Abstract
Much of the work on the basic molecular biology of human adenoviruses has been carried out on a very limited number of the more than 60 serotypes, primarily the highly related species C viruses adenovirus type 5 (Ad5) and Ad2 and, to some extent, Ad12 of species A. Until recently, it has been widely assumed that insights obtained with these model viruses were representative of all human adenoviruses. Recent studies on the E3 ubiquitin ligase formed by the viral E1B55K and E4orf6 proteins with a cellular Cullin-based complex indicated that although all species form such a functional complex, significant variations exist in terms of complex composition and the substrates that are degraded. In the present report we conducted a comprehensive analysis of the localization of E1B55K products from representatives of six of the seven adenovirus species in the presence and the absence of the corresponding E4orf6 protein. We found that although in some species E1B55K localized in aggresomes, such was not always the case, suggesting that these structures are not necessary for the efficient degradation of substrates. In addition, differences were evident in the localization of E1B55K, although all forms readily associated with PML. Finally, Ad5 E1B55K was seen to localize in close proximity to Rab11, a marker for the endosomal recycling compartment, and both focused at the microtubule organizing center. These findings suggest that E1B55K from some species may employ the transport system utilized by the membrane recycling pathway to assemble aggresomes and the possibility that this structure might then affect recycling of cell surface components.
INTRODUCTION
One of the products of early region 1B of human adenoviruses, E1B55K, has been studied extensively, especially that produced by serotype 5 (Ad5), which historically has served as a model for all adenoviruses. E1B55K is known to have several functions during lytic infection, some of which require a product of early region 4, the E4orf6 protein (1–8). Much of the Ad5 E1B55K in infected cells associates with and serves as the substrate recruitment component for a Cullin-based E3 ubiquitin ligase formed by E4orf6 that ubiquitinates several target substrates leading to their degradation to optimize the infectious cycle (4, 6, 9, 10). The formation of this ligase complex is highly conserved in adenoviruses, although substrate specificity varies somewhat between different adenovirus species (11). In the absence of E4orf6, E1B55K has been found to promote the degradation of the multifunctional protein Daxx (12) and to bind to and repress the transcriptional activation activity of p53 (13–16). E1B55K also contributes to cell transformation in combination with products of early region 1A (E1A), a function that is probably largely related to its ability to inactivate p53 (13–16) but also involves p53-independent functions (17–19). E1B55K is SUMOylated at its SUMO conjugation motif (SCM) (14, 20, 21) and phosphorylated on the C-terminal region by casein kinase 2 (CK2) (22–24). Both posttranslational modifications are required for efficient repression of p53-mediated transcription, for the nucleocytoplasmic relocalization of p53, and thus for cellular transformation (25). Furthermore, these modifications are linked in that phosphorylation is required for efficient SUMO conjugation to occur (25). The E1B55K/E4orf6 complex readily shuttles between the nucleus and cytoplasm (26–28). While the localization of Ad5 E1B55K in productively infected human cells as detected by immunofluorescence (IF) microscopy is mostly nuclear (7, 29) or near the nucleus (7), its localization in E1A/E1B-transformed human or rodent cells differs significantly. Little E1B55K is present in the nucleus, whereas a major quantity is concentrated in a single cytoplasmic body near the nucleus (30, 31), with some in regions of cell-cell contact at the cell surface (31) and low levels throughout the cytoplasm (31). The cytoplasmic bodies, which are evident in many infected human cells as well, were recently characterized as aggresomes, which localize near the microtubule organizing center (MTOC) region (32). In lytic infection the expression of E4orf6, as well as of E4orf3, is responsible for the relocalization of much of the E1B55K to the nucleus (33), but a portion can remain in these aggresomes (21).
Aggresome formation is a normal process that cells use to eliminate aggregates that can form when proteins fold only partially or incorrectly. When hydrophobic domains of such misfolded proteins are exposed they tend to aggregate if not properly controlled by chaperone proteins. Multiple aggregated particles are formed throughout the cytoplasm, presumably at sites of translation at polysomes (34), that are then rapidly transported toward the MTOC by dynein-dependent retrograde transport on microtubules (34, 35) where they are sequestered in a single structure called the aggresome. The aggresome is thus an aggregate of aggregates (36). If retrograde transport is inhibited by treatment with microtubule depolymerizing drugs such as nocodazole, or by the overexpression of p50/dynamitin, the aggresomal particles remain distributed throughout the cytoplasm (34, 35). Most aggresomes appear either as a single sphere or as an extended ribbon, depending in part on the nature of the aggregated proteins (36). Aggresomes are sometimes surrounded by a “vimentin cage” resulting from the collapse of the predominant intermediate filament (35, 36), although this formation is not universal or essential for aggresome formation (36; Ron Kopito, personal communication).
In the present study we have examined the localization of E1B55K and found a considerable variability among different adenovirus species. In fact, the localization of E1B55K of Ad5 and Ad12, the two most studied model serotypes, is quite distinct from the general pattern exhibited by representatives of other adenovirus species. We also found that the ability of E1B55K to localize in aggresomal structures is not conserved in all adenovirus species, suggesting that this structure is not required for efficient degradation of the nuclear substrates of the E4orf6/E1B55K E3 ubiquitin ligase complex. It was noted, however, that E1B55K proteins of all adenovirus species were able to bind PML and in most cases colocalized with PML in nuclear foci.
MATERIALS AND METHODS
Cells and cell lines.
Human small cell carcinoma H1299 cells (ATCC CRL-5803), carrying a deletion of the p53 gene (37), were cultured in Dulbecco modified Eagle medium (DMEM) containing 0.292-mg l-glutamine (Gibco)/ml supplemented with 10% fetal calf serum (FCS; PAA).
Plasmids and antisera.
cDNAs encoding FLAG-E4orf6 and HA-E1B55K proteins from each serotype were described previously (11). cDNA encoding GFP-Rab11 (38) was obtained from Morag Park (McGill University). Hemagglutinin (HA) epitopes were detected using anti-HA mouse monoclonal HA.11 (BabCO) (Western blotting or IF) or with rat monoclonal 3F10 (Roche) (IF), FLAG epitopes with rabbit anti-FLAG (Sigma-Aldrich), actin with mouse monoclonal antibody C4 (Millipore), PML with mouse monoclonal antibody ab6263 (Abcam), and γ-tubulin with mouse monoclonal antibody ab11316 (Abcam).
Indirect immunofluorescence.
Cells grown on glass coverslips in six-well dishes were transfected for 24 h with up to 1 μg of plasmid DNAs expressing HA-E1B55K of different serotypes. Some wells were treated with 1.5 μM nocodazole (Sigma). Cells were then fixed for 15 min with 4% paraformaldehyde (Electron Microscopy Science) and permeabilized for 15 min with 0.5% Triton X-100 in phosphate-buffered saline (PBS). For γ-tubulin experiments, cells were fixed with methanol at −20°C for 15 min with no permeabilization step. After washes in PBS, immunofluorescence was performed with appropriate antibodies for 2 h at room temperature in a humidity chamber. The secondary antibodies used were antibodies conjugated to Alexa 594 and Alexa 488 dyes (Molecular Probes). DAPI (4′,6′-diamidino-2-phenylindole) dye was added with the secondary antibodies. Coverslips were washed and mounted in Immu-Mount (Thermo Scientific). In all cases, we have analyzed cells expressing low to intermediate levels of protein to avoid potential localization artifacts related to overexpression. Images were obtained using an LSM3 confocal microscope with a ×63 objective using the LSM 4.2 Image browser software. Images were cropped using Adobe Photoshop CS3, the brightness adjusted, and then assembled with Adobe Illustrator CS3.
Coimmunoprecipitations.
Subconfluent cells grown in 100-mm dishes were treated with a mixture of DNA and 25-kDa linear polyethyleneimine (Polysciences Inc., Eppelheim, Germany). One hour before transfection, the medium was removed and replaced by fresh DMEM. The transfection solution was prepared by incubating a mixture of DNA, polyethyleneimine, and DMEM in a ratio of 1:10:100 for 30 min at room temperature. After application of the transfection solution, cells were incubated for 8 h before replacing the medium with standard culture medium. Protein extracts were prepared in full radioimmunoprecipitation assay lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5 mM EDTA, 1 mM dithiothreitol [DTT], 0.1% sodium dodecyl sulfate, 1% Nonidet P-40, 0.1% Triton X-100, 0.5% sodium deoxycholate) containing 1% (vol/vol) phenylmethylsulfonyl fluoride, 0.1% (vol/vol) aprotinin, 1 μg of leupeptin/ml, 1 μg of pepstatin/ml, 1 mM DTT, 25 mM iodacetamide, and 25 mM N-ethylmaleimide. For immunoprecipitations, protein A-Sepharose (Upstate) was coupled with PML antibodies for 1 h at 4°C. The antibody-coupled protein A-Sepharose was added to precleared extracts and rotated for 2 h at 4°C. Proteins bound to the antibody-coupled protein A-Sepharose were precipitated by centrifugation, washed three times, boiled for 3 min at 95°C in 2× Laemmli buffer, and analyzed by immunoblotting. For immunoblotting of steady-state protein levels/immunoprecipitation solutions, equal amounts of total protein were separated by SDS-PAGE, transferred to polyvinylidene difluoride membranes (Millipore) blocked using 5% skim milk. Primary antibodies were added on membranes for 2 to 3 h at room temperature. Membranes were washed with PBS containing 0.1% Tween 20 and the secondary antibody added for 45 min at room temperature. Detection was performed using the Western Lightning Chemiluminescence Reagent Plus (Perkin-Elmer).
RESULTS
The subcellular localization of E1B55K of Ad5 is not representative of all adenovirus species.
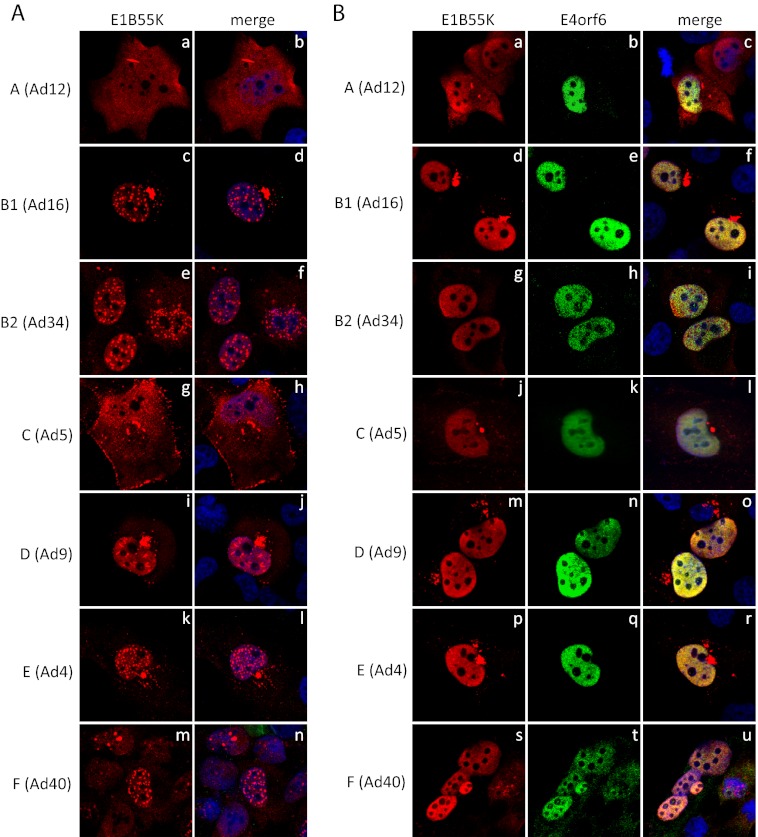
All human adenoviruses are predicted to encode similar E1B products, the largest of which in all serotypes is a highly similar species of about 55 kDa (C. Y. Cheng et al., unpublished data). For simplicity in the present report all will be referred to as E1B55K. It is widely believed that all E1B55K products exhibit largely common biological activities. For example, although there is some heterogeneity in composition and substrate specificity, we have found that representatives of all species form functional Cullin-based E3 ubiquitin ligase complexes with the E4orf6 protein (11). In the case of Ad5 it is likely that a major portion of the E1B55K in infected cells is present in this complex (4). The subcellular localization of E1B55K of Ad5 has been most extensively studied. Expressed alone in the absence of other viral proteins Ad5 E1B55K is mostly cytoplasmic with usually a juxtanuclear concentration within what has been determined to be an aggresome (32), and a variable but usually low level in the nucleus (31). After coexpression with E4orf6, Ad5 E1B55K becomes concentrated in the nucleus but a significant quantity remains with the aggresome. These results suggest that interactions with E4orf6 enhance transport to and/or retention in or inhibition of export from the nucleus (33). To determine whether this pattern of localization of E1B55K is conserved, H1299 cells grown on coverslips were transfected with plasmid DNAs expressing HA-E1B55K products from serotypes representing six of the seven adenovirus species and cells were fixed and stained using an HA-specific antibody. As shown in Fig. 1Ag and h, with Ad5 (species C) E1B55K the usual pattern was apparent, a mostly cytoplasmic localization with cytoplasmic dots, the aggresome, and a low (and in multiple fields of cells, not shown, variable) level of nuclear accumulation. A significant amount of Ad5 E1B55K was also evident at the plasma membrane. Figure 1Aa and b shows that the localization of Ad12 E1B55K corresponds well with results from previous studies (39, 40) in that it was evident throughout the cell with oval shaped aggresome-like structures being apparent, although unlike Ad5 some variation in the number of such structures was evident, ranging from 0 to even 3 or 4 in some cells, with 0 to 1 being most common (not shown). As seen previously (39), cells expressing only low levels of E1B55K exhibited mostly a nuclear localization (data not shown), which is also the predominant location of Ad12 E1B55K in transformed cells or DNA-transfected rat cells (31, 41). The localizations of E1B55K from the other serotypes were found to be quite similar, but all differed significantly from those of both the Ad5 and Ad12 proteins. All exhibited a mostly nuclear localization with intense nuclear dots. For Ad16 (Fig. 1Ac and d) and Ad34 (Fig. 1Ae and f), these dots were of a spherical nature. In the case of Ad16 (Fig. 1Ac and d), Ad9 (Fig. 1Ai and j), and Ad4 (Fig. 1Ak and l) E1B55K was also present in an aggresome-like structure near the nucleus. This structure was seen infrequently with Ad34 (Fig. 1Ae and f) but practically never with Ad40 (Fig. 1Am and n).
Fig 1.
Localization of E1B55K of different serotypes and effect of E4orf6 expression. (A) H1299 cells were transfected with plasmid DNAs expressing HA-tagged E1B55K of different serotypes, fixed in paraformaldehyde, and stained for HA and DNA (with DAPI). (B) H1299 cells were transfected with pairs of plasmid DNAs expressing HA-tagged E1B55K and FLAG-tagged E4orf6 of different serotypes, fixed in paraformaldehyde and stained for HA, FLAG, and DNA (with DAPI). Panels are as indicated in the figure. Letters next to each adenovirus serotype shown in parentheses refer to the HAdV species.
As described earlier, in the case of Ad5, E1B55K is present largely in the nucleus in the presence of the E4orf6 product. To determine whether a similar pattern exists with other serotypes, H1299 cells grown on coverslips were transfected with plasmid DNAs expressing HA-E1B55K and the corresponding FLAG-E4orf6 from the seven serotypes and examined by IF using HA- and FLAG-specific antibodies. Figure 1B (middle panels) shows that as seen before (11), in all cases E4orf6 protein localized exclusively in the nucleus. With Ad5 (Fig. 1Bj) the presence of E4orf6 resulted in full nuclear localization of E1B55K, as found previously. Figure 1B also shows that with the exception of Ad12, the expression of E4orf6 caused the loss of the nuclear dots of E1B55K from all other serotypes. Such was not the case with Ad12. Figures 1Ba and c show that even in the presence of Ad12 E4orf6 protein, Ad12 E1B55K was present in both the nucleus and cytoplasm with aggresome-like structures evident. This difference could occur for several reasons (see Discussion). Nevertheless, two major questions issue from the observations presented in Fig. 1 in terms of the nature of the intracellular structures containing E1B55K. The first is whether or not the presence of E1B55K in true aggresomes is conserved among all adenovirus species. The second concerns the composition of the nuclear dots containing E1B55K in many of the serotypes examined.
The nuclear dots containing E1B55K are associated with PML bodies.
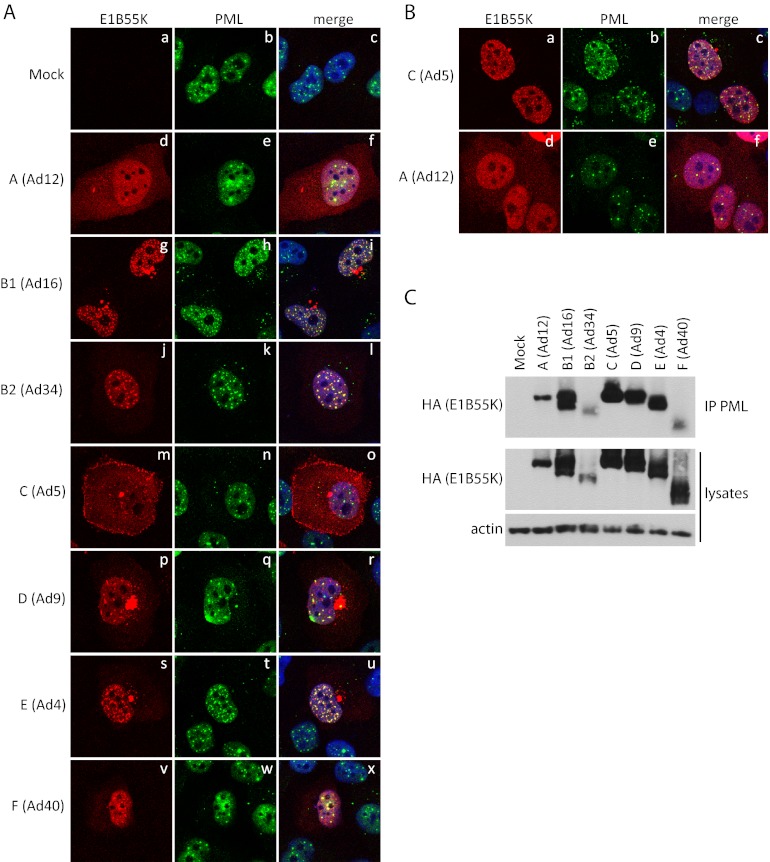
It seemed possible that the nuclear dots containing E1B55K could represent PML bodies, which are dense nuclear structures numbering from 10 to 20 per nucleus (42–44). They are composed of several proteins (45) among which PML is a key component (46, 47). The bodies are involved in a broad range of key cellular processes, including the innate immune response (reviewed in reference 48). Incoming Ad5 genomes localize adjacent to the PML bodies early in infection (49) and at least three viral proteins, E1A-13S (50), E4orf3 (51), and E1B55K (52) specifically interact with them. To address this question, colocalization of E1B55K with PML bodies was assessed in H1299 cells transfected with plasmid DNAs expressing HA-E1B55K proteins of various serotypes as in Fig. 1A by IF microscopy using antibodies against either HA or PML. As seen in Fig. 2A, the pattern of E1B55K from the various serotypes was similar to that shown in Fig. 1a; however, a clear colocalization of E1B55K and PML was seen for Ad4, Ad9, Ad16, Ad34, and Ad40. Unlike these serotypes the E1B55K proteins of Ad5 and Ad12 are not concentrated in the nucleus and thus colocalization was somewhat more difficult to determine. Nevertheless, the few E1B55K dots seen for Ad12 also colocalized with PML. To enhance the presence of Ad5 and Ad12 E1B55K in the nucleus, cells expressing HA-E1B55K of these serotypes were treated with leptomycin B (LMB) for 6 h before fixing. As shown previously, this treatment induces nuclear relocalization of Ad5 E1B55K and, as seen in Fig. 2Ba to c, a clear colocalization with PML was evident (52). LMB treatment also increased the level of Ad12 E1B55K in the nucleus and in some cells faint nuclear dots were present that also colocalized with PML (Fig. 2Bd to f). Ad5 E1B55K was shown previously to coimmunoprecipitate with PML (52). To determine if this interaction is conserved among adenovirus species, as suggested by the findings in Fig. 1, immunoprecipitates were prepared using anti-PML antibodies and the presence of HA-E1B55K was examined by Western blotting with anti-HA antibodies. Figure 2C shows that with all serotypes E1B55K interacted with PML.
Fig 2.
E1B55K of all serotypes colocalize and bind PML. H1299 cells were transfected with plasmid DNAs expressing HA-tagged E1B55K of different serotypes. (A) Transfected cells were fixed in paraformaldehyde and stained for HA, PML, and DNA (with DAPI). (B) Transfected cells were treated for the last 6 h with LMB, fixed in paraformaldehyde and stained for HA, PML, and DNA (with DAPI). (C) Transfected cells were lysed and immunoprecipitated with antibodies against PML and immunoblotted with antibodies against HA. Whole-cell lysates were also immunoblotted with HA and actin antibodies. Panels are as indicated in the figure.
Aggresome formation by E1B55K is not conserved in all adenovirus species.
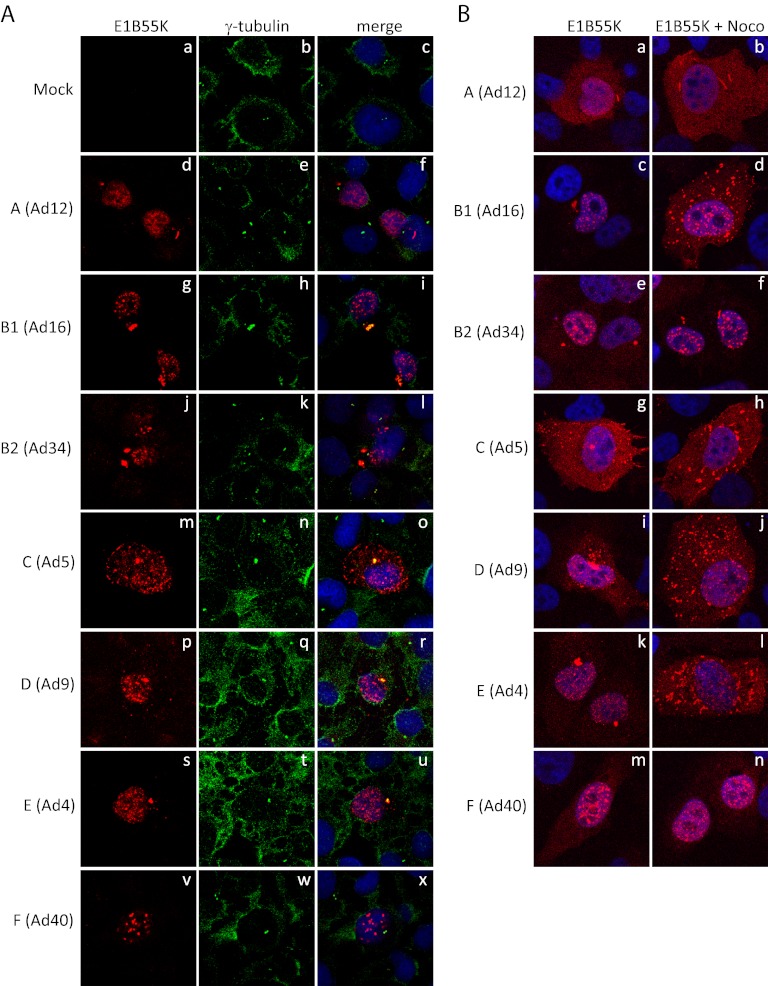
It was reported previously that Ad5 E1B55K assembles in an aggresome structure at the periphery of the nucleus (32). Major hallmarks of aggresomes are their location at or near microtubule organizing centers (MTOC), and the requirement of retrograde transport for their assembly (34, 35). To determine whether localization of E1B55K in aggresome structures is conserved with all serotypes, coimmunofluorescence studies of E1B55K proteins with the MTOC marker γ-tubulin were performed. H1299 cells were transfected with E1B55K-expressing plasmid DNAs as in Fig. 1A, they were fixed with methanol and stained with both anti-HA and anti-γ-tubulin antibodies. As seen in Fig. 3A, a colocalization between γ-tubulin and E1B55K from Ad4, Ad5, Ad9, and Ad16 was evident; however, the oval-shaped aggresome-like structures formed by Ad12 E1B55K did not colocalize with γ-tubulin, and in the case of the few cells that contain aggresome-like structures formed by Ad34 E1B55K, no colocalization with γ-tubulin was evident. No such structures were formed by Ad40 E1B55K. (Note that localization of γ-tubulin should be primarily at the MTOC in interphase cells, as we have observed in our studies; however, with the relatively weak signal obtained using the present antibody, an increased background was present in the image.)
Fig 3.
Formation of the aggresome structure is not conserved among serotypes. H1299 cells were transfected with plasmid DNAs expressing HA-tagged E1B55K of different serotypes. (A) Transfected cells were fixed in methanol and stained for HA, γ-tubulin, and DNA (with DAPI). (B) Transfected cells were treated for the last 6 h with nocodazole, fixed in paraformaldehyde, and stained for HA and DNA (with DAPI). Panels are as indicated in the figure.
To assemble into aggresomes, it is known that smaller aggregates utilize the dynein-dependent retrograde transport on microtubules (34, 35) and such transport is inhibited by nocodazole, which interferes with the polymerization of microtubules (34, 35). Previous studies have already shown that nocodazole treatment of Ad5 E1B55K-expressing cells disrupts the formation of aggresome structures (21, 32). To confirm that the aggresome-like structures formed by the other serotypes are indeed aggresomes, we looked at the pattern of E1B55K localization following nocodazole treatment. HA-E1B55K-expressing H1299 cells, as in Fig. 1A, were treated for the last 6 h before fixation for IF. As shown in Fig. 3B, treatment with nocodazole resulted in the dispersion of the aggresome structures of Ad4, Ad5, Ad9, and Ad16 into small aggregates throughout the cytoplasm; however, nocodazole treatment had no effect on the structures for Ad12, Ad34, and Ad40. In total, these studies suggested that E1B55K from several, but not all species are present in aggresomes.
E1B55K localizes in close proximity to the ERC.
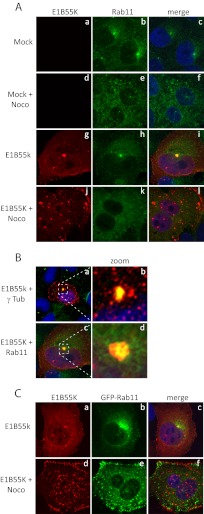
Certain other structures are also known to localize near the MTOC, one of these being the endosomal recycling compartment (ERC) (53). The ERC was of interest to us for two reasons. First, the formation of ERCs involves the same retrograde transport system used in the formation of aggresomes (54, 55). Second, one of the targets of the E4orf6/E1B55K ligase of some adenovirus species is a membrane protein, integrin α3 (56), making it possible that the ligase may interact with elements of the ERC. To determine whether E1B55K also colocalizes in the ERC, colocalization studies were conducted using the known ERC marker, Rab11 (57–59). Since both the aggresomes and ERCs are known to localize near the MTOC, E1B55K from a representative serotype that forms an aggresome, Ad5, was chosen for these experiments to provide at least an initial insight into this question. Control H1299 cells and those expressing Ad5 HA-E1B55K were fixed with paraformaldehyde and stained with antibodies recognizing E1B55K and endogenous Rab11. As seen in Fig. 4A, and as expected, Rab11 was localized largely in a dot-like structure near the nucleus (panels a to c). As expected (53, 60), treatment with nocodazole for the last 6 h before fixing resulted in the loss of these structures (Fig. 4Ad to f). Colocalization of Rab11 and E1B55K at the dot near the nucleus was clearly apparent (Fig. 4Ag to i), and this dot was absent following treatment with nocodazole (Fig. 4Aj to l). The image present in Fig. 4Ai is also shown in Fig. 4Bc with the small area denoted by the dashed square enlarged to show the colocalization in more detail. Figure 4Bd indicates clearly that E1B55K colocalized with the most intense part of the Rab11 signal but that Rab11 was present more diffusely in the surrounding area as well. Similarly, the image present Fig. 3Ao is also shown in Fig. 4Ba with the small area denoted by the dashed square enlarged. It shows that E1B55K strongly colocalizes with γ-tubulin but is also present as well in the immediate surrounding area. A similar colocalization between Ad5 E1B55K and Rab11 was also seen in cells expressing high levels of exogenous GFP-Rab11 (Fig. 4Ca to c). In this study, it was also possible to see that treatment with nocodazole disrupted the ERC, but several smaller dots of GFP-Rab11 were evident throughout the cytoplasm, mostly concentrated near the plasma membrane (Fig. 4Ce). E1B55K also was seen to collect in several cytoplasmic dots that did not overlap with the GFP-Rab11 dots but were quite close to the plasma membrane (Fig. 4Cd to f). At present, it is still unclear how Ad5 E1B55K at least partially colocalizes with the ERC, since we have been unable to demonstrate that E1B55K coimmunoprecipitates with Rab11 (data not shown); however, in addition to Rab11, several other proteins are present in ERC, and one of these proteins may bind E1B55K and target it to the ERC. Thus, further studies are planned, including those with other serotypes, to examine this possibility.
Fig 4.
Ad5 E1B55K colocalizes with the endosomal recycling compartment (ERC). H1299 cells were transfected with plasmid DNAs expressing HA-tagged E1B55K of different serotypes (A) or both E1B55K and GFP-Rab11 (C), fixed in paraformaldehyde and stained for HA, Rab11, and DNA (with DAPI) (A) or for HA and DNA (with DAPI) (C). (B) Sections from the merged panels of Fig. 4Ai and Fig. 3Ao (indicated by a dashed square in panels a and c) are magnified in panels b and d. Panels a and b show E1B55K in red, γ tubulin in green, and DAPI in blue. Panels c and d show E1B55K in red, Rab11 in green, and DAPI in blue.
DISCUSSION
The majority of studies on human adenoviruses have been performed using two highly related serotypes of the C species, Ad2 and Ad5, although a limited number have used Ad12, a member of the A species. It is only recently that researchers have taken an interest in serotypes representing all or several species. Our recent study on the E4orf6 protein indicated that significant functional differences exist between the serotypes (11). Another study on E4orf3 also showed significant variability among different serotypes in its subcellular localization and influence on the PML-nuclear bodies (61). The results of the present study suggest that such is also the case with E1B55K. The subcellular localization of Ad5 E1B55K when expressed alone or in E1A/E1B-transformed cells has been reported as mostly cytoplasmic with often the formation of an aggresome (21, 30–32); however, neither of these characteristics was conserved among the different serotypes representing six of the seven adenovirus species (we have not yet studied Ad52, the only serotype in species G). In fact, the majority displayed a nuclear E1B55K localization with intense nuclear dots or spheres. In this respect, E1B55K of both Ad5 and Ad12 differed, with Ad12 showing a mostly whole-cell localization with sometimes the presence of oval-shaped structures. The subcellular localization of a shuttling protein normally depends on the net rate difference between import and export. In the case of E1B55K a nuclear export signal (NES) has been identified (26), but not a nuclear localization signal (NLS) (26, 33). Figure 5 shows that the NES is well-conserved among serotypes except for Ad12 and Ad40; however, this dissimilarity alone cannot explain the differences in localization as Ad12 and Ad40 E1B55K proteins localize very differently. No obvious NLS motifs are evident in E1B55K of any serotype, so it probably must enter the nucleus by binding to another nuclear protein. In the case of Ad5, clearly E4orf6 as well as E4orf3 (62) can serve that function, but possibly some cellular protein may be involved with the E1B55K proteins of serotypes showing a nuclear localization when expressed alone.
Fig 5.
Partial alignment of E1B55K from different serotypes. A proportion of the sequences of E1B55K of different serotypes were aligned using the CLUSTAL W program. The portions contain the region containing the nuclear export sequence (NES) and SUMO conjugation motif (SCM).
In almost all cases, we found that the expression of E4orf6 resulted in the same effect: the presence of E1B55K spread evenly throughout the nucleus. In the case of Ad5 the presence of E4orf6 clearly resulted in a major relocalization of E1B55K from the cytoplasm to the nucleus. For most of the other serotypes the presence of E4orf6 resulted in the disappearance of E1B55K in the brightly staining nuclear dots. The exception in this respect was Ad12 for which expression of E4orf6 did not appear to have any significant effect. The lack of relocalization of Ad12 E1B55K by E4orf6 could be partly due to the observation that they seem to bind with somewhat less affinity than do the Ad5 proteins (data not shown). We have also noted that the levels of expression of Ad12 E4orf6 are usually somewhat lower than with most other serotypes and thus perhaps less E1B55K may be retained in the E4orf6 complex. However, other explanations may be more likely. Ad12 E1B55K localization is known to be largely nuclear during infection (63), thus making it possible that a viral protein other than E4orf6 may be responsible in the case of Ad12 for relocalization of E1B55K to the nucleus. In this context, Ad5 E1B55K is known to interact with the E4orf3 protein (62), and this protein might serve such a role more predominantly with Ad12.
Ad5 E1B55K was shown to coimmunoprecipitate with PML and to colocalize with PML in nuclear foci, at least when concentrated in the nucleus (52). This aspect of Ad5 E1B55K was found to be fully conserved in all of the serotypes tested. Interestingly, although previous studies indicated that SUMOylation of Ad5 E1B55K was not necessary for coimmunoprecipitation with PML, it was shown to be necessary for colocalization with PML in nuclear dots (52). This situation is probably different with at least some of the other serotypes, in particular with Ad40. Figure 5 shows that E1B55K protein from this serotype, as well as that of Ad12, does not contain a conserved SUMO conjugation motif (SCM). However, analysis by immunofluorescence indicated that Ad40 E1B55K clearly forms bright nuclear foci that colocalize with PML, demonstrating that at least with this serotype, conjugation by SUMO may not be required for colocalization with PML bodies.
As shown previously, Ad5 E1B55K protein exists in an aggresome (32). It has also been shown in general that aggresomes also recruit chaperone proteins (34, 64) and proteasomal components (34, 65, 66), presumably to help the clearance of aggregated proteins (36). It is possible that E1B55K has a tendency to misfold and thus aggregates and is transported to the aggresome and the presence of an unoccupied interaction site for E4orf6 could participate in this process. However, evidence exists to suggest that E1B55K is largely folded correctly and functional (32), in part because it is known to be able to recruit p53 and Mre11 to aggresomes (31, 32; Cheng et al., unpublished), and the aggresome can still form in the presence of E4orf6 in both cells expressing these proteins from transfected plasmid DNAs, as shown in Fig. 1B, and cells infected with wild-type virus (21, 32; unpublished observations). Thus, E1B55K may participate actively rather than passively in the aggresome formation process. This possibility combined with the presence of a high concentration of proteasomes at the MTOC and the localization of E4orf6 and Cul5 with E1B55K in the aggresome in 293 cells transfected with plasmid DNAs expressing E4orf6 (32) has led some to conclude that substrates of the E4orf6/E1B55K ligase complex are first brought to the aggresome before being degraded (67), although this idea was not a conclusion drawn in the original study. However, our results indicated that the formation of an aggresome is not conserved in three of the seven serotypes studied. In both Ad12 and Ad34, what were thought to be “aggresome-like” structures do not exhibit the known characteristics of aggresomes and in fact Ad40 does not form such structures at all. In both Ad12 and Ad34, the structures are not formed near the microtubule organizing center (MTOC) (Fig. 3) (40), and in the case of Ad12 they are not even necessarily near the nucleus. In addition, inhibition of microtubule polymerization by addition of nocodazole did not disrupt the formation of these structures, as should be the case with aggresomes. Interestingly, both Ad12 and Ad40 E1B55K/E4orf6 ligase complexes were shown to degrade p53 and Mre11 as efficiently as Ad5 (11). Thus, these results suggest that although some degradation of substrates of the viral E3 ligase complex probably takes place in aggresomes due to close proximity to an abundance of the ubiquitin machinery (32), this pathway is clearly not necessary for efficient degradation of substrates. This finding further suggests that it is unlikely that efficient protein degradation alone is the reason for the evolution of the ability of E1B55K to actively form an aggresome.
The MTOC is also the location of another structure, the endosomal recycling compartment (ERC), at least in several cell types (53). The ERC is a component of the membrane protein (and some lipid) recycling pathway (reviewed in references 68, 69, and 70). Briefly, after internalization, membrane proteins are delivered to early endosomes, also called sorting endosomes. Proteins destined for degradation are then transported to late endosome structures, but the majority of the cargos are returned to the plasma membrane in one of two pathways. In the short pathway, they are directly returned to the plasma membrane from the sorting endosomes. In the long pathway, they are first moved to the ERC (sometimes also called perinuclear recycling compartment), before returning to the plasma membrane. A major marker of the ERC is Rab11 (71), which with one of its interacting species, Rab11-FIP3, binds to the dynein motor complex to deliver the ERC to the MTOC area (54). Blocking the retrograde transport with nocodazole causes dispersion of the ERC, although it does not affect the kinetics of recycling (55). Rab11 also controls the return of proteins from the ERC to the plasma membrane (70). In the present studies, we have shown that E1B55K localizes quite close to a marker of the ERC, Rab11. It is not surprising that E1B55K has both the hallmarks of an aggresome and colocalizes with the ERC since both structures use the same retrograde transport mechanism to accumulate at the MTOC region. There is good evidence that E1B55K accumulated in this region is still functional and thus not misfolded, making it unlikely that the usual pathway that depends on misfolded proteins is used for assembly of the E1B55K aggresome. Thus, E1B55K may bind directly to proteins involved in retrograde transport or use the recycling pathway. In either case the final localization should be the same. The presence of E1B55K could have consequences in terms of the normal process of membrane protein recycling. In a screen for E4orf6/E1B55K ligase complex substrates, we showed that integrin α3 is degraded in response to the E4orf6/E1B55K ligase (56). Although we could detect efficient colocalization of E1B55K and integrin α3 in the aggresome structure (56), we have never been able to coimmunoprecipitate these two proteins. Of some interest, in recent continuing studies we have found that the levels of several integrin species were reduced at the cell surface following expression of Ad5 E4orf6 and E1B55K (F. Dallaire et al., unpublished results). Although the E1B55K-containing structure has all the hallmarks of an aggresome, it may use the recycling pathway to accumulate at the MTOC, and in the process disrupt normal protein recycling. This process might clarify how E1B55K can accumulate in this area of the cell while not actually being misfolded and thus inactive. It should be noted, however, that in a EM micrograph of E1B55K in the juxtanuclear structure, lipid bilayers were not readily observable (31), although the existence of such structures was not specifically addressed in a comprehensive way in this study. Thus, whereas Ad5 E1B55K and Rab11 may localize in the same general juxtanuclear region driven through a common transport system, the former seems unlikely to be present in or on membranous structures. Continuing research is currently being actively pursued on these issues.
ACKNOWLEDGMENTS
We thank Morag Park for supplying the GFP-Rab11 plasmid produced by Stephen Ferguson (38). We thank Sabrina Schreiner for critical reading of the manuscript.
Image collection for this manuscript was performed in the McGill University Life Sciences Complex Imaging Facility. Purchase of equipment in the facility was made possible with funding from the Canadian Foundation for Innovation (CFI) and the Ministère du Développement économique, innovation et exportation Québec (MDEIE).
This study was supported by a grant from the Canadian Institutes of Health Research (P.E.B. and P.B.).
Footnotes
Published ahead of print 13 February 2013
REFERENCES
- 1. Babiss LE, Ginsberg HS, Darnell JE., Jr 1985. Adenovirus E1B proteins are required for accumulation of late viral mRNA and for effects on cellular mRNA translation and transport. Mol. Cell. Biol. 5:2552–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gonzalez RA, Flint SJ. 2002. Effects of mutations in the adenoviral E1B 55-kilodalton protein coding sequence on viral late mRNA metabolism. J. Virol. 76:4507–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Halbert DN, Cutt JR, Shenk T. 1985. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J. Virol. 56:250–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Harada JN, Shevchenko A, Pallas DC, Berk AJ. 2002. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 76:9194–9206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pilder S, Moore M, Logan J, Shenk T. 1986. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol. Cell. Biol. 6:470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Querido E, Blanchette P, Yan Q, Kamura T, Morrison M, Boivin D, Kaelin WG, Conaway RC, Conaway JW, Branton PE. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sarnow P, Sullivan CA, Levine AJ. 1982. A monoclonal antibody detecting the adenovirus type 5-E1b-58Kd tumor antigen: characterization of the E1b-58Kd tumor antigen in adenovirus-infected and -transformed cells. Virology 120:510–517 [DOI] [PubMed] [Google Scholar]
- 8. Weinberg DH, Ketner G. 1986. Adenoviral early region 4 is required for efficient viral DNA replication and for late gene expression. J. Virol. 57:833–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blanchette P, Branton PE. 2009. Manipulation of the ubiquitin-proteasome pathway by small DNA tumor viruses. Virology 384:317–323 [DOI] [PubMed] [Google Scholar]
- 10. Blanchette P, Cheng CY, Yan Q, Ketner G, Ornelles DA, Dobner T, Conaway RC, Conaway JW, Branton PE. 2004. Both BC-box motifs of adenovirus protein E4orf6 are required to efficiently assemble an E3 ligase complex that degrades p53. Mol. Cell. Biol. 24:9619–9629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheng CY, Gilson T, Dallaire F, Ketner G, Branton PE, Blanchette P. 2011. The E4orf6/E1B55K E3 ubiquitin ligase complexes of human adenoviruses exhibit heterogeneity in composition and substrate specificity. J. Virol. 85:765–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schreiner S, Wimmer P, Sirma H, Everett RD, Blanchette P, Groitl P, Dobner T. 2010. Proteasome-dependent degradation of Daxx by the viral E1B-55K protein in human adenovirus-infected cells. J. Virol. 84:7029–7038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cathomen T, Weitzman MD. 2000. A functional complex of adenovirus proteins E1B-55kDa and E4orf6 is necessary to modulate the expression level of p53 but not its transcriptional activity. J. Virol. 74:11407–11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Endter C, Kzhyshkowska J, Stauber R, Dobner T. 2001. SUMO-1 modification required for transformation by adenovirus type 5 early region 1B 55-kDa oncoprotein. Proc. Natl. Acad. Sci. U. S. A. 98:11312–11317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yew PR, Berk AJ. 1992. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature 357:82–85 [DOI] [PubMed] [Google Scholar]
- 16. Yew PR, Liu X, Berk AJ. 1994. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 8:190–202 [DOI] [PubMed] [Google Scholar]
- 17. Hartl B, Zeller T, Blanchette P, Kremmer E, Dobner T. 2008. Adenovirus type 5 early region 1B 55-kDa oncoprotein can promote cell transformation by a mechanism independent from blocking p53-activated transcription. Oncogene 27:3673–3684 [DOI] [PubMed] [Google Scholar]
- 18. Schreiner S, Wimmer P, Groitl P, Chen SY, Blanchette P, Branton PE, Dobner T. 2011. Adenovirus type 5 early region 1B 55K oncoprotein-dependent degradation of cellular factor Daxx is required for efficient transformation of primary rodent cells. J. Virol. 85:8752–8765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sieber T, Dobner T. 2007. Adenovirus type 5 early region 1B 156R protein promotes cell transformation independently of repression of p53-stimulated transcription. J. Virol. 81:95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Endter C, Hartl B, Spruss T, Hauber J, Dobner T. 2005. Blockage of CRM1-dependent nuclear export of the adenovirus type 5 early region 1B 55-kDa protein augments oncogenic transformation of primary rat cells. Oncogene 24:55–64 [DOI] [PubMed] [Google Scholar]
- 21. Kindsmuller K, Groitl P, Hartl B, Blanchette P, Hauber J, Dobner T. 2007. Intranuclear targeting and nuclear export of the adenovirus E1B-55K protein are regulated by SUMO1 conjugation. Proc. Natl. Acad. Sci. U. S. A. 104:6684–6689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ching W, Dobner T, Koyuncu E. 2012. The human adenovirus type 5 E1B 55-kilodalton protein is phosphorylated by protein kinase CK2. J. Virol. 86:2400–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Teodoro JG, Branton PE. 1997. Regulation of p53-dependent apoptosis, transcriptional repression, and cell transformation by phosphorylation of the 55-kilodalton E1B protein of human adenovirus type 5. J. Virol. 71:3620–3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Teodoro JG, Halliday T, Whalen SG, Takayesu D, Graham FL, Branton PE. 1994. Phosphorylation at the carboxy terminus of the 55-kilodalton adenovirus type 5 E1B protein regulates transforming activity. J. Virol. 68:776–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wimmer P, Blanchette P, Schreiner S, Ching W, Groitl P, Berscheminski J, Branton PE, Will H, Dobner T. 21 May 2012. Cross-talk between phosphorylation and SUMOylation regulates transforming activities of an adenoviral oncoprotein. Oncogene. doi:10.1038/onc.2012.187 [DOI] [PubMed] [Google Scholar]
- 26. Dobbelstein M, Roth J, Kimberly WT, Levine AJ, Shenk T. 1997. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a Rev-like signal sequence. EMBO J. 16:4276–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dosch T, Horn F, Schneider G, Kratzer F, Dobner T, Hauber J, Stauber RH. 2001. The adenovirus type 5 E1B-55K oncoprotein actively shuttles in virus-infected cells, whereas transport of E4orf6 is mediated by a CRM1-independent mechanism. J. Virol. 75:5677–5683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kratzer F, Rosorius O, Heger P, Hirschmann N, Dobner T, Hauber J, Stauber RH. 2000. The adenovirus type 5 E1B-55K oncoprotein is a highly active shuttle protein and shuttling is independent of E4orf6, p53, and Mdm2. Oncogene 19:850–857 [DOI] [PubMed] [Google Scholar]
- 29. Rowe DT, Graham FL, Branton PE. 1983. Intracellular localization of adenovirus type 5 tumor antigens in productively infected cells. Virology 129:456–468 [DOI] [PubMed] [Google Scholar]
- 30. Brown CR, Doxsey SJ, White E, Welch WJ. 1994. Both viral (adenovirus E1B) and cellular (hsp70, p53) components interact with centrosomes. J. Cell Physiol. 160:47–60 [DOI] [PubMed] [Google Scholar]
- 31. Zantema A, Fransen JA, Davis-Olivier A, Ramaekers FC, Vooijs GP, DeLeys B, Van der Eb AJ. 1985. Localization of the E1B proteins of adenovirus 5 in transformed cells, as revealed by interaction with monoclonal antibodies. Virology 142:44–58 [DOI] [PubMed] [Google Scholar]
- 32. Liu Y, Shevchenko A, Berk AJ. 2005. Adenovirus exploits the cellular aggresome response to accelerate inactivation of the MRN complex. J. Virol. 79:14004–14016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Goodrum FD, Shenk T, Ornelles DA. 1996. Adenovirus early region 4 34-kilodalton protein directs the nuclear localization of the early region 1B 55-kilodalton protein in primate cells. J. Virol. 70:6323–6335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Garcia-Mata R, Bebok Z, Sorscher EJ, Sztul ES. 1999. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J. Cell Biol. 146:1239–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Johnston JA, Ward CL, Kopito RR. 1998. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 143:1883–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Garcia-Mata R, Gao YS, Sztul E. 2002. Hassles with taking out the garbage: aggravating aggresomes. Traffic 3:388–396 [DOI] [PubMed] [Google Scholar]
- 37. Mitsudomi T, Steinberg SM, Nau MM, Carbone D, D'Amico D, Bodner S, Oie HK, Linnoila RI, Mulshine JL, Minna JD. 1992. p53 gene mutations in non-small-cell lung cancer cell lines and their correlation with the presence of ras mutations and clinical features. Oncogene 7:171–180 [PubMed] [Google Scholar]
- 38. Dale LB, Seachrist JL, Babwah AV, Ferguson SS. 2004. Regulation of angiotensin II type 1A receptor intracellular retention, degradation, and recycling by Rab5, Rab7, and Rab11 GTPases. J. Biol. Chem. 279:13110–13118 [DOI] [PubMed] [Google Scholar]
- 39. Wienzek S, Roth J, Dobbelstein M. 2000. E1B 55-kilodalton oncoproteins of adenovirus types 5 and 12 inactivate and relocalize p53, but not p51 or p73, and cooperate with E4orf6 proteins to destabilize p53. J. Virol. 74:193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao LY, Liao D. 2003. Sequestration of p53 in the cytoplasm by adenovirus type 12 E1B 55-kilodalton oncoprotein is required for inhibition of p53-mediated apoptosis. J. Virol. 77:13171–13181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Grand RJ, Parkhill J, Szestak T, Rookes SM, Roberts S, Gallimore PH. 1999. Definition of a major p53 binding site on Ad2E1B58K protein and a possible nuclear localization signal on the Ad12E1B54K protein. Oncogene 18:955–965 [DOI] [PubMed] [Google Scholar]
- 42. Dyck JA, Maul GG, Miller WH, Jr, Chen JD, Kakizuka A, Evans RM. 1994. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell 76:333–343 [DOI] [PubMed] [Google Scholar]
- 43. Koken MH, Puvion-Dutilleul F, Guillemin MC, Viron A, Linares-Cruz G, Stuurman N, de Jong L, Szostecki C, Calvo F, Chomienne C. 1994. The t(15;17) translocation alters a nuclear body in a retinoic acid-reversible fashion. EMBO J. 13:1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. 1994. Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell 76:345–356 [DOI] [PubMed] [Google Scholar]
- 45. Negorev D, Maul GG. 2001. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene 20:7234–7242 [DOI] [PubMed] [Google Scholar]
- 46. Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh ET, Strauss JF, III, Maul GG. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhong S, Muller S, Ronchetti S, Freemont PS, Dejean A, Pandolfi PP. 2000. Role of SUMO-1-modified PML in nuclear body formation. Blood 95:2748–2752 [PubMed] [Google Scholar]
- 48. Borden KL. 2002. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol. Cell. Biol. 22:5259–5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ishov AM, Maul GG. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134:815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Berscheminski J, Groitl P, Dobner T, Wimmer P, Schreiner S. 2013. The adenoviral oncogene E1A-13S interacts with a specific isoform of the tumor suppressor PML to enhance viral transcription. J. Virol. 87:965–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hoppe A, Beech SJ, Dimmock J, Leppard KN. 2006. Interaction of the adenovirus type 5 E4 Orf3 protein with promyelocytic leukemia protein isoform II is required for ND10 disruption. J. Virol. 80:3042–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wimmer P, Schreiner S, Everett RD, Sirma H, Groitl P, Dobner T. 2010. SUMO modification of E1B-55K oncoprotein regulates isoform-specific binding to the tumour suppressor protein PML. Oncogene 29:5511–5522 [DOI] [PubMed] [Google Scholar]
- 53. Lin SX, Gundersen GG, Maxfield FR. 2002. Export from pericentriolar endocytic recycling compartment to cell surface depends on stable, detyrosinated (glu) microtubules and kinesin. Mol. Biol. Cell 13:96–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Horgan CP, Hanscom SR, Jolly RS, Futter CE, McCaffrey MW. 2010. Rab11-FIP3 links the Rab11 GTPase and cytoplasmic dynein to mediate transport to the endosomal-recycling compartment. J. Cell Sci. 123:181–191 [DOI] [PubMed] [Google Scholar]
- 55. McGraw TE, Dunn KW, Maxfield FR. 1993. Isolation of a temperature-sensitive variant Chinese hamster ovary cell line with a morphologically altered endocytic recycling compartment. J. Cell Physiol. 155:579–594 [DOI] [PubMed] [Google Scholar]
- 56. Dallaire F, Blanchette P, Groitl P, Dobner T, Branton PE. 2009. Identification of integrin alpha3 as a new substrate of the adenovirus E4orf6/E1B 55-kilodalton E3 ubiquitin ligase complex. J. Virol. 83:5329–5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Casanova JE, Wang X, Kumar R, Bhartur SG, Navarre J, Woodrum JE, Altschuler Y, Ray GS, Goldenring JR. 1999. Association of Rab25 and Rab11a with the apical recycling system of polarized Madin-Darby canine kidney cells. Mol. Biol. Cell 10:47–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schlierf B, Fey GH, Hauber J, Hocke GM, Rosorius O. 2000. Rab11b is essential for recycling of transferrin to the plasma membrane. Exp. Cell Res. 259:257–265 [DOI] [PubMed] [Google Scholar]
- 59. Ullrich O, Reinsch S, Urbe S, Zerial M, Parton RG. 1996. Rab11 regulates recycling through the pericentriolar recycling endosome. J. Cell Biol. 135:913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lapierre LA, Dorn MC, Zimmerman CF, Navarre J, Burnette JO, Goldenring JR. 2003. Rab11b resides in a vesicular compartment distinct from Rab11a in parietal cells and other epithelial cells. Exp. Cell Res. 290:322–331 [DOI] [PubMed] [Google Scholar]
- 61. Stracker TH, Lee DV, Carson CT, Araujo FD, Ornelles DA, Weitzman MD. 2005. Serotype-specific reorganization of the Mre11 complex by adenoviral E4orf3 proteins. J. Virol. 79:6664–6673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Leppard KN, Everett RD. 1999. The adenovirus type 5 E1b 55K and E4 Orf3 proteins associate in infected cells and affect ND10 components. J. Gen. Virol. 80(Pt 4):997–1008 [DOI] [PubMed] [Google Scholar]
- 63. Schughart K, Bause E, Esche H. 1985. Structure and expression of adenovirus type 12 E1B 58K protein in infected and transformed cells: studies using antibodies directed against a synthetic peptide. Virus Res. 3:41–56 [DOI] [PubMed] [Google Scholar]
- 64. Wigley WC, Fabunmi RP, Lee MG, Marino CR, Muallem S, DeMartino GN, Thomas PJ. 1999. Dynamic association of proteasomal machinery with the centrosome. J. Cell Biol. 145:481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fabunmi RP, Wigley WC, Thomas PJ, DeMartino GN. 2000. Activity and regulation of the centrosome-associated proteasome. J. Biol. Chem. 275:409–413 [DOI] [PubMed] [Google Scholar]
- 66. Lee HJ, Shin SY, Choi C, Lee YH, Lee SJ. 2002. Formation and removal of alpha-synuclein aggregates in cells exposed to mitochondrial inhibitors. J. Biol. Chem. 277:5411–5417 [DOI] [PubMed] [Google Scholar]
- 67. Fleisig HB, Liang H, Nagarajan L. 2010. Adenoviral oncoprotein E1B55K mediates colocalization of SSBP2 and PML in response to stress. J. Mol. Signal. 5:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Caswell PT, Norman JC. 2006. Integrin trafficking and the control of cell migration. Traffic 7:14–21 [DOI] [PubMed] [Google Scholar]
- 69. Caswell PT, Vadrevu S, Norman JC. 2009. Integrins: masters and slaves of endocytic transport. Nat. Rev. Mol. Cell. Biol. 10:843–853 [DOI] [PubMed] [Google Scholar]
- 70. Maxfield FR, McGraw TE. 2004. Endocytic recycling. Nat. Rev. Mol. Cell. Biol. 5:121–132 [DOI] [PubMed] [Google Scholar]
- 71. Grant BD, Donaldson JG. 2009. Pathways and mechanisms of endocytic recycling. Nat. Rev. Mol. Cell. Biol. 10:597–608 [DOI] [PMC free article] [PubMed] [Google Scholar]