Background: Regulation of ubiquitin ligases is the key control step in the post-translational attachment of ubiquitin.
Results: The ubiquitin-like protein SUMO post-translationally modifies the yeast ubiquitin ligase Rsp5p.
Conclusion: Rsp5p activity is regulated by the SUMO ligase Siz1p and in turn regulates Siz1p SUMO ligase activity.
Significance: Regulation of the ubiquitylation and SUMOylation machinery involves complex interactions.
Keywords: Intracellular Trafficking, Post-translational Modification, SUMO, Ubiquitin Ligase, Yeast
Abstract
The post-translational modifiers ubiquitin and small ubiquitin-related modifier (SUMO) regulate numerous critical signaling pathways and are key to controlling the cellular fate of proteins in eukaryotes. The attachment of ubiquitin and SUMO involves distinct, but related, machinery. However, it is now apparent that many substrates can be modified by both ubiquitin and SUMO and that some regulatory interaction takes place between the respective attachment machinery. Here, we demonstrate that the Saccharomyces cerevisiae ubiquitin ligase Rsp5p, a member of the highly conserved Nedd4 family of ubiquitin ligases, is SUMOylated in vivo. We further show that Rsp5p SUMOylation is mediated by the SUMO ligases Siz1p and Siz2p, members of the conserved family of PIAS SUMO ligases that are, in turn, substrates for Rsp5p-mediated ubiquitylation. Our experiments show that SUMOylated Rsp5p has reduced ubiquitin ligase activity, and similarly, ubiquitylated Siz1p demonstrates reduced SUMO ligase activity leading to respective changes in both ubiquitin-mediated sorting of the manganese transporter Smf1p and polySUMO chain formation. This reciprocal regulation of these highly conserved ligases represents an exciting and previously unidentified system of cross talk between the ubiquitin and SUMO systems.
Introduction
Post-translational modification by ubiquitin and the related homologue, Small Ubiquitin-related MOdifier (SUMO),2 dramatically changes the cellular fate of the modified protein. Ubiquitylation and SUMOylation reaction cascades are very similar, requiring an activating enzyme (E1), a conjugating enzyme (E2), and, in most cases, a ligase (E3) that dictates substrate specificity. Whereas both types of modifications have been well studied individually, it has become clear in recent years that ubiquitin and SUMO are closely interlinked. For example, it has been suggested that in some pathways, such as proliferating cell nuclear antigen regulation (1, 2), NF-κB transcription factor activation (3), and huntingtin-caused neurotoxicity (4), SUMO may act as a ubiquitin antagonist. The discovery of a conserved family of Really Interesting New Gene (RING) domain ubiquitin ligases termed SUMO-targeted Ubiquitin Ligases (STUBLs) has identified a different form of cross-talk between ubiquitin and SUMO. STUBLs bind to SUMOylated substrates via a SUMO interaction motif and subsequently target them for ubiquitylation (5, 6). Recently, a novel form of ligase activity, SUMO-regulated Ubiquitin Ligase (SRUBL) has been described in the enzyme BRCA1. In this case the activity of the RING-type ubiquitin ligase BRCA1, which is implicated in breast and ovarian cancer, is SUMO-regulated. The PIAS (Protein Inhibitors of Activated STATs) family of SUMO ligases control SUMO modification of BRCA1, and this modification enhances ubiquitin ligase activity (7).
The PIAS SUMO ligases are highly conserved in evolution with four family members, Siz1p, Siz2p, Mms21p, and Zip3p, identified in the yeast Saccharomyces cerevisiae. PIAS SUMO ligases resemble RING domain ubiquitin ligases and form a scaffold to bring SUMO-charged E2 together with the substrate (8, 9). It has been suggested that in yeast Siz1p is largely responsible for the majority of SUMO conjugation and numerous substrates of Siz1p have been reported mostly related to DNA repair and regulation of cell division (10–13). In contrast, the related protein Siz2p regulates telomere position and telomerase activity (14, 15), and although Siz1p and Siz2p are believed to have unique substrates, SUMOylation of many proteins can be stimulated by either ligase (16).
To date most SUMO ligases that have been identified are of the RING-type. However, with respect to ubiquitin ligation, both RING and HECT (Homologous to E6-AP Carboxyl Terminus) ligases have been identified. Unlike RING ligases, HECT ubiquitin ligases transiently accept ubiquitin from the E2 before transferring it to the substrate protein (3). One of the best studied examples of HECT ubiquitin ligases is the Nedd4 family, which is involved in a diverse range of cellular functions (17, 18). In humans, nine Nedd4 ligases have been identified, with Nedd4-2 shown to be involved in the regulated endocytosis of the renal epithelial sodium channel ENaC, misregulation of which causes hereditary hypertension (19). Members of the Nedd4 family are also involved in numerous other important cellular processes (18), including budding of retroviruses (20), regulation of bone morphogenetic protein signaling (21), and regulation of insulin-like growth factor 1 receptor abundance during development (22). Nedd4 family ligases contain a lipid- and protein-interacting C2 domain, several substrate interaction WW domains, and a catalytic HECT domain, which forms a thioester bond with ubiquitin before transferring it to the substrate (17).
S. cerevisiae contains a single Nedd4 ligase, Rsp5p, which is essential for cell viability (17, 24). Arguably the best studied role for Rsp5p is in ubiquitin-mediated endocytosis (23–25). Plasma membrane permeases, such as the manganese transporter Smf1p, the permease Gap1p, and many others, are ubiquitylated by Rsp5p in response to an excess of nutrients and then sorted via multivesicular bodies to the vacuole (a degradative compartment equivalent to the mammalian lysozome). As with other Nedd4 family ligases, Rsp5p generally interacts with substrates via its WW domains binding to short X1PX2Y (PY) motifs in the interacting protein, where X1 is often, but limited to, a proline or leucine residue. However, some substrates do not contain PY motifs and instead utilize adaptor proteins that contain PY motifs, such as Bsd2p, Tre1/2p, Ear1p, Ssh4p, and the arrestin-like Art proteins (26–29). Rsp5p-mediated ubiquitylation is also implicated in many other pathways, such as DNA repair (30), control of RNA polymerase stability (31, 32), regulation of substrate recruitment to the proteasome (33), and mitochondrial inheritance (34).
In this study we report the observation that Rsp5p is SUMOylated, largely by the PIAS SUMO ligase Siz1p. We demonstrate that SUMOylation leads to suppression of Rsp5p ligase activity and associated changes in ubiquitin-mediated endocytosis. Furthermore, we show that Siz1p and Siz2p are potential substrates for Rsp5p-mediated ubiquitylation and that mutations in Siz1p and Siz2p that alter their ability to interact with Rsp5p result in reduced SUMO ligase activity. These data represent a novel reciprocal regulation feedback loop between two important and conserved ligase families and shed further light on the complex regulatory interactions that takes place between ubiquitin and SUMO modification pathways.
EXPERIMENTAL PROCEDURES
Yeast Strains, Media, and Plasmids
Yeast strains siz1Δ and siz2Δ were derived from the BY4742 strain obtained from the Open Biosystems (Thermo Scientific) knock-out collection (MAT α his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 YDR409w::kanMX4 for siz1Δ and YOR156c::kanMX4 for siz2Δ). Strains producing Myc-Rsp5p, TAP-Rsp5p, and its variants were generated by plasmid complementation of a rsp5Δ strain as described previously (26). The siz1Δsiz2Δ strain was produced by replacing the SIZ2 gene in siz1Δ with Schizosaccharomyces pombe HIS5. The EN44 strain was made by inserting a natMX cassette just upstream of the RSP5 start codon (28).
Full-length ORFs of RSP5, SIZ1, SIZ2, UBC9, AOS1, UBA2, and UBC1 were PCR-amplified from yeast genomic DNA, whereas SMT3, and His8-FLAG-tagged SMT3 and ubiquitin ORFs were synthesized by Eurofins. The synthetic SMT3 sequence encoded a “mature” form of Smt3p with a C-terminal Gly-Gly motif (removing the last three C-terminal amino acids). All mutations described throughout the study were introduced by QuikChange site-directed mutagenesis protocol (Stratagene). PY motif mutants were produced by point mutation of indicated tyrosine residues to alanines. For Rsp5p SUMOylation site mutations, modified lysines were substituted for arginine. WW domain mutants of GST-Rsp5p were described previously (26). For protein expression in yeast ORFs of RSP5, SIZ1, SIZ2, SMT, His-FLAGSmt3, and His-FLAG-ubiquitin were subcloned into pYCYlac111 vector derivatives under the control of the TPI promoter with HA, Myc, or FLAG N-terminal epitope tags. The TAP-Bsd2p construct was described previously (26). For bacterial expression and protein purification, SIZ1, SIZ2, UBC9, and SMT3 ORFs were subcloned into pET-30a(+), pET-15a(+) (Novagen), or pGEX-6P-2 (GE Healthcare). For bacterial growth, 2XYT medium was used (Melford); for growing yeast, Synthetic Complete with appropriate amino acid drop-outs (Sigma) was utilized. Tests of cadmium sensitivity were performed with serial 10-fold dilutions of stationary phase cells as described in Ref. 35 using 50 μm CdCl2. For cycloheximide chase experiments yeast were grown to log phase in the appropriate amino acid drop-out media, and samples were taken for immunoblotting at 0, 0.5, 1, and 2 h following the addition of 100 μg ml−1 cycloheximide (Melford).
Protein Purification, Mass Spectrometry, and Immunoprecipitation
His-tagged Siz1p(1–465), Siz2p, and their PY mutants, cytBsd2p, Ubc9p, Ubc1p, Smt3p, and GST-tagged Rsp5p with its WW domain mutants were produced recombinantly in BL21-CodonPlusTM Escherichia coli (Stratagene) and purified by standard methods using HIS-Select® nickel affinity gel (Sigma) for His-tagged proteins or glutathione-Sepharose 4B (GE Healthcare) for GST-tagged proteins. GST-free Rsp5p was prepared using PreScission Protease (GE Healthcare). A heterodimer of Aos1/Uba2 was His-purified as described previously (36). For tandem affinity purification (TAP) of TAP-Rsp5p and TAP-Bsd2p, 2 liters of log phase cultures of yeast, producing the protein of interest, were resuspended in lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 5 mm MgCl2, EDTA-free protease inhibitors mixture (Roche Applied Science), 10% glycerol, 1 mm DTT, 1% Triton X-100 for TAP-Bsd2p or for TAP-Rsp5p 0.1% OPTG with 1% CHAPS) and lysed in a high pressure microfluidizer (Avestin) at 25,000 kilopascals. Subsequent purification was performed as described previously (26). Proteins were eluted with SDS-PAGE loading buffer and analyzed by SDS-PAGE with silver staining or immunoblotting.
TAP-Rsp5p bands were excised from a silver-stained 7.5% SDS-PAGE and in-gel digested with trypsin (Sigma) as described previously (37). The fragmented peptides were analyzed by LC-MS/MS in Agilent LC-MS, comprising a 1100 Series LC and SL Ion Trap Mass Selective Detector. The results were analyzed using an error-tolerant search for variable modifications including SUMOylation, EQIGG (K), using the Mascot search engine (Matrix Sciences). Prediction of possible SUMOylation sites was made using SUMOsp.
Immunoprecipitation and Detection of Proteins
FLAG-Smt3p, HA-tagged Siz1p, Siz2p, and their PY motif mutants were produced in appropriate yeast strains. Typically, 500 ml of a log phase culture was resuspended in lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 5 mm MgCl2, EDTA-free protease inhibitors mixture (Roche Applied Science), 20 mm N-ethylmaleimide, 10% glycerol, 1% Triton X-100) and lysed as above. Lysates were incubated overnight at 4 °C with anti-FLAG-M2 magnetic beads (Sigma) or EZviewTM red anti-HA affinity gel (Sigma). Anti-GFP antibody GSN149 (Sigma) conjugated with DynabeadsTM (Invitrogen) or Sepharose CL-4B (Sigma) was used as a negative control. All immunoprecipitants were washed with lysis buffer, and the proteins were eluted with SDS-PAGE sample buffer and analyzed by immunoblotting. For purification under denaturing conditions 100-ml yeast cultures for His-FLAG-ubiquitin or 500-ml cultures for His-FLAGSmt3 were grown to log phase and cells lysed by vortexing with glass beads for 5 min (ubiquitin) or using a microfluidizer as described above (Smt3) in a denaturing buffer (ubiquitin) (6 m guanidine HCl, 100 mm NaH2PO4·H2O, 10 mm Tris, 1% Triton X-100, 1 mm PMSF, pH 8.0). Cleared lysate were then incubated with HIS-Select resin for 2 h, and the resin was washed twice with 10 resin volumes of denaturing lysis buffer, twice with 10 resin volumes of nondenaturing wash buffer (50 mm NaH2PO4·H2O, 300 mm NaCl, 20 mm imidazole, pH 8.0) before bound proteins were eluted with SDS sample buffer.
For immunoblotting of cell lysates, samples were prepared using an adaptation of the alkaline lysis method (2, 6). Proteins were detected with anti-FLAG-M2 antibody (Sigma), anti-HA (Sigma), anti-Nedd4 (Abcam), anti-Bsd2 (26), anti-S-tag antibody (Novagen), anti-panubiquitin (Dako), anti-calmodulin-binding protein (TAP tag; Millipore), anti-Myc 9E10 (Sigma), anti-GST (Sigma), and anti-GAPDH (Sigma). Membranes were boiled in deionized water before incubation with anti-ubiquitin antibodies. For in vitro ubiquitylation and SUMOylation assays, secondary antibodies were IRDye® 680LT or IRDye® 800 goat anti-mouse/anti-rabbit (LI-COR), visualized using a LI-COR Biosciences Odyssey infrared scanner. For other experiments, horseradish peroxidase (HRP)-conjugated donkey anti-mouse or anti-rabbit secondary antibodies (Sigma) were used and detected with Immobilon HRP substrate (Millipore).
In Vitro Ubiquitylation and SUMOylation Assays
30-μl volume ubiquitylation assays were performed in 50 mm Tris, pH 7.4, 10 mm MgCl2, and 10 mm ATP with 100 ng of Uba1p (BostonBiochem), 100 ng of Ubc1p, 1 μg of methylated ubiquitin (Enzo Life Sciences), 500 ng of GST-Rsp5p or mutants, and 500 ng of either cytBsd2p, Siz1p(1–465), Siz2p, or their mutants. SUMOylation reactions were carried out in the Tris/MgCl2/ATP buffer described above with the addition of 80 ng of Aos1p/Uba2p, 50 ng of Ubc9p, and 1 μg of reductively methylated Smt3p (38). 500 ng of GST-RanGap1p(241–360) (Enzo Life Sciences) was used for Fig. 4E. Reactions were incubated at 30 °C and terminated by the addition of 30 μl of SDS loading buffer. Consecutive ubiquitylation/SUMOylation reactions for Figs. 3D and 4E were performed with an initial step incubated for 2 h in a 60-μl volume. Twice the concentration described above of E1 and E2 was used with respect to ubiquitin or SUMO-conjugating machinery. 15 μl of each reaction was used for GST-RanGap1p SUMOylation as described above. For Fig. 4E GST-Rsp5p was SUMOylated with Siz1p(1–465), without (negative control) or with methylated Smt3p, and 15 μl of each sample was used for ubiquitylation of cytBsd2p.
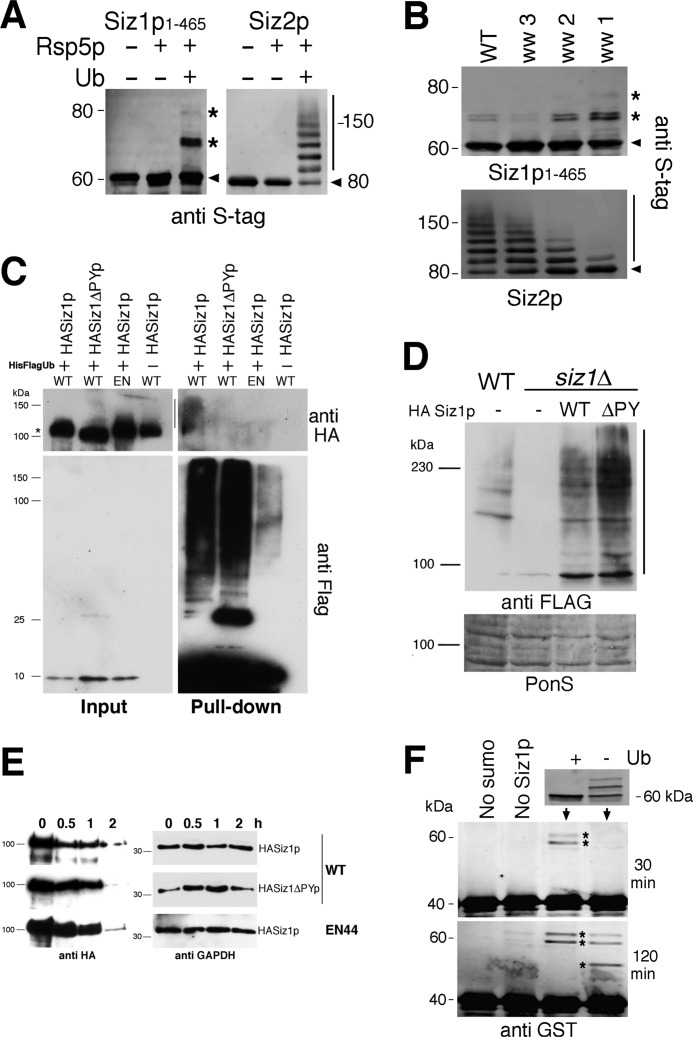
FIGURE 4.
A, in vitro ubiquitylation assays with recombinant Rsp5p and Siz1p(1–465) and Siz2p (10% SDS-PAGE). Arrowheads indicate unmodified Siz proteins; asterisks and bars indicate major modified forms of Siz1p(1–465) and Siz2, respectively. B, in vitro ubiquitylation of recombinant Siz1p(1–465) and Siz2p with either wild-type (WT) Rsp5p or variants in which only the indicated WW domain remains functional. Asterisks and bars indicate major modified forms of Siz1p(1–465) and Siz2, respectively. C, co-purification under denaturing conditions of His-FLAG-tagged ubiquitin with HASiz1p. Wild-type and an EN44 strain (EN) were co-transformed with HASiz1p or a version that lacks PY motifs (HASiz1ΔPYp) along with His-FLAG-tagged ubiquitin. Cells were grown to log phase, and cell lysates were prepared under denaturing conditions using a 6 m guanidine HCl buffer and pulldown assays carried out using His-select resin. Immunoblotting was performed using anti-HA (upper panels) and anti-FLAG antibodies (lower panels) on input (left panels) and pulldown (right panels) samples. Asterisk indicates position of HASiz1p, and bar shows position of modified HASiz1p. D, upper panel, total cell lysates expressing FLAG-Smt3p in a wild type yeast (WT) or siz1Δ strain were immunoblotted using anti-FLAG antibodies alongside total cell extracts from cells co-expressing an empty FLAG vector (−), HA-Siz1p (WT), or HA-Siz1pΔPY (ΔPY) as indicated. Lower panel, protein loading as assessed by Ponceau S staining (PonS). E, cycloheximide chase experiments with WT and an EN44 strain expressing HASiz1p or a version that lacks PY motifs (HASiz1ΔPYp). Cells were grown to log phase, and samples were taken at the time intervals indicated following the addition of 100 μg ml−1 cycloheximide. Immunoblotting was performed on the same samples using either anti-HA or anti-GAPDH antibodies. F, upper panel, recombinant Siz1p(1–465) (as visualized using anti-S-tag antibodies on 12% SDS-PAGE) subjected to in vitro ubiquitylation reaction with Rsp5p (+) with a negative control that contained all the ubiquitylation components except ubiquitin (−). Lower panels, modified and unmodified Siz1p(1–465) used in an in vitro SUMOylation reaction performed for 30 and 120 min using GST-RanGap1p(241–360) as a substrate. Asterisks indicate positions of SUMOylated GST-RanGap1p(241–360).
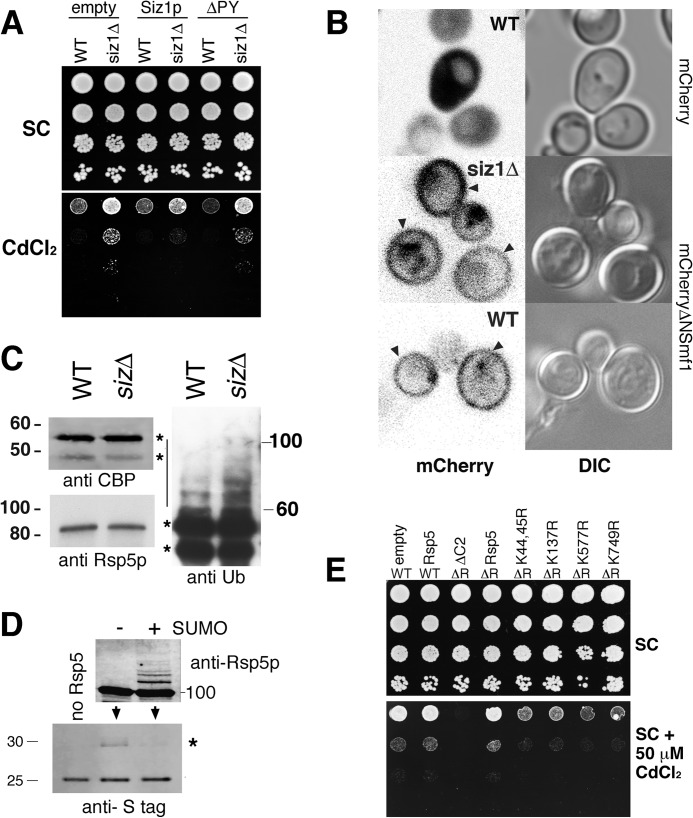
FIGURE 3.
A, wild-type yeast (WT) and a siz1Δ strain were transformed with empty FLAG (empty), FLAG-Siz1p (Siz1p), or FLAG-Siz1pΔPY (ΔPY) as indicated. Serial 10-fold dilutions of stationary phase cultures were replica-plated onto synthetic complete media without or with 50 μm CdCl2 (upper panel and lower panel, respectively) and incubated for 3 days at 30 °C. B, confocal imaging of mCherry and mCherry-tagged ΔNSmf1p and differential interference contrast (DIC) images of WT and siz1Δ yeast strains grown to log phase in metal-depleted media are shown. Arrowheads indicate mCherryΔNSmf1p signal at cell periphery. C, TAP-Bsd2p was expressed in WT yeast or siz1Δsiz2Δ strain (sizΔ) and TAP-purified. Purified proteins were subjected to SDS-PAGE followed by immunoblotting with anti-calmodulin-binding protein (CBP), anti-Rsp5, and anti-ubiquitin antibodies. Asterisks indicate cleaved (lower band) and uncleaved (upper band) forms of TAP-Bsd2p; bar indicates co-purifying HMW ubiquitin conjugates. D, upper panel shows recombinant Rsp5p (as visualized using anti-Rsp5 antibodies) subjected to in vitro SUMOylation reaction with Siz1p(1–465) (+) with a negative control that contained all the SUMOylation components except SUMO (−). Modified and unmodified Rsp5p were then used in an in vitro ubiquitylation reaction (lower panel) using the cytoplasmic portion of Bsd2p visualized using anti-Bsd2 antibodies. Asterisk indicates position of monoubiquitylated Bsd2p. E, cadmium sensitivity assays show lysine-to-arginine point mutations in SUMOylation sites identified in MS/MS analysis. Serial 10-fold dilutions were grown as in A of WT or an rsp5Δ strain (ΔR) expressing an empty plasmid, WT Rsp5 (Rsp5), an N-terminal deletion of Rsp5 that lacks the C2 domain (ΔC2) or full-length Rsp5 with lysine-to-arginine point mutations at the amino acid positions specified.
Microscopy
Yeast containing mCherry alone or mCherry-ΔNSmf1p were grown on metal-depleted media as described previously (26) and grown to log phase before imaging. Fluorescent and differential interference contrast imaging of log phase cells in water was performed using a Leica SP5 confocal microscope. Single channel image data were imported into Photoshop CS4 software (Adobe), converted into an 8-bit grayscale image, and then inverted using the standard Photoshop function. Brightness and contrast on the inverted images were then altered to the same arbitrary gray shade before a composite figure was generated.
RESULTS
Rsp5p Is SUMOylated in Vivo
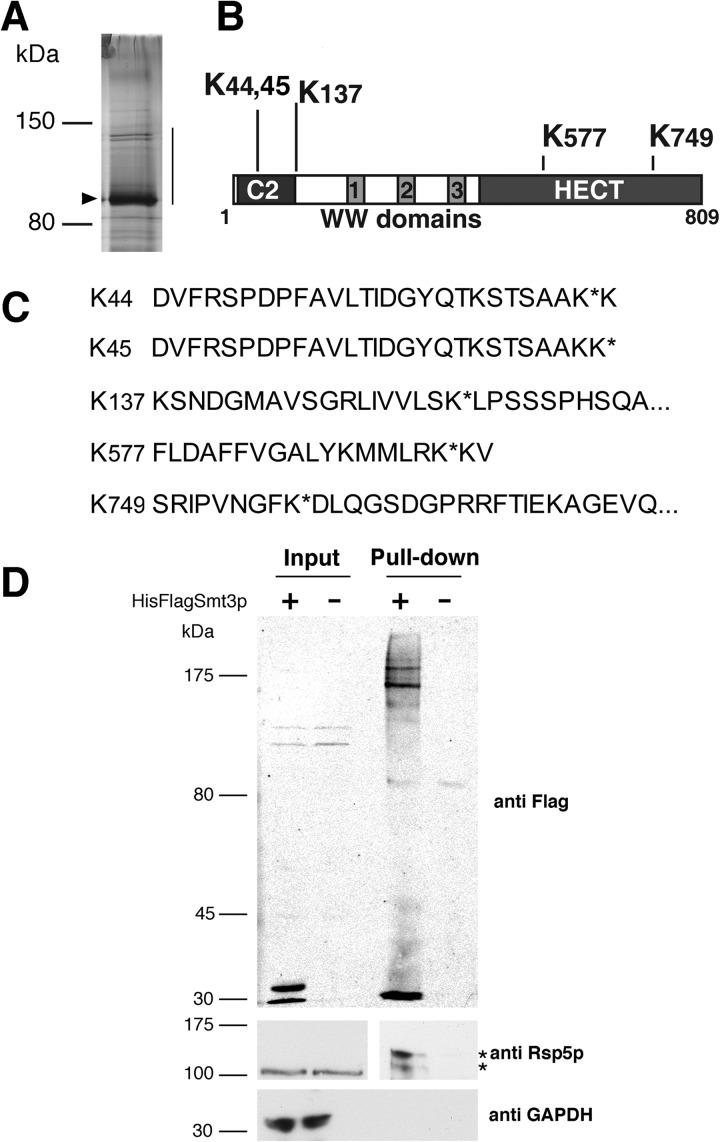
Previous work has shown that some ubiquitin ligases are regulated through post-translational modifications such as ubiquitylation and phosphorylation. To examine the possibility that Rsp5p is regulated by post-translational modifications, we purified Rsp5p using TAP and employed mass spectroscopy to identify any post-translational modifications and the modified residues. For this purpose, the RSP5 gene was substituted for a plasmid encoded N-terminally TAP-tagged RSP5 driven by the RSP5 promoter. TAP-Rsp5p protein was purified, resolved on SDS-PAGE (Fig. 1A), in-gel digested with trypsin and analyzed with MS/MS. The tryptic peptides were compared with the Mascot database for variable modifications of Rsp5p, including the attachment of Smt3p (the yeast SUMO homologue) to lysine residues (a residual EQIGG tryptic fragment on the modified lysine). Such analyses identified five peptides that carried modification by Smt3p (Fig. 1, B and C). This was particularly surprising because Rsp5p does not contain a consensus SUMOylation sequence, FKXE/D (39), although it has previously been identified as a low probability Smt3p substrate in a genome-wide survey (40). Two of the SUMOylated lysines, Lys44 and Lys45, were located within the C2 domain of Rsp5p. One SUMOylated residue, Lys137, was just outside of the C2 domain and two other lysines, Lys577 and Lys749, were found within the catalytic HECT domain (Fig. 1B). It is also interesting to note that ubiquitylated peptides modified at the Lys45 residue were also identified in our analysis. In support of the observation that Rsp5p is SUMOylated in vivo, His-FLAG-Smt3p was expressed in a Tap-Rsp5p strain and subjected to pulldown assays under denaturing conditions with His-select resin. Two major high molecular weight (HMW) bands of TAP-Rsp5 co-precipitated with His-FLAGSmt3p from the lysate representing Rsp5p/Smt3p conjugates (Fig. 1D).
FIGURE 1.
A, TAP-Rsp5p was purified from yeast and resolved on a 7.5% SDS-PAGE, and proteins were visualized by silver staining. Arrowhead indicates Rsp5p, bar indicates region of gel excised for MS/MS analysis. B, schematic diagram of Rsp5p shows conserved domains and position of SUMOylation residues. C, the LC-MS/MS peptide sequences in which SUMOylation was observed are listed (modified residues indicated with asterisk). D, His-FLAG-Smt3p (+) or an empty vector (−) was transformed into a Tap-Rsp5p strain, and total cell lysates were subjected to pulldown assays in the presence of 6 m guanidine hydrochloride with His-Select resin. Input and pulldown samples were immunoblotted with anti-FLAG antibodies for Smt3p, anti-Rsp5p, and anti-GAPDHp antibodies. Asterisks indicate two major species of Rsp5p that pull down with His-FLAGSmt3p.
Rsp5p Interacts with the SUMO Ligases Siz1p and Siz2p
To investigate the mechanism of Rsp5p SUMOylation, known components of the yeast SUMOylation machinery were scrutinized for the presence of putative Rsp5p-interacting PY motifs. We identified two proteins, Siz1p and Siz2p (known SUMO ligases), which contain numerous putative PY motifs. Siz1p contained three (LPVY100, SPFY183, IPEY638) and Siz2p five putative motifs (LPKY110, LPPY144, SPFY159, GPNY609, and LPPY653) that could potentially mediate the interaction with Rsp5p (Fig. 2A).
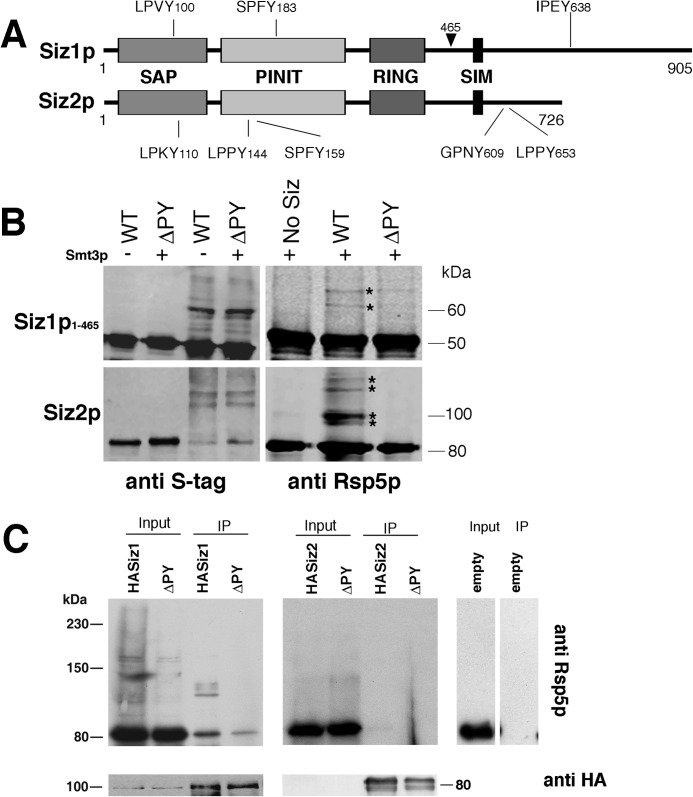
FIGURE 2.
A, schematic diagram of Siz1p and Siz2p shows conserved domains and position of putative PY motifs. Position of Siz1p truncation (amino acid 465) used in recombinant proteins is also indicated with an arrowhead. B, in vitro SUMOylation reactions were performed in the presence (+) or absence (−) of Smt3p using either wild-type (WT) Siz1p1–465 and Siz2p recombinant proteins or versions lacking PY motifs (ΔPY) in the presence of recombinant Rsp5p. Reactions were subjected to immunoblotting with anti-S-tag antibodies (for recombinant Siz proteins) (left panels) or anti-Rsp5 antibodies (right panels). Asterisks indicate positions of Rsp5p SUMO conjugates. C, full-length HA-tagged Siz1p and Siz2p, versions lacking PY motifs (ΔPY), or an empty HA plasmid (empty) was expressed in siz1Δ, siz2Δ, or wild-type strains, respectively. Total protein was extracted (Input) which was then subjected to IP with anti-HA antibodies followed by SDS-PAGE and immunoblotting with anti-Rsp5 (upper panels) and anti-HA antibodies (lower panels).
To study the interplay of Rsp5p with these SUMO ligases we first produced recombinant proteins and tested their interaction using in vitro SUMOylation assays. It should be noted that, as has been reported previously (9, 41), we were unable to produce full-length recombinant Siz1p and instead used a C-terminally truncated form (amino acids 1–465) lacking the SUMO interaction motif and the third putative PY motif (IPEY638) (Fig. 2A). In addition, we also produced altered forms of recombinant Siz1p(1–465) and Siz2p, in which the putative PY motifs had been inactivated by replacing the conserved tyrosine with an alanine (26).
In our assays (Fig. 2B) both recombinant Siz1p(1–465) and Siz2p demonstrated clear auto-SUMOylation activity. Interestingly, mutation of all putative PY motifs in Siz1p(1–465) and Siz2p had no effect on the level of this auto-SUMOylation (Fig. 2B, left panels). SUMO modification of recombinant Rsp5p was observed only when Siz1p(1–465) or Siz2p was present in the reactions, and was significantly attenuated (particularly with respect to Siz2p) when PY motifs were absent (Fig. 2B, right panels). Thus, in vitro Siz1p and Siz2p are capable of facilitating the modification of Rsp5p in a PY motif-dependent manner.
Because an interaction between Rsp5p and Siz1p and Siz2p could be demonstrated in vitro we decided to examine the interaction of these proteins in vivo using immunoprecipitation (IP) studies. To this end we generated modified yeast strains in either a siz1Δ or siz2Δ genetic background, expressing HA epitope-tagged full-length Siz1p and Siz2p (and ΔPY variants), respectively. Total protein extracts from these strains were then subjected to IP with anti-HA antibodies and subsequent immunoblotting with anti-HA and anti-Rsp5 antibodies (Fig. 2C).
HA-Siz1p (and ΔPY form) was readily detectable in total cell lysates as was endogenous Rsp5p (Fig. 2C, left panels), although it is interesting to note that HMW conjugates of Rsp5p were more abundant in a HA-Siz1p strain compared with the ΔPY variant. Following anti-HA IP, Rsp5p (and some HMW conjugates) co-precipitated with HA-Siz1p (Fig. 2C, right panels). In contrast, there was much reduced (although still detectable) co-precipitation of Rsp5p with the ΔPY variant of HA-Siz1p, and no HMW conjugates were observed (Fig. 2C, left panels). Similarly, HA-Siz2p (and its ΔPY form) could be immunoprecipitated from total protein extracts. However in this case, there was no change in the abundance of Rsp5p HMW conjugates between the wild-type and ΔPY forms of HA-Siz2p. Furthermore, no significant co-precipitation with Rsp5p was observed (Fig. 2C, right panels). Therefore, although Siz1p and Siz2p both have the potential to interact with and modify Rsp5p in a largely PY motif-dependent fashion, only the interaction with Siz1p is robust enough to be revealed in IP studies in vivo.
Siz1p Regulates the Manganese Transport Pathway
Because the role of Rsp5p in the ubiquitin-mediated endocytosis of the yeast manganese transporter Smf1p is well documented (26, 28, 42), and a clear interaction of Rsp5p and Siz1p was observed we decided to test a null siz1Δ strain for sensitivity to cadmium. Cadmium enters yeast cells via the manganese transporter Smf1p (43), and previous work has shown that when Rsp5p-mediated ubiquitylation of Smf1p is disrupted, Smf1p endocytosis from the plasma membrane is inhibited, and as a consequence cells become more sensitive to cadmium (26, 28, 42). Interestingly, we found that a strain lacking the SIZ1 gene (but not SIZ2, data not shown) exhibited a degree of resistance to cadmium compared with wild-type but showed no general change in growth on media lacking cadmium (Fig. 3A). To check whether the cadmium resistance phenotype of siz1Δ could be mediated via Rsp5p, a complementation assay with strains expressing either the HA-Siz1p or the ΔPY variant was performed. As expected, expression of HA-Siz1p could complement the cadmium resistance of siz1Δ, whereas overexpression of the ΔPY mutant failed to do so (Fig. 3A).
To investigate whether the cadmium resistance phenotype of siz1Δ could be due to an inability of Smf1p to reach the plasma membrane we examined the localization of an mCherry-tagged N-terminal deletion of Smf1p in wild-type and siz1Δ strains. ΔNSmf1p has a deletion of the N terminus of Smf1p, producing a functional transporter that undergoes ubiquitin-mediated metal-dependent but not stress-induced endocytosis (26). Interestingly, significant quantities of mCherry-ΔNSmf1p were observed at the cell periphery in both wild-type and siz1Δ strains when grown in metal-depleted media (Fig. 3B). Therefore, the cadmium resistance phenotype observed in siz1Δ is not simply due to an inability of Smf1p to reach the plasma membrane.
Because Smf1p reaches the plasma membrane we wondered whether the cadmium resistance observed in a siz1Δ strain could instead be due to changes in ubiquitin-mediated endocytosis. Previous work has shown that Bsd2p acts as an adaptor protein with Rsp5p in the regulated endocytosis of Smf1p and is itself ubiquitylated by Rsp5p (26, 42). To investigate Rsp5p-mediated ubiquitylation, Bsd2p was TAP-purified as described previously (26, 44) from wild-type yeast cells or strain lacking SIZ1. Purification of TAP-Bsd2p, expressed in both strains, resulted in similar yields with two major bands being identified, likely to be the cleaved and uncleaved forms of the TAP-tagged protein (Fig. 3C). Immunoblotting with anti-Rsp5 antibodies demonstrated that approximately equivalent quantities of Rsp5p co-precipitated with Bsd2p (Fig. 3C), suggesting that the lack of Siz1p did not affect the interaction of Rsp5p with its adaptor. In contrast, immunoblotting with anti-ubiquitin antibodies showed an increase in the abundance of Bsd2-ubiquitin conjugates in strains lacking SIZ1 compared with wild-type (Fig. 3C). Similar results with anti-ubiquitin antibodies were obtained if the TAP-Bsd2p was denatured with SDS prior to IP (data not shown).
Because in vivo Bsd2p showed increased ubiquitylation in a siz1Δ strain despite there being no apparent change in the level of steady-state interaction with Rsp5p, we decided to investigate directly the effect of Rps5p SUMOylation on its ligase activity. To this end we performed an in vitro SUMOylation reaction using recombinant Siz1p(1–465) to modify Rsp5p (Fig. 3D, upper panel). A negative control reaction was also performed alongside, which included all of the components of the SUMOylation machinery but lacking the addition of recombinant SUMO. This activity of modified and unmodified Rsp5p was then measured in an in vitro ubiquitylation reaction using the cytoplasmic portion of Bsd2p, which has been shown previously to contain all of the elements required for interaction with Rsp5p (26). In our assays SUMOylated Rsp5p showed less ubiquitin ligase activity toward Bsd2p compared with the unmodified control (Fig. 3D, lower panel). Thus, our in vitro data are consistent with our in vivo observations and support the hypothesis that SUMOylated Rsp5p exhibits lower activity toward Bsd2p and presumably Smf1p, thereby negatively regulating the manganese transport pathway.
Previous work has shown the importance of the Rsp5p C2 domain in the regulation of endocytosis (45). Therefore we decided to investigate the effects of removing all the SUMOylation sites we had identified by MS/MS within Rsp5, including a double mutant (K44R/K45R) removing the SUMOylation sites within the C2 domain. Lysine to arginine point mutations were made in TAP-tagged RSP5 plasmid constructs which were subsequently transformed into an RSP5-null strain and their ability to grow on cadmium-containing media assessed (Fig. 3E). All of the TapRsp5p constructs examined were able to complement the null phenotype of a rsp5Δ strain with no difference in growth on media lacking cadmium being observed between wild-type Rsp5p, the lysine point mutations, or a wild-type strain in which both endogenous Rsp5p and TapRsp5p were expressed (Fig. 3E, upper panel). Interestingly, overexpression of Rsp5p did not increase resistance to cadmium as might be expected, and all of the SUMOylation site mutations tested showed slightly reduced resistance to cadmium compared with the wild-type protein, although in all cases growth was significantly higher than that observed with a rsp5Δ strain expressing a version of Rsp5p that lacked its C2 domain (Fig. 3E, lower panel).
The Siz SUMO Ligases Are Rsp5p Substrates
Because we could detect a clear interaction with Siz1 (and Siz2 in vitro) with Rsp5p we wondered whether these proteins could be modified by Rsp5p. To this end we performed an in vitro ubiquitylation assay with recombinant Rsp5p, Siz1p(1–465), and Siz2p. Ubiquitylation of Siz1p(1–465) and Siz2p was only observed when Rsp5p was present, with Siz2p showing more extensive modification than Siz1p(1–465) (Fig. 4A). Interestingly, Siz1p(1–465) and Siz2p interact differentially with Rsp5p, with Siz1p(1–465) showing a clear interaction with the WW1 and WW2 but not the WW3 domains of Rsp5p (Fig. 4B, upper panel). In contrast, Siz2 shows a distinct preference for the WW3 domain of Rsp5p, with reduced interaction with WW2 and WW1 (Fig. 4B, lower panel).
Given that Siz1p showed a robust interaction with Rsp5p in vivo and that Rsp5p could modify Siz1p in vitro we decided to examine the ubiquitylation status of Siz1p in cells. We expressed full-length HASiz1p and its ΔPY variant in wild-type strain and in EN44, which contains an insertion in the upstream regulatory region of RSP5 resulting in reduced levels of Rsp5p (28). The HA-tagged Siz1 proteins were co-expressed with a His8-FLAG-tagged ubiquitin construct and co-precipitation studies were performed under denaturing conditions using HIS-Select resin. Under denaturing conditions HASiz1p co-precipitated with ubiquitin in extracts from wild-type strains in a PY motif-dependent fashion (Fig. 4C). In addition, no co-precipitation be detected between HASiz1p and ubiquitin in an EN44 strain and in a wild-type strain that did not express His-tagged ubiquitin (Fig. 4C). These results clearly demonstrate that Siz1p is ubiquitylated in an Rsp5p-dependent manner.
To investigate how the interaction between Siz1p and Rsp5p could affect its function in vivo, FLAG-Smt3p was expressed in wild-type yeast and null siz1Δ strains (Fig. 4D). As observed previously (10), deletion of SIZ1 caused a dramatic reduction in the abundance of FLAG-Smt3p HMW conjugates. Interestingly, on complementation with the wild-type Siz1p construct an increase in FLAG-Smt3p HMW conjugates was observed compared with wild-type cells, possibly due to overexpression of HA-Siz1p from the TPI1 promoter. Similarly, the ΔPY construct complemented the siz1Δ phenotype (Fig. 4D). However, the ΔPY form resulted in an increased abundance of FLAG-Smt3p conjugates, despite being expressed at a level similar to HA-Siz1p (Fig. 2C). Taken together, these data suggest that the interaction of Rsp5p with Siz1p, likely leading to its ubiquitylation, suppresses SUMO ligase activity.
One possibility to explain how Rsp5p-mediated modification of Siz1p results in reduced activity in vivo is that modification of Siz1 results in ubiquitin-mediated degradation. To address this possibility we used cycloheximide chase experiments to investigate the stability of HASiz1p and a version that lacks PY motifs in wild-type and EN44 strains. Following treatment with cycloheximide all of the Siz1p constructs tested were relatively unstable, with the vast majority of protein being degraded within 2 h (Fig. 4E, left panel). In contrast in the same samples the abundance of the metabolic enzyme GAPDH remained relatively unchanged in the same 2-h period (Fig. 4E, right panel). The presence or absence of Rsp5-interacting PY motifs had no apparent effect on Siz1p stability, and no change in stability was observed in the EN44 strain which has significantly reduced levels of Rsp5p. These results demonstrate that it is unlikely that proteolysis following Rsp5-mediated ubiquitylation of Siz1p is a significant mechanism for regulating the activity of the SUMO ligase.
Because Rsp5-mediated ubiquitylation does not appear to regulate Siz1p through a degradatory mechanism, it is therefore likely that ubiquitylation of Siz1p is directly altering its SUMO ligase activity. To validate this hypothesis, we decided to investigate how Siz1p ubiquitylation by Rsp5p affects SUMOylation of a well described in vitro substrate, RanGap1p (9, 46). Siz1p(1–465) was first ubiquitylated in vitro with Rsp5p alongside a negative control that included all the components of the ubiquitylation machinery but lacking ubiquitin. (Fig. 4F). Nonubiquitylated and ubiquitylated Siz1p(1–465) were then tested in a SUMOylation reaction with GST-RanGap1(241–360). Using short incubation times (30 min), at the limiting concentration of Ubc9p used, no SUMOylated GST-RanGap1(241–360) was observed with Ubc9p alone, but distinct SUMOylated products were observed after the addition of unmodified Siz1p(1–465) (Fig. 4F). In contrast, SUMOylated products were not observed with Siz1p(1–465) that had already been previously ubiquitylated (Fig. 4F). Interestingly, using a longer incubation time (120 min) a third lower molecular weight band corresponding to SUMOylated GST-RanGap1(241–360) was seen running at an approximate size equivalent to mono-SUMOylated protein (Fig. 4E). These in vitro assays demonstrate that Siz1p ubiquitylation by Rsp5p fundamentally alters its SUMO ligase activity.
DISCUSSION
In this study we have established that Rsp5p is SUMOylated in vivo and identified multiple lysine residues that are modified. To date, all of the reported ubiquitin ligases that undergo SUMOylation belong to the RING family of ligases, and all of them, apart from BRCA1, contain one or more SUMO interaction motifs (7, 47). Here, we show the first example of a HECT type ubiquitin ligase that is SUMO-regulated. The SUMOylated lysine residues identified were mostly located within and adjacent to the C2 domain with some within the catalytic HECT domain. Individual mutation of all the SUMO-modified lysine residues we identified did not result in any noticeable increase in the activity of Rsp5p against Smf1p as measured in cadmium sensitivity assays as might be expected. However, due to the limits of mass spectroscopy we cannot rule out that other, potentially more important, SUMOylation sites may exist in Rsp5p. There are 39 lysine residues in Rsp5p any of which could be subject to SUMOylation. In addition, all of these residues are also potential sites for other post-translational modifications such as ubiquitylation. Indeed, in our MS/MS analysis we identified Lys45 as a site for both SUMOylation and ubiquitylation, and there are many examples typified by the regulation of proliferating cell nuclear antigen (1, 2) where SUMOylation and ubiquitylation can have antagonistic activity making interpretation of lysine point mutations very difficult. In addition, overexpression of Rsp5p does not decrease sensitivity to cadmium, indicating that other, more complex mechanisms for regulating ubiquitin ligase activity are operating in yeast cells. When the same lysine residue can be modified by different ubiquitin-like proteins, leading to distinct, possibly antagonistic outcomes, the interpretation of site-directed mutagenesis experiments needs careful consideration and highlights the limitations of this commonly used approach.
Interestingly, we found that SUMOylation of Rsp5p is important in controlling the ubiquitin-mediated endocytosis of the manganese transporter Smf1p. Our first thought was that SUMOylation of Rsp5p might alter the affinity of the protein to adaptors, such as Bsd2p, because previous studies have tended to characterize SUMOylation as regulating protein-protein interactions (48). However, in this case the interaction of Bsd2p with Rsp5p in the absence of the SUMO ligase Siz1p was not noticeably different, suggesting that SUMOylation instead alters the ligase activity of Rsp5p with a concomitant effect on Smf1p endocytosis. A role for SUMO in the regulation of receptors and channels at the plasma membrane has been previously suggested. For example, the SUMO ligase PIAS3 was identified as a potassium channel-associated protein (49, 50), and the first membrane protein that was reported to be SUMOylated was the potassium channel K2P1, which when not SUMOylated exhibits constitutive channel activity (51, 52). Further plasma membrane proteins targeted for SUMOylation include the metabotropic glutamate receptor 8 (mGluR8) and the GluR6 subunit of kainate receptor (53, 54). Our results show that in addition to these intriguing direct roles for SUMO in the regulation of plasma membrane proteins, the role for SUMOylation in ubiquitin-mediated regulation of these proteins must now be considered.
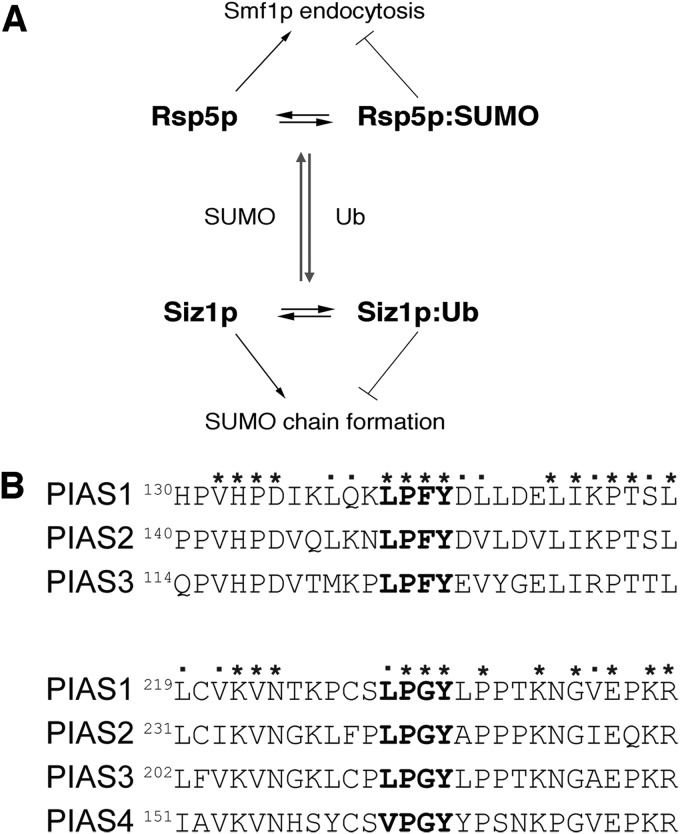
Our studies suggest a novel mechanism of reciprocal regulation between a ubiquitin ligase of the Nedd4 family and members of the PIAS family of SUMO ligases (Fig. 5A). The PIAS SUMO ligases, Siz1p and Siz2p, were identified as candidate SUMO ligases for Rsp5p modification as they contained putative PY motifs that could mediate their interaction with Rsp5p (Fig. 2A), although it should be noted that our results suggest that other non-PY Rsp5p interaction motifs may be present within Siz1p (Fig. 2C). Interestingly, Siz1p PY motifs are not essential for ligase activity as a Siz1p mutant that lacked all putative PY motifs was able to auto-SUMOylate and to complement a siz1Δ-null strain with respect to SUMO chain formation. The consequence of the interaction between Rsp5p and Siz1p and Siz2p appears to be reciprocal modification, in that Rsp5p, as well as being SUMOylated by these ligases, also ubiquitylates them. With respect to Siz1p the consequence of this interaction is the ubiquitin-mediated suppression of SUMO ligase activity. Such Rsp5p-mediated regulation of Siz1p activity is likely to have a huge impact, as Siz1p is responsible for the majority of SUMO conjugation in yeast cells (10).
FIGURE 5.
A, schematic diagram of reciprocal modification and regulation between Rsp5p and Siz1p. Covalent attachment (gray arrows) of ubiquitin (Ub) or SUMO alters activity of the modified ligase and has a concomitant effect on downstream processes such as Smf1p endocytosis and SUMO chain formation. B, human PIAS family of SUMO ligases with conserved PY motifs. ClustalW alignment (using MegAlign software, DNASTAR®Lasergene) shows the regions of protein sequences of human PIAS ligases (NCBI) with putative PY motifs highlighted in bold. Conserved residues within the regions are shown with asterisks, similar residues are highlighted with square dots.
We could not find much evidence for the interaction between Rsp5p and Siz2p in vivo, despite Siz2p having more potential Rsp5p binding motifs than Siz1p and the observation that it was a better Rsp5-modifiying partner for in vitro experiments (Figs. 2B and 4, A and B). Overexpression of Siz2p and its PY motif-lacking form in cells did not reveal any noticeable changes in the profile or quantity of HMW Rsp5p conjugates, and we could not precipitate any endogenous Siz2p-Rsp5p complex (Fig. 2C). However, despite these observations we cannot entirely rule out a role for Siz2p in the SUMOylation of Rsp5p. It should also be noted that S. cerevisiae has at least two other SUMO ligases (MMS21p and Zip3p) and that although neither of these proteins contains any recognizable Rsp5p interaction domains we cannot exclude the possibility that these ligases may be regulated by Rsp5p in a manner similar to Siz1p.
Interestingly, the PIAS family of SUMO ligases in humans contains two highly conserved putative PY motifs in PIAS1, PIAS2, and PIAS3 and one site in PIAS4 (Fig. 5B) (55). Thus, it is certainly possible that mammalian Nedd4 family ubiquitin ligases and PIAS family SUMO ligases might regulate each other as they do in yeast. Previous studies have identified some cross-talk between mammalian Nedd4 ligases and SUMO. For example, the androgen receptor, a ligand-controlled transcription factor deregulated in prostate cancer, is known to undergo SUMOylation by PIAS-like hZimp1, which contains at least four putative PY motifs (56). Interestingly, Nedd4-1 is involved in regulating androgen receptor stability through ubiquitylation via its interaction partner PMEPA1 (57). In addition, Nedd4-1 targets α-synuclein to the endosomal-lysosomal pathway whereas SUMOylation of α-synuclein stabilizes the protein and promote its solubility (58, 59). Recently, it has been shown that SUMO1 modification of the C2 domain in PTEN controls its association with the plasma membrane in mammalian cells (60). Such a mode of regulation may also exist with Nedd4 ligases, although our observation that SUMOylation inhibits Rsp5p in vitro suggests that a more direct inhibition of enzyme activity is involved.
This study sheds further light on the complex regulatory interactions that take place between the post-translational modifiers ubiquitin and SUMO. We show for the first time that a member of the HECT family of ubiquitin ligases is regulated through SUMOylation and that the role of SUMO in ubiquitin-mediated endocytosis may need to be reevaluated. Due to the highly conserved nature of ubiquitin and SUMO conjugation machinery, it seems likely that the reciprocal regulation we have observed in S. cerevisiae may exist in higher eukaryotes. Given the importance of Nedd4 and PIAS family ligases in many essential cellular processes and in several human diseases this is certainly worthy of further investigation.
Acknowledgments
We thank Dr. Hugh Pelham and Dr. Mike Lewis for the EN44 strain, Dr. Robin Maytum for advice on MS/MS, and Profs. Conrad Mullineaux and Ralf Stanewsky for critical reading of the manuscript.
This work was supported by the Biotechnology and Biological Sciences Research Council.
- SUMO
- small ubiquitin-related modifier
- HECT
- homologous to E6-AP carboxyl terminus
- HMW
- high molecular weight
- IP
- immunoprecipitation
- MS/MS
- tandem MS
- PIAS
- protein inhibitors of activated STATs
- RING
- really interesting new gene
- TAP
- tandem affinity purification.
REFERENCES
- 1. Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S. (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141 [DOI] [PubMed] [Google Scholar]
- 2. Stelter P., Ulrich H. D. (2003) Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425, 188–191 [DOI] [PubMed] [Google Scholar]
- 3. Huang L., Kinnucan E., Wang G., Beaudenon S., Howley P. M., Huibregtse J. M., Pavletich N. P. (1999) Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade. Science 286, 1321–1326 [DOI] [PubMed] [Google Scholar]
- 4. Steffan J. S., Agrawal N., Pallos J., Rockabrand E., Trotman L. C., Slepko N., Illes K., Lukacsovich T., Zhu Y. Z., Cattaneo E., Pandolfi P. P., Thompson L. M., Marsh J. L. (2004) SUMO modification of Huntingtin and Huntington's disease pathology. Science 304, 100–104 [DOI] [PubMed] [Google Scholar]
- 5. Perry J. J., Tainer J. A., Boddy M. N. (2008) A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem. Sci. 33, 201–208 [DOI] [PubMed] [Google Scholar]
- 6. Nagai S., Davoodi N., Gasser S. M. (2011) Nuclear organization in genome stability: SUMO connections. Cell. Res. 21, 474–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Morris J. R., Boutell C., Keppler M., Densham R., Weekes D., Alamshah A., Butler L., Galanty Y., Pangon L., Kiuchi T., Ng T., Solomon E. (2009) The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 462, 886–890 [DOI] [PubMed] [Google Scholar]
- 8. Hochstrasser M. (2001) SP-RING for SUMO: new functions bloom for a ubiquitin-like protein. Cell 107, 5–8 [DOI] [PubMed] [Google Scholar]
- 9. Yunus A. A., Lima C. D. (2009) Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol. Cell 35, 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Johnson E. S., Gupta A. A. (2001) An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106, 735–744 [DOI] [PubMed] [Google Scholar]
- 11. Pfander B., Moldovan G. L., Sacher M., Hoege C., Jentsch S. (2005) SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436, 428–433 [DOI] [PubMed] [Google Scholar]
- 12. Takahashi Y., Kikuchi Y. (2005) Yeast PIAS-type Ull1/Siz1 is composed of SUMO ligase and regulatory domains. J. Biol. Chem. 280, 35822–35828 [DOI] [PubMed] [Google Scholar]
- 13. Parker J. L., Bucceri A., Davies A. A., Heidrich K., Windecker H., Ulrich H. D. (2008) SUMO modification of PCNA is controlled by DNA. EMBO J. 27, 2422–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hang L. E., Liu X., Cheung I., Yang Y., Zhao X. (2011) SUMOylation regulates telomere length homeostasis by targeting Cdc13. Nat. Struct. Mol. Biol. 18, 920–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ferreira H. C., Luke B., Schober H., Kalck V., Lingner J., Gasser S. M. (2011) The PIAS homologue Siz2 regulates perinuclear telomere position and telomerase activity in budding yeast. Nat. Cell Biol. 13, 867–874 [DOI] [PubMed] [Google Scholar]
- 16. Reindle A., Belichenko I., Bylebyl G. R., Chen X. L., Gandhi N., Johnson E. S. (2006) Multiple domains in Siz SUMO ligases contribute to substrate selectivity. J. Cell Sci. 119, 4749–4757 [DOI] [PubMed] [Google Scholar]
- 17. Huibregtse J. M., Scheffner M., Beaudenon S., Howley P. M. (1995) A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. U.S.A. 92, 2563–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rotin D., Kumar S. (2009) Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 10, 398–409 [DOI] [PubMed] [Google Scholar]
- 19. Staub O., Gautschi I., Ishikawa T., Breitschopf K., Ciechanover A., Schild L., Rotin D. (1997) Regulation of stability and function of the epithelial Na+ channel (ENaC) by ubiquitination. EMBO J. 16, 6325–6336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Strack B., Calistri A., Accola M. A., Palu G., Gottlinger H. G. (2000) A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. U.S.A. 97, 13063–13068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhu H., Kavsak P., Abdollah S., Wrana J. L., Thomsen G. H. (1999) A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature 400, 687–693 [DOI] [PubMed] [Google Scholar]
- 22. Cao X. R., Lill N. L., Boase N., Shi P. P., Croucher D. R., Shan H., Qu J., Sweezer E. M., Place T., Kirby P. A., Daly R. J., Kumar S., Yang B. (2008) Nedd4 controls animal growth by regulating IGF-1 signaling. Sci. Signal. 1, ra5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hein C., Springael J. Y., Volland C., Haguenauer-Tsapis R., André B. (1995) NPl1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol. Microbiol. 18, 77–87 [DOI] [PubMed] [Google Scholar]
- 24. Kölling R., Hollenberg C. P. (1994) The ABC-transporter Ste6 accumulates in the plasma membrane in a ubiquitinated form in endocytosis mutants. EMBO J. 13, 3261–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lauwers E., Erpapazoglou Z., Haguenauer-Tsapis R., André B. (2010) The ubiquitin code of yeast permease trafficking. Trends Cell Biol. 20, 196–204 [DOI] [PubMed] [Google Scholar]
- 26. Sullivan J. A., Lewis M. J., Nikko E., Pelham H. R. (2007) Multiple interactions drive adaptor-mediated recruitment of the ubiquitin ligase rsp5 to membrane proteins in vivo and in vitro. Mol. Biol. Cell 18, 2429–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lin C. H., MacGurn J. A., Chu T., Stefan C. J., Emr S. D. (2008) Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell 135, 714–725 [DOI] [PubMed] [Google Scholar]
- 28. Nikko E., Sullivan J. A., Pelham H. R. (2008) Arrestin-like proteins mediate ubiquitination and endocytosis of the yeast metal transporter Smf1. EMBO Rep. 9, 1216–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Léon S., Erpapazoglou Z., Haguenauer-Tsapis R. (2008) Ear1p and Ssh4p are new adaptors of the ubiquitin ligase Rsp5p for cargo ubiquitylation and sorting at multivesicular bodies. Mol. Biol. Cell 19, 2379–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Erdeniz N., Rothstein R. (2000) Rsp5, a ubiquitin-protein ligase, is involved in degradation of the single-stranded-DNA binding protein rfa1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 20, 224–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huibregtse J. M., Yang J. C., Beaudenon S. L. (1997) The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin-protein ligase. Proc. Natl. Acad. Sci. U.S.A. 94, 3656–3661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harreman M., Taschner M., Sigurdsson S., Anindya R., Reid J., Somesh B., Kong S. E., Banks C. A., Conaway R. C., Conaway J. W., Svejstrup J. Q. (2009) Distinct ubiquitin ligases act sequentially for RNA polymerase II polyubiquitylation. Proc. Natl. Acad. Sci. U.S.A. 106, 20705–20710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Isasa M., Katz E. J., Kim W., Yugo V., González S., Kirkpatrick D. S., Thomson T. M., Finley D., Gygi S. P., Crosas B. (2010) Monoubiquitination of RPN10 regulates substrate recruitment to the proteasome. Mol. Cell 38, 733–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fisk H. A., Yaffe M. P. (1999) A role for ubiquitination in mitochondrial inheritance in Saccharomyces cerevisiae. J. Cell Biol. 145, 1199–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stimpson H. E., Lewis M. J., Pelham H. R. (2006) Transferrin receptor-like proteins control the degradation of a yeast metal transporter. EMBO J. 25, 662–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Werner A., Moutty M. C., Möller U., Melchior F. (2009) Performing in vitro sumoylation reactions using recombinant enzymes. Methods Mol. Biol. 497, 187–199 [DOI] [PubMed] [Google Scholar]
- 37. Wilm M., Shevchenko A., Houthaeve T., Breit S., Schweigerer L., Fotsis T., Mann M. (1996) Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 379, 466–469 [DOI] [PubMed] [Google Scholar]
- 38. Hershko A., Heller H. (1985) Occurrence of a polyubiquitin structure in ubiquitin-protein conjugates. Biochem. Biophys. Res. Commun. 128, 1079–1086 [DOI] [PubMed] [Google Scholar]
- 39. Rodriguez M. S., Desterro J. M., Lain S., Midgley C. A., Lane D. P., Hay R. T. (1999) SUMO-1 modification activates the transcriptional response of p53. EMBO J. 18, 6455–6461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wohlschlegel J. A., Johnson E. S., Reed S. I., Yates J. R. (2006) Improved identification of SUMO attachment sites using C-terminal SUMO mutants and tailored protease digestion strategies. J. Proteome Res. 5, 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Takahashi Y., Toh-E. A., Kikuchi Y. (2003) Comparative analysis of yeast PIAS-type SUMO ligases in vivo and in vitro. J. Biochem. 133, 415–422 [DOI] [PubMed] [Google Scholar]
- 42. Hettema E. H., Valdez-Taubas J., Pelham H. R. (2004) Bsd2 binds the ubiquitin ligase Rsp5 and mediates the ubiquitination of transmembrane proteins. EMBO J. 23, 1279–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu X. F., Culotta V. C. (1999) Post-translation control of Nramp metal transport in yeast: role of metal ions and the BSD2 gene. J. Biol. Chem. 274, 4863–4868 [DOI] [PubMed] [Google Scholar]
- 44. Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Séraphin B. (2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24, 218–229 [DOI] [PubMed] [Google Scholar]
- 45. Dunn R., Klos D. A., Adler A. S., Hicke L. (2004) The C2 domain of the Rsp5 ubiquitin ligase binds membrane phospholipids and directs ubiquitination of endosomal cargo. J. Cell Biol. 165, 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mahajan R., Delphin C., Guan T., Gerace L., Melchior F. (1997) A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88, 97–107 [DOI] [PubMed] [Google Scholar]
- 47. Geoffroy M. C., Hay R. T. (2009) An additional role for SUMO in ubiquitin-mediated proteolysis. Nat. Rev. Mol. Cell Biol. 10, 564–568 [DOI] [PubMed] [Google Scholar]
- 48. Bergink S., Jentsch S. (2009) Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458, 461–467 [DOI] [PubMed] [Google Scholar]
- 49. Wible B. A., Yang Q., Kuryshev Y. A., Accili E. A., Brown A. M. (1998) Cloning and expression of a novel K+ channel regulatory protein, KChAP. J. Biol. Chem. 273, 11745–11751 [DOI] [PubMed] [Google Scholar]
- 50. Wible B. A., Wang L., Kuryshev Y. A., Basu A., Haldar S., Brown A. M. (2002) Increased Ka+ efflux and apoptosis induced by the potassium channel modulatory protein KChAP/PIAS3β in prostate cancer cells. J. Biol. Chem. 277, 17852–17862 [DOI] [PubMed] [Google Scholar]
- 51. Rajan S., Plant L. D., Rabin M. L., Butler M. H., Goldstein S.A. (2005) Sumoylation silences the plasma membrane leak K+ channel K2P1. Cell 121, 37–47 [DOI] [PubMed] [Google Scholar]
- 52. Plant L. D., Dementieva I. S., Kollewe A., Olikara S., Marks J. D., Goldstein S. A. (2010) One SUMO is sufficient to silence the dimeric potassium channel K2P1. Proc. Natl. Acad. Sci. U.S.A. 107, 10743–10748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tang Z., El Far O., Betz H., Scheschonka A. (2005) Pias1 interaction and sumoylation of metabotropic glutamate receptor 8. J. Biol. Chem. 280, 38153–38159 [DOI] [PubMed] [Google Scholar]
- 54. Martin S., Nishimune A., Mellor J. R., Henley J. M. (2007) SUMOylation regulates kainate receptor-mediated synaptic transmission. Nature 447, 321–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gupta R., Kus B., Fladd C., Wasmuth J., Tonikian R., Sidhu S., Krogan N. J., Parkinson J., Rotin D. (2007) Ubiquitination screen using protein microarrays for comprehensive identification of Rsp5 substrates in yeast. Mol. Syst. Biol. 3, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sharma M., Li X., Wang Y., Zarnegar M., Huang C. Y., Palvimo J. J., Lim B., Sun Z. (2003) hZimp10 is an androgen receptor co-activator and forms a complex with SUMO-1 at replication foci. EMBO J. 22, 6101–6114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li H., Xu L. L., Masuda K., Raymundo E., McLeod D. G., Dobi A., Srivastava S. (2008) A feedback loop between the androgen receptor and a NEDD4-binding protein, PMEPA1, in prostate cancer cells. J. Biol. Chem. 283, 28988–28995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tofaris G. K., Kim H. T., Hourez R., Jung J. W., Kim K. P., Goldberg A. L. (2011) Ubiquitin ligase Nedd4 promotes α-synuclein degradation by the endosomal-lysosomal pathway. Proc. Natl. Acad. Sci. U.S.A. 108, 17004–17009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Krumova P., Meulmeester E., Garrido M., Tirard M., Hsiao H. H., Bossis G., Urlaub H., Zweckstetter M., Kügler S., Melchior F., Bähr M., Weishaupt J. H. (2011) Sumoylation inhibits α-synuclein aggregation and toxicity. J. Cell Biol. 194, 49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Huang J., Yan J., Zhang J., Zhu S., Wang Y., Shi T., Zhu C., Chen C., Liu X., Cheng J., Mustelin T., Feng G. S., Chen G., Yu J. (2012) SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane. Nat. Commun. 3, 911–923 [DOI] [PubMed] [Google Scholar]