Abstract
Phages are recognized as the most abundant and diverse entities on the planet. Their diversity is determined predominantly by their dynamic adaptation capacities when confronted with different selective pressures in an endless cycle of coevolution with a widespread group of bacterial hosts. At the end of the infection cycle, progeny virions are confronted with a rigid cell wall that hinders their release into the environment and the opportunity to start a new infection cycle. Consequently, phages encode hydrolytic enzymes, called endolysins, to digest the peptidoglycan. In this work, we bring to light all phage endolysins found in completely sequenced double-stranded nucleic acid phage genomes and uncover clues that explain the phage-endolysin-host ecology that led phages to recruit unique and specialized endolysins.
INTRODUCTION
Bacteriophages (phages) are the most abundant living entities on Earth and can be found in every conceivable habitat. Phages outnumber bacteria by an estimated 10-fold, accounting for a total population size of 1031 phage particles able to infect 108 species (1–4). As natural predators of bacteria, these viruses can kill 50% of the bacteria produced every day, information which demonstrates their ecological importance and impact on bacterial evolution (4, 5).
Phages are tremendously diversified, with genome sizes from as low as 17 kbp up to 0.5 Mbp, and constitute a reservoir of great genetic diversity, which is supported by the high frequency of novel genes found in newly characterized phage genomes. The percentage of genes carried in a newly sequenced genome that present homology to genes deposited in existing databases is usually low, in the range of 20% to 40% (5, 6). This high diversification in the phage population can be attributed to the high number of different bacterial hosts available for phage infection and the ability of phages to rapidly evolve in order to circumvent bacterial resistance mechanisms (7, 8). Even so, phages able to infect a common host may exhibit great diversity, with little or no identifiable nucleotide sequence similarity between them (9). The phage genomes are typically mosaic, with individual genes, or groups of genes, shared among unrelated genomes as a result of horizontal and vertical gene transfer during phage evolution, particularly as a result of recombination events between temperate phages (10–13).
The morphology of the phage particles and the type of nucleic acid form the basis of their classification, a responsibility attributed to the International Committee on the Taxonomy of Viruses (ICTV). However, ICTV's classification method is now being rectified to integrate genomic and proteomic homology (14). The major order of phages is the Caudovirales, with 96% of all reported phages (15). Caudovirales virions possess double-stranded DNA (dsDNA) genomes enclosed in heads with icosahedral symmetry and with tails that vary in length. These tailed phages are subdivided into three families: Siphoviridae (representing 61%), with long, flexible, and noncontractile tails, Myoviridae (representing 25%), having long, rigid, and contractile tails, and Podoviridae (representing 14%), with short and noncontractile tails. The remaining nontailed phages belong to a small and highly variable group, with single-stranded DNA (ssDNA), ssRNA, or dsRNA (3).
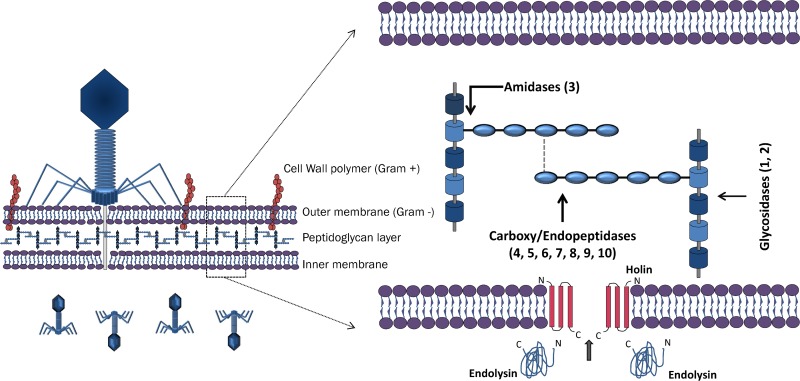
Phage replication demands a strategy for progeny release and dispersion to enable infection of new hosts. With the exception of the few known filamentous phages, which continuously extrude their progeny across the cell wall without killing their host cells, all phages lyse their bacterial hosts by compromising the structural integrity of the peptidoglycan (PG) layer (16–18). PG (or murein), a heteropolymer consisting of glycan strands cross-linked by peptides, forms an exoskeleton that withstands the internal cytoplasmic osmotic pressure, providing structural integrity to the cell (19–21). Consequently, compromising the PG will lead to an unsustainable internal osmotic pressure, ultimately resulting in the burst of the cell and progeny release. All double-stranded nucleic acid (dsNA) genome phages degrade the PG through an essential genome-encoded enzyme—the endolysin (17, 18). The muralytic activity of endolysins allows their classification according to the bond of the PG on which the enzymes act. At least four types of endolysins have already been identified: (i) lysozymes and (ii) transglycosidases act on the glycosidic bond that links the amino sugars in the cell wall, and (iii) amidases and (iv) endopeptidases act on the amide and peptide bonds of the cross-linking oligopeptide stems and interpeptide bridges (Fig. 1) (22–24).
Fig 1.
Schematic representation of how phage endolysins gain access to the PG through the most common holin-endolysin lytic system. A generalized PG structure illustrates all cleavage sites found: 1, N-acetyl-β-d-muramidase (LYSO, MURA, SLT, TRANG, GH19, GH25, and GH108); 2, N-acetyl-β-d-glucosaminidase (GH19 and GLUCO); 3, N-acetylmuramoyl-l-alanine amidase (AMI-2, AMI-3, AMI02-C, and CHAP); 4, l-alanoyl-d-glutamate (LD) endopeptidase (VANY); 5, c-d-glutamyl-m-diaminopimelic acid (DL) peptidase (NLPC-P60 and PET-C39-2); 6, d-Ala-m-DAP (DD) endopeptidase (PET-M23 and PET-M15-4); 7, m-DAP-m-DAP (LD) endopeptidase (PET-M23 and PET-M15-4); 8, d-alanyl-glycyl endopeptidase (CHAP); 9, d-alanyl-d-alanine peptidase (CHAP); 10, m-diaminopimelic acid-d-alanine (PET-U40). PET-M15-3, YKUD, and NLPD are peptidases with an unknown cleavage site.
An important issue is the regulation of the endolysin-mediated lysis event that has evolved individually to optimize phage fitness. The holin-endolysin system (lacking secretory signals), known as the lambda paradigm, is thought to be universal in almost all dsDNA phages, with some exceptions (23). In this system, endolysins accumulate in the intracellular space due to the bacterial inner membrane. Subsequently, a small hydrophobic membrane-spanning protein called holin is expressed at a genetically predetermined time and accumulates in clusters producing homo-oligomeric pores in the inner membrane, thereby exposing the PG layer to the endolysin (17, 25, 26). Some endolysins, such as the Bacillus cereus phage TP21-L, Oenococcus oeni phage fOg44, and Lactobacillus plantarum phage Øg1e endolysins, present different intrinsic signal sequences that allow them to pass the cytoplasmic membrane and reach the PG in a holin-independent manner (27, 28). The referred lytic systems are reported exclusively for Caudovirales phages, and not much is known about lytic systems present in cassettes of phages outside this order. A rare and interesting case is the Tectiviridae phage PRD1, in which the endolysin is associated with the viral membrane (29). The capacity of endolysins from a Gram-positive (G+) background to lyse bacteria when added externally was already tested in vitro, leading to the complete death of a streptococcus culture in a few seconds (30). The unique ability of endolysins to rapidly kill bacteria in a species-specific manner renders them promising as antibacterial and biocontrol agents and rapidly caught attention of many researchers that envisage applications in fermentations, food preservation, biotechnology, and medicine (23, 31). Exogenous action of endolysins on Gram-negative (G−) bacteria is still restricted due to the presence of an impermeable outer membrane, thus representing one of the most important challenges in endolysin therapy.
Although in recent years, endolysins have proven to be an interesting alternative to antibiotics, their sequences are poorly classified. PG hydrolases constitute an enormous pool of protein sequences that contain a broad range of lytic domains involved in PG digestion. To date, phage endolysins have been annotated mostly according to their enzymatic catalytic family; however, only a few researchers have tried to analyze their specific classes. Here we describe a bioinformatic analysis of all endolysins found in completely sequenced dsNA phage genomes, unraveling all the selected evolutionary markers behind the phage-endolysin-host ecology association.
MATERIALS AND METHODS
Computer analysis.
All computer analyses were performed on Intel-based PCs with the Windows 7 operating system with a Virtual Machine (Oracle VM Virtual Box) running the Ubuntu 12.05 operating system for sequence alignment using Clustal Omega (version 1.1.0) (32).
Sequence database.
To perform the study of the endolysin evolution/comparison, a sequence database was created by gathering phages assigned as completely sequenced at the NCBI (http://www.ncbi.nlm.nih.gov/pubmed). For the 890 retrieved phages, a query of the genome was performed to encounter any identified endolysin during the sequencing project. Putative endolysins were submitted to a BLASTP analysis (33) to identify new undiscovered proteins. The BLASTP was executed using all standard parameters with an E value cutoff of 1 × 10−5.
Endolysin motif search.
We considered correct endolysin identification (characterization of its type, catalytic characterization, and binding domains) of the upmost importance, and therefore, a set of online search platforms was used to accurately determine the protein cleavage and reaction mechanism. All proteins from the database (723 putative endolysins found in the 890 phages) were screened by the HHpred webserver (http://toolkit.tuebingen.mpg.de/hhpred) using Pfam, InterProScan, and COG with an E value cutoff of 1 × 10−5 and at least 80% query coverage. The multiple-sequence-alignment (MSA) generation method used was HHblits (34). Signal peptides were identified using SignalP (35) and PrediSi (36). To conclude the analysis, the signal-arrest-release (SAR) motif was queried. For that, we searched for transmembrane domains located at the N terminus using SOSUI (37), TMHMM (38), Phobius (39), Octopus (40), and Topcons (41). Endolysins for which a transmembrane domain was identified by at least one of the mentioned softwares and which present a high content (40% to 60%) of Gly/Ala and 0 to 2 basic residues (most of which are Lys) (42) were thus annotated as presenting a SAR domain.
Protein cladogram.
To understand the relation between our selected proteins and to comprehend phage evolution, a phylogenetic study was performed using our protein database. For that, a cladogram was constructed using putative endolysin sequences of only completely sequenced dsNA phage genomes. All these protein sequences were retrieved from our database in FASTA format, aligned using Clustal Omega (beta version for proteins only, v1.1.0) with the -o output.aln -MAC-RAM = 30000 –v –v parameters. The conversion to the PHYLIP format was done using ClustalX multiple alignment software (version 2.0). The phylogenetic construction was obtained using the Phylogeny Inference Package (PHYLIP v3.68) (43). The phylogenic analysis was performed using the following algorithms: maximum likelihood (ML), neighbor joining (NJ), and parsimony (P). These methods were applied to the PHYLIP sequence base using PROML, PROTDIST+NEIGHBOR, and PROTPARS, with SEQBOOT programmed to 100 replicates to bootstrap and number seed variation (the trees were compared to address similarity). Bootstrap values were calculated using CONSENSE. In all methods, the jumble number used was 10 and the bootstrap values were all above 80%. Although more than one method was used to construct the trees, the ML method was selected since it chooses those trees that maximize the probability of observing the data. Phylogenetic trees obtained from PHYLIP were represented and arranged, without being altered, using the FigTree software (v1.3.1) (44).
RESULTS AND DISCUSSION
Outline.
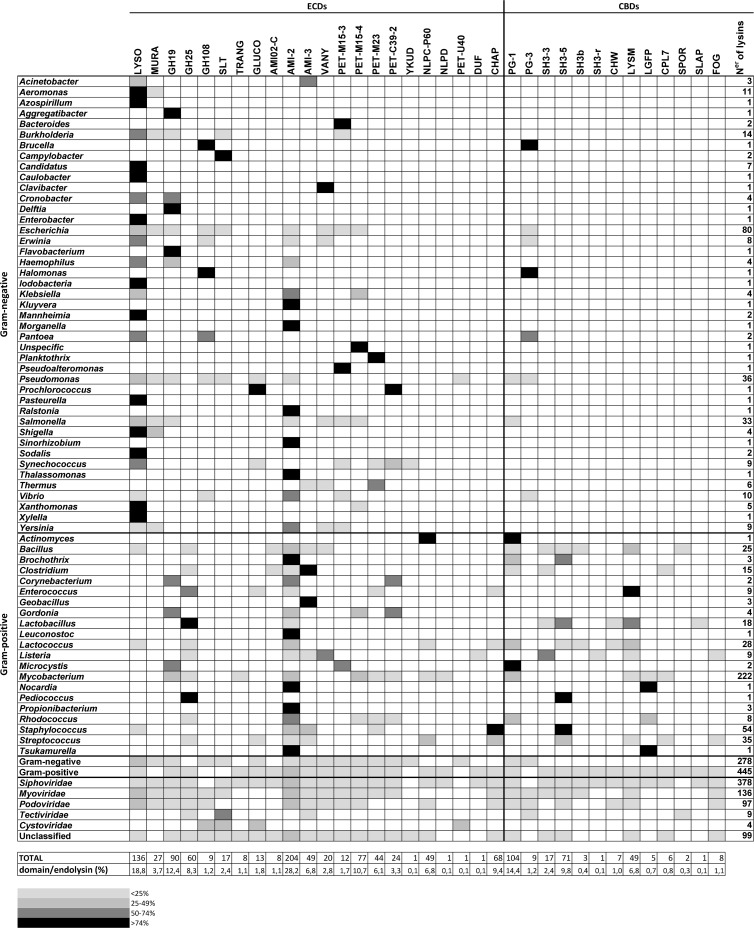
To study phage-host-endolysin ecology, a comprehensive list of all endolysins from dsNA phages with completed genome sequences deposited in the NCBI database was compiled. To sort a given open reading frame (ORF) as a probable endolysin, two criteria were followed: (i) the presence of a putative catalytic domain and (ii) the absence of additional domains with extraneous function. The latter allowed distinguishing of endolysins from structural lysins, also known as exolysins. For each encoded endolysin, the following information was gathered: (i) the presence and position of the enzymatic catalytic domain (ECD)/cell binding domain (CBD), (ii) the phage family type, (iii) the corresponding host Gram reaction, (iv) the host genus, and (v) the presence of signal peptides/transmembrane domains. The data set, altogether, includes a total of 890 complete phage sequences from which 723 putative endolysins were identified. Endolysins were identified in 136 Myoviridae, 378 Siphoviridae, 97 Podoviridae, nine Tectiviridae (dsDNA), four Cystoviridae (dsRNA), and 99 unclassified phages (see Table S1 in the supplemental material). Despite their conserved biological function, phage endolysins are enzymatically and architecturally tremendously diverse and hugely vary in length and size. With a remarkable 24 different ECDs and 13 CBD types (Table 1), these specialized enzymes comprehend 89 different types of architectural organizations (Fig. 2) and belong to phages infecting 64 different bacterial genera (Fig. 3), range from 72 to 578 amino acid residues with unique peptides/transmembrane signals (Fig. 4), and are evolutionarily distant from each other (Fig. 5).
Table 1.
Identified putative endolysin catalytic and binding domainsa
| Predicted catalytic or binding domain | Conserved domain(s) | Phage example | HHpred probability (%)/E value |
|---|---|---|---|
| Predicted catalytic domains | |||
| Phage_lysozyme (LYSO) | PF00959/IPR002196 | Burkholderia phage Bcep176 | 100.0/3.1E−34 |
| Muramidase (MURA) | COG4678 | Enterobacteria phage lambda | 100.0/6.2E−52 |
| Glyco_hydro_19 (GH19) | PF00182/IPR000726 | Pseudomonas phage F10 | 100.0/6.8E−42 |
| Glyco_hydro_25 (GH25) | PF01183/IPR002053 | Clostridium phage phiCTP1 | 100.0/4.9E−48 |
| Glyco_hydro_108 (GH108) | PF05838/IPR008565 | Vibrio phage VP882 | 100.0/1.5E−37 |
| SLT | PF01464/IPR008258 | Pseudomonas phage B3 | 99.9/2.9E−23 |
| Transglycosylase (TRANG) | PF06737/IPR010618 | Mycobacterium phage Blue7 | 100.0/8.3E−31 |
| Glucosaminidase (GLUCO) | PF01832/IPR002901 | Bacillus phage Bam35c | 100.0/6.8E−32 |
| Amidase02_C (AMI02-C) | PF12123/IPR021976 | Clostridium phage phiCD38-2 | 98.2/1.2E−06 |
| Amidase_5 (AMI-5) | PF05382/IPR008044 | Streptococcus phage 858 | 99.9/3.0E−26 |
| Amidase_3 (AMI-3) | PF01520/IPR002508 | Listeria phage 2389 | 100.0/6.4E−44 |
| Amidase_2 (AMI-2) | PF01510/IPR002502 | Gordonia phage GTE7 | 100.0/2.4E−29 |
| NlpD (NLPD) | COG0739 | Mycobacterium phage Timshel | 97.8/2E−05 |
| VanY (VANY) | PF02557/IPR003709 | Yersinia phage PY100 | 99.9/1.2E−24 |
| Peptidase_U40 (PET-U40) | PF10464/IPR19505 | Pseudomonas phage phi-6 | 100.0/2E−138 |
| Peptidase_M15_3 (PET-15-3) | PF08291/IPR013230 | Bacteroides phage B40-8 | 100.0/1.1E−38 |
| Peptidase_M15_4 (PET-15-4) | PF13539 | Salmonella phage ST64T | 99.7/8.2E−17 |
| Peptidase_M23 (PET-M23) | PF01551/IPR016047 | Thermus phage P23-45 | 100.0/2E−28 |
| YkuD (YKUD) | PF03734/IPR005490 | Synechococcus phage S-CBS4 | 99.7/7.1E−18 |
| NLPC_P60 (NLPC-P60) | PF00877/IPR000064 | Mycobacterium phage Dori | 99.8/2.9E−19 |
| Peptidase_C39_2 (PET-C39-2) | PF13529 | Rhodococcus phage REQ3 | 99.8/1.8E−17 |
| CHAP | PF05257/IPR007921 | Staphylococcus phage phi13 | 99.8/9.4E−22 |
| DUF3597 (DUF) | PF12200/IPR022016 | Listeria phage A188 | 99.5/7.6E−15 |
| Predicted binding domains | |||
| PG_binding_3 (PG-3) | PF09374/IPR018537 | Vibrio phage VP882 | 99.9/7.9E−25 |
| LysM (LYSM) | PF01476/IPR018392 | Enterococcus phage phiEf11 | 99.0/1.4E−10 |
| LysM (LYSM) | smart00257 | Lactobacillus phage Lb338-1 | 99.5/5.0E−14 |
| SH3_3 (SH3-3) | PF08239/IPR013247 | Lactobacillus phage Lv-1 | 97.6/5.3E−05 |
| SH3_5 (SH3-5) | PF08460/IPR013667 | Staphylococcus phage phi2958PVL | 99.5/1.5E−14 |
| PG_binding_1 (PG-1) | PF01471/IPR002477 | Mycobacterium phage Hertubise | 99.5/8.7E−14 |
| ChW (CHW) | smart00728 | Lactobacillus phage A2 | 99.3/2.3E−12 |
| ChW (CHW) | PF07538/IPR006637 | Lactococcus phage 949 | 99.2/7.1E−12 |
| Cpl-7 (CPL7) | PF08230/IPR013168 | Streptococcus phage SMP | 99.5/2.4E−14 |
| LGFP | PF08310/IPR013207 | Nocardia phage NBR1 | 99.7/1.6E−17 |
| SH3-related (SH3-r) | SUPFAM0051050 | Listeria phage A500 | 100.0/4.4E−54 |
| FOG | COG5263 | Listeria phage B054 | 99.8/4.0E−19 |
| SH3b | smart00287 | Lactococcus phage P087 | 97.5/4.4E−05 |
| SPOR | PF05036/IPR007730 | Bacillus phage AP50 | 99.1/1.8E−10 |
| SLAP | PF03217/IPR004903 | Bacillus phage 0305phi8-36 | 98.9 /3.0E−08 |
The acronyms used in all catalytic and binding domains are given in parentheses. Domain acronyms: Glyco_hydro, glycoside hydrolase; SLT, soluble lytic transglycosylase; CHAP, cysteine-, histidine-dependent amidohydrolases/peptidases; DUF, domain of unknown function; LYSM, lysin motif; CHW, clostridial hydrophobic with conserved W (Trp); SPOR, sporulation-related domain; SLAP, bacterial surface layer protein. Databases: PF, Pfam; COG, clusters of orthologous groups of proteins; IPR, InterProScan; Smart, Simple Modular Architecture Research Tool. AMI-5 is further considered NLPC-P60 (E values differ at most by 5 log).
Fig 2.
Diversity of ECDs/CBDs found at the N and C termini of the four major endolysin classes centrally located (from all 723 putative endolysins): N-acetylmuramidases (blue scale), N-acetylglucosaminidases (purple), N-acetylmuramoyl-l-alanine, and carboxy/endopeptidases (green scale). CHAP (orange) can act as an amidase or peptidase. These four major classes have been centrally located to show all ECDs (with colors mentioned above) and CBDs (gray scale) found attached to their N and C termini. For the correct interpretation of the schematic representation, for all endolysins found with a CHAP domain, a GLUCO at their N terminus was observed. In some proteins, a GLUCO, AMI-2, and AMI-3 domain at the C terminus was identified.
Fig 3.
Nature and repartition of PGH domains derived from 727 endolysins.
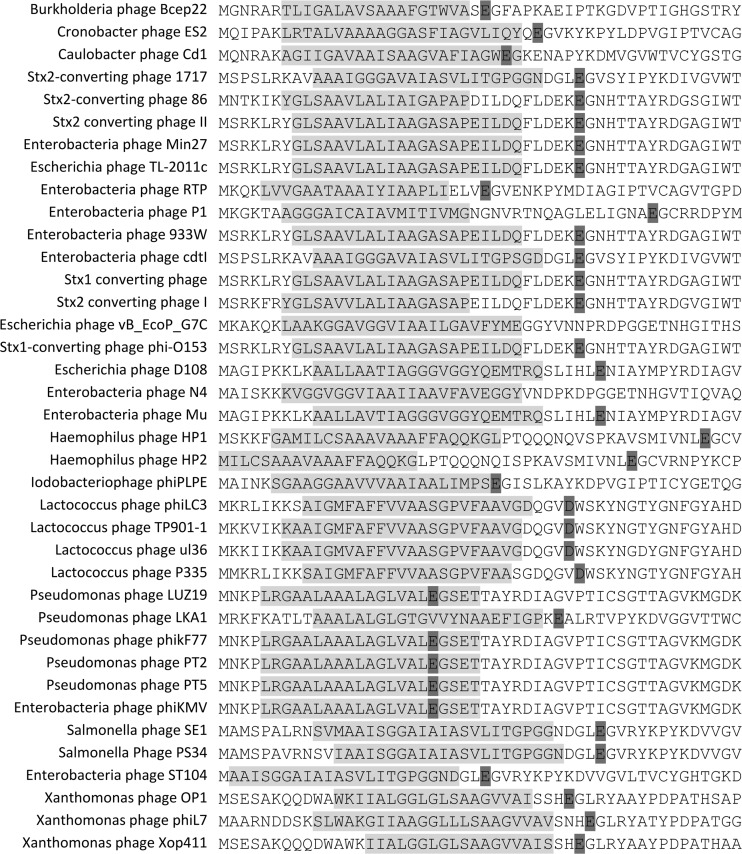
Fig 4.
Phage endolysins predicted to be secreted via the signal-arrest-release system (SAR). Only the first 50 residues are shown. Transmembrane domains are highlighted in light gray, and the predicted catalytic residue of the endolysin is highlighted in dark gray.
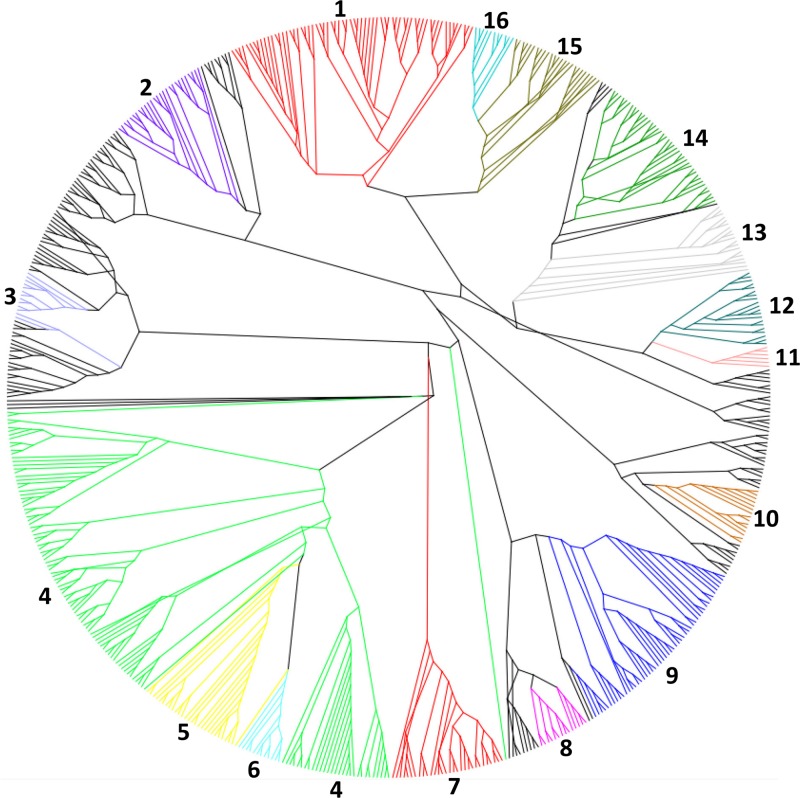
Fig 5.
Radial cladogram of bacteriophage endolysins of all characterized phages. The cladogram was constructed using PHYLIP v3.68 with a neighbor-joining algorithm. The resulting cladogram was represented and arranged, without being altered, using the FigTree software (version 1.3.1). Highlighted areas are numbered and correspond to the more-conserved clades. 1, Mycobacterium/Siphoviridae/AMI-2; 2, G+/GH25; 3, Lactococcus/Siphoviridae/AMI-2; 4, G−/LYSO; 5, G−/MURA; 6, Streptococcus/Siphoviridae/NLPC-P60; 7, G−/Podoviridae/AMI-2; 8, G−/Myoviridae/VANY; 9, Staphylococcus/Siphoviridae; 10, G−/GH19; 11, Escherichia/Tectiviridae/SLT; 12, Mycobacterium/Siphoviridae/NLPC-P60/GH25; 13, Mycobacterium/Siphoviridae/PET-M15-4/PG1; 14, Mycobacterium/Siphoviridae/PETM-15-4/GH19; 15, Mycobacterium/Siphoviridae/PET-M23/AMI-2; 16, Mycobacterium/Siphoviridae/PET-C39-2/GH19.
Endolysin-based lysis diversity among dsNA phages.
The complexity and variety of more than 100 different reported eubacterial PG chemotypes have driven an evolutionary pressure in phages to refine their lytic cassette in order to compromise the host cell wall (44). Toward this end, bacteriophages have acquired a huge diversity of PG hydrolases (PGHs), varying in type, number, and organization of their catalytic and binding domains, all illustrated in Fig. 2.
Endolysin ECDs.
Phage glycosidase PGHs (EC 3.2.1.X) are a group of enzymes belonging to the lysozyme-like superfamily that catalyze the glycolytic cleavage of the O-glycosidic bond of the bacterial PG. Glycosidase hydrolases (GHs), including GH24, GH25, and GH108 (EC 3.2.1.17), are a structurally diverse set of phage endolysin members with the same lysozyme activity cleaving the MurNAc-N-GlcNAc bond of the carbohydrate backbone. GH24 is the most predominant PGH, generally containing globular muramidase (MURA) or phage_lysozyme (LYSO) domains, strictly present in lytic cassettes of phages infecting G− bacteria. A few exceptions are the modular endolysins derived from Synechococcus phages S-CAM8, S-CBS3 (LYSO/PET15-3), and S-CBS1 (LYSO/PET39-2). GH25 endolysins, on the other hand, are acquired by Siphoviridae viruses targeting G+ organisms, mostly belonging to the Firmicutes family (e.g., Bacillus spp., Staphylococcus spp., Streptococcus spp.), that can be linked to several distinct C-terminal CBDs (SLAP, CPL-7, LYSM, SH3, and FOG). The GH108 class contains just nine endolysins with an enzymatic activity restricted to phages infecting only seven G− genera and has never been characterized in vitro. Other soluble lytic transglycosylase (SLT) and transglycosylase (TRANG) domains belong to the lytic transglycosylase group of enzymes. Although degrading the same PG covalent bond as the previous group, they are not hydrolases. SLT-encoding endolysins are globular and acquired by phages to target G− PG (found in Burkholderia, Campylobacter, Escherichia, and Pseudomonas phage endolysins), while the TRANG-type enzymes are modular and specialized in G+ PG (found only in the mycobacteriophages Gladiator, Da Vinci, Hammer, Trixie, Jeffabunny, Blue7, and Turbido). Pseudomonas aeruginosa phage phiKZ endolysin is an example of a (SLT) lytic transglycosylase for which activity has been biochemically confirmed (45). GH19 (EC 3.2.1.14) is a distinct class cleaving the glycosidic β-1,4 linkages of unbranched chains of N-acetylglucosamine polymers. This structure is uncommon in bacterial cell walls; therefore, one can speculate that GH19-identified endolysins can have glycosidase activity, acting either as N-acetylglucosaminidase or as N-acetylmuramidase. They are found mainly in Mycobacterium spp.-, Gordonia spp.-, and Corynebacterium spp.-infecting phages and have been recently characterized in Pseudomonas fluorescens OBP and Salmonella enterica serovar Enteritidis phage PVP-SE1 (46, 47). Finally, glucosaminidases (GLUCO) (EC.3.2.1.52) are core-specific lysosomal enzymes cleaving the GlcNAc-(β-1,4)-MurNAc bond. In phage endolysins, this domain is repeatedly found in Siphoviridae phage lytic cassettes that mostly target hosts within the Firmicutes family. Although this catalytic class is frequently present in Staphylococcus-infecting phages containing three domains in a cysteine-, histidine-dependent amidohydrolase/peptidase (CHAP)/amidase_2 (AMI-2)/GLUCO module, only the Streptococcus agalactiae prophage LambdaSa2 was shown to display β-d-N-acetylglucosaminidase activity (48).
Phage amidase PGHs (EC 3.5.1.28) and N-acetylmuramoyl-l-alanine amidases are endolysins composed of domains of the AMI-2, AMI-3, AMI-5, and AMI02-C types, cleaving the same PG bond between the N-acetylmuramoyl residues and l-amino acid residues. AMI-2, the most predominant domain (representing 28.22%) found in Mycobacterium-like and Staphylococcus-like endolysins, is usually located as a central domain. Escherichia phage T7 and Staphylococcus phage Twort endolysins are well-studied cases (49, 50). AMI-3 domains are predominant on G+-like endolysins. It is important to highlight that all Staphylococcus phage endolysins bearing AMI-3 have a CHAP/AMI-3/SH3-5 structure. Finally, AMI02-C is located at the C-terminal side and is merely present in some Bacillus and Clostridium-like endolysins. To our knowledge, no in vitro studies have yet been performed to confirm lytic activity of this domain.
Phage carboxy/endopeptidase PGHs form the larger group of ECDs that cleave short peptides that link the sugar polymers of the PG. A total of nine different peptidases (PET), PET-M23, NLPC-P60, NLPD, PET-U40, and PET-C39-2 endopeptidases and VANY, PET-M15-4, PET-M15-3, and YKUD domain carboxypeptidases, were found. PET-M23 is found in phage endolysins from several genera. It has been reported that the Bacillus subtilis prophage Sp-β endolysin (named CwlP) contains a related domain, and it has been shown to function as a dd-endopeptidase cleaving 4R3 d-Ala-meso-diaminopimelic acid (m-DAP) interpeptide linkages (51). The NLPC-P60 domain is described as a superfamily with a very different range of activities. Only found in G+-like endolysins, they can cleave N-acetylmuramate-l-alanine linkages, as was shown for the Streptococcus phage Dp-1 and Lactococcus lactis phage BK5-T endolysins (52, 53). More recently, NLPC-P60, found in some mycobacteriophage endolysins, was described as a cysteine proteinase cleaving the 4→3 linkage between d-Glu and m-DAP residues (54). PET-C39-2 was first identified in lysin A proteins (mycobacteriophage cell wall hydrolytic enzymes), cleaving the d-Glu–m-DAP linkages (54). This peptidase is also a cysteine proteinase and is present mainly in mycobacteriophages. VANY is described in the Pfam database as a d-alanyl-d-alanine carboxypeptidase, possibly due to the Enterococcus faecium vancomycin resistance protein VanY (55). Concerning its phage origin, this domain was identified in both G+- and G−-like endolysins, particularly in phages infecting Listeria, Bacillus, and Escherichia organisms. In opposition to what is described by Pfam, the Listeria phage endolysins Ply118 and Ply500 and the enterobacteriophage T5 endolysin classified as VANY carboxypeptidases act as l-alanyl-d-glutamate endopeptidases (56, 57). PET-M15-4 is a domain also described as d-alanyl-d-alanine carboxypeptidase; however, it has been predicted to display l-Ala-d-Glu activity (54). This domain was found in several mycobacteriophages and in some phages infecting G− organisms, such as Salmonella. PET-M15-3 peptidase domains are metallopeptidase domains belonging to the family M15A, found in only nine different endolysins, mainly from G−-infecting phages. The exact cleavage site has not yet been identified due to the lack of biochemical evidence. CHAP is a common domain found in phage endolysins identified in 87 proteins. This CHAP domain is strictly encoded in phages infecting G+ hosts, predominantly in phages infecting Streptococcus spp. and Staphylococcus spp. Of particular interest is their association with several other families of amidases, a connection which suggests that they might act in a cooperative manner to cleave multiple PG substrates. Reports have shown that CHAP can serve as a peptidase or amidase, acting as a d-alanyl-l-alanyl endopeptidase (in Streptococcus phage B30), as a d-alanyl-glycyl endopeptidase (in Staphylococcus phage phi11), or as an N-acetylmuramoyl-l-analine amidase (in Streptococcus phage C1). The latter endolysin (PlyC) is a unique multimeric protein composed of two separate gene products (PlyA and PlyB) (58–60). The remaining NLPD, PET-U40, and YKUD have been found in only one phage endolysin each, mycobacteriophage Timshel (PET-M15-4/NLPD), Cystoviridae Pseudomonas phage phi-6 (PET-U40), and Synechococcus phage S-CBS4 (GLUCO/YKUD) endolysins, respectively. NLPD and YKUD remain experimentally uncharacterized.
Phage endolysins with multicatalytic activities.
Through evolution, endolysins appear to have acquired certain substrate specificities by obtaining multiple ECDs found in bacteriophages infecting a range of 11 different genera but present predominantly in Staphylococcus and Mycobacterium-like phages. The latter have 26 different types of structures, 14 of which combine two ECDs. The most prevalent ones contain an N-terminal predicted peptidase, centrally located AMI-2, MURA, and TRANG, and a C-terminal PG-1. Less frequently, staphylococcus-like endolysins with multiple ECDs can contain up to three ECDs with alternating CHAP/AMI-3/SH3-5, CHAP/AMI-2/SH3-5, or CHAP/AMI-2/GLUCO modules. An unusual double amidase activity was found with an AMI-2/AMI-2C structure and an AMI-3/AMI-2C structure in six Bacillus phages and two Clostridium phages, respectively. In G−-like endolysins, the few multi-ECDs found belong to the Prochlorococcus marinus phage PSS2 and 4 Synechococcus phages with an N-terminal GH activity (MURA or GLUCO) and an additional peptidase activity (PET-C39-2, YKUD, or PET-M15-3). Another notable detail is that 150 (77%) of the 196 annotated multi-ECD phage endolysins belong to Siphoviridae viruses, mostly infecting G+ hosts. Studied examples including plural active lytic domains are the staphylococcal Φ11 (59), the group B streptococcal endolysin B30 (61), and the streptococcal lSa2 phage endolysin (48).
ECDs—evolutionary claims.
The structural distribution of the endolysin domains, as illustrated in Fig. 3, reflects the phages' different strategies to ensure the same biological function, i.e., to disrupt the PG. The acquirement of specific ECDs (Table 1) from a large pool of potential candidates may be influenced by three factors.
(i) The specific ECD substrate may be restricted to a few bacteria.
For instance, because Staphylococcus, Streptococcus, and Lactococcus have unique 3→4 glycine pentaglycine (Gly)5, dialanine (l-Ala-l-Ala), or l-Lys-d-Asp interpeptide bridges within the PG, their specific phages have acquired a CHAP domain that exclusively cleaves these links (62). LYSO activity is dominant in phages infecting G− species. This can be explained by the ability of many G+ pathogens to modify their glycan strands by N-glycosylation, de-N-acetylations, and/or O-acetylations that contribute to high levels of resistance to lysozyme (62).
(ii) The composition and length of the PG structure vary with growth phase and conditions.
As a result of generation of the cellular shape, its maintenance, and bacterial adaptation, bacterial PG dynamics continuously incorporate new material into the sacculus with different muropeptide compositions (63), meaning biosynthesis of new (and removal of old) substrates available for endolysin targeting. This would drive phages to a careful selection of a capable endolysin that efficiently digests all PG variants. Escherichia coli, Mycobacteria, and Streptococcus pneumoniae are known microorganisms in which muropeptides undergo several changes (63–65). Interestingly, their infecting phages represent the niche with lytic cassettes that span the greatest ECD diversity and multiplicity. Mycobacteriophage endolysins especially have an impressive 127 multifunctional ECD combinations, with preferred combinations of PEP-M13/AMI-2/PG-1, NLPC-P60/GH25, and PET-M15-4/GH19 modules. In some cases, ECDs within the same endolysin may act synergistically, cooperatively, or even alone.
(iii) Distinct ECDs guarantee a similar biological function.
Cleavage of a specific bacterial PG is not restricted to a single ECD but can be ensured by several ECDs with different cleavage sites. For instance, the MurNAc-N-GlcNAc glycosidase activity is performed exclusively by the LYSO-containing endolysins in the Xanthomonas phages, whereas at least four distinct domains (LYSO, MURA, GH108, and SLT) are responsible for this activity in Escherichia phage endolysins. The amidase cleavage of the d-lactyl moieties of MurNAc and l-alanine of the short stem peptide (NAM-amidase) is carried out by AMI-2 in Vibrio phage endolysins, while in Bacillus-like endolysins, it is performed by distinct AMI02-C, AMI-2, and AMI-3 domains. In general, ECDs are not global but, rather, are found in a restrictive number of bacterial genera. ECD variety among endolysins resulted from the historical battle between phage and bacterium, attaining a wealth of possible PGH sequences to reach the same final outcome: bacteriolysis.
Endolysin CBDs.
Endolysin modularity can also associate ECDs with a selected cell binding domain (CBD) (Table 1) to target different receptors (e.g., PG subunits, saccharides, proteins, lipoteichoic acid, choline, and PG itself). Notably, all CBDs are present mostly in Bacillus, Lactobacillus, Lactococcus, Mycobacterium, Staphylococcus, and Streptococcus phage endolysins (Fig. 3). Among the great variety of CBD motifs found, only PG-1, SH3, LYSM, and CPL-7 have been characterized in vitro (46, 66, 67). PG-1 is almost only restricted to phages infecting G+ genera, with a few exceptions detected in G−-like endolysins (Salmonella phage PVP-SE1 and Pseudomonas phages phiKZ, EL, 201phi21, and OBP), with a unique inverse rearrangement (one or more PG-1 CBDs at the N termini). PG-3 type has been acquired from phages with a rare modular arrangement (GH108/PG-3), infecting G− counterparts. SH3 domains commonly found in autolysins and phage endolysins often belong to the SH3b, SH3-3, or SH3-5 type. This CBD, shared by lysostaphin (a bacteriocin from Staphylococcus simulans bv. staphylolyticus) and its homologue, hydrolase ALE-1, has been uncovered to recognize pentaglycine cross bridges in PG (68, 69). Therefore, the fact that most of the SH3 domains are found in Staphylococcus-like endolysins is not surprising. The LysM domain is considered to be the CBD with the widest range of receptors, identified in more than 4,000 proteins of both prokaryotes and eukaryotes. It has been postulated that this domain binds to various types of PG and most likely recognizes the N-acetylglucosamine moiety (70). This is in agreement with our findings that show that LysM is always correlated to MurNAc-N-GlcNAc glycosidase activity (LYSO or GH25), with the exception of three endolysins, in which LysM is joined with an amidase domain (AMI-2 or AMI-3 type). Interestingly, the LysM domain is often found in double motifs, having a unique architecture of three motifs in the Lactobacillus phage Lb338-1 endolysin (GH25/LYSM/LYSM/LYSM). Endolysin modules containing the less abundant CPL-7 are GH25/Cpl-7/Cpl-7, NLPC-P60/GH25/Cpl-7, and NLPC-P60/Cpl-7/GLUCO, present in Clostridium phage (phiCTP1), Mycobacterium phages (Optimus and Baka), and Streptococcus phages (SMP and 315.3), respectively. Although CBD location is usually found at the N or C terminus, the aforementioned CBDs (LYSM, CPL-7, and PG-1) can also be located as central domains (e.g., mycobacteriophage Adjutor with NLPC-P60/PG-1/PG-1/GH19, Streptococcus phage SMP with NLPC/CPL-7/CPL-7/GLUCO, and Streptococcus phage phi-SsUD.1 with AMI-3/LYSM/LYSM/NLPC-P60). Concerning CHW (clostridial hydrophobic with conserved W), it has been stated that this protein family is almost exclusively limited to the Clostridium acetobutylicum bacterial species (71). Our results reveal only nine CHW domains detected in three Lactococcus and six Lactobacillus phage endolysins. Curiously, this domain is always present in endolysins attached to an AMI-2-type domain and bears multi-CHW motif copies. The Lactococcus phage A2 endolysin has an AMI-2/CHW/CHW/CHW arrangement, while Lactococcus phages 949, asccphi28, and KSY1 contain the AMI-2/CHW/CHW modules. The acquirement of other uncommon CBD motifs, like LGFP, SPOR, FOG, and SLAP, can represent alternative ways for tighter binding by directing the catalytic cavity into target muropeptides or changes in species specificity. Remarkably, FOG motifs are characterized by having six CBD (PF01473) repeats found only in six streptococcal phages (Cp-1, Dp-1, EJ-1, MM1, PH10, and SM1) and one Listeria phage (B054). To our knowledge, these CBDs have never been characterized in vitro in the big pool of phage PGHs.
CBDs—evolutionary claims.
An evolutionary relationship between S. pneumoniae autolysins and some streptococcal phage endolysins was originally proposed based on their significant nucleotide sequence similarities (72). It was suggested that modular endolysins may have evolved by the interchange of phage and bacterial genes encoding individual modules (72), gaining strong support after the creation of functional chimeric phage-bacterial enzymes, which consisted of one phage- and one bacterial-derived exolysin domain (73, 74). The recruitment and distribution of CBD through phage genomes (Fig. 3) can be correlated by different facts.
(i) Specific CBD receptors may be restricted to a few bacteria.
As the major role of endolysins is to enable progeny virions to be released from the host cell to ensure the effective release of phages, the CBDs may have evolved to target a unique and essential component of the host cell wall (75). This would explain the variability of this module and the highly selective lytic activity of endolysins. Therefore, in some cases, CBDs can confer highly specific ligand recognition to endolysins, resulting in a very narrow substrate spectrum of the full-length endolysin, often limited to the host species or strain (76) or, to a lesser extent, genus specific (67, 77). Well-studied examples of ligand recognition are the pneumococcal endolysins derived from phages Cp-1 (Cpl-1 endolysin), Dp-1 (PAL endolysin), and Ej-1 (Ej-1 endolysin). Their corresponding CBDs specifically bind choline, which is present only in teichoic acids of the pneumococcal cell wall and which is essential for bacterial viability (52, 72, 78). Also, the CBDs of the Listeria monocytogenes bacteriophage endolysins Ply118 and Ply500 can bind to different Listeria serovar groups, as visualized by fusions with green fluorescent protein (GFP), showing that they are correlated with the occurrence of somatic antigens related to these serovars (67).
(ii) Minimal disruption of normal flora.
The reason for the presence of a CBD typically seen in G+-like endolysins relies on the absence of an outer membrane in G+ bacteria. After external bacterial lysis, CBD maintains the endolysin tethered to the PG, making them unavailable for degrading any adjacent G+ PG from forthcoming potential hosts (bacterial threshold), thereby not compromising phage survival (79). The strong substrate specificity of the L. monocytogenes phage endolysins Ply118 and Ply500 of nanomolar affinities is in favor of this hypothesis, suggesting that they have evolved to bind irreversibly to their cell wall ligand (67). On the other hand, the presence of an outer membrane in G− bacteria that impairs direct contact of the endolysin from the outside eliminates the need for a CBD in endolysins from phages infecting these bacteria. In our in silico analysis, we observed a predominant CBD distribution among G+-infecting phages (Fig. 3). Lactobacillus, Lactococcus, and Bacillus genera represent the phage niche with the most diverse CBD collection (PG-1, SH3-3, SH3-5, SH3b, CHW, LYSM, and SPOR). Excellent examples are phage endolysins with 3 CBDs, observed only in Lactobacillus phage A2 (AMI-2/CHW/CHW/CHW) and Lactobacillus phage Lb338-1 (GH25/LYSM/LYSM/LYSM), or endolysins containing different types of CBDs, as is the case for the Lactobacillus phage phiAT3 endolysin (GH25/SH3-5/LYSM).
(iii) Catalytic activity regulator.
Rather than binding to the cell wall, CBD-associated domains sometimes can play determining roles in the enzymatic efficiency by allowing enough substrate to reach the catalytic site. Other carbohydrate hydrolases, such as xylanases and cellulases that cleave insoluble carbohydrate polymers, share a similar modular architecture (80), and their CBDs act by increasing enzyme-substrate proximity (81). Equilibrium association constants of CBDs of Listeria phage endolysins Ply118 and Ply500 place them in the same range as affinity-matured antibodies against bacterial cell surface antigens. Interestingly, the deletion of their CBDs rendered enzymes with no muralytic activity (67). Also, the activity of a C-terminally truncated Clostridium phage phi3626 endolysin was entirely abolished, emphasizing the important role that CBDs play in the enzyme's activity (82). As opposed to previous observations, structural analysis of PlyL indicates that the C-terminal CBD not only targets the endolysin to a specific ligand but also inhibits the catalytic activity in the absence of the cognate substrate. It has been hypothesized that substrate recognition by the CBD of PlyL disrupts interactions between the CBD and the ECD, relieving the inhibitory effect on the latter (83). These interdomain interactions are absent in the C-terminally truncated variant, converting the catalytic domain into a constitutively active domain. Therefore, B. subtilis, which is lacking the cognate target, is hydrolyzed more efficiently by the truncated form than by the full-length version. Consequently, the role of the CBD is variable, and therefore, each case must be examined individually.
(iv) Is it possible to lose nonessential domains?
Though a multidomain architecture is indicative of a modular enzyme, it does not guarantee its functionality as a modular protein. An analogy comparison can be done for phage endolysins, in which the presence of a CBD does not often constitute a prerequisite for enzyme activity. Deletion of the CBD of the L. monocytogenes endolysin PlyPSA only reduces the lytic activity (84). Also, the C-terminal domain of the Lactobacillus delbrueckii subsp. lactis bacteriophage LL-H endolysin can be removed without destroying the lytic activity (85). C-terminal deletions of the Staphylococcus aureus bacteriophage endolysins PlyTW and Ply187, L. monocytogenes bacteriophage endolysin Ply511, Bacillus amyloliquefaciens phage endolysin Morita2001, Bacillus anthracis prophage endolysin PlyL, and Bacillus cereus phage endolysin Ply21 even increase the muralytic activity (50, 83, 86–88). This pattern is also present in endolysins with dual lytic domains, for which, in theory, both should be equally active. Indeed, silent ECDs (almost devoid of activity) have been observed in several multi-ECD endolysins studied (streptococcal phage lambdaSa2 and B30 and staphylococcal phage phi11 endolysins) (89–91). Interestingly, in all these cases, the N-terminal ECD encodes the active domain with the highest activity.
Signal peptides and transmembrane domains within phage endolysins.
The presence of N-terminal signal sequences in endolysins suggests an alternative to the lambda paradigm and thus provides a mode of action different from that of the great majority of endolysins to access the PG at the end of the lytic phage cycle. Signal sequences may enable transport of the endolysin through the inner membrane to achieve the periplasm in a holin-independent manner by making use of the host Sec machinery (24, 27, 77).
Another alternative to the lambda paradigm is the SAR system, in which the endolysin possesses a noncleavable N-terminal type II signal anchor that remains part of the mature endolysin and stays embedded in the inner cell membrane in an inactive form (42, 92, 93). Consequently, these endolysins require pinholins to cause a membrane depolarization to release the SAR anchor enabling the endolysin to get access to the cell wall (92, 94, 95).
A SAR endolysin system alone is thought to be the most primitive phage-encoded lysis system, since, as a protein alone, it can still produce lysis of the cell without the need of any additional protein to achieve access to the PG. The pinholin-SAR endolysin system may thus constitute an intermediate step in the evolution of the canonical holin-endolysin two-component cell lysis system. Because the acquisition of a holin results in a more abrupt or saltatory time-determined lysis, this provides a selective advantage for phage maturation by preventing slow host cell deterioration. This system precedes the holin-endolysin system in which premature cell lysis is prevented by enabling the accumulation of large amounts of endolysin in the cytoplasm before holins oligomerize and allow contact between the endolysin and the PG.
From the created PGH database, 71 endolysins were predicted to present a signal peptide. The search for the presence of SAR domains revealed 103 endolysins presenting an N-terminal transmembrane domain, 38 of which have a high Gly/Ala content (40% to 60%) and fewer than three basic residues (42, 96). These endolysins were annotated as presenting a putative SAR sequence and are listed in Fig. 4. From the 38 endolysins containing predicted SAR sequences, roughly 85% are of the LYSO type, suggesting that this type of PGH is the most primitive. While phage endolysins presenting an N-terminal signal peptide are distributed randomly among all phage families infecting different hosts, phage endolysins with predicted SAR sequences are encoded only by phages infecting G− bacteria (mostly Enterobacteriaceae hosts), with the exception of four lactococcal phage endolysins.
Unusual genetic endolysin organization.
Usually, all phage endolysins, exolysins, and bacterial autolysins are products of single genes. However, within the studied genomes, nine phages were found to present two ORFs encoding endolysins presenting three uncommon features. First, the genomes from Gordonia phages GRU1 and GTE5 and from Streptococcus phage 315.6 contain two ORFs coding for an endolysin. However, they were considered a single product since the concatenated gene products present homology with single proteins from other systems. It was recently suggested that they were once encoded by a single gene or possibly fused in other systems (97). Second, the phages Enterococcus phiEf11, Microcystis Ma-LMM01, Streptococcus M102, Thermus P23-77, and Synechococcus CBS2 and SPM2 encode two predicted endolysins often found in adjacent genes and proximate to the putative holin ORF. Since no homology was found with single products, one possible scenario is that these phages can harbor two distinct endolysins with different cleavage sites that act simultaneously. The presence of a double lytic system in Lactococcus KSY1, containing two holins (ORF 53b and ORF 72) and two endolysins (ORF 53a and ORF 73), can reinforce this hypothesis (98). The third group reported here relates to endolysins formed as a product of two adjacent genes containing group I introns, as reported for the staphylococcal phage endolysins X2, G1, and 85 (99) or staphylokinase in phage phiNM3. Concerning these endolysins, there is no report of these ORFs encoding a functional protein besides their homology with a NUMOD4 motif and an HNH endonuclease or staphylokinase in phage phiNM3. These putative introns raise questions regarding their role in phage genome evolution. It has been suggested that introns can be involved in the process of exonization of intronic sequences or exon shuffling (100). In these cases, as introns are located between two different lytic domains, they probably enhance the chances of domain recombination in these regions, increasing the chances of exon shuffling.
Cladogram phage-host-endolysin relationship.
To comprehend the possible evolutionary position of phage endolysins, a maximum likelihood tree was constructed. By analyzing the cladogram (Fig. 5), we were able to perceive the great variety of endolysin domain structures, generally with conserved ECDs and little homology within the CBDs. Interestingly, there is no distinct clade organization, which shows that the tree is not conceived by just one factor, such as phage family, host, or ECD.
For tailed phages, the parameter that is apparently more important for the cladogram organization is the endolysins' domain architecture, which is visible on the highlighted clades—the more-robust clades.
Regarding nontailed phage endolysins, Escherichia-like endolysins from Tectiviridae contain an SLT catalytic domain and, as expected, they are in the same evolutionary clade. However, it was reported that, despite the amount of similar Tectiviridae genomes, one of the major differences was observed when comparing their lytic enzymes (101). Oppositely, Cystoviridae Pseudomonas-like endolysins appear not to be conserved, as they are dispersed over the tree.
Concluding remarks.
The ubiquity and huge genetic diversity found in phages are a result of both vertical and horizontal evolution. Through these processes, phages have acquired different lytic systems employing PG-degrading enzymes to fight a diversified and changeable bacterial cell wall. Therefore, endolysins can be seen as an important result of phage evolution to permit rapid adaptation to new environmental conditions. The fact that PG degradation leads to bacterial death has intensified a particular interest in the study of endolysins in an era in which the threat of antibiotic and multiresistant bacteria is increasing and solutions are scarce.
The work presented herein provides the first complete list of all known endolysins encoded by dsNA completely sequenced phage genomes as well their structure and predicted mechanisms of action. Novel endolysins and endolysin domains are here described, and further biological studies are needed to understand their molecular and biological basis. Through an increasing knowledge on protein design and with better molecular biology tools available, it is expected that more engineered endolysins can make the transition from the proof-of-principle stage to industrial and medical use.
The annotation of ECDs within endolysins is an important issue and thus far has been less than optimal. Simply calling all endolysins lysozymes as a generic term for PGHs has created historical misconception. Unfortunately, this older nomenclature persists. We intend to resolve previous issues in nomenclature and would like to encourage a universal annotation of phage endolysins and even the creation of an endolysin database.
Only then can we begin to understand and appreciate the fascinating meticulous evolutionary histories that individual phages have taken to lyse host cells and facilitate the virus egress at the end of its lytic cycle.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the Portuguese Foundation for Science and Technology in the scope of the projects PTDC/AGR-ALI/100492/2008 and PTDC/AGR-ALI/121057/2010. Hugo Oliveira, Luís D. R. Melo, and Sílvio B. Santos acknowledge the FCT grants SFRH/BD/63734/2009, SFRH/BD/66166/2009, and SFRH/BPD/75311/2010, respectively.
Footnotes
Published ahead of print 13 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03277-12.
REFERENCES
- 1. Skurnik M, Strauch E. 2006. Phage therapy: facts and fiction. Int. J. Med. Microbiol. 296:5–14 [DOI] [PubMed] [Google Scholar]
- 2. Rohwer F. 2003. Global phage diversity. Cell 113:141. [DOI] [PubMed] [Google Scholar]
- 3. Ackermann HW. 2007. 5500 phages examined in the electron microscope. Arch. Virol. 152:227–243 [DOI] [PubMed] [Google Scholar]
- 4. Brussow H, Hendrix RW. 2002. Phage genomics: small is beautiful. Cell 108:13–16 [DOI] [PubMed] [Google Scholar]
- 5. Suttle CA. 2005. Viruses in the sea. Nature 437:356–361 [DOI] [PubMed] [Google Scholar]
- 6. Comeau AM, Hatfull GF, Krisch HM, Lindell D, Mann NH, Prangishvili D. 2008. Exploring the prokaryotic virosphere. Res. Microbiol. 159:306–313 [DOI] [PubMed] [Google Scholar]
- 7. Sturino JM, Klaenhammer TR. 2006. Engineered bacteriophage-defence systems in bioprocessing. Nat. Rev. Microbiol. 4:395–404 [DOI] [PubMed] [Google Scholar]
- 8. Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8:317–327 [DOI] [PubMed] [Google Scholar]
- 9. Kwan T, Liu J, Dubow M, Gros P, Pelletier J. 2006. Comparative genomic analysis of 18 Pseudomonas aeruginosa bacteriophages. J. Bacteriol. 188:1184–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hendrix RW, Lawrence JG, Hatfull GF, Casjens S. 2000. The origins and ongoing evolution of viruses. Trends Microbiol. 8:504–508 [DOI] [PubMed] [Google Scholar]
- 11. Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. U. S. A. 96:2192–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Comeau AM, Bertrand C, Letarov A, Tetart F, Krisch HM. 2007. Modular architecture of the T4 phage superfamily: a conserved core genome and a plastic periphery. Virology 362:384–396 [DOI] [PubMed] [Google Scholar]
- 13. Simon MN, Davis RW, Davidson N. 1971. Heteroduplexes of DNA molecules of lambdoid phages: physical mapping of their base sequence relationships by electron microscopy, p 313–328 In Hershey AD. (ed), The bacteriophage lambda, vol 2 Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 14. Lavigne R, Seto D, Mahadevan P, Ackermann HW, Kropinski AM. 2008. Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using BLASTP-based tools. Res. Microbiol. 159:406–414 [DOI] [PubMed] [Google Scholar]
- 15. Ackermann HW. 2003. Bacteriophage observations and evolution. Res. Microbiol. 154:245–251 [DOI] [PubMed] [Google Scholar]
- 16. Russel M. 1995. Moving through the membrane with filamentous phages. Trends Microbiol. 3:223–228 [DOI] [PubMed] [Google Scholar]
- 17. Young I, Wang I, Roof WD. 2000. Phages will out: strategies of host cell lysis. Trends Microbiol. 8:120–128 [DOI] [PubMed] [Google Scholar]
- 18. Wang IN. 2006. Lysis timing and bacteriophage fitness. Genetics 172:17–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weidel W, Pelzer H. 1964. Bagshaped macromolecules—a new outlook on bacterial cell walls. Adv. Enzymol. Relat. Areas Mol. Biol. 26:193–232 [DOI] [PubMed] [Google Scholar]
- 20. Typas A, Banzhaf M, Gross CA, Vollmer W. 2012. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10:123–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mitchell P, Moyle J. 1956. Osmotic function and structure of bacteria. Symp. Soc. Gen. Microbiol. 6:150–180 [Google Scholar]
- 22. Young R. 1992. Bacteriophage lysis: mechanism and regulation. Microbiol. Rev. 56:430–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Loessner MJ. 2005. Bacteriophage endolysins—current state of research and applications. Curr. Opin. Microbiol. 8:480–487 [DOI] [PubMed] [Google Scholar]
- 24. Fischetti VA. 2010. Bacteriophage endolysins: a novel anti-infective to control Gram-positive pathogens. Int. J. Med. Microbiol. 300:357–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang IN, Smith DL, Young R. 2000. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54:799–825 [DOI] [PubMed] [Google Scholar]
- 26. Young R. 2002. Bacteriophage holins: deadly diversity. J. Mol. Microbiol. Biotechnol. 4:21–36 [PubMed] [Google Scholar]
- 27. Sao-Jose C, Parreira R, Vieira G, Santos MA. 2000. The N-terminal region of the Oenococcus oeni bacteriophage fOg44 lysin behaves as a bona fide signal peptide in Escherichia coli and as a cis-inhibitory element, preventing lytic activity on oenococcal cells. J. Bacteriol. 182:5823–5831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kakikawa M, Yokoi KJ, Kimoto H, Nakano M, Kawasaki K, Taketo A, Kodaira K. 2002. Molecular analysis of the lysis protein Lys encoded by Lactobacillus plantarum phage phig1e. Gene 299:227–234 [DOI] [PubMed] [Google Scholar]
- 29. Rydman PS, Bamford DH. 2002. The lytic enzyme of bacteriophage PRD1 is associated with the viral membrane. J. Bacteriol. 184:104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nelson D, Loomis L, Fischetti VA. 2001. Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proc. Natl. Acad. Sci. U. S. A. 98:4107–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oliveira H, Azeredo J, Lavigne R, Kluskens LD. 2012. Bacteriophage endolysins as a response to emerging foodborne pathogens. Trends Food Sci. Technol. 28:103–115 [Google Scholar]
- 32. Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 34. Remmert M, Biegert A, Hauser A, Soding J. 2012. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods 9:173–175 [DOI] [PubMed] [Google Scholar]
- 35. Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786 [DOI] [PubMed] [Google Scholar]
- 36. Hiller K, Grote A, Maneck M, Munch R, Jahn D. 2006. JVirGel 2.0: computational prediction of proteomes separated via two-dimensional gel electrophoresis under consideration of membrane and secreted proteins. Bioinformatics 22:2441–2443 [DOI] [PubMed] [Google Scholar]
- 37. Hirokawa T, Boon-Chieng S, Mitaku S. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378–379 [DOI] [PubMed] [Google Scholar]
- 38. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 39. Kall L, Krogh A, Sonnhammer EL. 2004. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338:1027–1036 [DOI] [PubMed] [Google Scholar]
- 40. Viklund H, Elofsson A. 2008. OCTOPUS: improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics 24:1662–1668 [DOI] [PubMed] [Google Scholar]
- 41. Hennerdal A, Elofsson A. 2011. Rapid membrane protein topology prediction. Bioinformatics 27:1322–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu M, Struck DK, Deaton J, Wang IN, Young R. 2004. A signal-arrest-release sequence mediates export and control of the phage P1 endolysin. Proc. Natl. Acad. Sci. U. S. A. 101:6415–6420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Felsenstein J. 2009. Phylogeny Inference Package (PHYLIP), version 3.6. Department of Genome Sciences, University of Washington, Seattle, WA [Google Scholar]
- 44. Schleifer K, Kandler O. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Paradis-Bleau C, Cloutier I, Lemieux L, Sanschagrin F, Laroche J, Auger M, Garnier A, Levesque RC. 2007. Peptidoglycan lytic activity of the Pseudomonas aeruginosa phage phiKZ gp144 lytic transglycosylase. FEMS Microbiol. Lett. 266:201–209 [DOI] [PubMed] [Google Scholar]
- 46. Walmagh M, Briers Y, Dos Santos SB, Azeredo J, Lavigne R. 2012. Characterization of modular bacteriophage endolysins from Myoviridae phages OBP, 201varphi2-1 and PVP-SE1. PLoS One 7:e36991 doi:10.1371/journal.pone.0036991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lai MJ, Lin NT, Hu A, Soo PC, Chen LK, Chen LH, Chang KC. 2011. Antibacterial activity of Acinetobacter baumannii phage varphiAB2 endolysin (LysAB2) against both gram-positive and gram-negative bacteria. Appl. Microbiol. Biotechnol. 90:529–539 [DOI] [PubMed] [Google Scholar]
- 48. Pritchard DG, Dong S, Kirk MC, Cartee RT, Baker JR. 2007. LambdaSa1 and LambdaSa2 prophage lysins of Streptococcus agalactiae. Appl. Environ. Microbiol. 73:7150–7154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cheng X, Zhang X, Pflugrath JW, Studier FW. 1994. The structure of bacteriophage T7 lysozyme, a zinc amidase and an inhibitor of T7 RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 91:4034–4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Loessner MJ, Gaeng S, Wendlinger G, Maier SK, Scherer S. 1998. The two-component lysis system of Staphylococcus aureus bacteriophage Twort: a large TTG-start holin and an associated amidase endolysin. FEMS Microbiol. Lett. 162:265–274 [DOI] [PubMed] [Google Scholar]
- 51. Sudiarta IP, Fukushima T, Sekiguchi J. 2010. Bacillus subtilis CwlP of the SP-β prophage has two novel peptidoglycan hydrolase domains, muramidase and cross-linkage digesting DD-endopeptidase. J. Biol. Chem. 285:41232–41243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sheehan MM, Garcia JL, Lopez R, Garcia P. 1997. The lytic enzyme of the pneumococcal phage Dp-1: a chimeric lysin of intergeneric origin. Mol. Microbiol. 25:717–725 [DOI] [PubMed] [Google Scholar]
- 53. Boyce JD, Davidson BE, Hillier AJ. 1995. Identification of prophage genes expressed in lysogens of the Lactococcus lactis bacteriophage BK5-T. Appl. Environ. Microbiol. 61:4099–4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Payne KM, Hatfull GF. 2012. Mycobacteriophage endolysins: diverse and modular enzymes with multiple catalytic activities. PLoS One 7:e34052 doi:10.1371/journal.pone.0034052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wright GD, Molinas C, Arthur M, Courvalin P, Walsh CT. 1992. Characterization of VanY, a dd-carboxypeptidase from vancomycin-resistant Enterococcus faecium BM4147. Antimicrob. Agents Chemother. 36:1514–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Loessner MJ, Wendlinger G, Scherer S. 1995. Heterogeneous endolysins in Listeria monocytogenes bacteriophages: a new class of enzymes and evidence for conserved holin genes within the siphoviral lysis cassettes. Mol. Microbiol. 16:1231–1241 [DOI] [PubMed] [Google Scholar]
- 57. Mikoulinskaia GV, Odinokova IV, Zimin AA, Lysanskaya VY, Feofanov SA, Stepnaya OA. 2009. Identification and characterization of the metal ion-dependent l-alanoyl-d-glutamate peptidase encoded by bacteriophage T5. FEBS J. 276:7329–7342 [DOI] [PubMed] [Google Scholar]
- 58. Nelson D, Schuch R, Chahales P, Zhu S, Fischetti VA. 2006. PlyC: a multimeric bacteriophage lysin. Proc. Natl. Acad. Sci. U. S. A. 103:10765–10770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Navarre WW, Ton-That H, Faull KF, Schneewind O. 1999. Multiple enzymatic activities of the murein hydrolase from staphylococcal phage phi11. Identification of a d-alanyl-glycine endopeptidase activity. J. Biol. Chem. 274:15847–15856 [DOI] [PubMed] [Google Scholar]
- 60. Baker JR, Liu C, Dong S, Pritchard DG. 2006. Endopeptidase and glycosidase activities of the bacteriophage B30 lysin. Appl. Environ. Microbiol. 72:6825–6828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pritchard DG, Dong S, Baker JR, Engler JA. 2004. The bifunctional peptidoglycan lysin of Streptococcus agalactiae bacteriophage B30. Microbiology 150:2079–2087 [DOI] [PubMed] [Google Scholar]
- 62. Vollmer W, Born P. 2009. Bacterial cell envelope peptidoglycan, p 15–28 In Moran AP. (ed), Microbial glycobiology—structures, relevance and applications. Elsevier Inc, London, United Kingdom [Google Scholar]
- 63. Glauner B, Holtje JV, Schwarz U. 1988. The composition of the murein of Escherichia coli. J. Biol. Chem. 263:10088–10095 [PubMed] [Google Scholar]
- 64. Pisabarro AG, de Pedro MA, Vazquez D. 1985. Structural modifications in the peptidoglycan of Escherichia coli associated with changes in the state of growth of the culture. J. Bacteriol. 161:238–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lavollay M, Arthur M, Fourgeaud M, Dubost L, Marie A, Veziris N, Blanot D, Gutmann L, Mainardi JL. 2008. The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by l,d-transpeptidation. J. Bacteriol. 190:4360–4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hu S, Kong J, Kong W, Guo T, Ji M. 2010. Characterization of a novel LysM domain from Lactobacillus fermentum bacteriophage endolysin and its use as an anchor to display heterologous proteins on the surfaces of lactic acid bacteria. Appl. Environ. Microbiol. 76:2410–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Loessner MJ, Kramer K, Ebel F, Scherer S. 2002. C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol. Microbiol. 44:335–349 [DOI] [PubMed] [Google Scholar]
- 68. Grundling A, Schneewind O. 2006. Cross-linked peptidoglycan mediates lysostaphin binding to the cell wall envelope of Staphylococcus aureus. J. Bacteriol. 188:2463–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lu JZ, Fujiwara T, Komatsuzawa H, Sugai M, Sakon J. 2006. Cell wall-targeting domain of glycylglycine endopeptidase distinguishes among peptidoglycan cross-bridges. J. Biol. Chem. 281:549–558 [DOI] [PubMed] [Google Scholar]
- 70. Buist G, Steen A, Kok J, Kuipers OP. 2008. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 68:838–847 [DOI] [PubMed] [Google Scholar]
- 71. Sullivan L, Paredes CJ, Papoutsakis ET, Bennett GN. 2007. Analysis of the clostridial hydrophobic with a conserved tryptophan family (ChW) of proteins in Clostridium acetobutylicum with emphasis on ChW14 and ChW16/17. Enzyme Microb. Technol. 42:29–43 [Google Scholar]
- 72. Garcia E, Garcia JL, Garcia P, Arraras A, Sanchez-Puelles JM, Lopez R. 1988. Molecular evolution of lytic enzymes of Streptococcus pneumoniae and its bacteriophages. Proc. Natl. Acad. Sci. U. S. A. 85:914–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Diaz E, Lopez R, Garcia JL. 1990. Chimeric phage-bacterial enzymes: a clue to the modular evolution of genes. Proc. Natl. Acad. Sci. U. S. A. 87:8125–8129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Diaz E, Lopez R, Garcia JL. 1991. Chimeric pneumococcal cell wall lytic enzymes reveal important physiological and evolutionary traits. J. Biol. Chem. 266:5464–5471 [PubMed] [Google Scholar]
- 75. Fischetti VA. 2003. Novel method to control pathogenic bacteria on human mucous membranes. Ann. N. Y. Acad. Sci. 987:207–214 [DOI] [PubMed] [Google Scholar]
- 76. Perez-Dorado I, Campillo NE, Monterroso B, Hesek D, Lee M, Paez JA, Garcia P, Martinez-Ripoll M, Garcia JL, Mobashery S, Menendez M, Hermoso JA. 2007. Elucidation of the molecular recognition of bacterial cell wall by modular pneumococcal phage endolysin CPL-1. J. Biol. Chem. 282:24990–24999 [DOI] [PubMed] [Google Scholar]
- 77. Loessner MJ, Maier SK, Daubek-Puza H, Wendlinger G, Scherer S. 1997. Three Bacillus cereus bacteriophage endolysins are unrelated but reveal high homology to cell wall hydrolases from different bacilli. J. Bacteriol. 179:2845–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Saiz JL, Lopez-Zumel C, Monterroso B, Varea J, Arrondo JL, Iloro I, Garcia JL, Laynez J, Menendez M. 2002. Characterization of Ejl, the cell-wall amidase coded by the pneumococcal bacteriophage Ej-1. Protein Sci. 11:1788–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Fischetti VA. 2008. Bacteriophage lysins as effective antibacterials. Curr. Opin. Microbiol. 11:393–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Khosla C, Harbury PB. 2001. Modular enzymes. Nature 409:247–252 [DOI] [PubMed] [Google Scholar]
- 81. Bolam DN, Ciruela A, McQueen-Mason S, Simpson P, Williamson MP, Rixon JE, Boraston A, Hazlewood GP, Gilbert HJ. 1998. Pseudomonas cellulose-binding domains mediate their effects by increasing enzyme substrate proximity. Biochem. J. 331:775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zimmer M, Vukov N, Scherer S, Loessner MJ. 2002. The murein hydrolase of the bacteriophage phi3626 dual lysis system is active against all tested Clostridium perfringens strains. Appl. Environ. Microbiol. 68:5311–5317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Low LY, Yang C, Perego M, Osterman A, Liddington RC. 2005. Structure and lytic activity of a Bacillus anthracis prophage endolysin. J. Biol. Chem. 280:35433–35439 [DOI] [PubMed] [Google Scholar]
- 84. Korndorfer IP, Danzer J, Schmelcher M, Zimmer M, Skerra A, Loessner MJ. 2006. The crystal structure of the bacteriophage PSA endolysin reveals a unique fold responsible for specific recognition of Listeria cell walls. J. Mol. Biol. 364:678–689 [DOI] [PubMed] [Google Scholar]
- 85. Vasala A, Valkkila M, Caldentey J, Alatossava T. 1995. Genetic and biochemical characterization of the Lactobacillus delbrueckii subsp. lactis bacteriophage LL-H lysin. Appl. Environ. Microbiol. 61:4004–4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Loessner MJ, Gaeng S, Scherer S. 1999. Evidence for a holin-like protein gene fully embedded out of frame in the endolysin gene of Staphylococcus aureus bacteriophage 187. J. Bacteriol. 181:4452–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Morita M, Tanji Y, Orito Y, Mizoguchi K, Soejima A, Unno H. 2001. Functional analysis of antibacterial activity of Bacillus amyloliquefaciens phage endolysin against Gram-negative bacteria. FEBS Lett. 500:56–59 [DOI] [PubMed] [Google Scholar]
- 88. Gaeng S, Scherer S, Neve H, Loessner MJ. 2000. Gene cloning and expression and secretion of Listeria monocytogenes bacteriophage-lytic enzymes in Lactococcus lactis. Appl. Environ. Microbiol. 66:2951–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Donovan DM, Foster-Frey J. 2008. LambdaSa2 prophage endolysin requires Cpl-7-binding domains and amidase-5 domain for antimicrobial lysis of streptococci. FEMS Microbiol. Lett. 287:22–33 [DOI] [PubMed] [Google Scholar]
- 90. Donovan DM, Foster-Frey J, Dong S, Rousseau GM, Moineau S, Pritchard DG. 2006. The cell lysis activity of the Streptococcus agalactiae bacteriophage B30 endolysin relies on the cysteine, histidine-dependent amidohydrolase/peptidase domain. Appl. Environ. Microbiol. 72:5108–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Horgan M, O'Flynn G, Garry J, Cooney J, Coffey A, Fitzgerald GF, Ross RP, McAuliffe O. 2009. Phage lysin LysK can be truncated to its CHAP domain and retain lytic activity against live antibiotic-resistant staphylococci. Appl. Environ. Microbiol. 75:872–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Xu M, Arulandu A, Struck DK, Swanson S, Sacchettini JC, Young R. 2005. Disulfide isomerization after membrane release of its SAR domain activates P1 lysozyme. Science 307:113–117 [DOI] [PubMed] [Google Scholar]
- 93. Briers Y, Peeters LM, Volckaert G, Lavigne R. 2011. The lysis cassette of bacteriophage varphiKMV encodes a signal-arrest-release endolysin and a pinholin. Bacteriophage 1:25–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Park T, Struck DK, Dankenbring CA, Young R. 2007. The pinholin of lambdoid phage 21: control of lysis by membrane depolarization. J. Bacteriol. 189:9135–9139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Pang T, Savva CG, Fleming KG, Struck DK, Young R. 2009. Structure of the lethal phage pinhole. Proc. Natl. Acad. Sci. U. S. A. 106:18966–18971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lee CN, Lin JW, Chow TY, Tseng YH, Weng SF. 2006. A novel lysozyme from Xanthomonas oryzae phage varphiXo411 active against Xanthomonas and Stenotrophomonas. Protein Expr. Purif. 50:229–237 [DOI] [PubMed] [Google Scholar]
- 97. Petrovski S, Tillett D, Seviour RJ. 2012. Genome sequences and characterization of the related Gordonia phages GTE5 and GRU1 and their use as potential biocontrol agents. Appl. Environ. Microbiol. 78:42–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Chopin A, Deveau H, Ehrlich SD, Moineau S, Chopin MC. 2007. KSY1, a lactococcal phage with a T7-like transcription. Virology 365:1–9 [DOI] [PubMed] [Google Scholar]
- 99. Becker SC, Foster-Frey J, Stodola AJ, Anacker D, Donovan DM. 2009. Differentially conserved staphylococcal SH3b_5 cell wall binding domains confer increased staphylolytic and streptolytic activity to a streptococcal prophage endolysin domain. Gene 443:32–41 [DOI] [PubMed] [Google Scholar]
- 100. Schmidt EE, Davies CJ. 2007. The origins of polypeptide domains. Bioessays 29:262–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Saren AM, Ravantti JJ, Benson SD, Burnett RM, Paulin L, Bamford DH, Bamford JK. 2005. A snapshot of viral evolution from genome analysis of the Tectiviridae family. J. Mol. Biol. 350:427–440 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.