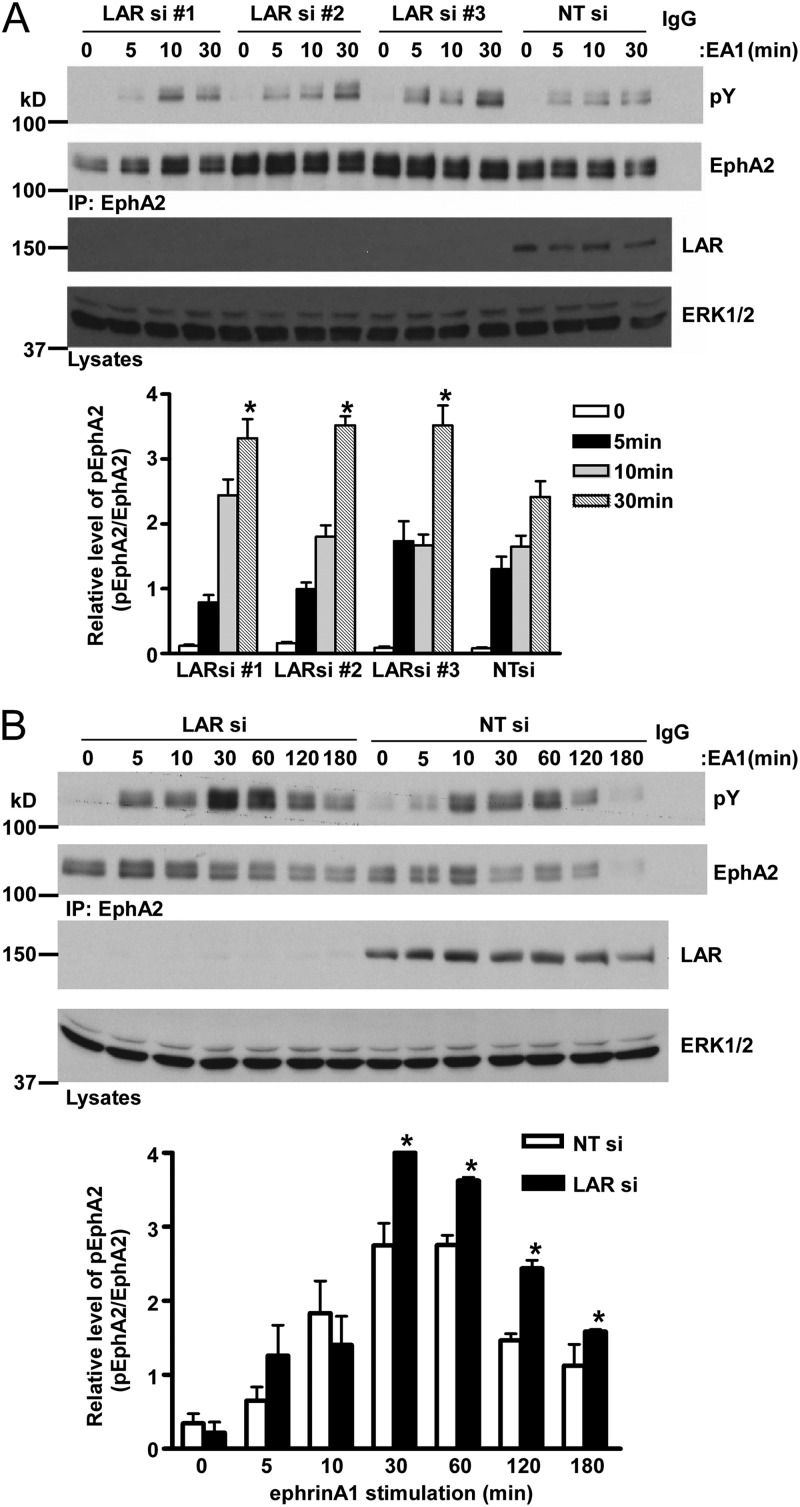
Notably, PTPRF, referred to herein as the leukocyte common antigen related (LAR) when knocked down, induced the highest-ranked hyper-tyrosyl-phosphorylated hit toward EphA2 (Fig. 1B and C). Although other PTPs, including PTPRO, have been shown to dephosphorylate EphA family members (33, 36), it is not known whether LAR acts specifically as an EphA2 phosphatase. Therefore, we set out to verify the siRNA RPTP-RTK screen by validating the effects of LAR knockdown on EphA2 tyrosyl phosphorylation using three distinct LAR siRNAs. MCF10A cells were transfected with LAR siRNAs or NT siRNA as a control. These cells were serum starved and then stimulated with ephrinA1. The EphA2 receptor was immunoprecipitated, and its phosphotyrosyl levels were determined. All siRNAs knocked down LAR and increased total EphA2 tyrosyl phosphorylation in response to ephrinA1 (Fig. 2A). The kinetics of EphA2 hyper-tyrosyl phosphorylation was most significant at the latest time point, 30 min (Fig. 2A). Therefore, we used an extended time of ephrinA1 stimulation to more carefully examine the kinetics of EphA2 tyrosyl dephosphorylation in response to LAR knockdown. When LAR was knocked down and MCF10A cells were stimulated with ephrinA1, EphA2 tyrosyl phosphorylation was enhanced and sustained beginning at 30 min for up to 180 min, compared with NT siRNA-transfected controls (Fig. 2B). These results demonstrate that suppression of LAR enhances EphA2 tyrosyl phosphorylation, suggesting that EphA2 serves as a LAR substrate.
Fig 2.
Depletion of LAR enhances EphA2 tyrosyl phosphorylation. (A) MCF10A cells were subjected to LAR knockdown with three different siRNAs. MCF10A cells were serum starved and stimulated with ephrinA1 (EA1) for the indicated times. EphA2 tyrosyl phosphorylation was determined by EphA2 immunoprecipitation followed by antiphosphotyrosyl immunoblotting. As a control, immunoblots were reprobed with anti-EphA2 antibodies to detect the levels of EphA2 expression. (B) MCF10A cells were subjected to LAR knockdown with siRNA #3 from panel A. EphA2 tyrosyl phosphorylation was determined in response to EA1. The graphs below at the bottoms of both panels represent densitometric quantitation of tyrosyl-phosphorylated EphA2 normalized to total EphA2. Data represent the mean ± standard errors of the means from three independent experiments. *, P < 0.05 between cells transfected with LAR siRNA and the NT siRNA control.
EphA2 is a direct LAR substrate.
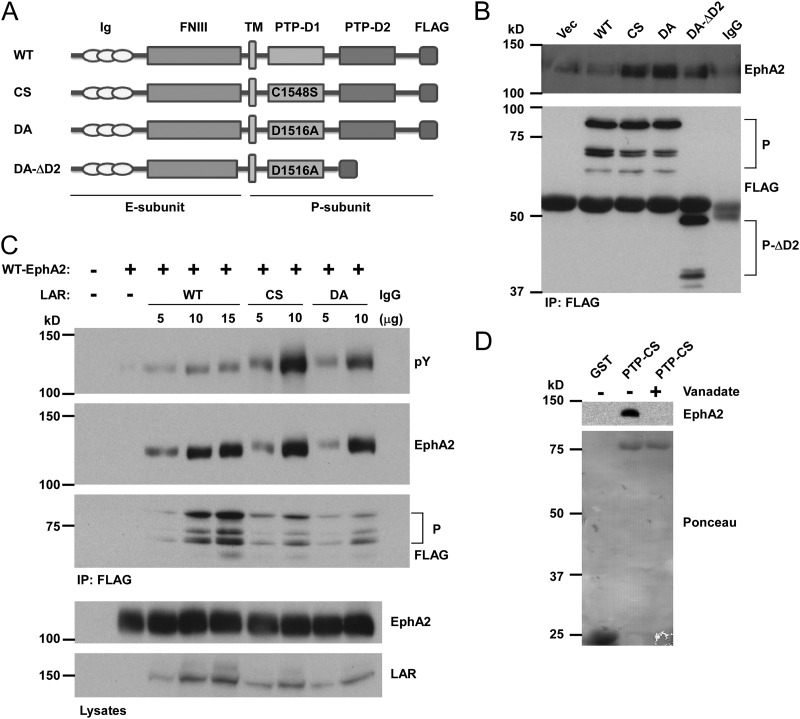
To determine whether EphA2 is a direct LAR substrate, we utilized LAR substrate-trapping mutants. PTP substrate-trapping mutants stably bind their cognate substrate but are unable to catalyze their dephosphorylation (37, 38). FLAG epitope-tagged substrate-trapping LAR mutants representing a change substitution at the conserved aspartic acid (D1516) residue in the active site of the PTP-D1 domain to alanine (A1516) (D/A) and a mutation of cysteine (C1548) to serine (S1548) (C/S) were generated (Fig. 3A). These mutants were transfected in to COS7 cells along with wild-type LAR. We found that both LAR substrate-trapping mutants formed stable interactions with endogenous tyrosyl-phosphorylated EphA2, whereas wild-type LAR interacted to a much lesser extent (Fig. 3B). In addition, the D/A mutant in context of a truncated D2 domain (Fig. 3B) bound EphA2 to levels greater than with wild-type LAR, albeit to a lower extent compared with the full-length substrate-trapping LAR mutants (Fig. 3B). We also performed substrate-trapping experiments in 293 cells that lack EphA2 expression. We again found that LAR substrate-trapping mutants exhibited increased EphA2 complex formation and that EphA2 was hyper-tyrosyl phosphorylated compared with wild-type EphA2 when coexpressed with wild-type LAR (Fig. 3C). These results strongly suggest that LAR complexes with EphA2 through its catalytic site, supporting the notion that LAR dephosphorylates EphA2.
Fig 3.
LAR substrate-trapping mutants define EphA2 as a LAR substrate. (A) Schematic structure of LAR substrate-trapping mutants. Shown are full-length FLAG-tagged wild-type LAR (WT) and substrate-trapping mutants CS (Cys1548 in the PTP-D1 domain was replaced by Ser1548), DA (Asp1516 in the PTP-D1 domain was replaced by Ala1516), and DA-ΔD2 (truncated PTP-D2 domain from the DA mutant). Ig, immunoglobulin-like domain; FNIII, fibronectin type III repeat domain; TM, transmembrane; PTP-D1, protein tyrosine phosphatase D1 domain; PTP-D2, PTP-pseudo-phosphatase D2 domain. (B) FLAG-tagged LAR substrate-trapping mutants were transfected into COS7 cells. Lysates were immunoprecipitated (IP) with FLAG antibodies and membranes probed with EphA2 and FLAG antibodies. FLAG immunoblots show LAR in its processed P-form (P) containing D1/D2 and deletion of D2-containing DA (P-ΔD2) form. Vec, vector alone. (C) FLAG-tagged wild-type LAR (WT) and substrate-trapping mutants of LAR along with wild-type EphA2 were cotransfected in to 293 cells. Lysates were immunoprecipitated with FLAG antibodies and immunoprecipitates were immunoblotted with either EphA2 or phosphotyrosine antibodies. Total cell lysates were blotted with EphA2 and LAR antibodies. (D) Wild-type EphA2 was transfected into 293 cells. Lysates were subjected to GST affinity precipitation with GST-PTP-CS (C1548S) in the absence or presence of vanadate (10 mM). The complexes were resolved, and membranes were immunoblotted with anti-EphA2 antibody and stained with Ponceau S for the detection of input purified protein.
If complex formation between wild-type EphA2 and the LAR substrate-trapping mutant is direct, this interaction should be disrupted by the PTP catalytic-site inhibitor vanadate. To test this, we performed affinity precipitation assays using purified LAR PTP domain substrate-trapping proteins that were incubated in either the absence or presence of vanadate, with lysates prepared from cells expressing wild-type EphA2. As expected, the substrate-trapping PTP domain mutant of LAR formed an enzyme-substrate complex with EphA2 in the absence of vanadate; in contrast, this complex was disrupted in the presence of vanadate (Fig. 3D). Collectively, these results demonstrate that LAR complexes with EphA2 through a direct PTP substrate-dependent mechanism, supporting the interpretation that LAR acts as an EphA2 phosphatase.
Colocalization of substrate-trapping LAR mutant with EphA2.
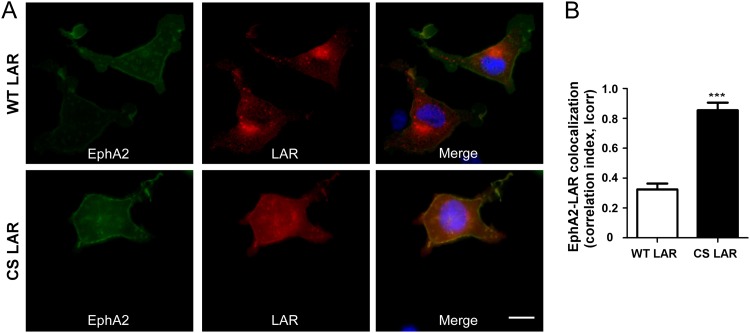
To further investigate LAR dephosphorylation of EphA2 in a cellular context, we examined whether the LAR substrate-trapping mutant exhibits increased EphA2 colocalization. When expressed in cells, a much greater proportion of the substrate-trapping LAR mutant than of wild-type LAR colocalized with wild-type EphA2 (Fig. 4A). The observation that the LAR substrate-trapping mutant colocalized to a higher degree than wild-type LAR was supported by quantitative assessment of colocalization (Fig. 4B). These data showed that the substrate-trapping LAR mutant had a correlation index that was approximately twice that of wild-type LAR (Fig. 4B). These results further support the conclusion that EphA2 forms a stable enzyme-substrate complex with LAR in a cellular context and suggest that LAR is appropriately localized in order to exert EphA2 tyrosyl dephosphorylation.
Fig 4.
Colocalization of EphA2 and substrate-trapping mutant of LAR. FLAG-tagged wild-type LAR (WT LAR) and substrate-trapping mutant LAR (CS-LAR) along with wild-type EphA2 were cotransfected into 293 cells. (A) Cells were fixed and immunostained with EphA2 (green), FLAG (red), and 4′,6-diamidino-2-phenylindole (DAPI) (blue). Scale bar = 10 μm. (B) Correlation indices were obtained as described in Materials and Methods. Data represent the mean correlation indices ± standard errors of the means from three independent experiments (n = ∼50 per experiment). ***, P < 0.001.
LAR dephosphorylates pY930 on the EphA2 receptor.
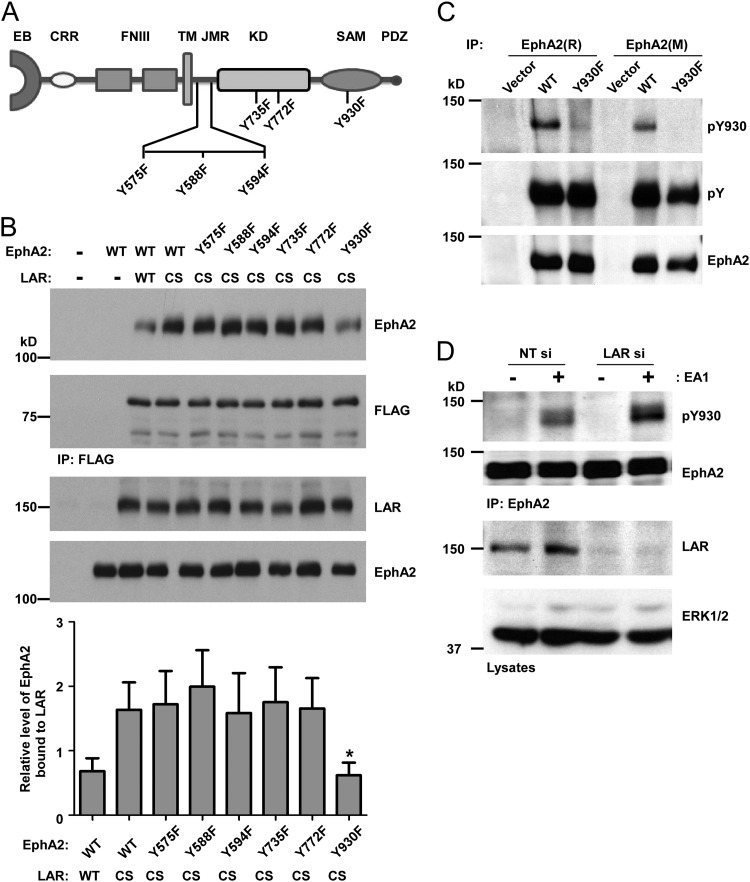
EphA2 has six major tyrosyl phosphorylation sites: Y575, Y588, and Y594 in the juxtamembrane region, Y735 and Y772 in the kinase domain, and Y930 in the SAM domain (Fig. 5A) (39). In order to identify the site of dephosphorylation by LAR on the EphA2 receptor, we performed LAR substrate-trapping experiments. We assessed which site of tyrosyl phosphorylation on EphA2 when mutated results in the failure to form a substrate-trapping complex with LAR. We mutated each of the tyrosyl residues on EphA2 to phenylalanine, and these tyrosyl phosphorylation site mutants were cotransfected in to 293 cells along with the LAR substrate-trapping mutant (CS-LAR). Substrate-trapping experiments revealed that Y930F-EphA2 was the least efficient at forming a substrate-trapping complex with CS-LAR (Fig. 5B), whereas all other tyrosyl phosphorylation site EphA2 mutants formed efficient substrate-trapped complexes to levels equivalent to that of wild-type EphA2 (Fig. 5B). These experiments identify Y930, which lies within the SAM domain of EphA2, as a major site of LAR dephosphorylation.
Fig 5.
Identification of tyrosine 930 on EphA2 as a target of LAR dephosphorylation. (A) Schematic structure of human EphA2 and depiction of major tyrosyl phosphorylation sites at Y575, Y588, and Y594 in the juxtamembrane region (JMR), Y735 and Y772 in the kinase domain (KD), and Y930 in the SAM domain (SAM). EB, ephrin binding domain; CRR, cysteine-rich region; FNIII, fibronectin type III repeat domain; TM, transmembrane; PDZ, PDZ domain-binding site. (B) Wild-type EphA2 or the indicated EphA2 mutants were coexpressed with a vector alone, WT-LAR, or CS-LAR in 293 cells. Lysates were immunoprecipitated with FLAG antibody and immune complexes were immunoblotted with either EphA2 or FLAG antibodies. Total cell lysates were immunoblotted with either EphA2 or LAR antibodies (bottom two panels). The graph at the bottom presents the densitometric quantitation of EphA2 bound to LAR normalized to FLAG. Data represent the means ± standard errors of the means from five independent experiments. *, P < 0.05 between CS-LAR/EphA2-WT and with CS-LAR/EphA2-Y930F. (C) 293 cells were transfected with a vector alone, wild-type EphA2 (WT), or the EphA2-Y930F mutant. At 48 h posttransfection, lysates were immunoprecipitated with rabbit EphA2(R) or mouse EphA2(M) antibody, and pY930 on EphA2 was detected by immunoblotting with EphA2 pY930-specific (pY930) antibodies, reprobed with antiphosphotyrosine, and then reprobed using EphA2 antibody as a control. (D) MCF10A cells were subjected to LAR knockdown. MCF10A cells were serum starved and stimulated with ephrinA1 (EA1) for 30 min. Phosphorylation of tyrosine 930 on EphA2 was determined by EphA2 immunoprecipitation followed by anti-pY930 immunoblotting and reprobing with EphA2 antibody. Total cell lysates were blotted with ERK1/2 and LAR antibodies.
To further evaluate the ability of LAR to dephosphorylate phosphotyrosyl 930 on EphA2, we generated EphA2 phosphotyrosyl 930 (pY930)-specific antibodies. We transfected 293 cells with vector alone, wild-type EphA2, or the EphA2-Y930F mutant; cell lysates were immunoprecipitated with either a rabbit [EphA2(R)] or mouse [EphA2(M)] EphA2 antibody and immunoblotted with both antiphosphotyrosine and anti-pY930 antibodies (Fig. 5C). Although the total content of EphA2 tyrosyl phosphorylation remained unchanged in EphA2-Y930F-expressing cells, analysis of pY930 using anti-pY930 EphA2 antibodies showed immunoreactivity only in cells expressing wild-type EphA2 and not cells expressing the EphA2-Y930F mutant (Fig. 5C). These results establish the specificity of anti-pY930 EphA2 antibodies to tyrosyl-phosphorylated 930 on EphA2. Next, we showed that depletion of LAR by siRNA enhances the level of pY930 on EphA2 in response to ephrinA1 (Fig. 5D). These results support the conclusion that LAR dephosphorylates phosphotyrosyl 930 on EphA2.
LAR regulates Nck1-EphA2 complex formation.
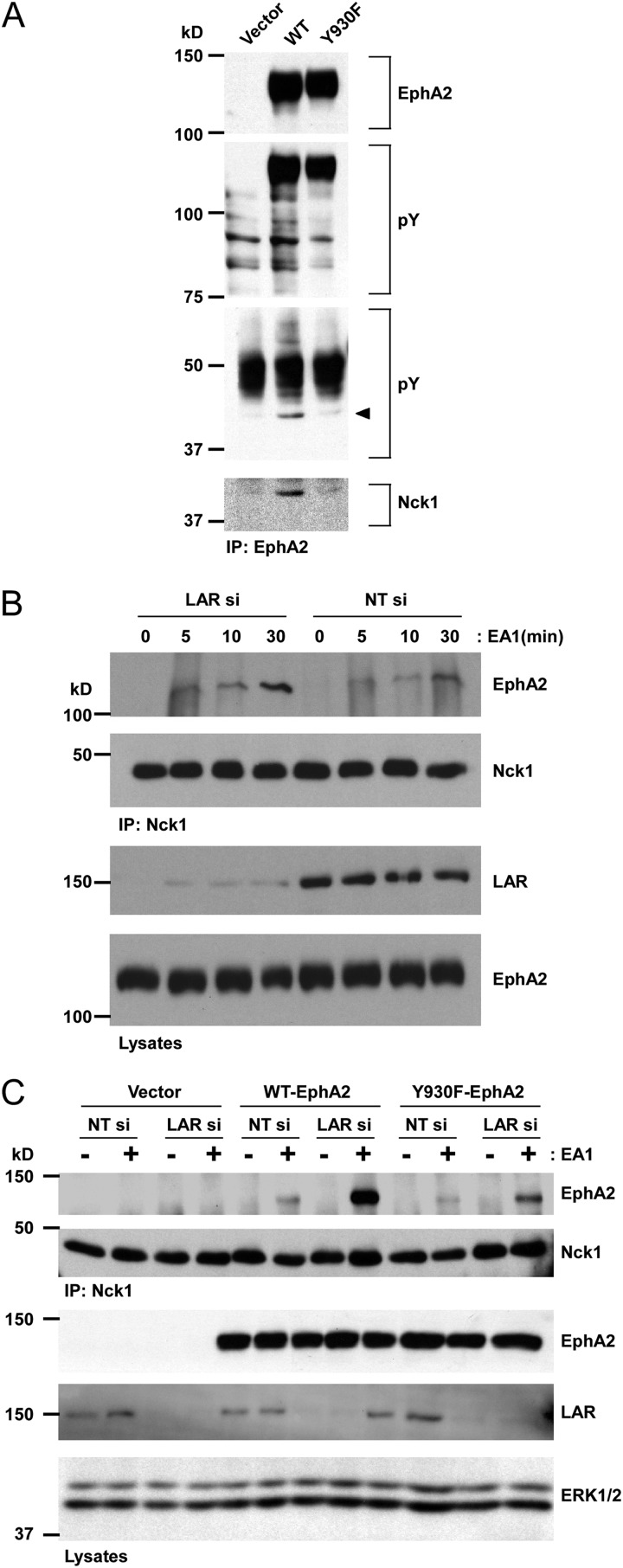
To explore the role of LAR Y930 dephosphorylation in EphA2 signaling, we sought to identify EphA2-interacting proteins that were dependent upon phosphorylation of this residue. Previous studies had suggested that when overexpressed, Vav3 and p85 can interact with EphA2 in a manner dependent upon several phosphotyrosyl residues (39). However, in MCF10A and 293 cells, we were unable to detect under endogenous conditions EphA2 interactions with either Vav3 or p85 (data not shown). Interestingly, we noted that Y930 on EphA2 was similar to the binding site for the Src homology 2 (SH2) domains of SHP-2, which has been shown to interact with EphA2 (40). However, we were not able to detect SHP-2 in a complex with EphA2 either under endogenous conditions or when overexpressed (data not shown). Because we were unable to identify using a candidate approach EphA2-interacting proteins that were dependent upon phosphorylation of Y930, we performed a nonbiased survey for potential tyrosyl-phosphorylated EphA2-interacting proteins that bound in a manner dependent upon Y930 phosphorylation. We transfected 293 cells with a vector alone, wild-type EphA2, or the EphA2-Y930F mutant and assessed the differential association of tyrosyl-phosphorylated EphA2-associated proteins (Fig. 6A). We observed reduced tyrosyl phosphorylation of an ∼45-kDa EphA2-interacting protein in EphA2-Y930F mutant-expressing cells (Fig. 6A). Several studies have shown that the adaptor protein Nck1, which is ∼45 kDa, interacts with tyrosyl-phosphorylated Eph family members through its SH2 domains and that this interaction plays an important role in cytoskeleton-mediated signaling, including cell migration (41–45). When EphA2 immune complexes were immunoblotted with anti-Nck1 antibodies, we detected the presence of Nck1 in wild-type EphA2 complexes. In contrast, Nck1 complex formation with EphA2 was markedly diminished in cells expressing the EphA2-Y930F mutant. Conversely, when we depleted LAR expression with siRNA in MCF10A cells, we found that Nck1-EphA2 complex formation was increased following stimulation with ephrinA1 (Fig. 6B). These experiments demonstrated that in MCF10A cells, LAR also negatively regulates the interaction between Nck1 and EphA2, presumably by dephosphorylating pY930 on EphA2.
Fig 6.
LAR attenuates EphA2-Nck1 interaction through EphA2 Y930. (A) 293 cells were transfected with a vector alone, wild-type EphA2 (WT), or the EphA2-Y930F mutant. At 48 h posttransfection, lysates were immunoprecipitated with EphA2 antibodies and phosphotyrosine was detected by immunoblotting with antiphosphotyrosine antibodies. EphA2 and Nck1 were reprobed using EphA2 and Nck1 antibodies. (B) MCF10A cells were transfected with either LAR siRNA (LAR si) or nontargeting siRNA (NT si) control. Cells were serum starved and then stimulated with ephrinA1 (EA1) for the indicated times. Cell lysates were incubated with Nck1 antibodies, and Nck1-bound EphA2 was detected by immunoblotting. The membrane was reprobed with Nck1 antibodies, and total cell lysates were immunoblotted with EphA2 and LAR antibodies. (C) Either LAR si or NT si was cotransfected in to 293 cells along with a vector alone, wild-type EphA2, or the EphA2-Y930F mutant. Cells were serum starved and then stimulated with EA1 for 30 min, lysates were immunoprecipitated with Nck1 antibodies, and EphA2 was detected by immunoblotting. The membrane was reprobed with Nck1 antibodies as a control. Total cell lysates were blotted with EphA2, ERK1/2, and LAR antibodies.
To confirm that LAR negatively regulates EphA2-Nck1 complex formation specifically through Y930 on EphA2, we cotransfected 293 cells with a vector alone, wild-type EphA2, or the EphA2-Y930F mutant along with either nontargeting siRNA or LAR siRNA. We found that in response to ephrinA1, Nck1 formed a complex with the EphA2-Y930F mutant to a lesser extent than with wild-type EphA2 (Fig. 6C). However, we were still able to detect Nck1 in a complex with the mutated EphA2 receptor, suggesting the likely participation of EphA2 Y594 in EphA2-Nck1 interactions, as reported previously (43). These results demonstrate that LAR negatively regulates EphA2-Nck1 complex formation by controlling phosphorylation on Y930 of EphA2.
LAR regulates EphA2-mediated cell migration through tyrosine 930 dephosphorylation.
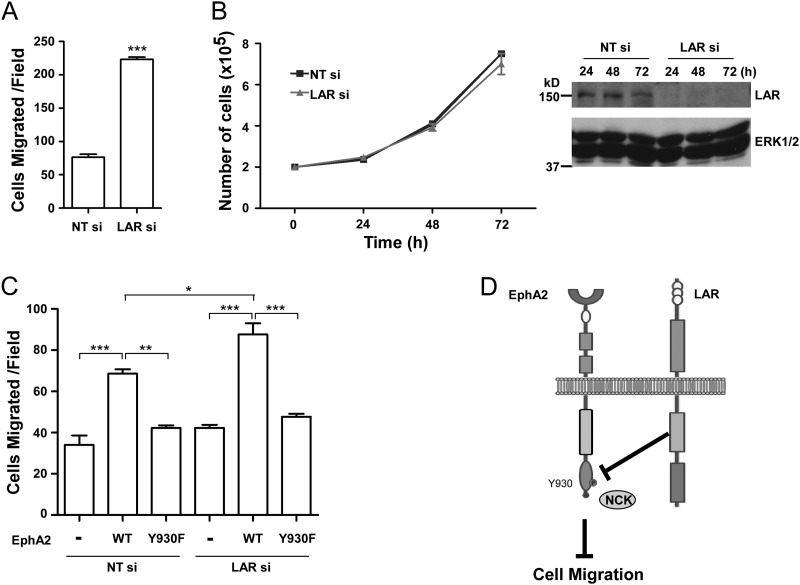
The EphA2 receptor regulates cell migration; in some cases, ligand activation of EphA2 promotes cell migration (46), and in others, it inhibits it (47, 48). The finding that Nck1 and EphA2 interact through phosphotyrosyl 930 (Fig. 6) prompted us to test the contribution of this site to EphA2-mediated cell migration. First, we asked whether knockdown of LAR, which leads to the enhanced binding of Nck1 to EphA2, would promote cell migration. When LAR was knocked down in MCF10A cells, we found that cell migration was significantly enhanced (Fig. 7A). This was unlikely to be a result of increased cell proliferation, since we found no differences in proliferation of LAR siRNA- and NT siRNA-treated cells (Fig. 7B).
Fig 7.
LAR regulates EphA2-mediated cell migration. (A) MCF10A cells were examined for cell migration 48 h following LAR siRNA knockdown. Data represent the number of cells migrated per field (means ± standard errors of the means from three independent experiments; ***, P; < 0.001). (B) MCF10A cells were transfected with either LAR siRNA (LAR si) or nontargeting siRNA (NT si). Twenty-four hours posttransfection, MCF10A cells were plated in six-well dishes. At 24-h intervals, cells were counted and lysates were prepared and immunoblotted with the indicated antibodies. (C) Either LAR siRNA (LAR si) or nontargeting siRNA (NT si) was cotransfected into 293 cells along with a vector alone, wild-type EphA2 (WT), or the EphA2-Y930F mutant. 293 cells were examined for cell migration. Data represent the means ± standard errors of the means from three independent experiments. *, P < 0.05; **, P, < 0.01; ***, P < 0.001. (D) LAR attenuates EphA2-mediated cell migration. LAR sets a threshold for EphA2-mediated effects on cell migration by regulating the levels of Nck1 associated with EphA2 through Y930 phosphorylation.
Next, we tested whether LAR affects cell migration through dephosphorylation of phosphotyrosyl 930 on the EphA2 receptor. When 293 cells were transiently transfected with wild-type EphA2 receptor along with nontargeting siRNA, cell migration was significantly enhanced (Fig. 7C). In contrast, expression of the Y930F mutant of the EphA2 receptor failed to promote cell migration (Fig. 7C). These results indicated that the EphA2 Y930 positively regulates cell migration. In order to determine whether LAR regulates cell migration by controlling EphA2 Y930 phosphorylation, we next asked whether LAR knockdown-induced cell migration is impaired in cells expressing the Y930F mutant of the EphA2 receptor. Cell migration was potentiated in wild-type EphA2-expressing cells in which LAR had been knocked down compared with that in wild-type EphA2-expressing cells alone (Fig. 7C). The ability of LAR when knocked down to promote cell migration was completely abrogated upon expression of the EphA2-Y930F mutant (Fig. 7C). Collectively, these results imply that LAR negatively regulates cell migration by selectively dephosphorylating EphA2 on Y930, thereby uncoupling Nck1 from the EphA2 receptor.
DISCUSSION
Aberrant RTK activation underlies the mechanism driving a number of human diseases, such as cancer. Therefore, elucidating how RTKs are inactivated is an important contribution toward a complete understanding of both physiological and pathophysiological processes that stem from RTK signaling. Although much progress has been made toward identifying PTPs involved in RTK dephosphorylation, a large proportion of RTKs are dephosphorylated by PTPs of unknown identity. In this report, we describe a novel approach to identify RPTPs that exhibit phosphatase activity toward RTKs. We employed an siRNA RPTP-RTK screen that revealed that 12 of the expressed RPTPs in MCF10A cells exert distinct effects of both hypo- and hyper-tyrosyl phosphorylation on selected RTKs throughout the RTKome. These results demonstrate that RPTPs exert a unique fingerprint of RTK regulation, which represents an extensive level of cross talk between the RPTPome and the RTKome. Although it has been appreciated that RPTPs can dephosphorylate RTKs, the data presented here reveal a comprehensive picture of the potential RPTP-RTK relationships. RTKs that were hyper-tyrosyl phosphorylated in the siRNA RPTP-RTK screen represented putative RPTP substrates. Consequently, the results of this screen uncover putative RTKs that may serve as novel RPTP substrates. Finally, as a proof of principle, we validated a RPTP-RTK hit from this screen and demonstrated a novel functional relationship between LAR and EphA2, whereby LAR selectively dephosphorylates EphA2 to control cell migration.
The findings that several silenced RPTPs that are known to be RTK phosphatases exhibited hyper-tyrosyl phosphorylation of their cognate RTK substrates supported the validity of the RPTP-RTK screen. For example, we found that HER2 was hyper-tyrosyl phosphorylated when PTPRN2 was knocked down, consistent with the demonstration that downregulation of PTPRN2 increases tyrosyl phosphorylation of HER2 (49). PTPRJ dephosphorylates VEGFR2 (25), and in the screen, downregulation of PTPRJ resulted in the hyper-tyrosyl phosphorylation of VEGFR2, supporting the observation that PTPRJ acts as a VEGFR2 phosphatase. PTPRJ knockdown also leads to hyper-tyrosyl phosphorylation of VEGFR1 and VEGFR3 in the screen. It is conceivable that both VEGFR1 and VEGFR3 are also PTPRJ substrates, since PTPRJ dephosphorylates pY1054/1059 within the VEGFR2 kinase loop, where both of these sites are conserved in VEGFR1 and VEGFR3. Knockdown of PTPRO induced hyper-tyrosyl phosphorylation of EphA2, consistent with the report that when PTPRO was overexpressed, it reduced EphA2 tyrosyl phosphorylation (33). PTPRO has recently been shown to dephosphorylate ErbB2 (HER2) (50), and indeed, ErbB2 was among the highest-ranked hyper-tyrosyl-phosphorylated proteins in this screen. Finally, LAR has been shown to dephosphorylate the insulin (34) and IGF-1 receptors (35), and we found that when LAR was knocked down, both the insulin and IGF-1 receptors were hypertyrosyl phosphorylated. Collectively, these results support the validity of the siRNA RPTP-RTK screen as a strategy to identify RPTPs that dephosphorylate RTKs.
Interestingly, knockdown of RPTPs also leads to the hypo-tyrosyl phosphorylation of several RTKs, such as ROR2, epidermal growth factor receptor (EGFR), and HER2. Clearly, RTK hypo-tyrosyl phosphorylation upon RPTP knockdown is not consistent with the interpretation that these RTKs are RPTP substrates. Rather, it is likely that RPTP knockdown results in the inactivation of a tyrosine kinase(s) whose substrate is that of an RTK present in this array. Several studies have shown that RPTPs can activate tyrosine kinases. For example, PTPε (PTPRE) knockout mice show decreased Syk kinase activity (51). In addition, PTPα (PTPRA) activates Src and Fyn (52, 53) and positively regulates phosphorylation of focal adhesion kinase in an integrin-dependent manner (54). Hence, hypo-tyrosyl-phosphorylated RTKs likely represent an indirect mode of regulation from an RPTP.
The results from the siRNA RPTP-RTK screen identified several novel RPTP-RTK substrates, one of which was EphA2 as a LAR substrate. Multiple lines of evidence are consistent with the interpretation that EphA2 serves as a physiological LAR substrate. First, knockdown of LAR using siRNAs distinct from those employed in the screen resulted in the hyper-tyrosyl phosphorylation of EphA2. Second, the LAR D1 substrate-trapping mutant formed a stable enzyme complex with tyrosyl-phosphorylated EphA2, and this complex formation was disrupted by the PTP catalytic site inhibitor vanadate. Third, we observed increased colocalization of the LAR substrate-trapping mutant and EphA2 in intact cells. Fourth, mutation of Y930 to a nonphosphorylatable residue disrupted the interaction between the LAR substrate-trapping mutant and EphA2. Concomitantly, LAR knockdown by siRNA resulted in the direct hyper-tyrosyl phosphorylation of Y930. Collectively, these results satisfy the criteria of PTP-substrate assignment between LAR and EphA2.
In order to define the functional consequences of LAR dephosphorylating EphA2 at Y930, we sought to identify potential EphA2-interacting proteins that were dependent upon Y930 phosphorylation. We identified the adaptor protein Nck1 as an EphA2-interacting protein that requires Y930 phosphorylation in order to complex with EphA2. Knockdown of LAR resulted in increased EphA2-Nck1 interactions, and the EphA2-Y930F mutant failed to complex with Nck1. Nck1 has been shown to bind EphA3 through its SH2 domains to phosphotyrosyl residue 602 on EphA3, and this interaction controls EphA-mediated cell migration (42). Moreover, Nck1 has also been reported to be involved in cytoskeleton-mediated cellular function (41–45). The functional consequences of the interaction between Nck1 and EphA2 (pY930) suggest that LAR controls EphA2-Nck1 interactions, which, in turn, play an essential role in cell migration. Interestingly, our data suggest that Nck1 forms a complex with EphA2 at Y930, in addition to Y594, as reported previously (43). The sequence surrounding Y930 is not strictly consistent with the SH2 binding domain consensus in Nck1 as seen in Y594, suggesting that Nck1 may complex with EphA2 indirectly. Nevertheless, our data clearly define an additional mode of Nck1-mediated signaling downstream of the EphA2 receptor.
Like other RPTPs, LAR is involved in cytoskeleton-mediated functions (35, 55, 56). Our finding that LAR negatively regulates EphA2-Nck1 interactions also suggested that LAR controls cell migration through an EphA2-mediated pathway. Indeed, knockdown of LAR, which resulted in enhanced cell migration, was completely impaired in its ability to promote cell migration when the Y930F mutant of EphA2 receptor was coexpressed. Collectively, these results demonstrate that LAR serves as a specific EphA2 phosphatase in the control of cell migration (Fig. 7D). It is noteworthy that genetic evidence in Caenorhabditis elegans also supports a functional relationship between LAR and EphA/ephrins in morphogenesis (57). Therefore, the data presented here offer molecular insight into the functional relationships between Eph/ephrins and LAR in lower organisms.
In summary, we have developed an siRNA RPTP-RTK screen that identified both known and unknown RPTP-RTK relationships. This information sets the foundation from which the mechanisms of RTK dephosphorylation can be further explored. The identification of LAR as a novel EphA2 phosphatase in the control of cell migration has provided proof of principle for the validity of this screen. The identification of RPTP-RTK substrates may open up new avenues for therapeutics against diseases that are caused by aberrant RTK signaling by identifying RPTPs as targets to indirectly modulate site-specific RTK functions.
Supplementary Material
ACKNOWLEDGMENTS
We thank David Stern, Ruey-Hwa Chen, and Paola Chiarugi for reagents and cell lines and Kyunghee Kim for assistance with data analysis. We are grateful to Murim Choi and Lisa Mijung Chung for bioinformatics assistance and Baehoon Kim for assistance with microscopic analyses.
A.M.B. was supported by NIH grant R01 GM099801, and H.L. was supported by a Leslie H. Warner Postdoctoral Fellowship.
Footnotes
Published ahead of print 28 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01708-12.
REFERENCES
- 1. Soulsby M, Bennett AM. 2009. Physiological signaling specificity by protein tyrosine phosphatases. Physiology (Bethesda) 24: 281– 289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tonks NK. 2006. Protein tyrosine phosphatases: from genes, to function, to disease. Nat. Rev. Mol. Cell Biol. 7: 833– 846 [DOI] [PubMed] [Google Scholar]
- 3. Elchebly M, Payette P, Michaliszyn E, Cromlish W, Collins S, Loy AL, Normandin D, Cheng A, Himms-Hagen J, Chan CC, Ramachandran C, Gresser MJ, Tremblay ML, Kennedy BP. 1999. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science 283: 1544– 1548 [DOI] [PubMed] [Google Scholar]
- 4. Bentires-Alj M, Neel BG. 2007. Protein-tyrosine phosphatase 1B is required for HER2/Neu-induced breast cancer. Cancer Res. 67: 2420– 2424 [DOI] [PubMed] [Google Scholar]
- 5. Chan G, Kalaitzidis D, Neel BG. 2008. The tyrosine phosphatase Shp2 (PTPN11) in cancer. Cancer Metastasis Rev. 27: 179– 192 [DOI] [PubMed] [Google Scholar]
- 6. Julien SG, Dube N, Hardy S, Tremblay ML. 2011. Inside the human cancer tyrosine phosphatome. Nat. Rev. Cancer 11: 35– 49 [DOI] [PubMed] [Google Scholar]
- 7. Klaman LD, Boss O, Peroni OD, Kim JK, Martino JL, Zabolotny JM, Moghal N, Lubkin M, Kim YB, Sharpe AH, Stricker-Krongrad A, Shulman GI, Neel BG, Kahn BB. 2000. Increased energy expenditure, decreased adiposity, and tissue-specific insulin sensitivity in protein-tyrosine phosphatase 1B-deficient mice. Mol. Cell. Biol. 20: 5479– 5489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ostman A, Hellberg C, Bohmer FD. 2006. Protein-tyrosine phosphatases and cancer. Nat. Rev. Cancer 6: 307– 320 [DOI] [PubMed] [Google Scholar]
- 9. Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. 2004. Protein tyrosine phosphatases in the human genome. Cell 117: 699– 711 [DOI] [PubMed] [Google Scholar]
- 10. Andersen JN, Mortensen OH, Peters GH, Drake PG, Iversen LF, Olsen OH, Jansen PG, Andersen HS, Tonks NK, Moller NP. 2001. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol. Cell. Biol. 21: 7117– 7136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bixby JL. 2001. Ligands and signaling through receptor-type tyrosine phosphatases. IUBMB Life 51: 157– 163 [DOI] [PubMed] [Google Scholar]
- 12. den Hertog J, Ostman A, Bohmer FD. 2008. Protein tyrosine phosphatases: regulatory mechanisms. FEBS J. 275: 831– 847 [DOI] [PubMed] [Google Scholar]
- 13. Jiang G, den Hertog J, Hunter T. 2000. Receptor-like protein tyrosine phosphatase alpha homodimerizes on the cell surface. Mol. Cell. Biol. 20: 5917– 5929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Toledano-Katchalski H, Tiran Z, Sines T, Shani G, Granot-Attas S, den Hertog J, Elson A. 2003. Dimerization in vivo and inhibition of the nonreceptor form of protein tyrosine phosphatase epsilon. Mol. Cell. Biol. 23: 5460– 5471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bilwes AM, den Hertog J, Hunter T, Noel JP. 1996. Structural basis for inhibition of receptor protein-tyrosine phosphatase-alpha by dimerization. Nature 382: 555– 559 [DOI] [PubMed] [Google Scholar]
- 16. Jiang G, den Hertog J, Su J, Noel J, Sap J, Hunter T. 1999. Dimerization inhibits the activity of receptor-like protein-tyrosine phosphatase-alpha. Nature 401: 606– 610 [DOI] [PubMed] [Google Scholar]
- 17. Felberg J, Johnson P. 1998. Characterization of recombinant CD45 cytoplasmic domain proteins. Evidence for intramolecular and intermolecular interactions. J. Biol. Chem. 273: 17839– 17845 [DOI] [PubMed] [Google Scholar]
- 18. Streuli M, Krueger NX, Thai T, Tang M, Saito H. 1990. Distinct functional roles of the two intracellular phosphatase like domains of the receptor-linked protein tyrosine phosphatases LCA and LAR. EMBO J. 9: 2399– 2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Blanchetot C, den Hertog J. 2000. Multiple interactions between receptor protein-tyrosine phosphatase (RPTP) alpha and membrane-distal protein-tyrosine phosphatase domains of various RPTPs. J. Biol. Chem. 275: 12446– 12452 [DOI] [PubMed] [Google Scholar]
- 20. Blanchetot C, Tertoolen LG, Overvoorde J, den Hertog J. 2002. Intra- and intermolecular interactions between intracellular domains of receptor protein-tyrosine phosphatases. J. Biol. Chem. 277: 47263– 47269 [DOI] [PubMed] [Google Scholar]
- 21. Lemmon MA, Schlessinger J. 2010. Cell signaling by receptor tyrosine kinases. Cell 141: 1117– 1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li S, Depetris RS, Barford D, Chernoff J, Hubbard SR. 2005. Crystal structure of a complex between protein tyrosine phosphatase 1B and the insulin receptor tyrosine kinase. Structure 13: 1643– 1651 [DOI] [PubMed] [Google Scholar]
- 23. Kulas DT, Goldstein BJ, Mooney RA. 1996. The transmembrane protein-tyrosine phosphatase LAR modulates signaling by multiple receptor tyrosine kinases. J. Biol. Chem. 271: 748– 754 [DOI] [PubMed] [Google Scholar]
- 24. Kulas DT, Zhang WR, Goldstein BJ, Furlanetto RW, Mooney RA. 1995. Insulin receptor signaling is augmented by antisense inhibition of the protein tyrosine phosphatase LAR. J. Biol. Chem. 270: 2435– 2438 [DOI] [PubMed] [Google Scholar]
- 25. Chabot C, Spring K, Gratton JP, Elchebly M, Royal I. 2009. New role for the protein tyrosine phosphatase DEP-1 in Akt activation and endothelial cell survival. Mol. Cell. Biol. 29: 241– 253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Debnath J, Muthuswamy SK, Brugge JS. 2003. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30: 256– 268 [DOI] [PubMed] [Google Scholar]
- 27. Lee H, Gaughan JP, Tsygankov AY. 2008. c-Cbl facilitates cytoskeletal effects in v-Abl transformed fibroblast through Rac1- and Rap1-mediated signaling. Int. J. Biochem. Cell Biol. 40: 1930– 1943 [DOI] [PubMed] [Google Scholar]
- 28. Lee H, Tsygankov AY. 2010. c-Cbl regulates glioma invasion through matrix metalloproteinase 2. J. Cell. Biochem. 111: 1169– 1178 [DOI] [PubMed] [Google Scholar]
- 29. Mercan F, Lee H, Kolli S, Bennett AM. 2013. Novel role for SHP-2 in nutrient-responsive control of S6 kinase 1 signaling. Mol. Cell. Biol. 33: 293– 306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jaskolski F, Mulle C, Manzoni OJ. 2005. An automated method to quantify and visualize colocalized fluorescent signals. J. Neurosci. Methods 146: 42– 49 [DOI] [PubMed] [Google Scholar]
- 31. Frangioni JV, Neel BG. 1993. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem. 210: 179– 187 [DOI] [PubMed] [Google Scholar]
- 32. Mercan F, Bennett AM. 2010. Analysis of protein tyrosine phosphatases and substrates. Curr. Protoc. Mol. Biol. 91: 18.16.1–18.16.17 doi:10.1002/0471142727.mb1816s91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shintani T, Ihara M, Sakuta H, Takahashi H, Watakabe I, Noda M. 2006. Eph receptors are negatively controlled by protein tyrosine phosphatase receptor type O. Nat. Neurosci. 9:761– 769 [DOI] [PubMed] [Google Scholar]
- 34. Ahmad F, Considine RV, Goldstein BJ. 1995. Increased abundance of the receptor-type protein-tyrosine phosphatase LAR accounts for the elevated insulin receptor dephosphorylating activity in adipose tissue of obese human subjects. J. Clin. Invest. 95: 2806– 2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Niu XL, Li J, Hakim ZS, Rojas M, Runge MS, Madamanchi NR. 2007. Leukocyte antigen-related deficiency enhances insulin-like growth factor-1 signaling in vascular smooth muscle cells and promotes neointima formation in response to vascular injury. J. Biol. Chem. 282: 19808– 19819 [DOI] [PubMed] [Google Scholar]
- 36. Parri M, Buricchi F, Taddei ML, Giannoni E, Raugei G, Ramponi G, Chiarugi P. 2005. EphrinA1 repulsive response is regulated by an EphA2 tyrosine phosphatase. J. Biol. Chem. 280: 34008– 34018 [DOI] [PubMed] [Google Scholar]
- 37. Flint AJ, Tiganis T, Barford D, Tonks NK. 1997. Development of “substrate-trapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proc. Natl. Acad. Sci. U. S. A. 94: 1680– 1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tiganis T, Bennett AM. 2007. Protein tyrosine phosphatase function: the substrate perspective. Biochem. J. 402: 1– 15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fang WB, Brantley-Sieders DM, Hwang Y, Ham AJ, Chen J. 2008. Identification and functional analysis of phosphorylated tyrosine residues within EphA2 receptor tyrosine kinase. J. Biol. Chem. 283: 16017– 16026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miao H, Burnett E, Kinch M, Simon E, Wang B. 2000. Activation of EphA2 kinase suppresses integrin function and causes focal-adhesion-kinase dephosphorylation. Nat. Cell Biol. 2: 62– 69 [DOI] [PubMed] [Google Scholar]
- 41. Holland SJ, Gale NW, Gish GD, Roth RA, Songyang Z, Cantley LC, Henkemeyer M, Yancopoulos GD, Pawson T. 1997. Juxtamembrane tyrosine residues couple the Eph family receptor EphB2/Nuk to specific SH2 domain proteins in neuronal cells. EMBO J. 16: 3877– 3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hu T, Shi G, Larose L, Rivera GM, Mayer BJ, Zhou R. 2009. Regulation of process retraction and cell migration by EphA3 is mediated by the adaptor protein Nck1. Biochemistry 48: 6369– 6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Miura K, Nam JM, Kojima C, Mochizuki N, Sabe H. 2009. EphA2 engages Git1 to suppress Arf6 activity modulating epithelial cell-cell contacts. Mol. Biol. Cell 20: 1949– 1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stein E, Huynh-Do U, Lane AA, Cerretti DP, Daniel TO. 1998. Nck recruitment to Eph receptor, EphB1/ELK, couples ligand activation to c-Jun kinase. J. Biol. Chem. 273: 1303– 1308 [DOI] [PubMed] [Google Scholar]
- 45. Vindis C, Teli T, Cerretti DP, Turner CE, Huynh-Do U. 2004. EphB1-mediated cell migration requires the phosphorylation of paxillin at Tyr-31/Tyr-118. J. Biol. Chem. 279: 27965– 27970 [DOI] [PubMed] [Google Scholar]
- 46. Brantley-Sieders DM, Caughron J, Hicks D, Pozzi A, Ruiz JC, Chen J. 2004. EphA2 receptor tyrosine kinase regulates endothelial cell migration and vascular assembly through phosphoinositide 3-kinase-mediated Rac1 GTPase activation. J. Cell Sci. 117: 2037– 2049 [DOI] [PubMed] [Google Scholar]
- 47. Miao H, Li DQ, Mukherjee A, Guo H, Petty A, Cutter J, Basilion JP, Sedor J, Wu J, Danielpour D, Sloan AE, Cohen ML, Wang B. 2009. EphA2 mediates ligand-dependent inhibition and ligand-independent promotion of cell migration and invasion via a reciprocal regulatory loop with Akt. Cancer Cell 16: 9– 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Taddei ML, Parri M, Angelucci A, Onnis B, Bianchini F, Giannoni E, Raugei G, Calorini L, Rucci N, Teti A, Bologna M, Chiarugi P. 2009. Kinase-dependent and -independent roles of EphA2 in the regulation of prostate cancer invasion and metastasis. Am. J. Pathol. 174: 1492– 1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhu JH, Chen R, Yi W, Cantin GT, Fearns C, Yang Y, Yates JR, III, Lee JD. 2008. Protein tyrosine phosphatase PTPN13 negatively regulates Her2/ErbB2 malignant signaling. Oncogene 27: 2525– 2531 [DOI] [PubMed] [Google Scholar]
- 50. Yu M, Lin G, Arshadi N, Kalatskaya I, Xue B, Haider S, Nguyen F, Boutros PC, Elson A, Muthuswamy LB, Tonks NK, Muthuswamy SK. 2012. Expression profiling during mammary epithelial cell three-dimensional morphogenesis identifies PTPRO as a novel regulator of morphogenesis and ErbB2-mediated transformation. Mol. Cell. Biol. 32: 3913– 3924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. De Franceschi L, Biondani A, Carta F, Turrini F, Laudanna C, Deana R, Brunati AM, Turretta L, Iolascon A, Perrotta S, Elson A, Bulato C, Brugnara C. 2008. PTPepsilon has a critical role in signaling transduction pathways and phosphoprotein network topology in red cells. Proteomics 8: 4695– 4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Su J, Muranjan M, Sap J. 1999. Receptor protein tyrosine phosphatase alpha activates Src-family kinases and controls integrin-mediated responses in fibroblasts. Curr. Biol. 9: 505– 511 [DOI] [PubMed] [Google Scholar]
- 53. Ponniah S, Wang DZ, Lim KL, Pallen CJ. 1999. Targeted disruption of the tyrosine phosphatase PTPalpha leads to constitutive downregulation of the kinases Src and Fyn. Curr. Biol. 9: 535– 538 [DOI] [PubMed] [Google Scholar]
- 54. Zeng L, Si X, Yu WP, Le HT, Ng KP, Teng RM, Ryan K, Wang DZ, Ponniah S, Pallen CJ. 2003. PTP alpha regulates integrin-stimulated FAK autophosphorylation and cytoskeletal rearrangement in cell spreading and migration. J. Cell Biol. 160: 137– 146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Müller T, Choidas A, Reichmann E, Ullrich A. 1999. Phosphorylation and free pool of beta-catenin are regulated by tyrosine kinases and tyrosine phosphatases during epithelial cell migration. J. Biol. Chem. 274: 10173– 10183 [DOI] [PubMed] [Google Scholar]
- 56. Debant A, Serra-Pages C, Seipel K, O'Brien S, Tang M, Park SH, Streuli M. 1996. The multidomain protein Trio binds the LAR transmembrane tyrosine phosphatase, contains a protein kinase domain, and has separate rac-specific and rho-specific guanine nucleotide exchange factor domains. Proc. Natl. Acad. Sci. U. S. A. 93: 5466– 5471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Harrington RJ, Gutch MJ, Hengartner MO, Tonks NK, Chisholm AD. 2002. The C. elegans LAR-like receptor tyrosine phosphatase PTP-3 and the VAB-1 Eph receptor tyrosine kinase have partly redundant functions in morphogenesis. Development 129: 2141– 2153 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.