Abstract
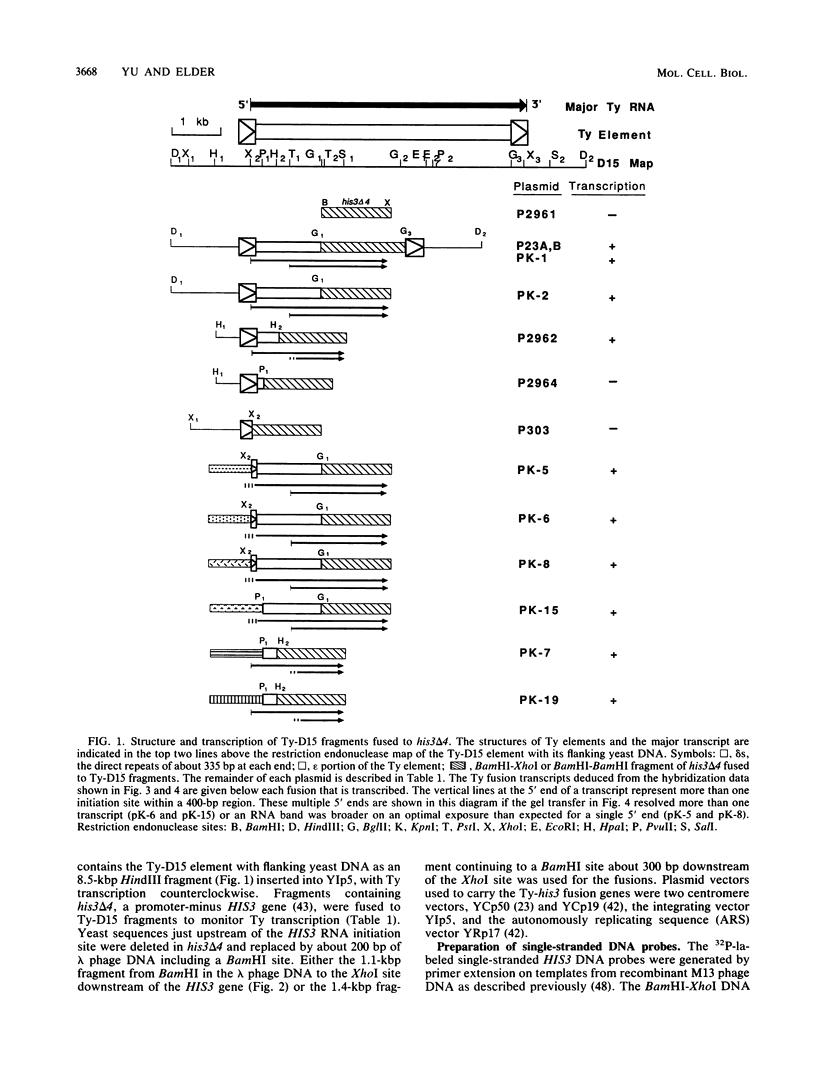
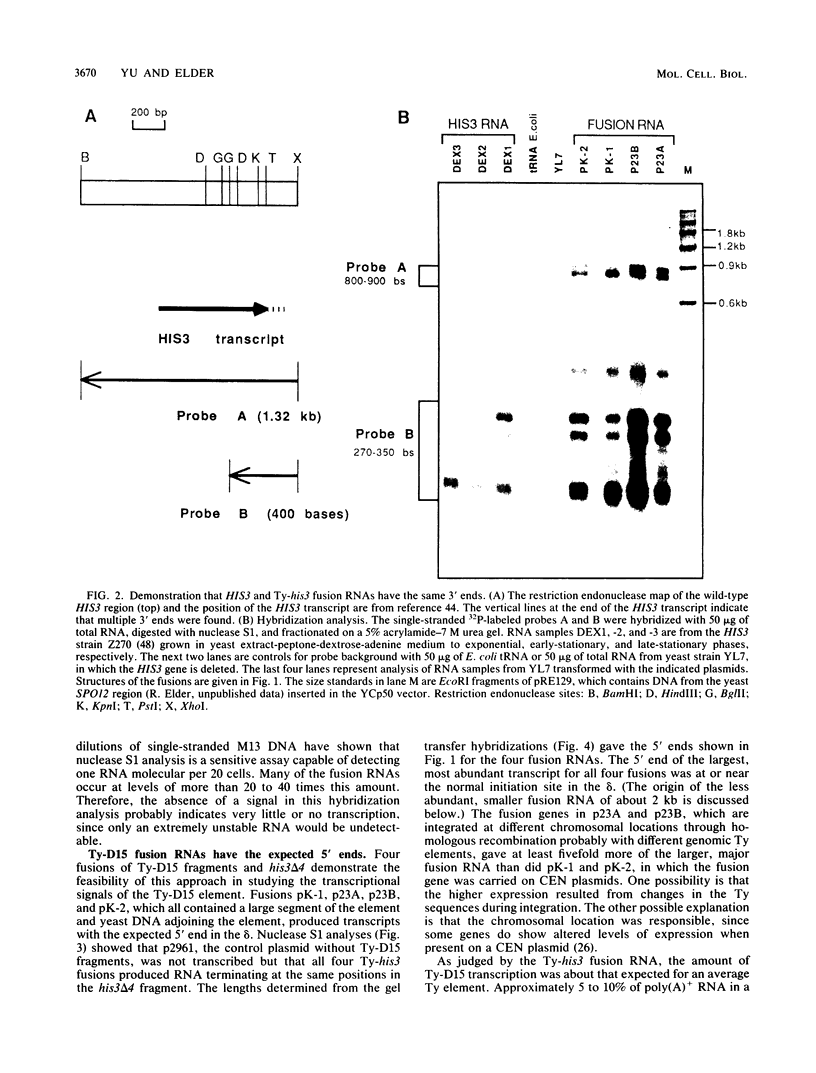
The major transcript of the yeast transposable element Ty1 has its 5' end in one delta and the 3' end in the opposite delta, the direct repeats of about 335 base pairs (bp) at each end of the element. The transcriptional initiation signals of the Ty-D15 element that give rise to this transcript were found to have a number of unusual characteristics. The 5' delta by itself, which contained the initiation site for Ty transcription, gave no detectable transcription. A region internal to the transcript in a translated part of the element and about 140 bp downstream of the 5' delta was essential for initiation of the major Ty transcript. This internal activating region (IAR) had several interesting properties. When the portion of the delta upstream of the initiation site was replaced with DNA fragments that did not by themselves act as promoters, initiation directed by the IAR still occurred at about the same position, 200 to 400 bp upstream of the IAR. If fragments containing the IAR were inverted, transcription could still occur. When 468 or 636 bp was inserted between the delta and the IAR, initiations occurred near the normal delta initiation site and in the inserted DNA. Therefore, the location and properties of transcription signals for Ty-D15 differ considerably from those expected for a yeast gene transcribed by RNA polymerase II.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arndt K. T., Styles C., Fink G. R. Multiple global regulators control HIS4 transcription in yeast. Science. 1987 Aug 21;237(4817):874–880. doi: 10.1126/science.3303332. [DOI] [PubMed] [Google Scholar]
- Arrigo S., Yun M., Beemon K. cis-acting regulatory elements within gag genes of avian retroviruses. Mol Cell Biol. 1987 Jan;7(1):388–397. doi: 10.1128/mcb.7.1.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Eichinger D., Castrillon D., Fink G. R. The Saccharomyces cerevisiae genome contains functional and nonfunctional copies of transposon Ty1. Mol Cell Biol. 1988 Apr;8(4):1432–1442. doi: 10.1128/mcb.8.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Garfinkel D. J., Styles C. A., Fink G. R. Ty elements transpose through an RNA intermediate. Cell. 1985 Mar;40(3):491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- Bowen B. A., Fulton A. M., Tuite M. F., Kingsman S. M., Kingsman A. J. Expression of Ty-lacZ fusions in Saccharomyces cerevisiae. Nucleic Acids Res. 1984 Feb 10;12(3):1627–1640. doi: 10.1093/nar/12.3.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Struhl K. Saturation mutagenesis of a yeast his3 "TATA element": genetic evidence for a specific TATA-binding protein. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2691–2695. doi: 10.1073/pnas.85.8.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare J., Farabaugh P. Nucleotide sequence of a yeast Ty element: evidence for an unusual mechanism of gene expression. Proc Natl Acad Sci U S A. 1985 May;82(9):2829–2833. doi: 10.1073/pnas.82.9.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Company M., Adler C., Errede B. Identification of a Ty1 regulatory sequence responsive to STE7 and STE12. Mol Cell Biol. 1988 Jun;8(6):2545–2554. doi: 10.1128/mcb.8.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Company M., Errede B. Cell-type-dependent gene activation by yeast transposon Ty1 involves multiple regulatory determinants. Mol Cell Biol. 1987 Sep;7(9):3205–3211. doi: 10.1128/mcb.7.9.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coney L. R., Roeder G. S. Control of yeast gene expression by transposable elements: maximum expression requires a functional Ty activator sequence and a defective Ty promoter. Mol Cell Biol. 1988 Oct;8(10):4009–4017. doi: 10.1128/mcb.8.10.4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen B. R., Lomedico P. T., Ju G. Transcriptional interference in avian retroviruses--implications for the promoter insertion model of leukaemogenesis. Nature. 1984 Jan 19;307(5948):241–245. doi: 10.1038/307241a0. [DOI] [PubMed] [Google Scholar]
- Elder R. T., Loh E. Y., Davis R. W. RNA from the yeast transposable element Ty1 has both ends in the direct repeats, a structure similar to retrovirus RNA. Proc Natl Acad Sci U S A. 1983 May;80(9):2432–2436. doi: 10.1073/pnas.80.9.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder R. T., St John T. P., Stinchcomb D. T., Davis R. W., Scherer S., Davis R. W. Studies on the transposable element Ty1 of yeast. I. RNA homologous to Ty1. II. Recombination and expression of Ty1 and adjacent sequences. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):581–591. doi: 10.1101/sqb.1981.045.01.075. [DOI] [PubMed] [Google Scholar]
- Errede B., Cardillo T. S., Sherman F., Dubois E., Deschamps J., Wiame J. M. Mating signals control expression of mutations resulting from insertion of a transposable repetitive element adjacent to diverse yeast genes. Cell. 1980 Nov;22(2 Pt 2):427–436. doi: 10.1016/0092-8674(80)90353-0. [DOI] [PubMed] [Google Scholar]
- Errede B., Company M., Ferchak J. D., Hutchison C. A., 3rd, Yarnell W. S. Activation regions in a yeast transposon have homology to mating type control sequences and to mammalian enhancers. Proc Natl Acad Sci U S A. 1985 Aug;82(16):5423–5427. doi: 10.1073/pnas.82.16.5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errede B., Company M., Hutchison C. A., 3rd Ty1 sequence with enhancer and mating-type-dependent regulatory activities. Mol Cell Biol. 1987 Jan;7(1):258–265. doi: 10.1128/mcb.7.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton A. M., Mellor J., Dobson M. J., Chester J., Warmington J. R., Indge K. J., Oliver S. G., de la Paz P., Wilson W., Kingsman A. J. Variants within the yeast Ty sequence family encode a class of structurally conserved proteins. Nucleic Acids Res. 1985 Jun 11;13(11):4097–4112. doi: 10.1093/nar/13.11.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton A. M., Rathjen P. D., Kingsman S. M., Kingsman A. J. Upstream and downstream transcriptional control signals in the yeast retrotransposon, TY. Nucleic Acids Res. 1988 Jun 24;16(12):5439–5458. doi: 10.1093/nar/16.12.5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel D. J., Boeke J. D., Fink G. R. Ty element transposition: reverse transcriptase and virus-like particles. Cell. 1985 Sep;42(2):507–517. doi: 10.1016/0092-8674(85)90108-4. [DOI] [PubMed] [Google Scholar]
- Hall B. D., Clarkson S. G., Tocchini-Valentini G. Transcription initiation of eucaryotic transfer RNA genes. Cell. 1982 May;29(1):3–5. doi: 10.1016/0092-8674(82)90083-6. [DOI] [PubMed] [Google Scholar]
- Hauber J., Nelböck-Hochstetter P., Feldmann H. Nucleotide sequence and characteristics of a Ty element from yeast. Nucleic Acids Res. 1985 Apr 25;13(8):2745–2758. doi: 10.1093/nar/13.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerome J. F., Jaehning J. A. mRNA transcription in nuclei isolated from Saccharomyces cerevisiae. Mol Cell Biol. 1986 May;6(5):1633–1639. doi: 10.1128/mcb.6.5.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M., Davis R. W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X. B., Clare J. J., Farabaugh P. J. The upstream activation site of a Ty2 element of yeast is necessary but not sufficient to promote maximal transcription of the element. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8520–8524. doi: 10.1073/pnas.84.23.8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marczynski G. T., Jaehning J. A. A transcription map of a yeast centromere plasmid: unexpected transcripts and altered gene expression. Nucleic Acids Res. 1985 Dec 9;13(23):8487–8506. doi: 10.1093/nar/13.23.8487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J., Kingsman A. J., Kingsman S. M. Ty, an endogenous retrovirus of yeast? Yeast. 1986 Sep;2(3):145–152. doi: 10.1002/yea.320020302. [DOI] [PubMed] [Google Scholar]
- Mellor J., Malim M. H., Gull K., Tuite M. F., McCready S., Dibbayawan T., Kingsman S. M., Kingsman A. J. Reverse transcriptase activity and Ty RNA are associated with virus-like particles in yeast. Nature. 1985 Dec 12;318(6046):583–586. doi: 10.1038/318583a0. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Shore D. Transcriptional regulation in the yeast life cycle. Science. 1987 Sep 4;237(4819):1162–1170. doi: 10.1126/science.3306917. [DOI] [PubMed] [Google Scholar]
- Nonet M., Scafe C., Sexton J., Young R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol. 1987 May;7(5):1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osley M. A., Hereford L. Identification of a sequence responsible for periodic synthesis of yeast histone 2A mRNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7689–7693. doi: 10.1073/pnas.79.24.7689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathjen P. D., Kingsman A. J., Kingsman S. M. The yeast ROAM mutation--identification of the sequences mediating host gene activation and cell-type control in the yeast retrotransposon, Ty. Nucleic Acids Res. 1987 Sep 25;15(18):7309–7324. doi: 10.1093/nar/15.18.7309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder G. S., Coney L. R., Pearlman R. E., Rose A. B. Control of yeast gene expression by transposable elements. Basic Life Sci. 1986;40:545–555. doi: 10.1007/978-1-4684-5251-8_42. [DOI] [PubMed] [Google Scholar]
- Roeder G. S., Rose A. B., Pearlman R. E. Transposable element sequences involved in the enhancement of yeast gene expression. Proc Natl Acad Sci U S A. 1985 Aug;82(16):5428–5432. doi: 10.1073/pnas.82.16.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Hong G. F., Hill D. F., Petersen G. B. Nucleotide sequence of bacteriophage lambda DNA. J Mol Biol. 1982 Dec 25;162(4):729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- Scherer S., Davis R. W. Recombination of dispersed repeated DNA sequences in yeast. Science. 1980 Sep 19;209(4463):1380–1384. doi: 10.1126/science.6251545. [DOI] [PubMed] [Google Scholar]
- Scherer S., Mann C., Davis R. W. Reversion of a promoter deletion in yeast. Nature. 1982 Aug 26;298(5877):815–819. doi: 10.1038/298815a0. [DOI] [PubMed] [Google Scholar]
- Scherer S. Regulated yeast promoters produced by DNA rearrangements selected in vivo. J Mol Biol. 1986 Oct 5;191(3):355–365. doi: 10.1016/0022-2836(86)90132-4. [DOI] [PubMed] [Google Scholar]
- Silverman S. J., Fink G. R. Effects of Ty insertions on HIS4 transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Jul;4(7):1246–1251. doi: 10.1128/mcb.4.7.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcomb D. T., Mann C., Davis R. W. Centromeric DNA from Saccharomyces cerevisiae. J Mol Biol. 1982 Jun 25;158(2):157–190. doi: 10.1016/0022-2836(82)90427-2. [DOI] [PubMed] [Google Scholar]
- Struhl K. Deletion mapping a eukaryotic promoter. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4461–4465. doi: 10.1073/pnas.78.7.4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Nucleotide sequence and transcriptional mapping of the yeast pet56-his3-ded1 gene region. Nucleic Acids Res. 1985 Dec 9;13(23):8587–8601. doi: 10.1093/nar/13.23.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987 May 8;49(3):295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- Wang H. T., Frackman S., Kowalisyn J., Esposito R. E., Elder R. Developmental regulation of SPO13, a gene required for separation of homologous chromosomes at meiosis I. Mol Cell Biol. 1987 Apr;7(4):1425–1435. doi: 10.1128/mcb.7.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson V. M., Cox D., Young E. T., Russell D. W., Smith M. Characterization of transposable element-associated mutations that alter yeast alcohol dehydrogenase II expression. Mol Cell Biol. 1983 Jan;3(1):20–31. doi: 10.1128/mcb.3.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson V. M., Young E. T., Ciriacy M. Transposable elements associated with constitutive expression of yeast alcohol dehydrogenase II. Cell. 1981 Feb;23(2):605–614. doi: 10.1016/0092-8674(81)90156-2. [DOI] [PubMed] [Google Scholar]
- Winston F., Dollard C., Malone E. A., Clare J., Kapakos J. G., Farabaugh P., Minehart P. L. Three genes are required for trans-activation of Ty transcription in yeast. Genetics. 1987 Apr;115(4):649–656. doi: 10.1093/genetics/115.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Durbin K. J., Fink G. R. The SPT3 gene is required for normal transcription of Ty elements in S. cerevisiae. Cell. 1984 Dec;39(3 Pt 2):675–682. doi: 10.1016/0092-8674(84)90474-4. [DOI] [PubMed] [Google Scholar]