Background: Expression of the G protein-coupled receptor LGR5 demarcates adult tissue stem cells in the intestine, stomach, hair follicle, and mammary epithelium.
Results: LGR5 is rapidly and constitutively internalized to the trans-Golgi network at steady state.
Conclusion: Internalization occurs through a potential phosphorylation domain within the C-terminal tail.
Significance: An understanding of LGR5 trafficking dynamics is expected to clarify its role in signaling and stem cell biology.
Keywords: Cell Biology, Cell Surface Receptor, G Protein-coupled Receptor (GPCR), Stem Cells, Trafficking, Trans-Golgi Network
Abstract
LGR5 is a Wnt pathway associated G protein-coupled receptor (GPCR) that serves as a molecular determinant of stem cells in numerous tissues including the intestine, stomach, hair follicle, eye, and mammary gland. Despite its importance as a marker for this critical niche, little is known about LGR5 signaling nor the biochemical mechanisms and receptor determinants that regulate LGR5 membrane expression and intracellular trafficking. Most importantly, in cells LGR5 is predominantly intracellular, yet the mechanisms underlying this behavior have not been determined. In this work we elucidate a precise trafficking program for LGR5 and identify the motif at its C terminus that is responsible for the observed constitutive internalization. We show that this process is dependent upon dynamin GTPase activity and find that wild-type full-length LGR5 rapidly internalizes into EEA1- and Rab5-positive endosomes. However, LGR5 fails to rapidly recycle to the plasmid membrane through Rab4-positive vesicles, as is common for other GPCRs. Rather, internalized LGR5 transits through Rab7- and Rab9-positive vesicles, co-localizes in vesicles with Vps26, a retromer complex component that regulates retrograde trafficking to the trans-Golgi network (TGN) and reaches a steady-state distribution in the TGN within 2 h. Using mutagenesis, particularly of putative phosphorylation sites, we show that the amino acid pair, serine 861 and 864, is the principal C-tail determinant that mediates LGR5 constitutive internalization. The constitutive internalization of LGR5 to the TGN suggests the existence of novel biochemical roles for its Wnt pathway related, but ill defined signaling program.
Introduction
LGR53 was originally cloned in 1998 and found to be a member of the leucine-rich repeat-containing G protein-coupled receptor (LGR) family (1). The LGR family comprises three subfamilies, the most notable being the glycoprotein hormone subfamily comprised of the follicle-stimulating, thyroid-stimulating, and luteinizing hormone receptors (FSH, TSH, and LHR, respectively). The two other subfamilies contain Lgrs4–6 and Lgrs7/8. In addition to the prototypical 7-transmembrane bundle that all GPCRs share, LGR5 possesses a large N-terminal extracellular ectodomain, comprising 17 repetitive leucine-rich domains, a number which varies in the LGR family (2).
In 2007 Barker et al. discovered that LGR5 expression provides a key molecular determinant for identifying the intestinal epithelial stem cell (3). Using an LGR5-driven lineage tracing strategy, they found that epithelial cells of the small intestine and colon are derived from a corresponding LGR5+ cell located at the crypt base. The importance of LGR5 as a robust marker of stem cells has now been expanded to include other tissues such as the hair follicle (4), stomach (5), eye (6), and the mammary gland (7). In addition to marking the LGR5 lineage in vivo, LGR5 expression enables an ex vivo fractionation of single LGR5+ cells for growing fully differentiated intestinal organoids (8), stomach (5), and mammary gland (7). More recently, LGR5+ tumors in mouse models have been lineage traced and “retraced” to demonstrate that they can act as bona fide cancer stem cells, potentially explaining the increased expression of LGR5 found in cancer (9–11). LGR4- and LGR6-driven lineage-tracing systems have also been useful in identifying a range of additional cell types, which, respectively, identify cells with less restricted or more restricted expression patterns and cell lineages compared with LGR5 (12, 13).
The notion that LGR5 may be an instrumental regulator of critical physiology and an important therapeutic target has led to an explosion of genetic studies as well as renewed searches for its endogenous ligand. In 2011, R-spondins 1–4 were reported to be ligands for Lgrs 4, 5, and 6 (12, 14–17), and in these studies it was demonstrated that LGR5 binding of R-spondins led to a potentiation of Wnt/β-catenin signaling (12, 17). Despite LGR4–6 having stereotypical domains for coupling to G proteins and recruiting β-arrestin, no combination of LGR5/ligand has been able to activate these signaling pathways (12, 17). In addition to scaffolding GPCR-signaling proteins, β-arrestins also regulate GPCR membrane expression and their internalization through motifs found in the receptor intracellular loops and C-tails (18–21). LGR5 is poorly expressed at the plasma membrane in model cell systems, and a recent report indicates that LGR5 is constitutively internalized (15). Although the mechanisms underlying LGR5 endocytosis in general are unclear, its C-tail contains numerous putative serine regulatory motifs, including one, 872–875 (TSSS) canonically associated with G protein receptor kinase-dependent phosphorylation and high affinity receptor/β-arrestin binding, prolonged vesicular trafficking, and eventual plasma membrane recycling (18, 19). In contrast to the prototypical trafficking behaviors elucidated for most GPCRs with this domain, we find that LGR5 is constitutively internalized and rapidly trafficked to the TGN independent of the TSSS motif. Rather, we demonstrate the existence of a separate domain (Ser861/Ser864) responsible for initiating internalization of LGR5. Our identification of a β-arrestin-independent mechanism responsible for LGR5 constitutive internalization will facilitate untangling its distinctive signaling and trafficking behaviors. The presence of multiple and independent internalization domains suggest that proper trafficking of LGR5 either at steady state or following ligand occupancy is an essential aspect of its signaling competency. These findings raise the intriguing question of whether the potent β-arrestin binding domain in the LGR5 tail is a vestigial motif, or more provocatively, it could indicate the existence of another class of endogenous LGR5 ligands.
EXPERIMENTAL PROCEDURES
Plasmids, Cloning, Cell Lines, Transfection
A clone containing the open reading frame encoding full-length LGR5 (40008253) was purchased from Open Biosystems. LGR5 contains an N-terminal and cleavable signal sequence required for proper trafficking. The signal sequence of LGR5 (amino acids 1–21; MDTSRLGVLLSLPVL-LQLATG) was cloned upstream of an N-terminal 3×HA tag, and the remainder of LGR5 starting at amino acid 22 was cloned in-frame and immediately downstream of the 3×HA epitope in pEGFP-N3 to yield an N-terminally 3×HA-tagged receptor with an enhanced green fluorescent protein (EGFP) fused to the C-terminal tail. Truncations to the C-terminal tail of LGR5, tail swapping with the human vasopressin 2 receptor (V2R), and point mutations were generated using standard PCR-based cloning techniques, QuikChange mutagenesis, and overlap exchangePCR (22). Dynamin K44A and human V2R were available in the laboratory (23). EGFP-tagged Rab4 and Rab5 were available in the laboratory; and Rab7 (24), Rab9 (24), and Rab11 (24) were purchased from Addgene (12605, 12663, and 12674, respectively). HEK239 T/17 (HEK) cells were obtained from the ATCC (CRL-11268). HEK cells were cultured in the recommended media (1× DMEM (Mediatech/Cellgro 10-013-CV), 10% FBS (Sigma F2442), 1× Antibiotic-Antimycotic (Invitrogen 15240-062)) and transfected using a calcium phosphate protocol that was modified according to cell number and assay as described below (25).
Internalization Assays
Confocal
HEK cells were transfected and plated on 35-mm glass bottom dishes (MatTek Corporation, Ashland, MA) P35G-0-10C) that were previously treated with 75 μg/ml fibronectin for 1 h at room temperature. The next day, cells were placed on ice to block endocytosis and pulse-stained with a mouse monoclonal anti-HA antibody (1:500, hybridoma line available in the laboratory) or chicken anti-HA antibody (1:750 (Abcam Ab9111)) in staining medium (SM: clear MEM with supplements, 1× MEM (Invitrogen 51200), 10 mm Hepes (Invitrogen 15630), 1× GlutaMAX-1 (Invitrogen 35050)) for 45 min. Cells were washed four times with cold SM and either fixed or chased at 37 °C for the indicated chase times and then fixed. To detect labeled receptor, cells were permeabilized and blocked with 0.12% Triton X-100 and 5% BSA/PBS for 30 min and incubated with secondary antibody as indicated. As indicated in the text, cells expressing human V2R were also stimulated with arginine vasopressin at 0.1 IU/ml (Sigma V0377). Where indicated, cells were also stained with mouse anti-CI-M6PR* (1:500 (Abcam Ab2733)), rabbit anti-Trip230* (1:1000 (Abcam Ab72223)), rabbit anti-EEA1 (1:125 (Abcam Ab2300)), rabbit anti-Vps26 (1:1000 (Abcam Ab23892)), or sheep anti-TGN46 (1:800 (AbdSerotec AHP500GT)). Secondary antibodies utilized were goat anti-mouse-568 (1:1000 (Invitrogen A11004)), goat anti-chicken-568 (1:1000 (Invitrogen A11041)), goat anti-mouse-633 (1:1000 (Invitrogen A21050), or donkey anti-sheep-568 (1:1000 (Invitrogen A21099)). Cells were imaged using a Zeiss LSM 510 (Carl Zeiss MicroImaging) at 100× and a digital zoom of 2×, unless otherwise noted in the figure legends. The asterisk denotes antibodies that required cells to be fixed in methanol rather than 4% paraformaldehyde. For Figs. 4–7, MatTek 24-well plate glass bottomed dishes (P24G-0-10-F) were used to analyze the internalization dynamics of all of the constructs in the same batch analysis. Cells were imaged in a blinded manner, in which a minimum of three images was captured per well. As this experiment was performed on the same day under the same conditions, the same WT control was used for each figure to visually normalize each figure to the normal WT time course.
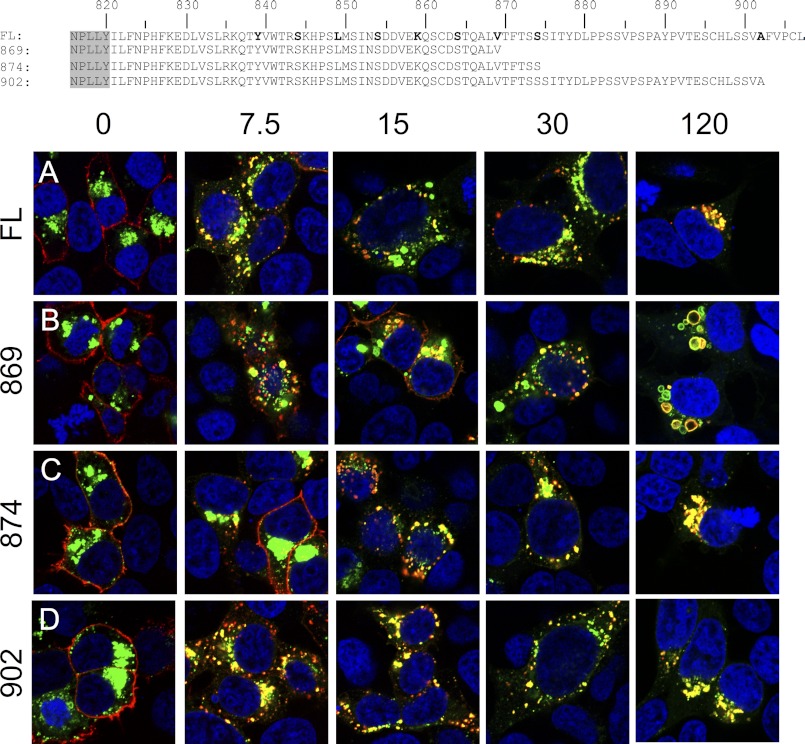
FIGURE 4.
Internalization of LGR5 is regulated by a motif between positions 834 and 869. Shown are primary amino acid sequences of the C-terminal tail for each construct (canonical GPCR NPXXY domain in gray). HEK 293T cells were transiently transfected with the indicated 3×HA N-terminally (red) and C-terminally EGFP (green)-tagged Lgr5 constructs: FL-LGR5 (A), 869 (B), 874 (C), or 902 (D). Cells were pulsed with a MαHA antibody for 45 min on ice, washed, chased for 0, 7.5, 15, 30, or 120 min at 37 °C, fixed, permeabilized, and stained with a GαM568 antibody (red). Merged 100× confocal images are presented (blue, nuclear counterstain).
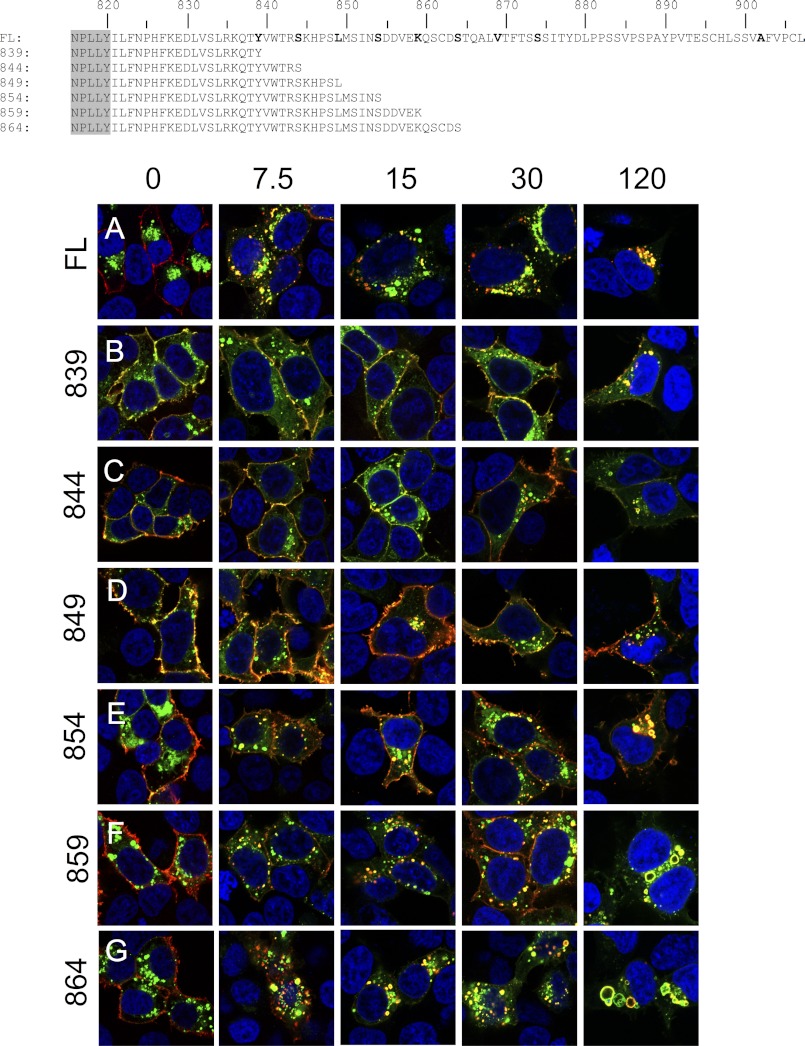
FIGURE 5.
Truncation analysis identifies a putative region regulating LGR5 internalization. Shown are primary amino acid sequences of the C-terminal tail for each construct (canonical GPCR NPXXY domain in gray). HEK 293T cells were transiently transfected with the indicated 3×HA N-terminally (red) and C-terminally EGFP (green)-tagged constructs: FL-LGR5 (A), 839 (B), 844 (C), 849 (D), 854 (E), 859 (F), or 864 (G). Cells were pulsed with a MαHA antibody for 45 min on ice, washed, chased for 0, 7.5, 15, 30, or 120 min at 37 °C, fixed, permeabilized, and stained with a GαM568 antibody (red). Merged 100× confocal images are presented (blue, nuclear counterstain).
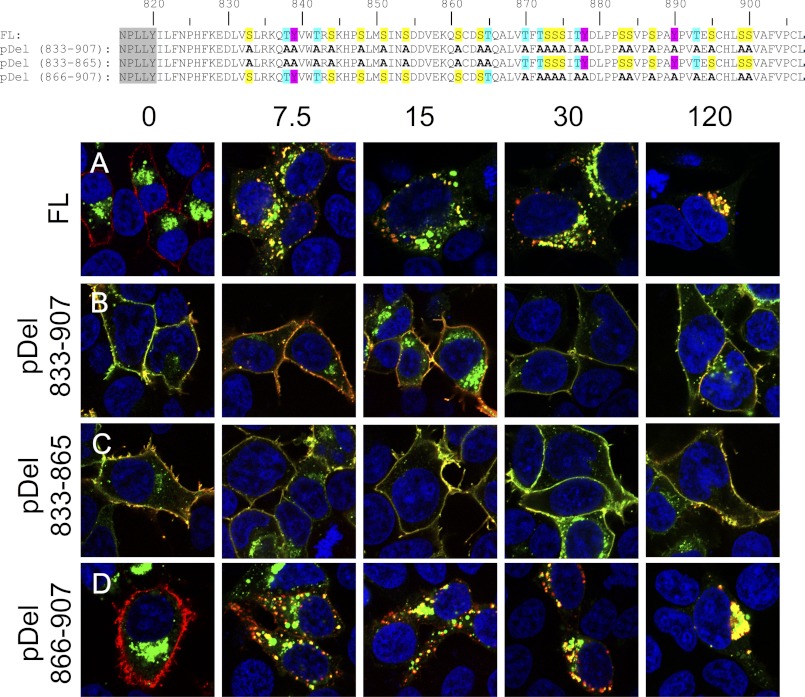
FIGURE 6.
Mutating putative phosphorylation sites inhibits internalization. Shown are primary amino acid sequences of the C-terminal tail for each construct (canonical GPCR NPXXY domain in gray). HEK 293T cells were transiently transfected with the indicated 3×HA N-terminally (red) and C-terminally EGFP (green)-tagged constructs: FL-LGR5 (A), pDel 833–907 (B), pDel 833–865 (C), and pDel 866–907 (D). Cells were pulsed with a MαHA antibody for 45 min on ice, washed, chased for 0, 7.5, 15, 30, or 120 min at 37 °C, fixed, permeabilized, and stained with a GαM568 antibody (red). Merged 100× confocal images are presented (blue, nuclear counterstain).
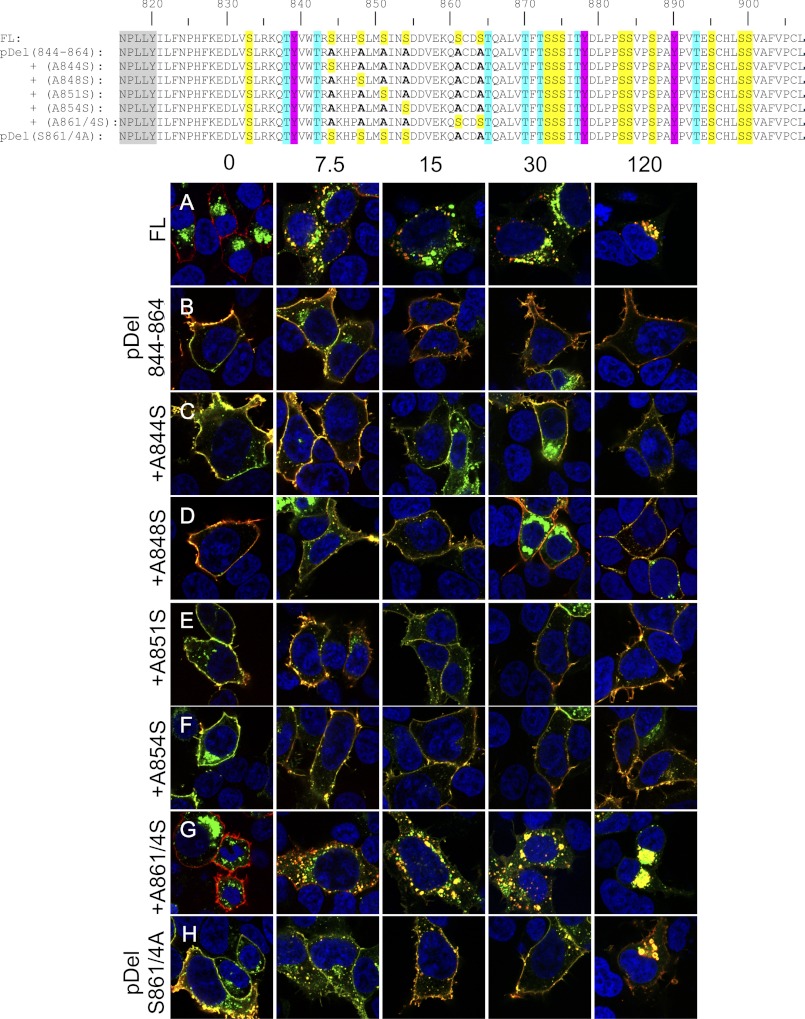
FIGURE 7.
Identification of the primary motif responsible for internalization of LGR5. Shown are the primary amino acid sequences of the C-terminal tail for each construct (canonical GPCR NPXXY domain in gray). HEK 293T cells were transiently transfected with the indicated 3×HA N-terminally (red) and C-terminally EGFP (green)-tagged constructs: FL-LGR5 (A), pDel 844–864 (B), +A844S (C), +A848S (D), +A851S (E), +A854S (F), +A861S/A864S (G), or S861A/S864A (H). Cells were pulsed with a MαHA antibody for 45 min on ice, washed, chased for 0, 7.5, 15, 30, or 120 min at 37 °C, fixed, permeabilized, and stained with a GαM568 antibody (red). Merged 100× confocal images are presented (blue, nuclear counterstain).
On-cell ELISA
On-cell ELISAs were performed according to published protocol with several modifications (26). Briefly, Corning Costar Tissue Culture-treated clear 24-well plates (Corning Costar 3526, Corning) were incubated with 100 μg/ml poly-d-lysine (Sigma P0899) for 6 h at room temperature or overnight at 4 °C. Plates were washed with H2O and air dried in a laminar flow hood. 5.25 × 106 HEK cells (100-mm plate) were transfected with 5 μg of receptor ± 5 μg of dynamin K44A or pcDNA3.1 empty vector and plated at 250,000 cells/well in the prepared 24-well plates. Cells were pulsed with primary mouse anti-HA antibody (1:500, 250 μl/well) for 45 min in SM on ice to block endocytosis, washed four times with 250 μl of cold SM, and chased at 37 °C for the indicated time provided in the text, fixed (4% paraformaldehyde), and then stained with goat anti-mouse-680 (Invitrogen) for 1 h. Each well was washed in PBS three times, aspirated, and imaged on a LI-COR Odyssey using the 700-nm channel and focal offset of 1.5. Untransfected stained cells were used to subtract background signal from each condition. Data were normalized within each receptor type to the amount of receptor present and time zero. Each experiment comprised three technical replicates for each time point and construct tested. A minimum of three independent experiments were performed and analyzed in GraphPad Prism (see “Statistical Analysis”).
Hierarchical Clustering and Heat Map Rendering
The normalized and average percentage surface expression values for each construct were log2 normalized across the entire time course, hierarchically clustered, and presented as a heat map using Tree view, and adapted from a previously published protocol for analysis of microarray data (27).
Statistical Analysis
Data were collected in Microsoft Excel and then transferred to GraphPad Prism (GraphPad Software). Two-way unmatched ANOVAs with a Bonferroni post hoc test were performed, and the results are summarized in Table 1.
TABLE 1.
Analysis of significant change in internalization relative to WT (two-way ANOVA, post hoc Bonferroni)
Results of each data set were analyzed separately (Fig. 8, A–E) and are presented together. For each data set, interaction p < 0.05, receptor p < 0.05, and time <0.05. +, p < 0.05; −, p > 0.05; NT, not tested.
| Receptor | Time |
||||
|---|---|---|---|---|---|
| 3.75 | 7.5 | 15 | 30 | 120 | |
| min | min | min | min | min | |
| +K44A | − | + | + | + | + |
| WT/V2R tail | + | + | + | + | + |
| 834 | + | + | + | + | + |
| 839 | NT | + | + | + | + |
| 844 | NT | + | + | + | + |
| 849 | NT | + | + | + | + |
| 854 | NT | + | + | + | − |
| 859 | NT | + | + | + | − |
| 864 | NT | − | − | − | − |
| 869 | NT | + | − | − | − |
| 874 | NT | − | − | − | − |
| 902 | NT | + | − | − | − |
| pDel 833–907 | NT | + | + | + | + |
| pDel 833–865 | NT | + | + | + | + |
| pDel 866–907 | NT | − | − | − | − |
| pDel 844–864 | NT | + | + | + | + |
| +A844S | NT | + | + | + | + |
| +A848S | NT | + | + | + | + |
| +A851S | NT | + | + | + | + |
| +A854S | NT | + | + | + | + |
| +A861S/A864S | NT | − | − | − | − |
| A861S/A864S | NT | + | + | − | − |
RESULTS
Internalization and Trafficking of LGR5
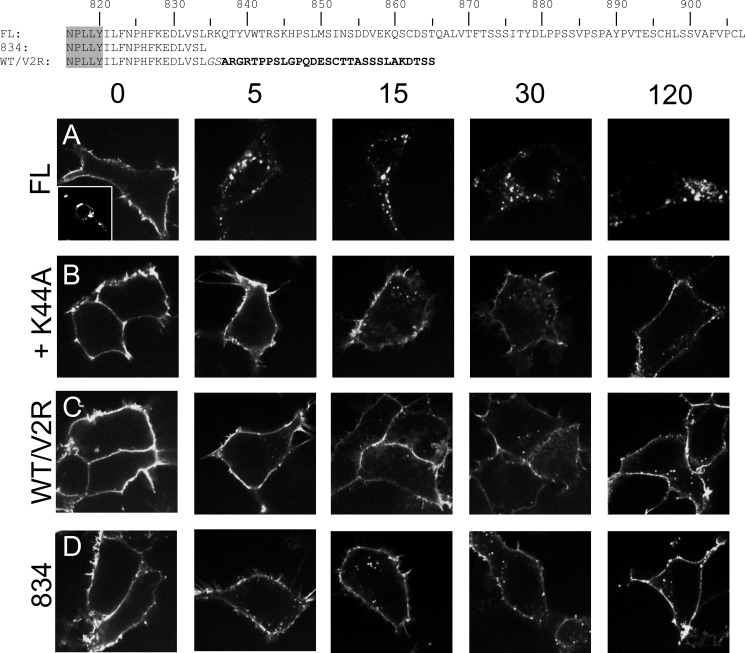
To enable fluorescence visualization of the LGR5 receptor in HEK293T cells at steady state, we constructed a chimera from full-length (FL, 1–907) FL-LGR5 by placing a 3×HA epitope tag at the N terminus and an EGFP moiety at the C terminus. Imaging for EGFP in transiently transfected cells revealed that LGR5 was expressed predominantly in intracellular vesicles in a perinuclear distribution (Fig. 1A, inset). To determine whether these perinuclear receptors first trafficked to the plasma membrane before internalizing, we performed antibody pulse-chase assays in live cells. After antibody labeling of plasma membrane receptor on ice at the 3×HA epitope, the cells were warmed and chased in serum-free medium for 0, 5, 15, 30, or 120 min before fixation, permeabilization, and labeling with a fluorescent secondary antibody (Fig. 1, A–D). Fluorescence imaging and analysis revealed that plasma membrane FL-LGR5 was rapidly internalized into small vesicles within 5 min and trafficked to a perinuclear compartment by 120 min (Fig. 1A). Importantly, as an unbiased confirmation for all confocal based internalization assays, we performed quantitative on-cell ELISAs to precisely measure receptor internalization for Figs. 1 and 4–7. These results are summarized in Table 1 and later in the paper in Fig. 8.
FIGURE 1.
The C-terminal tail of LGR5 regulates its constitutive internalization. Shown are primary amino acid sequences of the C-terminal tail for each construct (canonical GPCR NPXXY domain in gray and V2R tail in bold). HEK 293T cells were transiently transfected with the indicated 3×HA N-terminally epitope-tagged constructs: FL-WT LGR5 full-length (A), FL-LGR5 + dynamin K44A (B), WT/V2R tail (C), or Lgr5 with a truncation at amino acid position 834 (D). A, inset depicts a 3×HA FL-WT LGR5-EGFP fusion and imaged for native EGFP fluorescence. A–D, cells were pulsed with a MαHA antibody for 45 min on ice, washed, chased for 0, 5, 15, 30, or 120 min at 37 °C, fixed, permeabilized, and stained with a GαM-568 antibody (gray scale). 100× confocal images are presented.
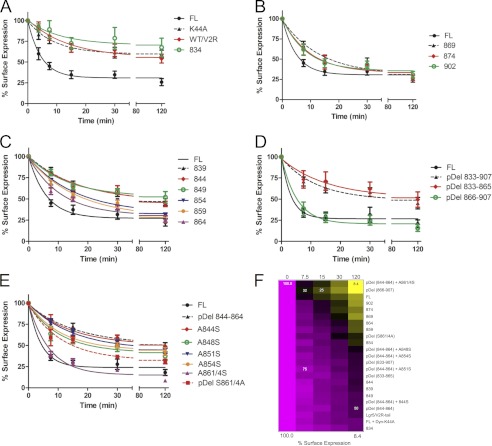
FIGURE 8.
Unbiased quantitative analysis of LGR5 internalization reveals the C-terminal motif responsible for internalization. HEK cells were transfected with the constructs utilized according to Fig. 1 (A), Fig. 4 (B), Fig. 5 (C), Fig. 6 (D), and Fig. 7 (E). Cells were pulse-chased at 37 °C with primary MSαHA antibody fixed and then stained with a GαM680 without permeabilization to assess the fraction of the receptor pulsed that remained on the surface following the chase. Cells were chased for (A) 0, 3.75, 7.5, 15, 30, or 120 min or (B–E) 0, 7.5, 15, 30, or 120 min. Cells were imaged on a LiCOR Odyssey and data normalized to the receptor on the cell surface at time 0 for each construct. F, data from each receptor construct were log2-transformed and normalized to the geometric average of the FL-LGR5 construct and presented as a heat map over the internalization time course (0, 7.5, 15, 30, and 120 min) where bright magenta indicates 100% cell surface expression and bright yellow indicates 8.4% cell surface expression. Reference values for cell surface expression and their correlation to color are indicated on the map.
The C-terminal Tail of LGR5 Is a Primary Modulator of Its Constitutive Internalization
To assess whether constitutive internalization of wild-type LGR5 could be clathrin-mediated, we co-transfected dominant negative dynamin I (K44A) with the receptor (23) and repeated the pulse-chase assay. As expected for clathrin-dependent GPCR internalization, surface LGR5 was extensively stabilized (Fig. 1B). Because GPCR internalization is regulated by G protein receptor kinase-dependent phosphorylation and β-arrestin recruitment to the C-terminal tail (28), we tested this paradigm for LGR5 by replacing its tail with one whose behavior is well characterized, the human V2R tail. V2R is normally stably expressed at the plasma membrane, and its tail also conferred stable plasma membrane expression of LGR5 (Fig. 1C) throughout an internalization time course. Finally, exploiting another known paradigm for stabilizing GPCR surface expression, we truncated the tail of LGR5 14 amino acids downstream of the conserved NPXXY domain at amino acid position 834 (29, 30) to allow for expression of a tailless receptor while preserving the stereotypical eighth α-helix (31). 834 LGR5 also displayed robust plasma membrane expression and very little constitutive internalization (Fig. 1D) as similarly demonstrated in Ref. 15. Collectively, these data demonstrate that the C-terminal tail of LGR5 is a primary modulator of its rapid internalization into the perinuclear compartment at steady state.
Endosome Trafficking of LGR5
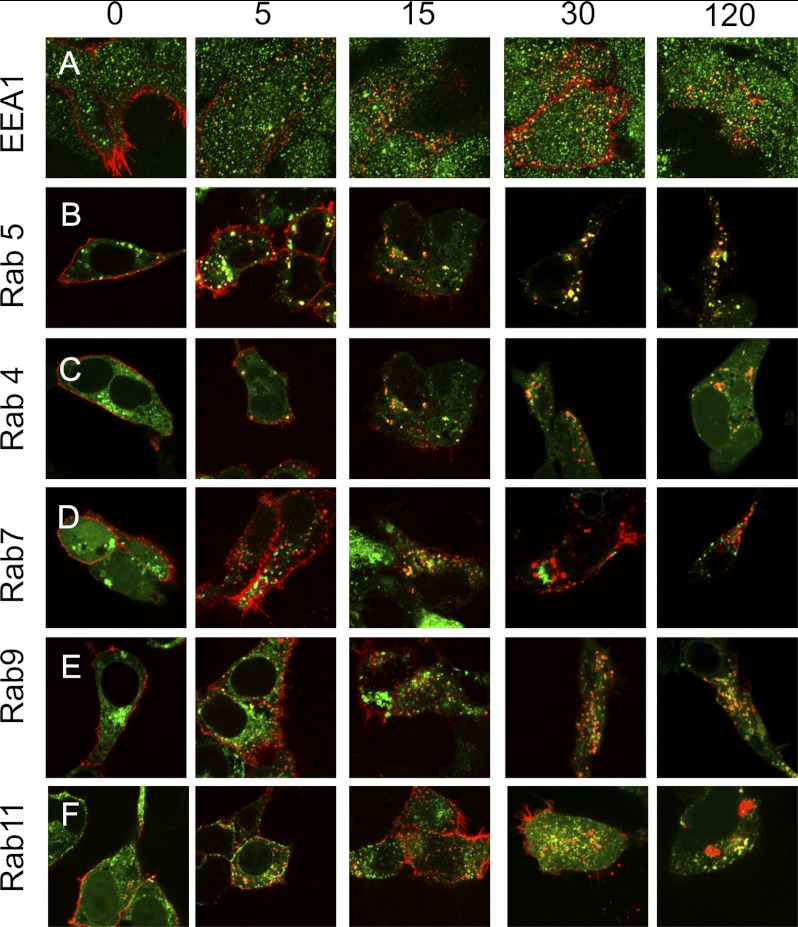
We further characterized the vesicular distribution of FL-LGR5 by analyzing its transit from early to late endosomes. Antibody pulse-chase experiments followed FL-Lgr5 distributions at 0, 5, 15, 30, and 120 min (Fig. 2). Receptors appear in red in these images. To simultaneously identify the corresponding endosome compartments, we also immunostained for specific markers. Fig. 2A demonstrates that the EEA1 (green) co-localizes with LGR5 from between 5 min and 120 min (yellow vesicles), suggesting that LGR5 is rapidly internalized from the plasma membrane into early endosomes (32). LGR5 also extensively co-localized from 5 to 120 min with a GFP-tagged Ras-associated protein-5 (Rab5) (green, Fig. 2B), another early endosome marker necessary for GPCR retrieval from clathrin coated pits into early endosomes. In contrast, much less co-localization with Rab4-GFP was found, a marker of fast recycling endosomes that is often used for rapid delivery of desensitized GPCRs back to the plasma membrane (Fig. 2C) (32). A previous report suggested that FL-LGR5 is degraded following its internalization (15). We correspondingly saw strong co-localization of FL-LGR5 with Rab7-GFP- (Fig. 2D) and Rab9-GFP (Fig. 2E)-tagged endosomes, evidence that LGR5 is trafficking to late endosomes (33). However, we also observed FL-LGR5 in recycling endosomes marked by a Rab11 GFP tag (Fig. 2F) (34). Rab11 has been shown to regulate the transit of cargo from early endosomes to the TGN (35) and delivery from the TGN back to the plasma membrane (36). Moreover, Rab7 and Rab9, in addition to their roles in proper trafficking of cargo to late endosomes and lysosomes, are both essential components of retrograde transport of cargo to the TGN (37–39). The observation that LGR5 is found predominantly in a perinuclear compartment together with its localization with Rab7, Rab9, and Rab11 endosomes led us to the test the hypothesis that a membrane population of LGR5 rapidly internalizes to the trans-Golgi network.
FIGURE 2.
LGR5 rapidly internalizes into early endosomes and transits through late endosomes and recycling endosomes. HEK 293T cells were transiently transfected with a 3×HA N-terminally epitope WT-LGR5 and stained for EEA1 (A, green) or co-transfected with EGFP (green)-tagged Rab5 (B), Rab4 (C), Rab7 (D), Rab9 (E), or Rab11 (F). Cells were pulsed with a MαHA antibody for 45 min on ice, washed, chased for 0, 5, 15, 30, or 120 min at 37 °C, fixed, permeabilized, and stained with a GαM568 antibody (red). Merged 100× confocal images are presented.
Internalized LGR5 Assumes a Trans-Golgi Network Fate
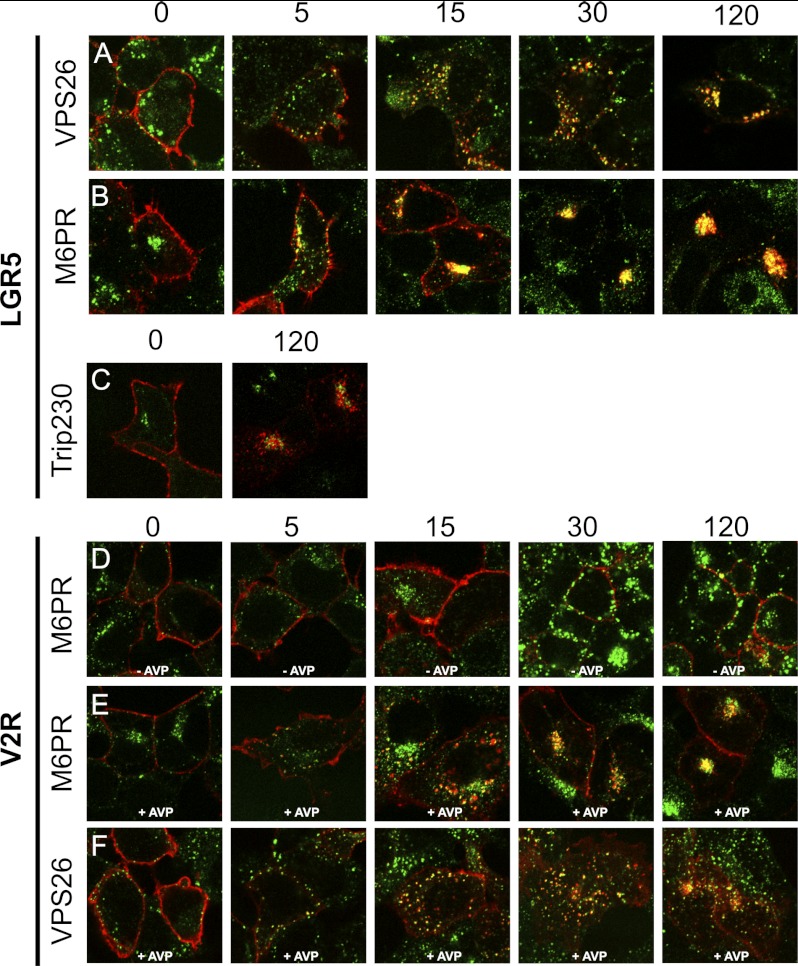
To test this hypothesis we performed an antibody pulse-chase experiment at 0, 5, 15, 30, and 120 min (Fig. 3) to determine whether LGR5 (in red) transits through endosomes positive for vacuolar protein sorting-associated protein 26 (Vps26), a critical component of the retromer complex that facilitates cargo transit to the TGN (40–42). A substantial degree of co-localization of LGR5 with Vps26 was found between 5 and 120 min (Fig. 3A). The CI-M6PR is a marker for perinuclear endosomes (39, 43). We observed co-localization of LGR5 with M6PR (Fig. 3B) by 5 min and extensive perinuclear co-localization in the TGN by 15, 30, and 120 min. To define the TGN more precisely, we also co-stained with another TGN marker, TGN46 (44, 45), and found extensive co-localization of both the steady-state population and internalized fraction of LGR5 with TGN46 (supplemental Fig. 1). LGR5 co-localization with the cis-Golgi marker Trip230 was unremarkable (46) (Fig. 3C). The constitutive activity at steady state of LGR5 is in stark contrast to the human V2R, which showed little to no appreciable internalization throughout a similar pulse-chase experiment, nor co localization with M6PR in the absence of ligand (Fig. 3D). However, when the V2R is stimulated by arginine vasopressin(0.1 IU/ml), we also saw co-localization of V2R with M6PR and Vps26 (Fig. 3, E and F, respectively). These data demonstrate that LGR5 is constitutively internalized through retromer positive endosomes and delivered to the TGN. Moreover, although no agonist was used in these experiments, LGR5 displays properties of an agonist-activated receptor.
FIGURE 3.
Constitutively internalized LGR5 internalizes into VPS26-positive endosomes and is deposited to the TGN. A–C, HEK 293T cells were transiently transfected with a 3×HA N-terminally epitope-tagged WT-LGR5. Cells were pulsed with an HA antibody for 45 min on ice, washed, chased for 0, 5, 15, 30, or 120 min at 37 °C, fixed, permeabilized, and stained with appropriate primary and secondary antibodies to visualize HA (A–C, red), VPS26 (A, green), M6PR (B, green), or Trip230 (C, green). D–F, HEK cells were transfected with a 3×HA N-terminally epitope-tagged human V2R, pulsed with an αHA antibody for 45 min on ice, washed, chased for 0, 5, 15, 30, or 120 min at 37 °C in the absence (−) or presence (+) of arginine vasopressin (0.1 IU/ml), fixed, permeabilized, and stained with appropriate primary and secondary antibodies to visualize HA (D–F, red), M6PR (D and E, green), or Vps26 (F, green). Merged 100× confocal images are presented.
Identification of the Protein Motif Responsible for the Constitutive Internalization of LGR5
An alignment of the C-terminal tails of LGR5 and V2R revealed a cluster of threonine and serine residues previously shown to be important for β-arrestin2-mediated internalization (TFTSS in LGR5, amino acids 870–875) (18, 19). In addition, several GPCRs contain PDZ binding domains, which regulate internalization in the last 4 amino acids of the C-tails (47). Therefore, we designed N-terminally HA-tagged, C-terminally EGFP-tagged, tail truncation mutants around these sites (869) and (874) to test the putative β-arrestin2 interaction motif and (902) to test the final 4 C-terminal amino acids. Similar to above, we performed antibody pulse-chase for each of these receptors to compare their behaviors with FL-LGR5 over the course of 0, 7.5, 15, 30, and 120 min (Fig. 4). Our experimental design allowed for simultaneous assessment of surface labeled receptor (red) to total receptor (EGFP-tagged, green). Compared with a FL-LGR5 (Fig. 4A), which displayed a rapid internalization by 7.5 min, the mutants exhibited only a subtle decrease in internalization: 869 (Fig. 4B), 874del (Fig. 4C), and 902 (Fig. 4D). These data suggest that the constitutive internalization motif is N-terminal to position 869, and the putative β-arrestin2 TFTSS domain is not significant for this process. Interestingly, we did notice at 120 min that 869 localized in markedly dilated vesicles instead of the tightly packed ones typical of the TGN (Fig. 4B). In the (874) at 120 min (Fig. 4C) this phenotype is lost, and the receptor returns to a perinuclear TGN distribution, suggesting that another motif is responsible for proper late trafficking of the internalized receptor.
An LGR5 Internalization Motif between Amino Acid Positions 854 and 864
To narrow the search for the internalization motif we reconstituted the C-terminal tail from position 834 to 864 in 5-amino acid segments and again performed antibody pulse-chase experiments with these truncation mutants for 0, 7.5, 15, 30, or 120 min. Compared with the FL-LGR5 (Fig. 5A), 839 (Fig. 5B), 844 (Fig. 5C), and 849 (Fig. 5D) had significant reductions in internalization rates and reductions in total receptor internalized. 854 (Fig. 5E) and 859 (Fig. 5F) also had significant reductions in internalization rates, but to a lesser extent than 839, 844, and 849. However, for both 854 and 859 the total amount of receptor internalized over a 120-min chase returned to amounts similar to FL-LGR5. These data indicate that the motif responsible for the rapid internalization of LGR5 is between amino acid positions 854 and 864. Intriguingly 859 (Fig. 5F) and 864 (Fig. 5G) both had vesicles that were dilated similarly to those of 869 (Fig. 4B) at 120 min, again suggesting the existence of another motif necessary for correct trafficking of LGR5 once internalized.
Internalization and Phosphorylation Motifs
The C-terminal tail of LGR5 contains 26 potential phosphorylation sites, several of which are located within amino acids 854–864. To first test the role that phosphorylation may play on internalization we made a construct where every potential phosphorylation site was mutated to alanine, pDel 833–907 (Fig. 6B). In an antibody pulse-chase experiment from 0, 7.5, 15, 30, and 120 min, confocal imaging demonstrated that this receptor possessed stable expression on the membrane compared with (Fig. 6A) FL-LGR5, throughout the 120-min time course. We hypothesized that if our above C-tail reconstitution analyses were correct, the putative phosphorylation sites within the first half of the C-tail should be the most important factor regulating internalization. To test this, we separately mutated the phosphorylation sites in each half of the tail. We generated two constructs, one with all potential phosphorylation sites mutated to alanine from positions 833 to 865 and WT sequence from 865–907 (pDel 833–865), and the other with WT sequence from position 833–865 and all putative phosphorylation sites mutated to alanine from position 865 to 907 (pDel 866–907). Our data demonstrate that pDel 833–865 (Fig. 6C) confers robust membrane expression and is resistant to internalization throughout the time course tested. In contrast, pDel 865–907 (Fig. 6D) is initially at the cell membrane but is rapidly internalized by 7.5 min and assumes a perinuclear distribution by 120 min, in a manner similar to FL-LGR5 (Fig. 6A). These data demonstrate that the required motif is within positions 833–865 and reinforces the concept that this process is functionally distinct from that regulated by the TSSS domain present at position 872.
Amino Acid Positions 861 and 864 Are Critical for the Rapid Internalization of LGR5
C-tail region 844–864 contains several putative G protein receptor kinase and casein kinase phosphorylation sites according to the phosphorylation prediction software GPS2.1 (48) (supplemental Table 1). We mutated all of them to alanine (pDel 844–864) and in pulse-chase experiments found that pDel 844–864 was present at the cell surface and robustly resistant to internalization (Fig. 7B). With pDel 844–864 as a template, we mutated each alanine within this region back to its WT residue. The resulting receptors: +A844S (Fig. 7C), +A848S (Fig. 7D), +A851S (Fig. 7E), and +A854S (Fig. 7F) all display robust surface expression and resistance to internalization over a 120-min time course, indicating that these residues are not critical to the internalization. In contrast, the +A861S/A864S possessed internalization dynamics almost identical to those of the WT FL-LGR5 (Fig. 7G). As a proof of principle that these two residues are critical to proper internalization dynamics we mutated only them in the WT receptor, and as expected this mutant, pDel S861A/S864A receptor, verified their importance by displaying robust surface expression and delayed internalization rates (Fig. 7H).
Quantitative Determination of LGR5 Internalization
We performed on-cell ELISAs to quantify precisely the internalization of LGR5 in an unbiased manner. From these experiments we confirmed the imaging data presented previously. We found that LGR5 is constitutively internalized and that this process is dependent upon clathrin-mediated endocytosis and the C-terminal tail of LGR5 (Fig. 8A). Internalization of LGR5 is independent of a PDZ domain or the TSSS domain present at position 872 (Fig. 8B). Rather, our data point to the existence of an additional internalization motif between positions 854 and 864 (Fig. 8C). We confirmed results from Fig. 6, which point to phosphorylation as a likely modulator of LGR5 internalization (Fig. 8D). Finally, we demonstrate the importance of amino acid positions 861 and 864 for proper internalization of LGR5 (Fig. 8E). Statistical analyses supporting these conclusions are presented in tabular form (Table 1). Collectively, these data indicate that amino acid positions 861 and 864 are critical for the rapid internalization of LGR5 and that serines at amino acid positions 844, 848, 851, and 854 may secondarily contribute to the dynamics of LGR5 internalization.
DISCUSSION
The cellular trafficking of GPCRs is an important process that regulates not only the complement of receptors at the plasma membrane but also dynamically controls the cellular responsiveness to activating ligands. A large number of GPCR sorting signals and interacting proteins that support the processing of newly synthesized receptors or their endocytosis, recycling, or degradation following their activation have been characterized (49). Endocytosis of GPCRs is usually regarded as an event by which an activated receptor is uncoupled from its cognate G protein and downstream effectors. This process that leads to desensitization of G protein-mediated signaling usually occurs through an agonist-dependent recruitment of adaptor molecules, like the arrestins, and removal of the receptor from the plasma membrane, typically through clathrin-mediated endocytosis (28). In certain cases, however, constitutively activated GPCRs have been shown to internalize in an agonist-independent fashion (21). Interestingly, in the case of LGR5, it is poorly expressed at the plasma membrane as a result of its efficient endocytosis in the apparent absence of agonist. Our data identify that the structural determinant underlying its constitutive internalization lies in two C-tail serine residues, 861 and 864, that are distinct from those required for arrestin-dependent internalization. We have also demonstrated similar findings in other cell lines including the human colon carcinoma cell line HCT-116 and human osteosarcoma cell line U2OS (data not shown), thereby to eliminate the possibility that this is a cell type-specific phenomenon. We further demonstrate that upon removal from the membrane, LGR5 undergoes retrograde trafficking in Rab5, 7, 9, and Vps26 endosomes en route to the TGN.
Retrograde trafficking of cargo is a tightly regulated process, which delivers cargo from early endosomes to the TGN. Cargo can be delivered to the TGN from the early endosomes directly by retromer or indirectly by retromer in a Rab7- and Rab9-dependent manner (37, 38), routes that TGN46 or CI-M6PR/furin, respectively, employ (38, 50). Our study demonstrates conclusively that LGR5 internalizes through Vps26 endosomes with minimal and transient co-localization with Rab7, 9, and 11, leading us to conclude that the bulk of LGR5 likely traffics directly from early endosomes to the TGN following an internalization route more reminiscent of TGN46 than M6PR or furin. However, previous work has indicated that a fraction of LGR5 is destined for lysosomal degradation (15); therefore, trafficking of LGR5 to Rab7- and Rab9-positive endosomes may reflect this indirect pathway.
Understanding the basic biochemical process through which LGR5 is rapidly internalized and trafficked to the TGN is necessary to more precisely characterize its role in stem and cancer cell biology. TGN trafficking of GPCRs is a relatively new area of study. Its importance to receptor signaling and regulation, however, as illustrated by the following four examples of GPCRs now known to traffic to the TGN, is gaining greater recognition. In the case of the parathyroid hormone receptor type 1 (PTHR), a recent study demonstrated that retromer and not arrestin is ultimately responsible for complete PTHR desensitization, and moreover, PTHR is able to co-immunoprecipitate components of the retromer complex (51). These findings add yet one more component to the fine-tuning of GPCR signaling and raise the interesting possibility that retromer, like arrestin, may also recruit and scaffold other signaling proteins. The CC chemokine receptor CCR5, binds to CCL3–5 and is the primary receptor responsible for HIV binding. Intriguingly, some of the best anti-HIV treatments available that reduce HIV/CCR5 binding induce CCR5 long term desensitization and trafficking to the TGN (52). The β1-adrenergic receptor through an arrestin-dependent mechanism and the G protein-coupled estrogen receptor are just two additional examples of an increasing cohort of GPCRs that redistribute to the TGN following activation for reasons that are not yet fully understood (31, 51).
A comprehensive heat map view of our findings (Fig. 8E) demonstrates the existence of a wide range of receptor- and mutation-specific internalization groupings. This analysis clearly demonstrates that the FL-LGR5 is rapidly internalized and behaves similarly to receptors with C-terminal tails extending past position 859 or those whose potential phosphorylation sites after position 866 were all mutated to alanine. These data also demonstrate that the constitutive internalization of LGR5 is independent of the threonine and serine clusters, which form a putative β-arrestin2 recruitment domain (18). Collectively, the data point to serine residues 861/864 as the most critical determinants of LGR5 constitutive internalization, as is confirmed by the gain- and loss-of-function mutants corresponding to those residues. Moreover, these data also suggest that serines 861/864 are necessary and sufficient for rapid internalization of LGR5. However, from our data it appears that even if these sites are mutated, internalization still ensues but at a much slower rate. The requirement or redundancy of multiple residues regulating internalization has been observed for other receptors in which the most important residues serve some necessary binding or priming function (53). Interestingly, a putative “dileucine” motif, 867LV, resides just after those two Ser residues, whose phosphorylation could enhance the ability of this motif to interact with clathrin-coated pit adaptor proteins in an agonist-independent fashion (54, 55). Our results suggest that phosphorylation at serines 861/864 may serve as an obligate priming event. The GPS2.1 group-based prediction system (48) indicated that LGR5 contains a myriad of putative phosphorylation sites that include motifs for casein kinase1/2 and G protein receptor kinases (supplemental Table 1), which could affect receptor desensitization and signaling (28, 56–58). Position 864 in particular is predicted to be a substrate of the G protein receptor kinase superfamily, and additional studies will be necessary to fully characterize the specific kinases involved.
Two critical and related questions are evident concerning agonist-dependent and constitutive LGR5 internalization. Both center on the determinants that identify LGR5 as a particular type of cargo. The serine/threonine LGR5 cluster at 872–875 TSSS is similar to those observed in other GPCRs, and these motifs are primarily associated with stable receptor/β-arrestin2 complexes that internalize through clathrin-coated pits (18, 19). Although in our study these residues do not appear to regulate the internalization of LGR5, these residues are also conserved and subject to agonist-dependent phosphorylation in the LGR family member FSHR. Importantly, when this domain is mutated, FSHR agonist-mediated arrestin recruitment and internalization are markedly attenuated (59). On this basis we propose that LGR5 ligand-independent internalization is separated functionally from ligand-dependent activation and internalization, which may be driven instead by its TSSS domain. Such a dichotomy, although rare, has been demonstrated for PAR1 receptor, which is constitutively internalized in an arrestin-independent manner yet upon its activation is desensitized through recruitment of arrestins (60, 61). For the PAR1 receptor, this ensures that a reserve population of receptors is ready for deployment to the cell surface following a one-time stimulus and degradation of an activated receptor population. For LGR5 it may signal a requirement to associate with other TGN proteins with which it shares common signaling pathway partners. However, the surprising finding that LGR5 does not recruit arrestin, even following nanomolar binding of R-spondins 1–4 (17), suggests either that ligand-mediated activation of LGR5 breaks the current rules of GPCR activation or that a separate class of endogenous LGR5 ligands exists that can initiate arrestin mediated desensitization. Characterizing the fundamental aspects of LGR5 activation will be important to understand how alterations in LGR5 internalization and trafficking impact receptor signaling, stem cell fate, or tumorigenesis.
Acknowledgments
We thank Vann Bennett, Caroline Ray, Tama Evron, and Yushi Bai for thoughtful discussions and advice.
This work was supported by the Susan G. Komen Foundation Grant KG080627 (to J. C. S. and H. K. L.); Duke Cancer Center Stewart Trust and Duke Cancer Center Cancer and the Environment (to J. C. S., L. K. R., L. S. B., and M. C. G.); and National Institute of Drug Abuse Grant P30 5P30DA29925 (to L. S. B. and M.G.C.).

This article contains supplemental Fig. 1 and Table 1.
- LGR5
- leucine-rich repeat-containing G protein-coupled receptor 5
- CI-M6PR
- cation-independent M6PR
- EEA1
- early endosomal antigen-1
- FL
- full-length
- GPCR
- G protein-coupled receptor
- M6PR
- mannose 6-phosphate receptor
- Rab
- Ras-associated protein
- SM
- staining medium
- TGN
- trans-Golgi network
- Trip230
- Golgi-microtubule-associated protein of 210 kDa
- V2R
- vasopressin 2 receptor
- Vps26
- vacuolar protein sorting-associated protein 26.
REFERENCES
- 1. Hsu S. Y., Liang S.-G., Hsueh A. J. (1998) Characterization of two LGR genes homologous to gonadotropin and thyrotropin receptors with extracellular leucine-rich repeats and a G protein-coupled, seven-transmembrane region. Mol. Endocrinol. 12, 1830–1845 [DOI] [PubMed] [Google Scholar]
- 2. Hsu S. Y., Kudo M., Chen T., Nakabayashi K., Bhalla A., van der Spek P. J., van Duin M., Hsueh A. J. (2000) The three subfamilies of leucine-rich repeat-containing G protein-coupled receptors (LGR): identification of LGR6 and LGR7 and the signaling mechanism for LGR7. Mol. Endocrinol. 14, 1257–1271 [DOI] [PubMed] [Google Scholar]
- 3. Barker N., van Es J. H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P. J., Clevers H. (2007) Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 [DOI] [PubMed] [Google Scholar]
- 4. Jaks V., Barker N., Kasper M., van Es J. H., Snippert H. J., Clevers H., Toftgård R. (2008) Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat. Genet. 40, 1291–1299 [DOI] [PubMed] [Google Scholar]
- 5. Barker N., Huch M., Kujala P., van de Wetering M., Snippert H. J., van Es J. H., Sato T., Stange D. E., Begthel H., van den Born M., Danenberg E., van den Brink S., Korving J., Abo A., Peters P. J., Wright N., Poulsom R., Clevers H. (2010) Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6, 25–36 [DOI] [PubMed] [Google Scholar]
- 6. Brzeszczynska J., Ramaesh K., Dhillon B., Ross J. A. (2012) Molecular profile of organ culture-stored corneal epithelium: LGR5 is a potential new phenotypic marker of residual human corneal limbal epithelial stem cells. Int. J. Mol. Med. 29, 871–876 [DOI] [PubMed] [Google Scholar]
- 7. de Visser K. E., Ciampricotti M., Michalak E. M., Tan D. W., Speksnijder E. N., Hau C. S., Clevers H., Barker N., Jonkers J. (2012) Developmental stage-specific contribution of LGR5+ cells to basal and luminal epithelial lineages in the postnatal mammary gland. J. Pathol. 228, 300–309 [DOI] [PubMed] [Google Scholar]
- 8. Sato T., Vries R. G., Snippert H. J., van de Wetering M., Barker N., Stange D. E., van Es J. H., Abo A., Kujala P., Peters P. J., Clevers H. (2009) Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 [DOI] [PubMed] [Google Scholar]
- 9. Barker N., Ridgway R. A., van Es J. H., van de Wetering M., Begthel H., van den Born M., Danenberg E., Clarke A. R., Sansom O. J., Clevers H. (2009) Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457, 608–611 [DOI] [PubMed] [Google Scholar]
- 10. Schepers A. G., Snippert H. J., Stange D. E., van den Born M., van Es J. H., van de Wetering M., Clevers H. (2012) Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science 337, 730–735 [DOI] [PubMed] [Google Scholar]
- 11. Leushacke M., Barker N. (2012) Lgr5 and Lgr6 as markers to study adult stem cell roles in self-renewal and cancer. Oncogene 31, 3009–3022 [DOI] [PubMed] [Google Scholar]
- 12. de Lau W., Barker N., Low T. Y., Koo B. K., Li V. S., Teunissen H., Kujala P., Haegebarth A., Peters P. J., van de Wetering M., Stange D. E., van Es J. E., Guardavaccaro D., Schasfoort R. B., Mohri Y., Nishimori K., Mohammed S., Heck A. J., Clevers H. (2011) Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 476, 293–297 [DOI] [PubMed] [Google Scholar]
- 13. Snippert H. J., Haegebarth A., Kasper M., Jaks V., van Es J. H., Barker N., van de Wetering M., van den Born M., Begthel H., Vries R. G., Stange D. E., Toftgård R., Clevers H. (2010) Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science 327, 1385–1389 [DOI] [PubMed] [Google Scholar]
- 14. Glinka A., Dolde C., Kirsch N., Huang Y. L., Kazanskaya O., Ingelfinger D., Boutros M., Cruciat C. M., Niehrs C. (2011) LGR4 and LGR5 are R-spondin receptors mediating Wnt/β-catenin and Wnt/PCP signalling. EMBO Rep. 12, 1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carmon K. S., Lin Q., Gong X., Thomas A., Liu Q. (2012) LGR5 interacts and cointernalizes with Wnt receptors to modulate Wnt/β-catenin signaling. Mol. Cell. Biol. 32, 2054–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gong X., Carmon K. S., Lin Q., Thomas A., Yi J., Liu Q. (2012) LGR6 is a high affinity receptor of R-spondins and potentially functions as a tumor suppressor. PLoS ONE 7, e37137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carmon K. S., Gong X., Lin Q., Thomas A., Liu Q. (2011) R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/β-catenin signaling. Proc. Natl. Acad. Sci. U.S.A. 108, 11452–11457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oakley R. H., Laporte S. A., Holt J. A., Barak L. S., Caron M. G. (1999) Association of β-arrestin with G protein-coupled receptors during clathrin-mediated endocytosis dictates the profile of receptor resensitization. J. Biol. Chem. 274, 32248–32257 [DOI] [PubMed] [Google Scholar]
- 19. Oakley R. H., Laporte S. A., Holt J. A., Barak L. S., Caron M. G. (2001) Molecular determinants underlying the formation of stable intracellular G protein-coupled receptor-β-arrestin complexes after receptor endocytosis. J. Biol. Chem. 276, 19452–19460 [DOI] [PubMed] [Google Scholar]
- 20. Wilbanks A. M., Laporte S. A., Bohn L. M., Barak L. S., Caron M. G. (2002) Apparent loss-of-function mutant GPCRs revealed as constitutively desensitized receptors. Biochemistry 41, 11981–11989 [DOI] [PubMed] [Google Scholar]
- 21. Barak L. S., Oakley R. H., Laporte S. A., Caron M. G. (2001) Constitutive arrestin-mediated desensitization of a human vasopressin receptor mutant associated with nephrogenic diabetes insipidus. Proc. Natl. Acad. Sci. U.S.A. 98, 93–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bryksin A. V., Matsumura I. (2010) Overlap extension PCR cloning: a simple and reliable way to create recombinant plasmids. BioTechniques 48, 463–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang J., Ferguson S. S., Barak L. S., Ménard L., Caron M. G. (1996) Dynamin and β-arrestin reveal distinct mechanisms for G protein-coupled receptor internalization. J. Biol. Chem. 271, 18302–18305 [DOI] [PubMed] [Google Scholar]
- 24. Choudhury A., Dominguez M., Puri V., Sharma D. K., Narita K., Wheatley C. L., Marks D. L., Pagano R. E. (2002) Rab proteins mediate Golgi transport of caveola-internalized glycosphingolipids and correct lipid trafficking in Niemann-Pick C cells. J. Clin. Invest. 109, 1541–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kingston R. E., Chen C. A., Okayama H. (2001) Calcium phosphate transfection. Curr. Protoc. Immunol. Chapter 10, Unit 10.13 [DOI] [PubMed] [Google Scholar]
- 26. Chang X.-B., Mengos A., Hou Y.-X., Cui L., Jensen T. J., Aleksandrov A., Riordan J. R., Gentzsch M. (2008) Role of N-linked oligosaccharides in the biosynthetic processing of the cystic fibrosis membrane conductance regulator. J. Cell Sci. 121, 2814–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Snyder J. C., Zemke A. C., Stripp B. R. (2009) Reparative capacity of airway epithelium impacts deposition and remodeling of extracellular matrix. Am. J. Respir. Cell Mol. Biol. 40, 633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gainetdinov R. R., Premont R. T., Bohn L. M., Lefkowitz R. J., Caron M. G. (2004) Desensitization of G protein-coupled receptors and neuronal functions. Annu. Rev. Neurosci. 27, 107–144 [DOI] [PubMed] [Google Scholar]
- 29. Probst W. C., Snyder L. A., Schuster D. I., Brosius J., Sealfon S. C. (1992) Sequence alignment of the G-protein coupled receptor superfamily. DNA Cell Biol. 11, 1–20 [DOI] [PubMed] [Google Scholar]
- 30. Barak L. S., Tiberi M., Freedman N. J., Kwatra M. M., Lefkowitz R. J., Caron M. G. (1994) A highly conserved tyrosine residue in G protein-coupled receptors is required for agonist-mediated β2-adrenergic receptor sequestration. J. Biol. Chem. 269, 2790–2795 [PubMed] [Google Scholar]
- 31. Cheng S. B., Quinn J. A., Graeber C. T., Filardo E. J. (2011) Down-modulation of the G protein-coupled estrogen receptor, GPER, from the cell surface occurs via a trans-Golgi-proteasome pathway. J. Biol. Chem. 286, 22441–22455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mu F. T., Callaghan J. M., Steele-Mortimer O., Stenmark H., Parton R. G., Campbell P. L., McCluskey J., Yeo J. P., Tock E. P., Toh B. H. (1995) EEA1, an early endosome-associated protein: EEA1 is a conserved α-helical peripheral membrane protein flanked by cysteine “fingers” and contains a calmodulin-binding IQ motif. J. Biol. Chem. 270, 13503–13511 [DOI] [PubMed] [Google Scholar]
- 33. Soldati T., Rancaño C., Geissler H., Pfeffer S. R. (1995) Rab7 and Rab9 are recruited onto late endosomes by biochemically distinguishable processes. J. Biol. Chem. 270, 25541–25548 [DOI] [PubMed] [Google Scholar]
- 34. Ullrich O., Reinsch S., Urbé S., Zerial M., Parton R. G. (1996) Rab11 regulates recycling through the pericentriolar recycling endosome. J. Cell Biol. 135, 913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wilcke M., Johannes L., Galli T., Mayau V., Goud B., Salamero J. (2000) Rab11 regulates the compartmentalization of early endosomes required for efficient transport from early endosomes to the trans-Golgi network. J. Cell Biol. 151, 1207–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen W., Feng Y., Chen D., Wandinger-Ness A. (1998) Rab11 is required for trans-Golgi network-to-plasma membrane transport and a preferential target for GDP dissociation inhibitor. Mol. Biol. Cell 9, 3241–3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rojas R., van Vlijmen T., Mardones G. A., Prabhu Y., Rojas A. L., Mohammed S., Heck A. J., Raposo G., van der Sluijs P., Bonifacino J. S. (2008) Regulation of retromer recruitment to endosomes by sequential action of Rab5 and Rab7. J. Cell Biol. 183, 513–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chia P. Z., Gasnereau I., Lieu Z. Z., Gleeson P. A. (2011) Rab9-dependent retrograde transport and endosomal sorting of the endopeptidase furin. J. Cell Sci. 124, 2401–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pfeffer S. R. (2009) Multiple routes of protein transport from endosomes to the trans-Golgi network. FEBS Lett. 583, 3811–3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bonifacino J. S., Rojas R. (2006) Retrograde transport from endosomes to the trans-Golgi network. Nat. Rev. Mol. Cell Biol. 7, 568–579 [DOI] [PubMed] [Google Scholar]
- 41. Seaman M. N. (2005) Recycle your receptors with retromer. Trends Cell Biol. 15, 68–75 [DOI] [PubMed] [Google Scholar]
- 42. Seaman M. N. (2004) Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J. Cell Biol. 165, 111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Duncan J. R., Kornfeld S. (1988) Intracellular movement of two mannose 6-phosphate receptors: return to the Golgi apparatus. J. Cell Biol. 106, 617–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mallet W. G., Maxfield F. R. (1999) Chimeric forms of furin and TGN38 are transported with the plasma membrane in the trans-Golgi network via distinct endosomal pathways. J. Cell Biol. 146, 345–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ghosh R. N., Mallet W. G., Soe T. T., McGraw T. E., Maxfield F. R. (1998) An endocytosed TGN38 chimeric protein is delivered to the TGN after trafficking through the endocytic recycling compartment in CHO cells. J. Cell Biol. 142, 923–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rios R. M., Tassin A. M., Celati C., Antony C., Boissier M. C., Homberg J. C., Bornens M. (1994) A peripheral protein associated with the cis-Golgi network redistributes in the intermediate compartment upon brefeldin A treatment. J. Cell Biol. 125, 997–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cao T. T., Deacon H. W., Reczek D., Bretscher A., von Zastrow M. (1999) A kinase-regulated PDZ-domain interaction controls endocytic sorting of the β2-adrenergic receptor. Nature 401, 286–290 [DOI] [PubMed] [Google Scholar]
- 48. Xue Y., Ren J., Gao X., Jin C., Wen L., Yao X. (2008) GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol. Cell. Proteomics 7, 1598–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. von Zastrow M., Williams J. T. (2012) Modulating neuromodulation by receptor membrane traffic in the endocytic pathway. Neuron 76, 22–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ganley I. G., Espinosa E., Pfeffer S. R. (2008) A syntaxin 10-SNARE complex distinguishes two distinct transport routes from endosomes to the trans-Golgi in human cells. J. Cell Biol. 180, 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Feinstein T. N., Wehbi V. L., Ardura J. A., Wheeler D. S., Ferrandon S., Gardella T. J., Vilardaga J. P. (2011) Retromer terminates the generation of cAMP by internalized PTH receptors. Nat. Chem. Biol. 7, 278–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Escola J. M., Kuenzi G., Gaertner H., Foti M., Hartley O. (2010) CC chemokine receptor 5 (CCR5) desensitization: cycling receptors accumulate in the trans-Golgi network. J. Biol. Chem. 285, 41772–41780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kouhen O. M., Wang G., Solberg J., Erickson L. J., Law P. Y., Loh H. H. (2000) Hierarchical phosphorylation of δ-opioid receptor regulates agonist-induced receptor desensitization and internalization. J. Biol. Chem. 275, 36659–36664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Windheim M., Hilgendorf A., Burgert H. G. (2004) Immune evasion by adenovirus E3 proteins: exploitation of intracellular trafficking pathways. Curr. Top. Microbiol. Immunol. 273, 29–85 [DOI] [PubMed] [Google Scholar]
- 55. Pitcher C., Höning S., Fingerhut A., Bowers K., Marsh M. (1999) Cluster of differentiation antigen 4 (CD4) endocytosis and adaptor complex binding require activation of the CD4 endocytosis signal by serine phosphorylation. Mol. Biol. Cell 10, 677–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tobin A. B., Butcher A. J., Kong K. C. (2008) Location, location, location: site-specific GPCR phosphorylation offers a mechanism for cell type-specific signalling. Trends Pharmacol. Sci. 29, 413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Torrecilla I., Spragg E. J., Poulin B., McWilliams P. J., Mistry S. C., Blaukat A., Tobin A. B. (2007) Phosphorylation and regulation of a G protein-coupled receptor by protein kinase CK2. J. Cell Biol. 177, 127–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Budd D. C., McDonald J. E., Tobin A. B. (2000) Phosphorylation and regulation of a Gq/11-coupled receptor by casein kinase 1α. J. Biol. Chem. 275, 19667–19675 [DOI] [PubMed] [Google Scholar]
- 59. Kara E., Crépieux P., Gauthier C., Martinat N., Piketty V., Guillou F., Reiter E. (2006) A phosphorylation cluster of five serine and threonine residues in the C terminus of the follicle-stimulating hormone receptor is important for desensitization but not for β-arrestin-mediated ERK activation. Mol. Endocrinol. 20, 3014–3026 [DOI] [PubMed] [Google Scholar]
- 60. Paing M. M., Stutts A. B., Kohout T. A., Lefkowitz R. J., Trejo J. (2002) β-Arrestins regulate protease-activated receptor-1 desensitization but not internalization or down-regulation. J. Biol. Chem. 277, 1292–1300 [DOI] [PubMed] [Google Scholar]
- 61. Chen C. H., Paing M. M., Trejo J. (2004) Termination of protease-activated receptor-1 signaling by β-arrestins is independent of receptor phosphorylation. J. Biol. Chem. 279, 10020–10031 [DOI] [PubMed] [Google Scholar]