Background: Zinc is required for catalysis and protein structure.
Results: Zinc-deficient Chlamydomonas lose carbonic anhydrases and cannot grow photoautotrophically in air. They also hyperaccumulate copper but are phenotypically copper-deficient and therefore require Crr1, the nutritional copper sensor.
Conclusion: Zinc deficiency impacts the carbon-concentrating mechanism and disrupts copper homeostasis.
Significance: Cross-talk exists between zinc and copper homeostasis pathways.
Keywords: Algae, Copper, Gene Regulation, Photosynthesis, Transport Metals, Zinc
Abstract
Zinc is an essential nutrient because of its role in catalysis and in protein stabilization, but excess zinc is deleterious. We distinguished four nutritional zinc states in the alga Chlamydomonas reinhardtii: toxic, replete, deficient, and limited. Growth is inhibited in zinc-limited and zinc-toxic cells relative to zinc-replete cells, whereas zinc deficiency is visually asymptomatic but distinguished by the accumulation of transcripts encoding ZIP family transporters. To identify targets of zinc deficiency and mechanisms of zinc acclimation, we used RNA-seq to probe zinc nutrition-responsive changes in gene expression. We identified genes encoding zinc-handling components, including ZIP family transporters and candidate chaperones. Additionally, we noted an impact on two other regulatory pathways, the carbon-concentrating mechanism (CCM) and the nutritional copper regulon. Targets of transcription factor Ccm1 and various CAH genes are up-regulated in zinc deficiency, probably due to reduced carbonic anhydrase activity, validated by quantitative proteomics and immunoblot analysis of Cah1, Cah3, and Cah4. Chlamydomonas is therefore not able to grow photoautotrophically in zinc-limiting conditions, but supplementation with 1% CO2 restores growth to wild-type rates, suggesting that the inability to maintain CCM is a major consequence of zinc limitation. The Crr1 regulon responds to copper limitation and is turned on in zinc deficiency, and Crr1 is required for growth in zinc-limiting conditions. Zinc-deficient cells are functionally copper-deficient, although they hyperaccumulate copper up to 50-fold over normal levels. We suggest that zinc-deficient cells sequester copper in a biounavailable form, perhaps to prevent mismetallation of critical zinc sites.
Introduction
Zinc is an essential nutrient required in abundance by organisms ranging from bacteria to humans. Over 300 known enzymes utilize zinc as a cofactor, and whole genome surveys estimate that 4–10% of all sequenced proteins from prokaryotes and eukaryotes contain zinc-binding domains (1). Excess zinc is believed to be toxic because it can compete for metal binding sites in other proteins and can indirectly generate damaging reactive oxygen species. Thus, intracellular zinc content must be regulated to ensure that zinc-containing proteins can function while excess zinc is avoided. The first level of control is at the step of assimilation, where intracellular zinc status controls the expression and presentation of low and high affinity transporters at the plasma membrane (2–7). Another level of control is by compartmentalization, where transporters can sequester zinc in the vacuole (in fungi), lysosome-related compartments (in Caenorhabditis elegans), or in zincosomes (in mammalian cells) in a situation of excess (8–10). Sequestered zinc can be mobilized by efflux transporters (11). The expression of each type of transporter is therefore critical for homeostasis, and there are multiple levels of control from transcription to trafficking to protein degradation (2, 12).
Transporters in two families are important for zinc metabolism, the ZIP (Zrt-, Irt-like protein) family, whose members function primarily to move either zinc or iron into the cytoplasm, and the CDF (cation diffusion facilitator) family, whose members function primarily to move zinc out of the cytoplasm (13) (reviewed in Ref. 14). In plants, members of the CDF family are also called MTPs (metal tolerance proteins) because their function in moving metals out of the cytoplasm is important for handling toxicity (15, 16). The pattern of expression of individual members of these families in response to the concentration and type of metal nutrient provides a clue to their physiological functions.
Photosynthetic microorganisms rely on the carbon-concentrating mechanism (CCM)4 to concentrate CO2 at the active site of ribulose-bisphosphate carboxylase/oxygenase, the enzyme that catalyzes the first step in the CO2 fixation pathway (reviewed in Refs. 17–20). Carbonic anhydrases are integral components in this pathway and contribute to the high productivity of some of these organisms. These enzymes generally rely on a zinc cofactor (to activate water) to catalyze the interconversion of bicarbonate and CO2. In a zinc-deficient marine environment, cobalt can replace zinc, or an alternate enzyme that uses a cadmium cofactor can substitute (21–23). The occurrence of these zinc-sparing mechanisms is indicative of the contribution of carbonic anhydrases to the cellular zinc quota, meaning the optimally desired zinc content of the cell in a zinc-replete medium (24).
We have developed Chlamydomonas reinhardtii as a reference organism for understanding pathways of trace metal metabolism and homeostasis in algae and in the plant lineage, especially with respect to the impact of deficiency on bioenergetic pathways in the mitochondria and chloroplast (14, 25). Chlamydomonas species have been found in various environmental niches distinguished by metal content, pH, and oxygen availability, suggesting that this genus has developed adaptive systems to deal with changing environmental conditions (26). In the laboratory, dilute Chlamydomonas cultures can reach stationary phase in 2–3 days in a simple salt-containing medium in which the trace elements are buffered by chelation with EDTA (27). The absence of serum or amino acid supplementation simplifies the provision of trace metal nutrients and the establishment of deficiency. We have been able to exploit this in previous work with Chlamydomonas cells experiencing iron, copper, or manganese deficiency, in which we noted that the assimilatory transporters are responsive to metal nutrition at the transcriptional level, including two genes encoding ZIP family members, which were named IRT1 and IRT2 because they responded to iron deficiency (28–31). Nevertheless, there are as many as a dozen members of the ZIP family in Chlamydomonas, and some of them are likely to be involved in zinc assimilation.
The interaction of zinc and copper homeostasis pathways is also likely in Chlamydomonas. Cu(I) is taken up by the CTR (copper transporter) family of transporters, whose members are components of the nutritional copper regulon in Chlamydomonas. A zinc-containing SBP (squamosa-promoter binding protein) domain transcription factor named Crr1 (copper response regulator) controls the expression of the CTR genes (28). Its functional homolog in Arabidopsis is SPL7 (32–34). Besides the CTR genes encoding the plasma membrane-localized assimilatory transporters, Crr1 regulates over 60 genes involved in acclimation to copper deficiency through associated copper-responsive elements, which are the target sites for the SBP domain (28, 35–37). The best characterized of these target genes is CYC6, encoding cytochrome (Cyt) c6, which is a heme-containing replacement of the usual copper-containing protein, plastocyanin, in the photosynthetic electron transfer chain. Therefore, although copper-deficient Chlamydomonas cells do not accumulate plastocyanin, they remain photosynthetically competent (38). Decreased plastocyanin abundance and transcriptional activation of CYC6 are classic markers for the copper deficiency state.
In this work, we use growth and the expression of a subset of genes for ZIP family transporters as sentinels of zinc status to establish zinc deficiency and zinc limitation in Chlamydomonas, which are achieved by serial transfer of replete cells to medium lacking supplemental zinc. Transcriptome and proteome surveys identify the carbonic anhydrases and hence the carbon concentrating mechanism as a pathway impacted by poor zinc nutrition. We note also an impact on copper homeostasis, and we suggest the existence of mechanisms that control the ratio of intracellular metal ions.
EXPERIMENTAL PROCEDURES
Culturing and Strains
C. reinhardtii strains CC-4532 (wild-type, 2137), crr1–2 (referred to subsequently as crr1), and a rescued strain crr1–2::CRR1 (referred to subsequently as CRR1) were cultured under 50–100 μmol m−2 s−1 continuous illumination (2:1, cool white/warm white light) in Tris acetate-phosphate (TAP) or Tris phosphate (TP) medium with trace element supplements described by Kropat et al. (27). Briefly, stock solutions of 25 mm EDTA-Na2, 28.5 μm (NH4)6Mo7O24, 0.1 mm Na2SeO3, 2.5 mm ZnSO4 in 2.75 mm EDTA, 6 mm MnCl2 in 6 mm EDTA, 20 mm FeCl3 in 22 mm EDTA, and 2 mm CuCl2 in 2 mm EDTA were made individually in Milli-Q-purified water and diluted 1:1000 in the final growth medium. For metal-free studies, all glassware was triple washed in 6 n hydrochloric acid followed by at least six rinses in Milli-Q-purified (Millipore) water. All media were made using Milli-Q water (39). For experiments other than the proteomic studies, cells were grown in nutrient-replete medium, followed by one transfer into zinc medium with no supplemental zinc, before inoculation into the experimental conditions. For the proteomic studies, cells were grown in replete medium and directly inoculated into the experimental conditions. For experiments involving CO2 supplementation, cultures were bubbled with filtered air (control) or a mixture of 1% CO2 with air. Cell density was measured by counting with a hemocytometer.
Fluorescence Rise and Decay Kinetics
For CC-4532, room temperature fluorescence rise and decay kinetics were analyzed using a FluorCam 700MF (Photon Systems Instruments). Approximately 50 μl of concentrated mid-log phase liquid culture was spotted onto the lid of a plastic Petri dish and dark-adapted for 10 min prior to a flash of saturating light and measurement of the Kautsky effect in continuous red light at 150 μmol m−2 s−1 PFD (photon flux density) with 100% actinic light and 60% activity. For measurements of crr1 and CRR1 cells in zinc-replete and zinc-limited conditions, room temperature kinetics were measured using a laboratory-built instrument as described by Joliot and Joliot (40).
Cell Size Determination
Cell size was determined with a Beckman Coulter laser diffraction particle size analyzer LS13 320. Approximately 100 ml of cultures in exponential phase were poured into the micro-liquid module with a magnetic stir bar included to keep the cells in suspension. If necessary, cultures were concentrated or diluted until the sample obscuration was between 8 and 12%.
Nucleic Acid Analysis
Total Chlamydomonas RNA was prepared as described by Quinn and Merchant (39). RNA quality was assessed on an Agilent 2100 Bioanalyzer and by hybridization to CBLP (also called RACK1) as described previously (39). A 915-bp EcoRI fragment from the cDNA cloned in pcf8-13 was used as the probe (41).
Quantitative RT-PCR
Genomic DNA was removed from the total RNA preparation by treatment with Turbo DNase (Ambion) according to the manufacturer's instructions with the following modifications. 3 units of enzyme was used per 10 μg of nucleic acid, and incubation at 37 °C was increased to 90 min. Complementary DNA, primed with oligo(dT), was generated with reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Amplification was carried out with reagents from the iQ SYBR Green Supermix qPCR kit (Bio-Rad). Each reaction contained the vendor's master mix, a 0.3 μm concentration of each primer, and cDNA corresponding to 20 ng of input RNA in the reverse transcriptase reaction. The reaction conditions for the Opticon 2 from MJ Research were as follows: 95 °C for 5 min, followed by cycles of 95 °C for 10 s, 65 °C for 30 s, and 72 °C for 30 s, up to 40 cycles. The fluorescence was measured at each cycle at 72 and 83 °C. The 2−ΔΔCT method was used to analyze the database on the fluorescence at 83 °C (42). Melting curves were performed after the PCR to assess the presence of a unique final product.
RNA-seq
RNAs were sequenced by Illumina on a GAIIx system for estimating transcript abundance. The reads were aligned using Bowtie (43) in single-end mode and with a maximum tolerance of three mismatches to the Au10.2 transcript sequences (see the Phytozome Web site), corresponding to the version 4.0 assembly of the Chlamydomonas genome. Expression estimates were obtained for each individual run in units of reads per kilobase of mappable transcript length per million mapped reads (RPKM; see Ref. 44) after normalization by the number of aligned reads and transcript mappable length. The transcript coverage across the genome was visualized on a local installation of the UCSC browser. Differential expression analysis was performed in R with the DESeq package (45), and p values were adjusted to control for false discovery rate with the Benjamini-Hochberg method (46). Sequence files are publicly available in the NCBI Gene Expression Omnibus (accession numbers GSE25622 and GSE41096).
Protein Isolation for Immunoblot Analysis
Chlamydomonas cultures were collected by centrifugation (1000 × g, 5 min) and washed twice with 10 mm sodium phosphate, pH 7.0. The total protein fraction was further subfractionated into soluble and membrane components as described by Howe and Merchant (47). Protein concentration of soluble fractions was determined using the Pierce BCA protein assay kit following the manufacturer's instructions. Samples were diluted to 4 μg/μl. Membrane fractions were normalized by resuspending them to a volume that was equivalent to the final soluble fraction volume.
Immunoblot Analysis
Protein samples were denatured by the addition of 5% β-mercaptoethanol and boiling for 10 min before separation on denaturing polyacrylamide gels and transferring in a semidry blotter onto nitrocellulose membranes in transfer buffer (25 mm Tris, 192 mm glycine, 0.0004% SDS (w/v), and 20% (v/v) methanol). The membrane was blocked overnight with 1% dried milk in Tris-buffered saline (10 mm Tris-Cl, 150 mm NaCl, pH 7.5) plus Tween 20 (0.05% (w/v)) before incubation in primary antiserum for 4–12 h. Gel concentrations (acrylamide monomer) and dilutions for each primary antibody were as follows: plastocyanin, 16%, 1:1000 dilution; Cyt c6, 16%, 1:1000 dilution; Cah1, 12%, 1:2000 dilution; Cah3, 12%, 1:2000; Cah4, 12%, 1:15,000 dilution; ferredoxin 15%, 1:10,000 dilution; and chloroplast ATP synthase, 10%, 1:20,000 dilution. Secondary goat anti-rabbit horseradish peroxidase (Pierce) was used at a 1:5000 dilution.
Sample Preparation and Analysis for Quantitative Proteomics
Soluble protein was extracted from CC-4532 cells growing in 2.5 μm zinc and the first round of 0 supplemental zinc. For each condition, label-free, data-independent quantitative liquid chromatography-tandem mass spectrometry (LC-MS/MS or “LC-MSE”) was performed as previously described with slight modifications (35, 48). In brief, cells were collected at late exponential phase from replete zinc conditions or from the first transfer to zinc-depleted medium by centrifugation. Cells were broken by slow freezing and thawing at −80 °C and 24 °C, respectively. Insoluble material was removed by centrifugation at 16,000 × g for 10 min at 4 °C followed by centrifugation at 253,000 × g for 20 min at 4 °C. Approximately 30 μg of protein (per lane) was separated by gel electrophoresis on 4–12% NuPage BisTris gels (Invitrogen) and visualized by staining with Coomassie Blue (Bio-Rad). Gel lanes were divided into ∼3-mm bands, and individual bands were subjected to in-gel trypsin digestion (sequencing grade modified trypsin; Promega). Digested peptides were extracted into a 50:50 water/acetonitrile solution containing 2.5% formic acid and lyophilized. Peptides were then resuspended into a 25 fmol/μl bovine serum albumin (BSA) digest and quantification standard (Waters). LC-MSE was performed using a nanoAcquity UPLC (Waters) system coupled to a quadrupole time-of-flight mass spectrometer (Waters Xevo QTOF). Protein Lynx Global Server (PLGS version 2.4; Waters) was used to process the LC-MS raw data and determine protein identification and quantification. The quantification of protein levels was achieved via the addition of an internal protein standard (BSA trypsin digest standard) to which the data set was normalized. Our criteria were that the difference in protein abundance between growth conditions must be statistically significant (p < 0.05 by Student's t test) and at least 2-fold or greater in magnitude in order to define a change in protein abundance (48).
Stoichiometric Measurements of Cytochrome/P700 and Photosystem I (PSI)/PSII
Spectroscopic measurements were performed using a JTS-10 spectrophotometer (Biologic, Claix, France). Light-induced absorption changes were measured as absorption of flashed light at discrete times. Changes in the amount of functional photosynthetic complexes were evaluated measuring the electrochromic shift spectral change, a shift in the pigment absorption bands that is linearly correlated to the number of light-induced charge separations within the reaction centers. Functional PSI and PSII content was estimated from changes in the amplitude of the fast phase of the electrochromic shift signal (at 520–546 nm) upon excitation with a saturating laser flash (520 nm, 5-ns duration). PSII contribution was calculated from the decrease in the signal amplitude upon the addition of DCMU (3-(3′,4′-dichlorophenyl)-1,1-dimethylurea) (20 μm) and hydroxylamine (2 mm) to irreversibly block PSII charge separation. Conversely, PSI was estimated as the fraction of the signal that was insensitive to these inhibitors (49). Cytochrome redox changes were calculated as the difference between the absorption at 554 nm and a base line drawn between 545 and 573 nm and corrected for the contribution of the electrochromic signal (50). P700 (the primary electron donor to PSI) was measured at 705 nm. To evaluate the relative cytochrome/P700 stoichiometry, measurements were performed in continuous saturating light (1100 μmol of photons m−2 s−1) and in the presence of saturating concentrations of DBMIB (2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone) (10 μm). This is required to ensure full inhibition of cytochrome b6 f reduction by light-generated plastoquinol and therefore a maximum oxidation of P700 and of the cytochrome.
Measurement of Intracellular Metal Content
Cells were collected by centrifugation at 1700 × g for 5 min. Pellets were washed once in 1 mm EDTA to remove cell surface-associated metals and once in Milli-Q water. The washed cell paste was overlaid with nitric acid corresponding to a final concentration of 24% in 1 ml and digested at 65 °C. To obtain a corresponding blank, the volume of the cell paste was replaced by deionized water and treated as described above. Total metal and phosphorous content was measured by inductively coupled plasma-MS (ICP-MS; Agilent 7500).
RESULTS
Identification of Four Distinct Zinc Nutrition States
Chlamydomonas grows in media containing a wide range of supplemental zinc concentrations (51). Elemental analysis of wild-type Chlamydomonas cells grown in standard TAP medium supplemented with Hutner's trace elements, which contains about 80 μm of EDTA-chelated zinc, indicated a zinc quota of ∼2–3 × 107 atoms/cell. We calculated that a minimum concentration of 0.85 μm zinc ions would be necessary to support zinc-replete growth to stationary phase (∼2 × 107 cells/ml). After allowing for variation in zinc quota in situations of altered physiology (e.g. low CO2 partial pressure when carbonic anhydrases would be induced), we chose 2.5 μm zinc ions in a revised micronutrient solution (27, 51, 52). Indeed, this concentration of zinc is more than sufficient to support normal growth and a typical intracellular trace metal (copper, iron, manganese) quota; when the zinc content of the medium is reduced to 0.25 μm, there is no impact on growth (Fig. 1A). Therefore, 2.5 μm zinc ions is considered metal replete for laboratory growth of Chlamydomonas.
FIGURE 1.
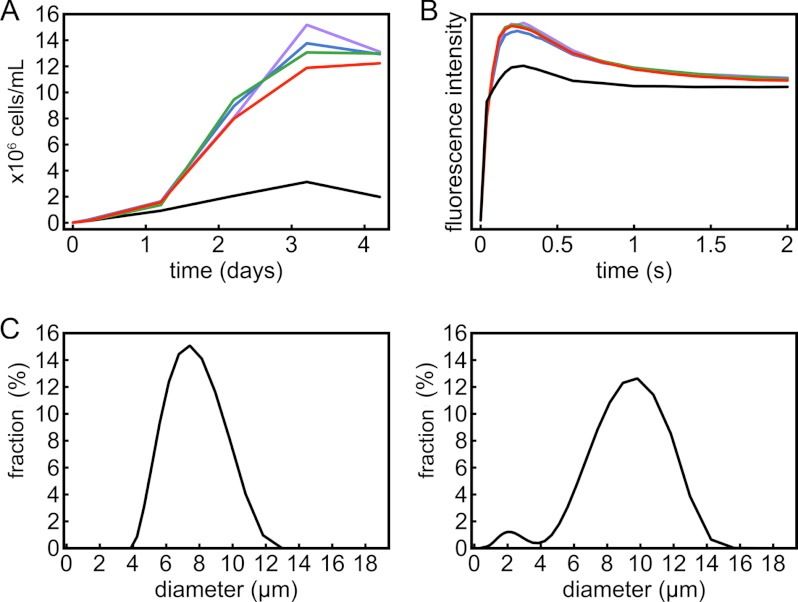
Phenotypes of zinc-deficient Chlamydomonas. Strain CC-4532 was grown in TAP medium with different amounts of supplemental zinc: 0 μm (black), 0.25 μm (blue), 2.5 μm (green), 80 μm (purple), and 250 μm (red). A, growth measured by counting cells; B, Kautsky fluorescence rise and decay kinetics; C, diameters of cells growing in medium containing 2.5 μm zinc (left) and 0 supplemental zinc (right). Growth curves represent the averages of biological triplicates from three separate inocula (S.D. values not shown for clarity). For fluorescence measurements and cell size determination, representative data from experimental triplicates are shown.
To generate zinc limitation, we inoculated cells (to a density of 105 cells/ml) from a replete culture into growth medium without any zinc supplementation (labeled as 0 in the figures). Although all medium constituents were prepared using high purity salts and all glassware was freshly acid-washed (39), there remained residual zinc in the medium, which we estimated at ∼10 nm based on ICP-MS analysis. When this culture (referred to as the first round in zinc-depleted medium) reached early stationary phase (∼1 × 107 cells/ml), the cells were used to reinoculate fresh “zinc-free” medium or medium with various amounts of supplemented zinc (Fig. 1A). A growth phenotype was identified in medium supplemented with no (0) or very low (10–25 nm; data not shown) zinc, and this correlated with a reduced intracellular zinc content of these cultures (varying from 25 to 50% of the zinc in the replete situation in individual experiments) (supplemental Fig. S1). The zinc content of cells transferred twice to the 0 zinc medium was only slightly less than or nearly the same as that of cultures transferred once to 0 zinc medium. Further subculturing was therefore not necessary, and we routinely used two transfers to 0 zinc medium as the base line for poor zinc nutrition. We also tried to generate zinc deficiency by reducing zinc bioavailability with the introduction of Zn(II)-specific chelators, like N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine into the medium, but this did not enhance the phenotype of cells in the second round of growth in 0 zinc medium. N,N,N′,N′-Tetrakis(2-pyridylmethyl)ethylenediamine was also not effective in generating a zinc-deficient (see below) or zinc-limited condition when it was added at higher concentrations to zinc-replete medium. Therefore, we concluded that, in the absence of a transporter-defective mutant, sequential transfer into defined medium with 0 supplemental Zn(II) is the only effective way to generate zinc limitation in Chlamydomonas. Analysis of the fluorescence induction curves shows that PSII activity is retained by cells growing in all concentrations of supplemental zinc, including those from cultures with no supplemental zinc, and that electron transfer between the two photosystems still operates under all of these conditions (Fig. 1B). Light microscopy indicated that zinc-limited cells are larger than the replete ones, and this is confirmed by measuring the size distribution of cells in batch culture. The average diameter of zinc-limited cells is 11 μm compared with the 9-μm size of zinc-replete cells. In addition, we noted the occurrence of smaller bodies of size ∼2.5 μm in the zinc-limited culture. Their identity is not known, but they may represent stress-induced cell fragmentation (Fig. 1C).
At the other extreme, zinc excess is defined as high zinc ion concentrations at which zinc starts to become toxic and the growth rate is inhibited relative to the maximum observed growth rates. In TAP medium, growth is inhibited when the medium contains more than 125–250 μm chelated (EDTA) zinc ions (Fig. 1A, red curve), and this defines the zinc excess situation.
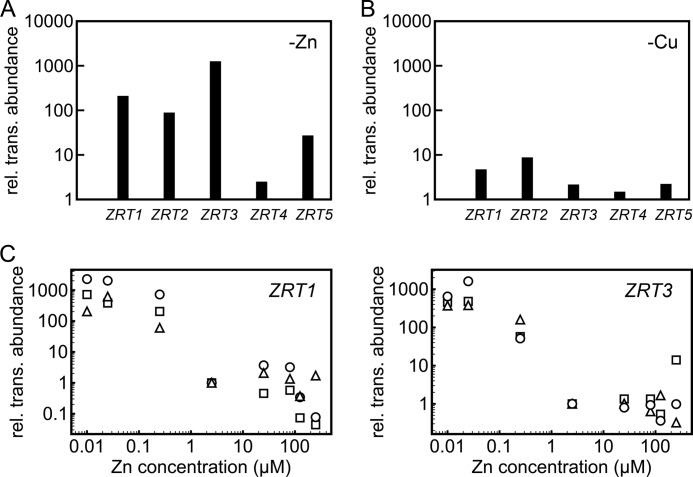
In previous work on iron nutrition, we distinguished the deficiency state from the limitation state. The iron deficiency state is characterized by the absence of a growth phenotype or other visual symptoms but the presence of a molecular signature, which in the case of iron nutrition is the expression of the iron assimilation pathway (31). Therefore, we sought to identify biomarkers for zinc deficiency. As putative zinc transporters, members of the ZIP family are excellent candidates for zinc assimilation proteins. Previously, 14 members of this family were identified in Chlamydomonas based on homology to Arabidopsis, yeast, and human sequences (14, 53). We assessed the patterns of mRNA accumulation for each of these as a function of zinc nutrition status to distinguish (a) which of these might respond to zinc nutrition and (b) whether their expression might be an early gauge or a sentinel of the zinc status. RNAs were isolated from cells transferred to medium containing the indicated amounts of zinc (after 1 round in 0 supplemental zinc) and analyzed by real-time RT-PCR for the expression of each of the ZIP genes. Among these, five zinc nutrition-responsive genes were identified and named ZRT1, ZRT2, ZRT3, ZRT4, and ZRT5 (for zinc-responsive transporter (Fig. 2, A and B). Based on the magnitude of the change, we chose ZRT1 and ZRT3 as sentinel genes and defined 25 nm as the boundary between zinc deficiency and zinc-limiting laboratory conditions (Fig. 2C). Thus, we established growth conditions to generate each of four states of zinc nutrition: 0–10 nm zinc (no added zinc) for zinc-limiting conditions, 25 nm zinc for deficient conditions, 2.5 μm zinc for replete conditions, and 250 μm for zinc-toxic conditions.
FIGURE 2.
Relative expression of ZIP family transporter genes identifies a subset responsive to zinc nutrition. RNA was isolated from cells grown in TAP medium containing various amounts of zinc or copper supplementation as described below. RNA abundance was assessed by real-time RT-PCR using the 2−ΔΔCT method. RNA abundance was normalized to CBLP abundance, and average CT values were calculated from technical triplicates. Each data point represents an independent RNA sample. A, RNA abundance in 0 zinc relative to 2.5 μm zinc supplementation. B, RNA abundance in 0 copper relative to 2 μm copper supplementation. C, ZRT1 (left) and ZRT3 (right) mRNA abundances in cells grown in medium containing 0, 0.025, 0.25, 2.5, 25, 80, 125, and 250 μm supplemental zinc. The zinc-deficient state was identified as the concentration of supplemental zinc at which ZRT1 (left) and ZRT3 (right) transcripts increased relative to 2.5 μm zinc supplementation (replete conditions).
Transcriptome Analysis of Cells Growing in Limited Versus Replete Zinc Conditions
To understand the basis for growth inhibition and to discover new zinc homeostasis factors, we used RNA-Seq for an exploratory transcriptome of zinc-limited CC-4532 (wild-type) cells. The cells were generated by transfer from the first round of growth in medium with no supplemental zinc into TAP medium supplemented or not with 2.5 μm Zn-EDTA. Cells were collected from all cultures at mid-exponential phase. RNA was prepared and validated for physiology by RT-PCR for sentinel gene expression and analyzed by sequencing of cDNA libraries on the Illumina platform. The reads were aligned to the Augustus 10.2 gene models on the version 4 assembly of the Chlamydomonas genome and were analyzed to quantify abundance of transcripts as described previously. Using a cut-off of 4-fold change and a p value of <0.05, we identified 533 genes that showed increased transcript abundance and 119 genes that showed decreased transcript abundance in zinc-limiting conditions relative to zinc-replete conditions (supplemental Data Set 1).
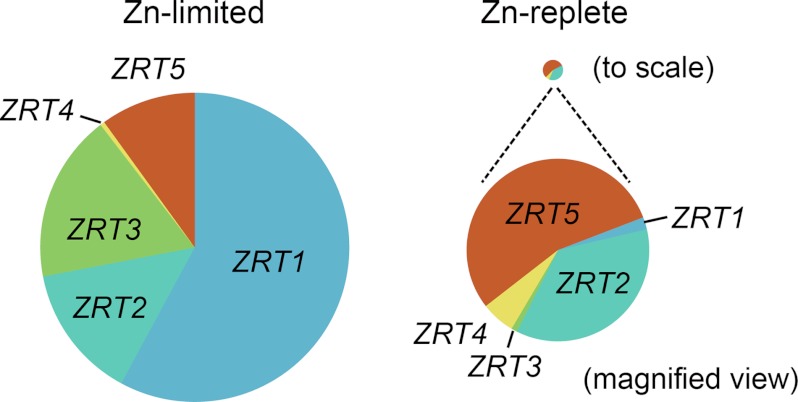
Total transcript abundance for the ZRT genes dramatically increased in zinc-limited conditions relative to zinc-replete conditions (Fig. 3). However, the proportion of each individual ZRT transcript in the total pool is different in the zinc-limited versus replete state. For instance, ZRT1 and ZRT3 are not prominent in replete cells but become more prominent in zinc-limited conditions, suggesting that they may encode the primary high affinity assimilative zinc transporters in a situation of deficiency. ZRT5 may encode a lower affinity form that can function with higher extracellular zinc availability.
FIGURE 3.
Abundance of Chlamydomonas ZRT transcripts. Pie charts show the contribution of each ZRT transcript during growth in zinc-limited (no supplemental zinc) and zinc-replete (2.5 μm zinc) media. The size of each pie chart represents the sum of all ZRT transcripts in that condition relative to the other.
Evident in the data set are genes belonging to three physiological categories: transcripts that were shown previously to increase in CO2-limited cells (54), transcripts that increase in copper-deficient cells (35), and transcripts encoding candidate zinc transporters and zinc homeostasis factors. The CO2-responsive genes HAP3 (Cre03.g177250.t1.1) and a gene encoding an unknown hypothetical protein (Cre10.g436450.t1.1) are among those whose transcripts increase most dramatically (∼103-fold), and the same is true for genes in the copper regulon, like CYC6 (Cre16.g651050.t1.1) encoding Cyt c6 and a gene encoding a hypothetical protein (Cre07.g352000.t1.1) (Table 1). The degree of regulation is as dramatic as that for the putative zinc transporters ZRT1 (Cre07.g351950.t1.2) and ZRT3 (Cre13.g573950.t1.2) and two genes encoding COG0523 domain-containing proteins that are implicated in zinc homeostasis (55). We named the two COG0523-domain proteins Zcp1 and Zcp2 (for zinc-responsive COG0523 domain-containing protein). Zcp2 corresponds to Cre02.g118400.t1.2, but Zcp1 is missing in the version 4 assembly of the Chlamydomonas genome. Nevertheless, when the reads are aligned to the version 3 assembly, the increase in transcript abundance for Zcp1 (Protein ID 117458) is dramatically evident.
TABLE 1.
Subsets of CO2-, Cu-, and Zn-responsive genes are highly up-regulated in Zn-limitation
Comparison of mRNA abundances among genes that are the most highly up-regulated in Zn-limited relative to Zn-replete photoheterotrophic wild-type cells identifies genes known previously to be regulated by CO2, Cu, and Zn. Protein ID Augustus 10.2, loci identified in the Augustus update 10.2 annotation of the JGI assembly version 4 for each gene. -Fold change is presented as the ratio of −Zn/+Zn. Differential expression statistics are provided in terms of p values.
| Protein ID Augustus 10.2 | Gene name | Defline | −Zinc | +Zinc | Change | p value |
|---|---|---|---|---|---|---|
| RPKM | RPKM | -fold | ||||
| Cre02.g118400.t1.2 | ZCP2 | Epressed hypothetical protein | 614 | 0.08 | 7 ×103 | 2 ×10−41 |
| Cre16.g651050.t1.1 | CYC6 | Cytochrome c6 | 1825 | 0.25 | 7 ×103 | 6 ×10−41 |
| Cre10.g436450.t1.1 | Expressed hypothetical protein | 114 | 0.03 | 4 ×103 | 5 ×10−33 | |
| Cre03.g177250.t1.1 | HAP3 | Haloperoxidase-like protein | 304 | 0.18 | 1 ×103 | 2 ×10−32 |
| Cre07.g351950.t1.2 | ZRT1 | Zinc nutrition-responsive transporter | 563 | 0.40 | 1 ×103 | 7 ×10−38 |
| Cre07.g352000.t1.1 | Expressed hypothetical protein | 4364 | 4.3 | 1 ×103 | 1 ×10−37 | |
| Cre13.g573950.t1.2 | ZRT3 | Zinc nutrition-responsive transporter | 171 | 0.19 | 1 ×103 | 2 ×10−29 |
The zinc responsiveness of two previously identified limiting CO2-inducible genes prompted us to compare the genes regulated by zinc starvation with those regulated by low CO2, including targets of the transcription factor Ccm1 (54) (supplemental Data Set 2). We found that 82 genes were similarly zinc-responsive and CO2-responsive, with 54 of them being predicted Ccm1 targets. Among extensively studied low CO2-induced genes that are also zinc-responsive are CAH4, CAH5, CCP1, HLA3, and LCIA. When we compared the genes regulated by zinc starvation with those regulated by copper starvation, we found a similar situation. There are 23 instances of genes responding similarly to deprivation of either micronutrient (supplemental Data Set 3), with 16 increasing in transcript abundance and 7 decreasing, and of the 23, 11 (or about half) are Crr1 targets, which is the same fraction as that of Crr1 targets among genes that respond to copper nutrition (35). The reason that we pick up only a fraction of the copper regulon and Crr1 targets is that the copper regulon is not as highly activated in zinc deficiency as it is in copper deficiency, and some of the transcript changes did not meet our criteria for inclusion. A few (eight) genes respond in an opposite fashion to zinc versus copper nutrition, and two of these are Crr1 targets, pointing to additional Crr1-independent controls at the transcriptional level for these two genes. When we checked the reproducibility of a subset of the changes in RNA abundance by real-time RT-PCR on RNAs isolated from triplicate cultures, we confirmed the findings (data not shown).
Therefore, we conclude that (a) there is a zinc nutrition-sensing regulatory pathway in Chlamydomonas that operates in part to control the abundance of transcripts for zinc transporters of the ZIP family and zinc homeostasis factors, and (b) there may be an impact of zinc starvation on copper homeostasis and CO2 assimilation.
When we used the Algal Functional Annotation Tool (10), with selected gene ontology terms from Arabidopsis thaliana to overcome the limitation of available annotations for Chlamydomonas, to identify metabolic functions or pathways that might be associated with the zinc-responsive genes identified from transcriptome profiling, we noted three meaningful (p value<0.01) biological processes for transcripts whose abundance increases: zinc and other ion transport (17 hits, scores range from 2 × 10−3 to 5 × 10−4), zinc and other ion homeostasis (4 hits, scores range from 8 × 10−3 to 2 × 10−4), and the response to arsenic (2 hits, score = 1 × 10−3). For transcripts that decrease, we found genes related to organelle and peroxisome organization, organic acid metabolism, fatty acid metabolism, and regulation of cell division and DNA replication, probably a consequence of the role of zinc ions as a structural cofactor in proteins involved in nucleic acid transactions.
Proteomic Analysis
To determine if changes in the transcript abundance of some genes were recapitulated at the level of the soluble proteome, we analyzed soluble protein samples by quantitative LC-MSE from cells growing in medium supplemented with 2.5 μm zinc or in an initial round of medium with no supplemental zinc (see also Ref. 48). We chose to use only one round of growth in medium lacking zinc in an attempt to distinguish between the primary effects of zinc deficiency versus more general stress markers, which are likely to be more prevalent after sustained growth (i.e. two transfers) in medium with 0 supplemental zinc. Among the proteins identified as the most abundant and differentially regulated in zinc-limiting conditions are the hypothetical protein Cre07.g352000.t1.1 and both Zcp1 and Zcp2. In addition, we identified and showed an increase in the abundance of Cgl78/Ycf54 and Fea1, whose transcripts increase in both zinc limitation and copper deficiency (supplemental Data Set 3).
Carbonic Anhydrases and CO2 Requirements during Photoautotrophic Growth
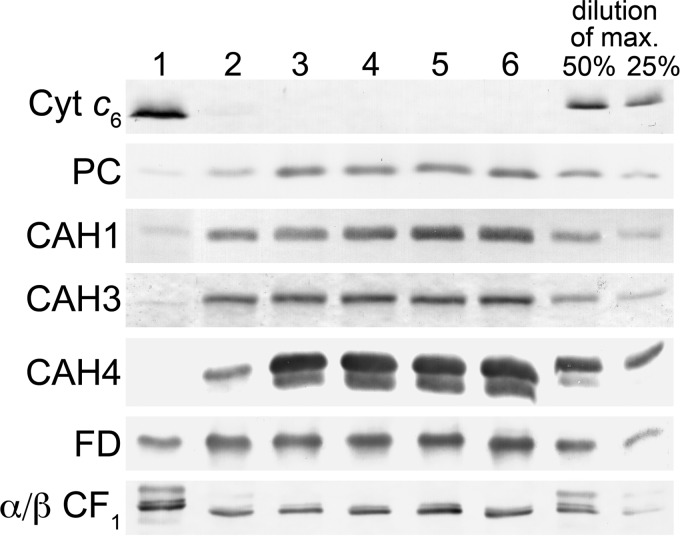
The carbon-concentrating mechanism allows algae and cyanobacteria to grow phototrophically at air levels of CO2, and carbonic anhydrases (CAs) are key enzymes in this mechanism (56–58). In Chlamydomonas, there are 12 genes predicted to encode zinc-containing carbonic anhydrases (59). The up-regulation of the CCM genes suggested to us that the activities of one or more of these enzymes might be compromised in zinc limitation, leading to a functional low CO2 phenotype despite the presence of acetate, which normally suppresses the low CO2-activated genes. Therefore, we surveyed the expression of the CAH genes that contribute to the bulk of the CA activity or that are known to be required for operation of the CCM. Cah1 is the major contributor to carbonic anhydrase activity in whole cell extracts in the presence of light and low (i.e. atmospheric levels) CO2, although no phenotype has been associated with its absence. Differences in transcript accumulation for CAH1 in response to zinc ion concentration were small but suggested a slight decrease in expression in response to decreasing zinc (supplemental Data Set 1). Immunoblot analysis revealed a more dramatic decrease in Cah1 accumulation in zinc-limiting medium (Fig. 4), and this result was recapitulated in proteomic studies (48). Cah3 is localized to the chloroplast and is believed to be the primary carbonic anhydrase required for maintenance of the CCM. Transcripts for CAH3 increase in zinc limitation, but again immunoblot analyses showed that Cah3 abundance is decreased in zinc deficiency. Two other carbonic anhydrases, Cah4 and Cah5 (nearly identical in sequence), are located in the mitochondria. The transcripts encoding these enzymes increase under zinc limitation, but immunoblot analysis revealed the decreasing abundance of Cah4 in zinc limitation as well. We conclude that zinc limitation has a major impact on carbonic anhydrase activity, as documented already for diatoms, which led to the hypothesis that the decreased growth rate under phototrophic conditions might result from impaired CCM.
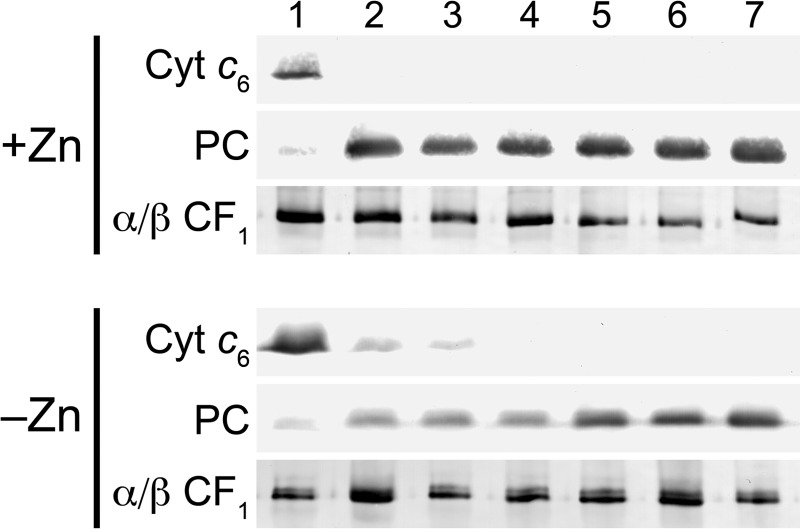
FIGURE 4.
Impact of zinc nutrition on abundance of select proteins in the photosynthetic apparatus. Total soluble and resuspended particulate fractions from Chlamydomonas cultures grown in TAP medium with the various supplemental zinc concentrations were separated on polyacrylamide gels under denaturing conditions. Lane 1, 0 zinc; lane 2, 0.25 μm zinc; lane 3, 2.5 μm zinc; lane 4, 25 μm zinc; lane 5, 80 μm zinc; lane 6, 125 μm zinc. The separated proteins were transferred to nitrocellulose membranes. Proteins of interest were detected by immunoblot analysis with antisera raised against Cyt c6, plastocyanin (PC), carbonic anhydrase 1 (CAH1), carbonic anhydrase 3 (CAH3), carbonic anhydrase 4 (CAH4), ferredoxin (FD), and the α and β subunits of the chloroplast ATP synthase (α/β CF1). For Cyt c6 and the α and β subunits of the chloroplast ATP synthase, dilutions of maximum (dilutions of max) are based on lane 1. For all other proteins, they are based on lane 6.
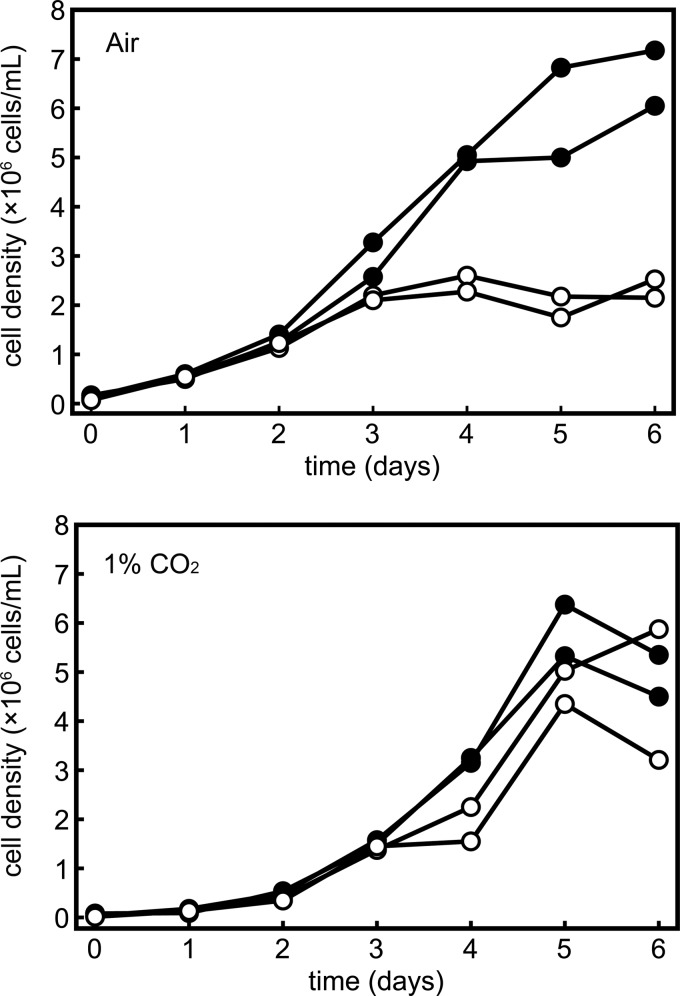
To test this hypothesis, we grew zinc-limited and zinc-replete cultures in TP medium, which lacks acetate as a carbon source, and bubbled the flasks with either air, representing limiting CO2 conditions, or air with 1% CO2, representing high CO2 conditions, where CA activity is not required. In air-bubbled flasks, zinc-limited cultures displayed limited growth relative to zinc-replete cultures (Fig. 5). When flasks were bubbled with 1% CO2, growth in zinc-limited cultures was comparable with growth in zinc-replete cultures, although overall growth in both cultures decreased slightly. We attribute the slight change in both cultures to a change in the pH of the medium caused by the introduction of 1% CO2. The zinc limitation phenotype in phototrophic medium is therefore rescued by high CO2, establishing a causal connection between CCM and the zinc limitation phenotype resulting from impaired CA activity. The importance of back-up CAs in diatoms for productivity in low zinc environments is evident.
FIGURE 5.
High CO2 suppresses the growth phenotype of zinc-limited photoautotrophically grown cells. Chlamydomonas cells (strain CC-4532) were preconditioned in TP medium and inoculated to a density of 105 cells/ml into TP medium bubbled with filtered air (top) or a mixture of 1% CO2 and air (bottom). Growth was monitored by counting with a hemocytometer. Duplicates of zinc-replete (closed symbols) and zinc-limited (open symbols) cultures are shown and represent one of two experimental duplicates.
Copper Deficiency and Crr1 Function in Zinc-limited Cells
The up-regulation of the Crr1 regulon and copper deficiency responses suggests that the zinc-deficient cells may be functionally copper-deficient, reminiscent of the connection between iron and copper, where copper-deficient cells are secondarily iron-deficient (60). To test this idea, we monitored the abundance of plastocyanin and Cyt c6 as a function of zinc concentration by immunoblotting (Fig. 4). Indeed, plastocyanin is decreased in zinc-deficient cells, and a concomitant increase in cytochrome c6 is evident. If excess copper is provided, we can restore plastocyanin content in proportion to the added copper, but much greater amounts of copper are required to maintain plastocyanin in zinc-limited cells compared with zinc-replete ones (Fig. 6). This supports the idea that zinc-limited cells are functionally copper-deficient. Therefore, we tested whether Crr1 might be required for Chlamydomonas to acclimate to zinc limitation. In zinc-limiting and toxic conditions, growth of crr1 mutant cultures was lower than growth of the complemented strain, CRR1 (Fig. 7A). Growth of the mutant was also slightly decreased in replete conditions, consistent with the role of Crr1 in all states of copper nutrition.
FIGURE 6.
Supplemental copper can restore plastocyanin abundance in zinc-limited cultures. Total soluble and particulate fractions were separated on a polyacrylamide gel and transferred as described in the legend to Fig. 5. Top, zinc-replete cultures express a constant level of plastocyanin (PC) over the range of 2–80 μm supplemental copper ion concentration. Bottom, zinc-limited cultures show a steady increase in plastocyanin with the increase in supplemental copper. Lane 1, 0 copper; lane 2, 2 μm copper; lane 3, 5 μm copper; lane 4, 10 μm copper; lane 5, 20 μm copper; lane 6, 40 μm copper; lane 7, 80 μm copper. Note that at 80 μm copper ion concentration, plastocyanin levels in zinc-limited cultures are still lower than plastocyanin levels in zinc-replete cultures growing with 2 μm supplemental copper.
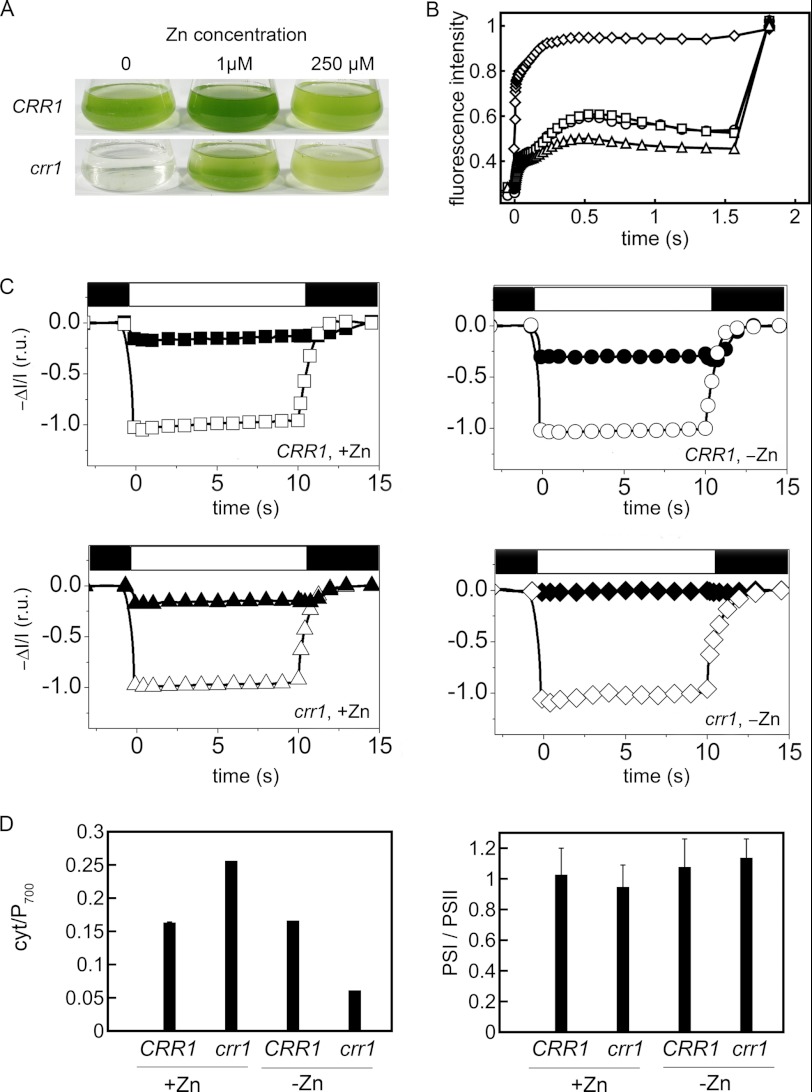
FIGURE 7.
Crr1 is required for acclimation to zinc deficiency. A, crr1 and CRR1 strains grown in TAP medium supplemented with 0, 1, and 250 μm zinc. Cells were inoculated at 105 cells/ml and photographed after 3 days of growth. B, Kautsky fluorescence rise and decay kinetics of strains growing in zinc-replete or zinc-limiting conditions normalized to maximum fluorescence intensity measured upon exposure to a saturating light pulse (1100 μmol of photons m−2 s−1). Squares, CRR1 in zinc-replete medium; circles, CRR1 in zinc-limited medium; diamonds, crr1 in zinc-limited medium; triangles, crr1 in zinc-replete medium. C, Cyt (solid symbols) and P700 (open symbols) redox changes of dark-adapted cells as measured by absorbance changes (same symbols as above); r.u., relative units. D, cytochrome/P700 and PSI/PSII ratios. In A–C, representative data are shown. For D, the error bars represent S.D. of three independent measurements.
Measurement of fluorescence rise and decay kinetics in crr1 versus CRR1 cells revealed that the mutant maintains a high steady state level of fluorescence emission in zinc limitation (Fig. 7B). This is probably due to the inability of the mutant to transfer electrons downstream of PSII to PSI in the photosynthetic electron transport chain, most probably as a result of the inability to up-regulate Cyt c6. To support this, we estimated the stoichiometry of Cyt c6 to P700 (PSI) in zinc-limited crr1 and CRR1 cultures by quantifying the ratio of redox-active Cyt/P700 using the electrochromic shift assay (Fig. 7C). For the complemented strain, Cyt/P700 ratios increased from 0.15 in zinc-replete conditions to 0.25 in zinc limitation, whereas the mutant showed a slight decrease in the Cyt/P700 ratio from 0.15 in zinc-replete conditions to ∼0.05 in zinc limitation (Fig. 7D). PSI/PSII ratios were ∼1:1 for both strains in both zinc-replete and zinc-limited conditions (Fig. 7D). These results support the idea that the increase in Cyt c6 is necessary to maintain photosynthesis in zinc-limited Chlamydomonas cells, even when copper is available in concentrations that should be sufficient to support plastocyanin maintenance.
Trace Metal Quota
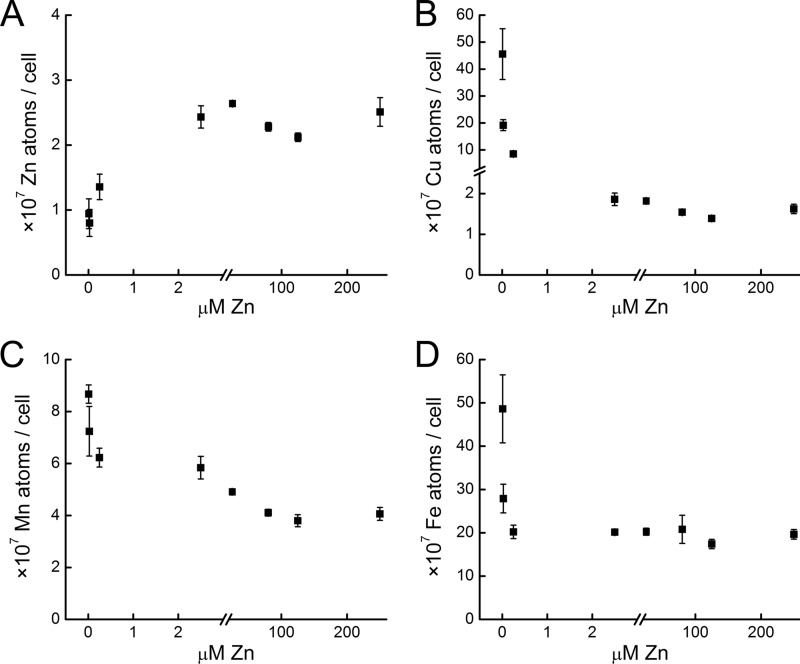
To validate the impact of zinc nutrition on copper content, we measured the metal content of Chlamydomonas cells over a range of supplemental zinc concentrations (Fig. 8). In the range of concentrations from 25 to 250 μm, the zinc quota is maintained at ∼3 × 107 atoms/cell with some variation that tracked changing external zinc concentrations. When zinc in the medium was less than 25 nm, the quota decreased to ∼1 × 107 atoms/cell, consistent with symptoms of zinc deficiency and limitation. In contrast, the abundance of iron and copper ions (and possibly also manganese ions) increased with decreasing zinc ion supplementation. This might be attributed to up-regulation of ZIP transporters and the relatively high concentrations of these divalent cations in the medium. In fact, we note that sentinel genes for iron assimilation are down-regulated in zinc deficiency, presumably because of higher intracellular iron content. Unexpectedly, given the molecular phenotype of the copper regulon (see above), the copper quota was increased most dramatically. Its intracellular abundance increased by over an order of magnitude from ∼1–2 × 107 atoms/cell in zinc-replete conditions to as much as 4 × 108 atoms/cell in zinc-limiting conditions. This contrasts strikingly with the tight regulation of copper content observed in previous work (28) and presents a conundrum with respect to the molecular phenotype. We conclude that normal copper homeostasis requires zinc.
FIGURE 8.
Zinc-deficient Chlamydomonas cells accumulate other metal ions. Intracellular metal ion concentrations of zinc (A), copper (B), manganese (C), and iron (D) were quantified as a function of supplemental zinc. Chlamydomonas strain CC-4532 was grown in TAP medium supplemented with 0, 0.025, or μm 0.25 zinc (Experiment 1) or with 2.5 25, 80, 125, or 250 μm zinc (Experiment 2). For analysis, cells were collected by centrifugation. The cell pellet was washed with 1 mm EDTA to remove extracellular bound metal and dissolved in 30% nitric acid by incubation at 65 °C. The metal content was measured by ICP-MS. Data points represent the average of experimental triplicates.
DISCUSSION
Although studies investigating acclimation strategies to zinc deficiency have been performed in yeast and other organisms (61), the impact of zinc deficiency on photosynthetic organisms has been underinvestigated. Here, we have defined and characterized distinct stages of zinc nutrition in Chlamydomonas because of an excellent infrastructure for understanding trace element homeostasis developed through our prior studies (25). We have distinguished a stage where growth is inhibited, and we refer to that as zinc limitation versus a stage where the zinc-sensing signal transduction pathway is operational to turn on assimilation genes but growth is not affected, and we refer to that as deficiency. As is the case for other organisms, excess zinc results in toxicity, which is also distinguished by growth inhibition (Fig. 1).
In Saccharomyces cerevisiae and Arabidopsis, a primary strategy for the regulation of zinc homeostasis involves the zinc-responsive expression of low and high affinity zinc transporters (62, 63). Therefore, these seemed like excellent candidates for targets of nutritional zinc signaling as well. A genome survey of ZIP and CDF family transporters (14) led to the identification of five putative Chlamydomonas zinc transporters whose transcripts accumulate in zinc-limiting conditions (Fig. 2). The regulation of these genes was used to identify zinc concentrations that induce a change in the abundance of the corresponding RNAs, and this corresponds to concentrations at which the zinc content is less than ∼2 × 107 atoms/cell, which defines the typical zinc quota for Chlamydomonas. Very low zinc content in the medium (zinc limitation) results in physiological stress, as suggested by the change in cell size (Fig. 1) as well as by the accumulation of Nile Red-staining bodies under zinc-limiting conditions (27).
Based on fluorescence measurements of photosynthetic ability, the impact of zinc limitation did not affect the photosynthetic apparatus per se (Fig. 1B). However, the known role of zinc in DNA-binding proteins and CAs participating in the CCM suggested that photosynthetic growth might be affected indirectly as a result of zinc deficiency. RNA-seq technology has been employed and rigorously evaluated in Chlamydomonas, showing this technology to be as quantitative as real-time PCR to evaluate changes in transcript abundance in response to external conditions (35). Therefore, we used it as an exploratory tool to discover pathways that might be impacted by poor zinc nutrition. We identified nearly 700 genes whose expression was significantly changed in zinc-limited versus -replete cells (supplemental Data Set 1).
Photoautotrophic Growth at Air Levels of CO2 and Carbonic Anhydrases
The finding that genes described/discovered in previous studies as being targets of the CCM pathway or Crr1 pointed to inorganic carbon assimilation and copper homeostasis, both of which affect photosynthetic performance, as being affected by poor zinc nutrition. In both cases, we substantiated this by rescuing the zinc starvation phenotype by provision of high CO2 or excess copper (Figs. 5 and 6, respectively). Consistent with an impact on the CCM pathway, we found that photoautotrophic growth of zinc-limited cells was inhibited relative to zinc-replete cells at air levels of CO2 (low CO2 conditions), but provision of 1% CO2 increased photoautotrophic growth rates in zinc-limited cells. The LHCSR1, LHCSR3.1, and LHCSR3.2 genes encoding a subtype of light-harvesting chlorophyll-binding protein are regulated by CO2 as well, but they are inversely regulated in response to zinc limitation relative to CO2 limitation (64). This suggests that these genes are under the control of multiple regulatory factors because they respond differently to low CO2 than they do to other nutrient deprivation regimes, including nitrogen and sulfur starvation (65).
The most obvious targets of poor zinc nutrition are the carbonic anhydrases, and we characterized their expression profiles relative to zinc nutrition. Transcripts for some carbonic anhydrases are increased in zinc limitation; however, immunoblots for Cah1, Cah3, and Cah4/5 revealed that these proteins decreased in zinc-limiting conditions. We could not detect Cah2 because its abundance is quite low (5%) relative to that of Cah1, and indeed it is not detected in cells grown in uninduced conditions (i.e. high CO2) (59). The discrepancy between transcript and protein accumulation may suggest post-transcriptional regulation of these genes or a compensatory up-regulation of the genes in response to decreased activity. It is possible that the proteins are indeed synthesized, but they might be degraded in the absence of the essential zinc cofactor, or they might be actively degraded as part of a zinc-sparing program (24, 48). Carbonic anhydrases are important for photosynthesis under low CO2 conditions based on the effect of enzyme inhibitors (acetazolamide and DBS) and based on the phenotype of a mutant lacking lumenal carbonic anhydrase Cah3 (66), and the expression of several CAH genes is increased in cells grown at air levels of CO2 as part of the CCM (59). The conditional CO2-repressed phenotype of zinc-deficient Chlamydomonas cells establishes the carbonic anhydrases as key targets of zinc deficiency and therefore, in an environmental context, emphasizes the importance of the ability of diatoms to substitute cobalt or cadmium in the active site of CAs (22, 67) to maintain photosynthetic growth in face of zinc deficiency.
Influence of Zinc on Copper Nutrition
The similar regulation of 23 copper-responsive genes, including 11 Crr1 targets, suggested a connection between copper and zinc metabolism. The fraction of Crr1 targets (50%) is about the same as the fraction of the responding genes in copper-deficient cells (35). Because most of these are genes that are highly responsive and highly expressed, we concluded that zinc-limited cells are signaling a slightly copper-deficient status. Nevertheless, when we measured intracellular metal content, we noted the opposite (i.e. severalfold higher copper content in zinc-deficient cells) (Fig. 8). We conclude therefore that cell-associated copper is somehow not accessible to the copper sensor. Further, the copper also seems to be inaccessible to plastocyanin. The biosynthetic pathway for plastocyanin is normal because when excess copper is added to zinc-deficient cells, this copper can be used for holoplastocyanin synthesis, and the expression of CYC6 is decreased. Therefore, cell-associated copper is generally not accessible for cuproprotein biosynthesis. The biosynthesis of Cyt c6 would, in this case, become essential, which may explain the non-photosynthetic phenotype of zinc-deficient crr1 cells (Fig. 7).
The advantage, if any, of copper sequestration is not known. The effects of zinc nutrition on organisms is known to be complicated by the fact that interactions exist between zinc and copper homeostasis pathways. In vivo, Drosophila metallothionein genes are induced by both copper and zinc, suggesting that metallothioneins bind both metals (68). Among humans, serum copper/zinc ratios were significantly higher in patients with malignant lung tumors than in healthy patients and may serve as a diagnostic test in lung cancer patients (69). The copper/zinc ratio is also significantly increased in disabled elderly patients relative to healthy elderly people and was associated with levels of systemic oxidative stress (70). In the case of patients with Wilson disease, zinc supplementation has been used to counteract copper hyperaccumulation. Mechanistic explanations for this effect are not readily available.
Crr1 and Ccm1 are both zinc-dependent transcription factors (37, 71). The maintenance of transcription factors Crr1 and Ccm1 in zinc limitation indicates that zinc remains available for their function although carbonic anhydrase function is compromised, indicative of hierarchical rules governing the allocation of zinc ions. This mechanism has already been documented previously for copper, manganese, and iron in Chlamydomonas (24, 72). It is tempting to speculate that the COG0523 domain proteins, Zcp1 and Zcp2, may play a role in metal allocation and sparing. The corresponding genes are as highly up-regulated (∼103-fold and ∼7 × 103-fold, respectively) and as highly expressed (∼6 × 102 RPKM) as are some of the genes encoding ZRT proteins (e.g. ZRT1 and ZRT3). These are excellent candidates for discovery of the mechanism of regulation or the zinc sensor in Chlamydomonas. There are also a number of proteins of unknown function that are also highly regulated by zinc deficiency, indicative of room for new discovery in zinc homeostasis.
Acknowledgments
We thank Rey Martin, Kara Velez and Esperanza del Rio for assistance during the early phases of the project; Joan Valentine at UCLA for use of the ICP-MS for metal measurements; Dr. James V. Moroney for providing antibodies to Cah1 and Cah4; Dr. Martin Spalding for providing an additional antibody for Cah1; and Dr. Joanna Porankiewicz for providing an antibody to Cah3.
This work was supported, in whole or in part, by National Institutes of Health Grant GM42143 (to S. M.) for work on the zinc-deficiency transcriptome. The proteomic analyses and expression of proteins for use as antigens were supported by the Institute of Genomics and Proteomics at UCLA (funded by the Office of Science (Biological and Environmental Research), United States Department of Energy, through Cooperative Agreement DE-FC02-02ER63421).

This article contains supplemental Fig. S1 and Data Sets 1–3.
- CCM
- carbon-concentrating mechanism
- CA
- carbonic anhydrase
- Cyt
- cytochrome
- TAP
- Tris acetate-phosphate
- TP
- Tris phosphate
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PSI and PSII
- photosystem I and II, respectively
- ICP
- inductively coupled plasma
- RPKM
- reads per kilobase of mappable transcript length per million mapped reads.
REFERENCES
- 1. Andreini C., Banci L., Bertini I., Rosato A. (2006) Zinc through the three domains of life. J. Proteome Res. 5, 3173–3178 [DOI] [PubMed] [Google Scholar]
- 2. Ehrensberger K. M., Bird A. J. (2011) Hammering out details. Regulating metal levels in eukaryotes. Trends Biochem. Sci. 36, 524–531 [DOI] [PubMed] [Google Scholar]
- 3. Eide D. J. (2009) Homeostatic and adaptive responses to zinc deficiency in Saccharomyces cerevisiae. J. Biol. Chem. 284, 18565–18569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ryu M. S., Lichten L. A., Liuzzi J. P., Cousins R. J. (2008) Zinc transporters ZnT1 (Slc30a1), Zip8 (Slc39a8), and Zip10 (Slc39a10) in mouse red blood cells are differentially regulated during erythroid development and by dietary zinc deficiency. J. Nutr. 138, 2076–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dainty S. J., Kennedy C. A., Watt S., Bähler J., Whitehall S. K. (2008) Response of Schizosaccharomyces pombe to zinc deficiency. Eukaryot Cell 7, 454–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhao H., Eide D. (1996) The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J. Biol. Chem. 271, 23203–23210 [DOI] [PubMed] [Google Scholar]
- 7. Zhao H., Eide D. (1996) The yeast ZRT1 gene encodes the zinc transporter protein of a high affinity uptake system induced by zinc limitation. Proc. Natl. Acad. Sci. U.S.A. 93, 2454–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Simm C., Lahner B., Salt D., LeFurgey A., Ingram P., Yandell B., Eide D. J. (2007) Saccharomyces cerevisiae vacuole in zinc storage and intracellular zinc distribution. Eukaryot. Cell 6, 1166–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Roh H. C., Collier S., Guthrie J., Robertson J. D., Kornfeld K. (2012) Lysosome-related organelles in intestinal cells are a zinc storage site in C. elegans. Cell Metab. 15, 88–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ballestín R., Molowny A., Marín M. P., Esteban-Pretel G., Romero A. M., Lopez-Garcia C., Renau-Piqueras J., Ponsoda X. (2011) Ethanol reduces zincosome formation in cultured astrocytes. Alcohol. Alcohol. 46, 17–25 [DOI] [PubMed] [Google Scholar]
- 11. Williams L. E., Pittman J. K., Hall J. L. (2000) Emerging mechanisms for heavy metal transport in plants. Biochim. Biophys. Acta 1465, 104–126 [DOI] [PubMed] [Google Scholar]
- 12. Kambe T., Andrews G. K. (2009) Novel proteolytic processing of the ectodomain of the zinc transporter ZIP4 (SLC39A4) during zinc deficiency is inhibited by acrodermatitis enteropathica mutations. Mol. Cell Biol. 29, 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Davis D. E., Roh H. C., Deshmukh K., Bruinsma J. J., Schneider D. L., Guthrie J., Robertson J. D., Kornfeld K. (2009) The cation diffusion facilitator gene cdf-2 mediates zinc metabolism in Caenorhabditis elegans. Genetics 182, 1015–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Blaby-Haas C. E., Merchant S. S. (2012) The ins and outs of algal metal transport. Biochim. Biophys. Acta 1823, 1531–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim D., Gustin J. L., Lahner B., Persans M. W., Baek D., Yun D. J., Salt D. E. (2004) The plant CDF family member TgMTP1 from the Ni/Zn hyperaccumulator Thlaspi goesingense acts to enhance efflux of Zn at the plasma membrane when expressed in Saccharomyces cerevisiae. Plant J. 39, 237–251 [DOI] [PubMed] [Google Scholar]
- 16. Podar D., Scherer J., Noordally Z., Herzyk P., Nies D., Sanders D. (2012) Metal selectivity determinants in a family of transition metal transporters. J. Biol. Chem. 287, 3185–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Badger M. R., Price G. D. (2003) CO2 concentrating mechanisms in cyanobacteria. Molecular components, their diversity and evolution. J. Exp. Bot. 54, 609–622 [DOI] [PubMed] [Google Scholar]
- 18. Giordano M., Beardall J., Raven J. A. (2005) CO2 concentrating mechanisms in algae. Mechanisms, environmental modulation, and evolution. Annu. Rev. Plant Biol. 56, 99–131 [DOI] [PubMed] [Google Scholar]
- 19. Roberts K., Granum E., Leegood R. C., Raven J. A. (2007) Carbon acquisition by diatoms. Photosynth. Res. 93, 79–88 [DOI] [PubMed] [Google Scholar]
- 20. Wang Y., Duanmu D., Spalding M. H. (2011) Carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii. Inorganic carbon transport and CO2 recapture. Photosynth. Res. 109, 115–122 [DOI] [PubMed] [Google Scholar]
- 21. Lane T. W., Morel F. M. (2000) A biological function for cadmium in marine diatoms. Proc. Natl. Acad. Sci. U.S.A. 97, 4627–4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lane T. W., Morel F. M. (2000) Regulation of carbonic anhydrase expression by zinc, cobalt, and carbon dioxide in the marine diatom Thalassiosira weissflogii. Plant Physiol. 123, 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yee D., Morel F. M. M. (1996) In vivo substitution of zinc by cobalt in carbonic anhydrase of a marine diatom. Limnol. Oceanogr. 41, 573–577 [Google Scholar]
- 24. Merchant S. S., Helmann J. D. (2012) Elemental economy. Microbial strategies for optimizing growth in the face of nutrient limitation. Adv. Microb. Physiol. 60, 91–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Merchant S. S., Allen M. D., Kropat J., Moseley J. L., Long J. C., Tottey S., Terauchi A. M. (2006) Between a rock and a hard place. Trace element nutrition in Chlamydomonas. Biochim. Biophys. Acta 1763, 578–594 [DOI] [PubMed] [Google Scholar]
- 26. Harris E. H. (1989) The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use, Academic Press, Inc., San Diego, CA: [DOI] [PubMed] [Google Scholar]
- 27. Kropat J., Hong-Hermesdorf A., Casero D., Ent P., Castruita M., Pellegrini M., Merchant S. S., Malasarn D. (2011) A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. Plant J. 66, 770–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Page M. D., Kropat J., Hamel P. P., Merchant S. S. (2009) Two Chlamydomonas CTR copper transporters with a novel Cys-Met motif are localized to the plasma membrane and function in copper assimilation. Plant Cell 21, 928–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Allen M. D., del Campo J. A., Kropat J., Merchant S. S. (2007) FEA1, FEA2, and FRE1, encoding two homologous secreted proteins and a candidate ferrireductase, are expressed coordinately with FOX1 and FTR1 in iron-deficient Chlamydomonas reinhardtii. Eukaryot. Cell 6, 1841–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Allen M. D., Kropat J., Tottey S., Del Campo J. A., Merchant S. S. (2007) Manganese deficiency in Chlamydomonas results in loss of photosystem II and MnSOD function, sensitivity to peroxides, and secondary phosphorus and iron deficiency. Plant Physiol. 143, 263–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. La Fontaine S., Quinn J. M., Nakamoto S. S., Page M. D., Göhre V., Moseley J. L., Kropat J., Merchant S. (2002) Copper-dependent iron assimilation pathway in the model photosynthetic eukaryote Chlamydomonas reinhardtii. Eukaryot. Cell 1, 736–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kropat J., Tottey S., Birkenbihl R. P., Depège N., Huijser P., Merchant S. (2005) A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl. Acad. Sci. U.S.A. 102, 18730–18735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yamasaki H., Hayashi M., Fukazawa M., Kobayashi Y., Shikanai T. (2009) SQUAMOSA promoter binding protein-like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 21, 347–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bernal M., Casero D., Singh V., Wilson G. T., Grande A., Yang H., Dodani S. C., Pellegrini M., Huijser P., Connolly E. L., Merchant S. S., Krämer U. (2012) Transcriptome sequencing identifies SPL7-regulated copper acquisition genes FRO4/FRO5 and the copper dependence of iron homeostasis in Arabidopsis. Plant Cell 24, 738–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Castruita M., Casero D., Karpowicz S. J., Kropat J., Vieler A., Hsieh S. I., Yan W., Cokus S., Loo J. A., Benning C., Pellegrini M., Merchant S. S. (2011) Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell 23, 1273–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Quinn J. M., Barraco P., Eriksson M., Merchant S. (2000) Coordinate copper- and oxygen-responsive Cyc6 and Cpx1 expression in Chlamydomonas is mediated by the same element. J. Biol. Chem. 275, 6080–6089 [DOI] [PubMed] [Google Scholar]
- 37. Sommer F., Kropat J., Malasarn D., Grossoehme N. E., Chen X., Giedroc D. P., Merchant S. S. (2010) The CRR1 nutritional copper sensor in Chlamydomonas contains two distinct metal-responsive domains. Plant Cell 22, 4098–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Merchant S., Bogorad L. (1986) Regulation by copper of the expression of plastocyanin and cytochrome c552 in Chlamydomonas reinhardtii. Mol. Cell Biol. 6, 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Quinn J. M., Merchant S. (1998) Copper-responsive gene expression during adaptation to copper deficiency. Methods Enzymol. 297, 263–279 [DOI] [PubMed] [Google Scholar]
- 40. Joliot P., Joliot A. (2002) Cyclic electron transfer in plant leaf. Proc. Natl. Acad. Sci. U.S.A. 99, 10209–10214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schloss J. A. (1990) A Chlamydomonas gene encodes a G protein β subunit-like polypeptide. Mol. Gen. Genet. 221, 443–452 [DOI] [PubMed] [Google Scholar]
- 42. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 43. Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 [DOI] [PubMed] [Google Scholar]
- 45. Anders S., Huber W. (2010) Differential expression analysis for sequence count data. Genome Biol. 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate. A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300 [Google Scholar]
- 47. Howe G., Merchant S. (1992) The biosynthesis of membrane and soluble plastidic c-type cytochromes of Chlamydomonas reinhardtii is dependent on multiple common gene products. EMBO J. 11, 2789–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hsieh S. I., Castruita M., Malasarn D., Urzica E., Erde J., Page M. D., Yamasaki H., Casero D., Pellegrini M., Merchant S. S., Loo J. A. (2013) The proteome of copper, iron, zinc, and manganese micronutrient deficiency in Chlamydomonas reinhardtii. Mol. Cell Proteomics 12, 65–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bailleul B., Cardol P., Breyton C., Finazzi G. (2010) Electrochromism: a useful probe to study algal photosynthesis. Photosynth. Res. 106, 179–189 [DOI] [PubMed] [Google Scholar]
- 50. Finazzi G., Büschlen S., de Vitry C., Rappaport F., Joliot P., Wollman F. A. (1997) Function-directed mutagenesis of the cytochrome b6f complex in Chlamydomonas reinhardtii. Involvement of the cd loop of cytochrome b6 in quinol binding to the Qo site. Biochemistry 36, 2867–2874 [DOI] [PubMed] [Google Scholar]
- 51. Harris E. H. (2009) Introduction to Chlamydomonas and Its Laboratory Use, Elsevier, San Diego, CA [Google Scholar]
- 52. Hutner S. H., Provasoli L., Schatz A., Haskins C. P. (1950) Some approaches to the study of the role of metals in the metabolism of microorganisms. Proc. Am. Philos. Soc. 94, 152–170 [Google Scholar]
- 53. Hanikenne M., Krämer U., Demoulin V., Baurain D. (2005) A comparative inventory of metal transporters in the green alga Chlamydomonas reinhardtii and the red alga Cyanidioschizon merolae. Plant Physiol. 137, 428–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fang W., Si Y., Douglass S., Casero D., Merchant S. S., Pellegrini M., Ladunga I., Liu P., Spalding M. H. (2012) Transcriptome-wide changes in Chlamydomonas reinhardtii gene expression regulated by carbon dioxide and the CO2-concentrating mechanism regulator CIA5/CCM1. Plant Cell 24, 1876–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Haas C. E., Rodionov D. A., Kropat J., Malasarn D., Merchant S. S., de Crécy-Lagard V. (2009) A subset of the diverse COG0523 family of putative metal chaperones is linked to zinc homeostasis in all kingdoms of life. BMC Genomics 10, 470 PMC2770081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tachiki A., Fukuzawa H., Miyachi S. (1992) Characterization of carbonic anhydrase isozyme CA2, which is the CAH2 gene product, in Chlamydomonas reinhardtii. Biosci. Biotechnol. Biochem. 56, 794–798 [DOI] [PubMed] [Google Scholar]
- 57. Villarejo A., Rolland N., Martínez F., Sültemeyer D. F. (2001) A new chloroplast envelope carbonic anhydrase activity is induced during acclimation to low inorganic carbon concentrations in Chlamydomonas reinhardtii. Planta 213, 286–295 [DOI] [PubMed] [Google Scholar]
- 58. Ynalvez R. A., Xiao Y., Ward A. S., Cunnusamy K., Moroney J. V. (2008) Identification and characterization of two closely related β-carbonic anhydrases from Chlamydomonas reinhardtii. Physiol. Plant. 133, 15–26 [DOI] [PubMed] [Google Scholar]
- 59. Moroney J. V., Ma Y., Frey W. D., Fusilier K. A., Pham T. T., Simms T. A., DiMario R. J., Yang J., Mukherjee B. (2011) The carbonic anhydrase isoforms of Chlamydomonas reinhardtii. Intracellular location, expression, and physiological roles. Photosynth. Res. 109, 133–149 [DOI] [PubMed] [Google Scholar]
- 60. Dancis A., Haile D., Yuan D. S., Klausner R. D. (1994) The Saccharomyces cerevisiae copper transport protein (Ctr1p). Biochemical characterization, regulation by copper, and physiologic role in copper uptake. J. Biol. Chem. 269, 25660–25667 [PubMed] [Google Scholar]
- 61. Rutherford J. C., Bird A. J. (2004) Metal-responsive transcription factors that regulate iron, zinc, and copper homeostasis in eukaryotic cells. Eukaryot. Cell 3, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Eide D. J. (2006) Zinc transporters and the cellular trafficking of zinc. Biochim. Biophys. Acta 1763, 711–722 [DOI] [PubMed] [Google Scholar]
- 63. Sinclair S. A., Krämer U. (2012) The zinc homeostasis network of land plants. Biochim. Biophys. Acta 1823, 1553–1567 [DOI] [PubMed] [Google Scholar]
- 64. Yamano T., Miura K., Fukuzawa H. (2008) Expression analysis of genes associated with the induction of the carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Physiol. 147, 340–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Toepel J., Albaum S. P., Arvidsson S., Goesmann A., la Russa M., Rogge K., Kruse O. (2011) Construction and evaluation of a whole genome microarray of Chlamydomonas reinhardtii. BMC Genomics 12, 579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sinetova M. A., Kupriyanova E. V., Markelova A. G., Allakhverdiev S. I., Pronina N. A. (2012) Identification and functional role of the carbonic anhydrase Cah3 in thylakoid membranes of pyrenoid of Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1817, 1248–1255 [DOI] [PubMed] [Google Scholar]
- 67. Lane T. W., Saito M. A., George G. N., Pickering I. J., Prince R. C., Morel F. M. (2005) Biochemistry. A cadmium enzyme from a marine diatom. Nature 435, 42. [DOI] [PubMed] [Google Scholar]
- 68. Zhang B., Egli D., Georgiev O., Schaffner W. (2001) The Drosophila homolog of mammalian zinc finger factor MTF-1 activates transcription in response to heavy metals. Mol. Cell Biol. 21, 4505–4514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dìez M., Cerdàn F. J., Arroyo M., Balibrea J. L. (1989) Use of the copper/zinc ratio in the diagnosis of lung cancer. Cancer 63, 726–730 [DOI] [PubMed] [Google Scholar]
- 70. Mezzetti A., Pierdomenico S. D., Costantini F., Romano F., De Cesare D., Cuccurullo F., Imbastaro T., Riario-Sforza G., Di Giacomo F., Zuliani G., Fellin R. (1998) Copper/zinc ratio and systemic oxidant load. Effect of aging and aging-related degenerative diseases. Free Radic. Biol. Med. 25, 676–681 [DOI] [PubMed] [Google Scholar]
- 71. Fukuzawa H., Miura K., Ishizaki K., Kucho K. I., Saito T., Kohinata T., Ohyama K. (2001) Ccm1, a regulatory gene controlling the induction of a carbon-concentrating mechanism in Chlamydomonas reinhardtii by sensing CO2 availability. Proc. Natl. Acad. Sci. U.S.A. 98, 5347–5352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Page M. D., Allen M. D., Kropat J., Urzica E. I., Karpowicz S. J., Hsieh S. I., Loo J. A., Merchant S. S. (2012) Fe sparing and Fe recycling contribute to increased superoxide dismutase capacity in iron-starved Chlamydomonas reinhardtii. Plant Cell 24, 2649–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]