Background: Regulation of hypoxia-inducible factor-1 (HIF-1) is focused on proteasomal degradation of the HIF-1α subunit.
Results: Pharmacological and genetic approaches establish that HIF-1α binds to effectors of chaperone-mediated autophagy (CMA) and is targeted for lysosomal degradation.
Conclusion: CMA targets HIF-1α for lysosomal degradation.
Significance: Lysosomal degradation of HIF-1α represents a novel mechanism of HIF-1 regulation and a potential therapeutic target.
Keywords: Autophagy, Hypoxia-inducible Factor, Lysosomes, Protein Degradation, Transcriptional Regulation
Abstract
Hypoxia-inducible factor-1 (HIF-1) is a heterodimeric transcription factor that mediates adaptive responses to hypoxia. We demonstrate that lysosomal degradation of the HIF-1α subunit by chaperone-mediated autophagy (CMA) is a major regulator of HIF-1 activity. Pharmacological inhibitors of lysosomal degradation, such as bafilomycin and chloroquine, increased HIF-1α levels and HIF-1 activity, whereas activators of chaperone-mediated autophagy, including 6-aminonicotinamide and nutrient starvation, decreased HIF-1α levels and HIF-1 activity. In contrast, macroautophagy inhibitors did not increase HIF-1 activity. Transcription factor EB, a master regulator of lysosomal biogenesis, also negatively regulated HIF-1 activity. HIF-1α interacts with HSC70 and LAMP2A, which are core components of the CMA machinery. Overexpression of HSC70 or LAMP2A decreased HIF-1α protein levels, whereas knockdown had the opposite effect. Finally, hypoxia increased the transcription of genes involved in CMA and lysosomal biogenesis in cancer cells. Thus, pharmacological and genetic approaches identify CMA as a major regulator of HIF-1 activity and identify interplay between autophagy and the response to hypoxia.
Introduction
Hypoxia-inducible factor-1 (HIF-1)2 is a transcription factor that is essential for mediating a broad repertoire of adaptive responses to reduced O2 availability. First identified in studies of erythropoietin gene transcriptional regulation (1), HIF-1 was subsequently shown to coordinate adaptive responses to hypoxia at both the cellular and systemic levels (2–4). To date, HIF-1 has been shown to regulate the expression of hundreds of target genes involved in angiogenesis, erythropoiesis, metabolism, autophagy, and other adaptive responses to hypoxia (5). Among these are GLUT1, encoding glucose transporter 1 (3, 6); PDK1, encoding pyruvate dehydrogenase kinase 1 (7, 8); VEGF, encoding vascular endothelial growth factor (9); LDHA, encoding lactate dehydrogenase A (10); and BNIP3, encoding Bcl-2/adenovirus E1B 19-kDa protein-interacting protein 3 (11). In addition, recent data indicate that HIF-1α has functions that are independent of gene transcription (12). The HIF-2α protein shares sequence similarity and functional overlap with HIF-1α, but its expression is restricted to certain cell types, and in some cases, it appears to mediate distinct biological functions (13).
In recent years, the mechanisms by which HIF-1 activity is regulated have been intensively studied. HIF-1 is a heterodimer composed of HIF-1α and HIF-1β subunits (2). O2-dependent proline hydroxylation marks HIF-1α for ubiquitination and proteasomal degradation (14–17), whereas asparagine hydroxylation blocks interaction of HIF-1α with the p300 coactivator (18, 19). During hypoxia, both proline and asparagine hydroxylation are inhibited, which provides a molecular basis for the observed increase in HIF-1α protein stability and HIF-1 transcriptional activity (20). Hydroxylase activity can be inhibited by iron chelators, such as desferrioxamine (DFX), and by competitive antagonists of α-ketoglutarate, such as dimethyloxalylglycine, because the hydroxylases contain Fe(II) in their catalytic centers and use α-ketoglutarate as a co-substrate (14). In recent years, proteins that regulate proteasomal degradation of HIF-1α in an O2-independent fashion have also been identified, including RACK1 (21), calcineurin (22), hypoxia-associated factor (23), CHIP/HSP70 (24), and SHARP1 (25).
Autophagy is a process by which proteins are degraded in lysosomes rather than proteasomes. Macroautophagy sequesters organelles and proteins in an autophagosome, which is then delivered to lysosomes, whereas chaperone-mediated autophagy (CMA) is a pathway for selective degradation of individual proteins (26) that is mediated by binding to HSC70 (heat shock cognate 70-kDa protein) (27), followed by unfolding and translocation of proteins through the lysosomal membrane by LAMP2A (lysosome-associated membrane protein type 2A) (28). The LAMP2 gene encodes two other splice variants, LAMP2B and LAMP2C, which have limited tissue distribution and unclear roles in lysosomal degradation (29, 30). Transcription factor EB (TFEB) coordinates lysosomal biogenesis and autophagy (31, 32). Lysosomes contain proteases in an acidic environment (pH ∼5.0 versus pH ∼7.2 in the cytosol) that is essential for their activity (33). The acidity of lysosomes is maintained by V-ATPase proton pumps. Various drugs, such as bafilomycin and chloroquine, have been used to block lysosomal degradation. Bafilomycin inhibits the activity of the V-ATPase proton pumps, whereas chloroquine is a weak alkaline compound that accumulates in and neutralizes the acidity of lysosomes (34).
Here, we report that HIF-1α is degraded in lysosomes via CMA. HIF-1α binds to key CMA effectors, including HSC70 and LAMP2A. Overexpression of either HSC70 or LAMP2A decreased HIF-1α protein levels and HIF-1 activity, whereas knockdown of HSC70 or LAMP2A had the opposite effect. Blocking lysosomal degradation using bafilomycin or chloroquine increased HIF-1 activity and HIF-1α protein levels, and the magnitude of this effect was comparable to the effect of hypoxia itself. Stimulation of lysosomal biogenesis by TFEB overexpression decreased HIF-1α protein levels and HIF-1 activity, and pharmacological agents that increase CMA, including digoxin, had a similar effect. Thus, we have identified a novel mechanism by which HIF-1α is degraded and that is independent of proteasome activity. In addition, we show that exposure to hypoxia leads to up-regulation of genes involved in CMA and lysosomal biogenesis.
EXPERIMENTAL PROCEDURES
Tissue Culture
293T, HeLa, Hep3B, mouse embryonic fibroblast (MEF), and human foreskin fibroblast cells were cultured in DMEM supplemented with 10% FBS and penicillin/streptomycin. Cells were maintained at 37 °C in a 5% CO2 and 95% air incubator. Cells were subjected to hypoxia by exposure to 1% O2, 5% CO2, and balance N2 at 37 °C in a modulator incubator chamber (Billups-Rothenberg).
Immunoprecipitation and Immunoblot Assays
Cells were lysed in PBS with 0.1% Tween 20, 1 mm DTT, protease inhibitor mixture, 1 mm Na3VO4, and 10 mm NaF, followed by gentle sonication. For immunoprecipitation assays, 30 μl of V5-agarose beads (Sigma) were incubated overnight with 2.5 mg of cell lysate at 4 °C. Beads were washed four times with lysis buffer. Proteins were eluted in SDS sample buffer and separated by SDS-PAGE. The following antibodies were used in immunoblot and immunoprecipitation assays: anti-LAMP2A and anti-lysosomal HSC70 (Abcam), anti-LAMP2A and anti-β-actin (Santa Cruz Biotechnology), anti-HIF-1α (BD Biosciences), anti-FLAG (Sigma), anti-HSC70 and anti-TFEB (Novus Biologicals), and anti-V5 (Invitrogen).
Immunofluorescence Assay
Cells were processed as described previously (35). Cells were plated on gelatin-coated glass-bottomed plates (Live Assay). For immunocytochemistry, samples were washed with ice-cold PBS, fixed with 4% paraformaldehyde for 20 min at room temperature, permeabilized with 0.05% Triton X-100 for 15 min, washed twice with PBS, and blocked with 10% goat serum and 1% AlbuMAX (Invitrogen) for 1 h. Samples were incubated with primary antibody for 1 h, washed, and incubated with Alexa Fluor-conjugated secondary antibody (Invitrogen) for 1 h. Samples were washed and mounted on microscope slides with a drop of SlowFade (Invitrogen) and sealed with medical adhesive (Hollister).
Luciferase Reporter Assay
HeLa or Hep3B cells were seeded onto 24-well plates at 20,000 cells/well, and 48 h after seeding, the cells were transfected with plasmid DNA using PolyJet (SignaGen). Reporters pSV-RL (10 ng) and p2.1 (120 ng) were cotransfected with expression vectors. Cells were lysed, and luciferase activities were determined with a multiwell luminescence reader (PerkinElmer Life Sciences) using the Dual-Luciferase reporter assay system (Promega).
Real-time Quantitative RT-PCR (RT-qPCR) Assay
Total RNA was extracted from 293T cells using TRIzol (Invitrogen) and treated with DNase I (Ambion). Total RNA (1 μg) was used for first-strand synthesis with the iScript cDNA synthesis system (Bio-Rad). qPCR was performed using iQ SYBR Green Supermix and the iCycler real-time PCR detection system (Bio-Rad). Expression of target mRNA relative to 18 S rRNA was calculated based on the threshold cycle (Ct) for amplification as 2−(ΔCt), where ΔCt = Ct(target) − Ct(18 S). The primer sequences used were as follows: 18 S rRNA, CGG CGA CGA CCC ATT CGA AC and GAA TCG AAC CCT GAT TCC CCG TC; ATG6 (mouse), AAT CTA AGG AGT TGC CGT TAT AC and CCA GTG TCT TCA ATC TTG CC; ATP6V0E1 (V-type proton ATPase subunit E1), CAT TGT GAT GAG CGT GTT CTG G and AAC TCC CCG GTT AGG ACC CTT A; ATP6V1H (V-type proton ATPase subunit H), GGA AGT GTC AGA TGA TCC CCA and CCG TTT GCC TCG TGG ATA AT; BNIP3, CTT CCA TCT CTG CTG CTC TC and GTA ATC CAC TAA CGA ACC AAG TC; cathepsin A, CAG GCT TTG GTC TTC TCT CCA and TCA CGC ATT CCA GGT CTT TG; cathepsin B, AGT GGA GAA TGG CAC ACC CTA and AAG AAG CCA TTG TCA CCC CA; GLUT1 (human), CGG GCC AAG AGT GTG CTA AA and TGA CGA TAC CGG AGC CAA TG; GLUT1 (mouse), GCT CTA CGT GGA GCC CTA and CAC ATC GGC TGT CCC TCG A; HIF-1α, CCA CAG GAC AGT ACA GGA TG and TCA AGT CGT GCT GAA TAA TAC C; LAMP1, GCG AGC TCC AAA GAA ATC AA and TGG ACC TGG GTG CCA CTA A; LAMP2, CTC TGC GGG GTC ATG GTG and CGC ACA GCT CCC AGG ACT; LDHA, ATC TTG ACC TAC GTG GCT TGG A and CCA TAC AGG CAC ACT GGA ATC TC; PDK1, ACC AGG ACA GCC AAT ACA AG and CCT CGG TCA CTC ATC TTC AC; and VEGF (human), CTT GCC TTG CTG CTC TAC and TGG CTT GAA GAT GTA CTG G.
shRNA Expression
The vector pLKO-TET was used for shRNA expression. Sequences used were as follows: ATG6, CAG TTT GGC ACA ATC AAT A; HSC70-A, GCC CGA TTT GAA GAA CTG AAT; HSC70-B, GCA ACT GTT GAA GAT GAG AAA; LAMP2A-A, GCC ATC AGA ATT CCA TTG AAT; LAMP2A-B, GAA GTG AAC ATC AGC ATG TAT; TFEB-A, CCC ACT TTG GTG CTA ATA GCT; and TFEB-B, GAA CAA GTT TGC TGC CCA CAT.
Statistical Analysis
All data are presented as means ± S.E., except where indicated otherwise. Differences between two conditions were analyzed using Student's t test.
RESULTS
Inhibition of Lysosomal Degradation Increases HIF-1α Levels
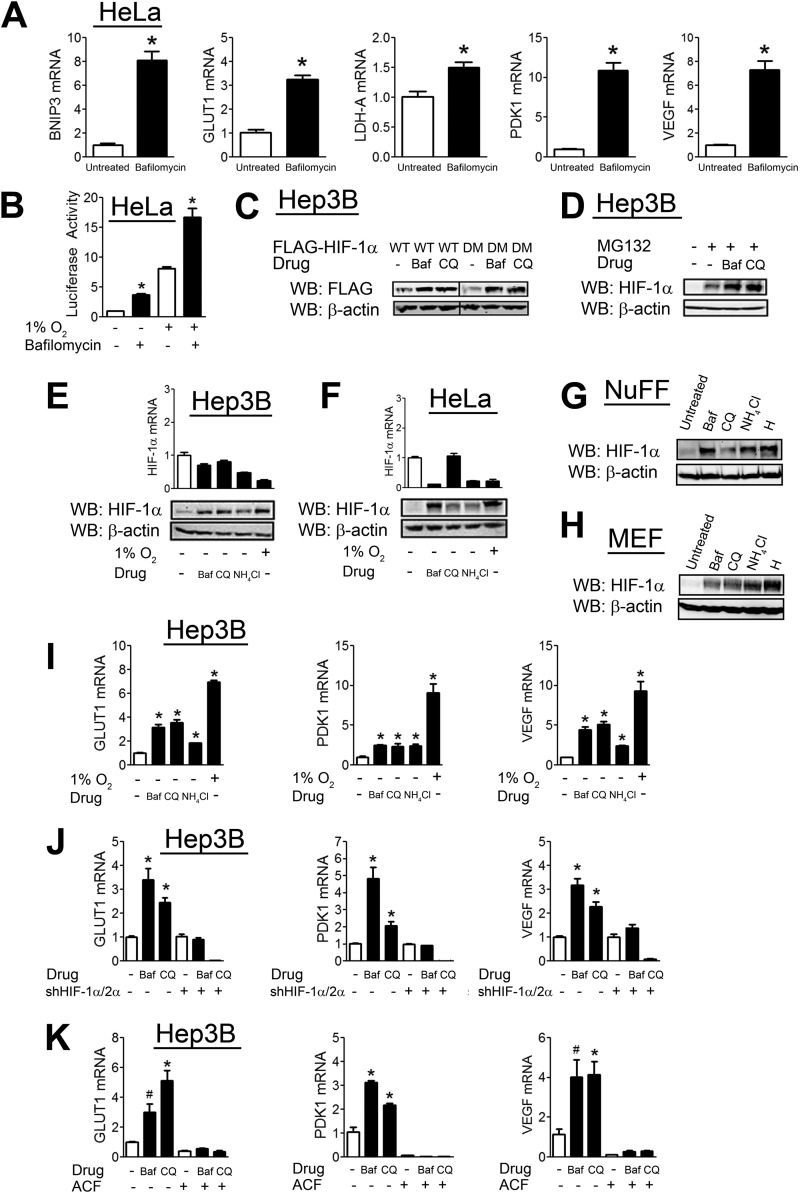
We determined the effect of bafilomycin on the expression of several HIF-1 target genes, including GLUT1, VEGF, PDK1, LDHA, and BNIP3. Bafilomycin treatment of HeLa human cervical carcinoma cells led to a significant increase in the expression of all HIF-1 target genes analyzed under non-hypoxic conditions (Fig. 1A). We further analyzed the effect of bafilomycin using a previously described HIF reporter assay (10). HeLa cells were cotransfected with p2.1, a reporter plasmid that contains a 68-bp hypoxia response element from the human ENO1 gene upstream of the SV40 promoter and firefly luciferase coding sequences, and pSV-RL, a control reporter that contains Renilla luciferase coding sequences downstream of the SV40 promoter only. The ratio of firefly to Renilla luciferase activity served as a measure of HIF-1 transcriptional activity. Bafilomycin increased HIF-1 activity under both hypoxic and non-hypoxic conditions (Fig. 1B). Bafilomycin, as well as another inhibitor of lysosomal degradation (chloroquine), led to an increase in FLAG-HIF-1α protein levels and had a similar effect on a double mutant HIF-1α construct with both proline hydroxylation sites (Pro-402 and Pro-564) mutated (Fig. 1C). To determine whether the effect of these drugs is independent of the proteasome, we analyzed the effect of bafilomycin and chloroquine in the presence of the proteasomal inhibitor MG132. Treatment with bafilomycin or chloroquine increased HIF-1α levels in the presence of MG132 (Fig. 1D), confirming that they operate through a proteasome-independent pathway.
FIGURE 1.
CMA inhibitors increase HIF-1α protein levels. A, HeLa cells were left untreated or treated with bafilomycin (10 nm) for 24 h, after which RNA was isolated, and RT-qPCR of the indicated HIF target genes was performed. B, HeLa cells were transfected with the HIF-1 reporter plasmid p2.1 and the Renilla luciferase reporter pSV-RL, exposed to 1% O2, and left untreated or treated with bafilomycin (10 nm) for 48 h. Afterward, cells were lysed, and reporter activity was measured. C, Hep3B cells were transfected with FLAG-HIF-1α expression vector that was either unmutated (WT) or harbored P402A/P564A mutations (double mutant (DM)). 24 h post-transfection, the cells were left untreated or treated with bafilomycin (Baf; 10 nm) or chloroquine (CQ; 50 μm) for an additional 8 h, after which cells were lysed, and lysates were probed with the indicated antibodies. WB, Western blot. D, Hep3B cells were left untreated or treated with the proteasomal inhibitor MG132 (10 μm) either alone or in combination with bafilomycin (10 nm) or chloroquine (50 μm) for 8 h. Afterward, cells were lysed, and lysates were probed with the indicated antibodies. Hep3B (E) and HeLa (F) cells were left untreated at 20% O2 (normoxia); treated with bafilomycin (10 nm), chloroquine (50 μm), or NH4Cl (10 mm); or left untreated and exposed to 1% O2 (hypoxia) for 8 h. Afterward, cells were lysed, and lysates were probed with the indicated antibodies. RNA was also isolated, and RT-qPCR of HIF-1α mRNA was performed. Normal human foreskin fibroblasts (NuFF; G) and MEFs (H) were left untreated at 20% O2; treated with bafilomycin (10 nm), chloroquine (50 μm), or NH4Cl (10 mm); or left untreated and exposed to 1% O2 (hypoxia (H)) for 8 h. Afterward, cells were lysed, and lysates were probed with the indicated antibodies. I, Hep3B cells were left untreated; treated with bafilomycin (10 nm), chloroquine (50 μm), or NH4Cl (10 mm); or exposed to 1% O2 for 24 h. Afterward, RNA was isolated, and RT-qPCR of the indicated HIF target genes was performed. J, Hep3B cells stably infected with empty shRNA vector or vectors against both HIF-1α and HIF-2α (shHIF-1α/2α) were left untreated or treated with bafilomycin (10 nm) or chloroquine (50 μm) for 24 h. Afterward, RNA was isolated, and RT-qPCR of the indicated HIF target genes was performed. K, Hep3B cells were treated with various combinations of bafilomycin (10 nm), chloroquine (50 μm), and the HIF-1 inhibitor acriflavine (ACF; 5 μm) for 24 h. Afterward, RNA was isolated, and RT-qPCR of the indicated HIF target genes was performed. Results are shown as means ± S.E. *, p < 0.01; #, p < 0.05.
We examined the effect on endogenous HIF-1α levels of inhibiting lysosomal degradation with bafilomycin, chloroquine, or ammonium chloride (NH4Cl). In Hep3B human hepatocellular carcinoma cells (Fig. 1E), HeLa cells (Fig. 1F), normal human foreskin fibroblasts (Fig. 1G), and MEFs (Fig. 1H), blocking lysosomal degradation led to an increase in HIF-1α levels that was comparable to the effect of hypoxia. However, we found no increase in HIF-1α mRNA levels upon treatment with these drugs in either Hep3B (Fig. 1E) or HeLa (Fig. 1F) cells, indicating that changes in transcription do not account for the increased HIF-1α levels.
We analyzed the effect of inhibiting lysosomal degradation on the expression of three HIF-1 target genes, GLUT1, PDK1, and VEGF, in Hep3B cells. Treatment with bafilomycin, chloroquine, or NH4Cl significantly increased the expression of all three HIF target genes (Fig. 1I). To demonstrate that the increased expression of these genes was HIF-dependent, we compared the effect of lysosomal inhibitors in Hep3B cells stably transfected with shRNAs targeting both HIF-1α and HIF-2α or with an empty shRNA vector. Whereas bafilomycin and chloroquine increased HIF target gene expression in the empty shRNA vector-transfected cells, there was no increase in the HIF-1α/2α shRNA-transfected cells treated with bafilomycin, and gene expression was even more severely inhibited in chloroquine-treated HIF-1α/2α shRNA-transfected cells (Fig. 1J). Similarly, we examined HIF target gene expression in the presence of acriflavine, which is an inhibitor of HIF subunit dimerization (36). Lysosomal inhibitors had no effect on HIF target gene expression in the presence of acriflavine (Fig. 1K). Taken together, the data presented in Fig. 1 indicate that the inhibition of lysosomal protein degradation leads to an increase in HIF-1α protein levels and HIF target gene expression that is independent of prolyl hydroxylation.
Inhibition of Macroautophagy Has No Effect on HIF-1α Levels
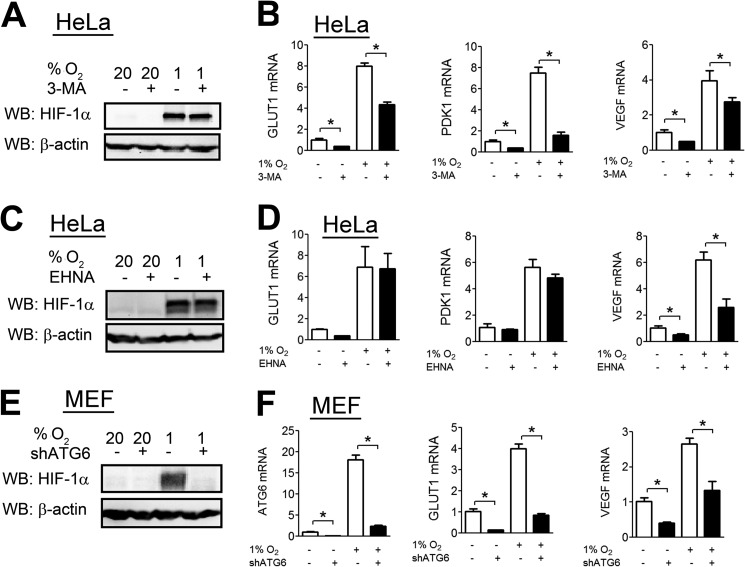
The inhibitors of lysosomal degradation utilized above block both CMA and macroautophagy. To determine whether macroautophagy contributes to HIF-1 regulation, we analyzed HIF-1α levels in HeLa cells treated with 3-methyladenine (3-MA), a well characterized and selective inhibitor of macroautophagy (37). 3-MA had no effect on HIF-1α levels (Fig. 2A). 3-MA treatment led to decreased expression of several HIF target genes, including GLUT1, PDK1, and VEGF (Fig. 2B). We determined HIF-1α levels upon treatment with a second macroautophagy inhibitor, erythro-9-(2-hydroxy-3-nonyl)adenine, which has a distinct mechanism of action (38). erythro-9-(2-Hydroxy-3-nonyl)adenine treatment did not increase HIF-1α levels (Fig. 2C) or HIF target gene expression (Fig. 2D).
FIGURE 2.
Macroautophagy does not contribute to HIF-1α degradation. A, HeLa cells were left untreated or treated with 3-MA (5 mm) at either 20% or 1% O2. 8 h later, cells were lysed, and lysates were probed with the indicated antibodies. WB, Western blot. B, HeLa cells were left untreated or treated with 3-MA (5 mm) at either 20% or 1% O2 for 24 h. Afterward, RNA was isolated and subjected to RT-qPCR against GLUT1, PDK1, and VEGF. C, HeLa cells were left untreated or treated with erythro-9-(2-hydroxy-3-nonyl)adenine (EHNA; 10 μm) at either 20% or 1% O2. 8 h later, cells were lysed, and lysates were probed with the indicated antibodies. D, HeLa cells were left untreated or treated with EHNA at 20% or 1% O2 for 24 h. Afterward, RNA was isolated and subjected to RT-qPCR against GLUT1, PDK1, and VEGF. E, MEFs stably transfected with empty shRNA vector or shRNA vector targeting ATG6 (shATG6) were exposed to either 20% or 1% O2 for 8 h. Afterward, cells were lysed, and lysates were probed with the indicated antibodies. F, MEFs stably transfected with empty shRNA vector or shRNA vector targeting ATG6 were exposed to 20% or 1% O2 for 24 h. Afterward, RNA was isolated and subjected to RT-qPCR against ATG6, GLUT1, and VEGF. Results are shown as means ± S.E. *, p < 0.01.
We also analyzed HIF-1α levels in previously described ATG6 knockdown MEFs (39). Atg6 encodes a protein that is essential for formation of the autophagosome during macroautophagy. Surprisingly, ATG6 knockdown led to a dramatic decrease in HIF-1α levels (Fig. 2E) and a corresponding decrease in HIF target gene expression (Fig. 2F). Taken together, the results from pharmacological and genetic approaches indicate that macroautophagy does not contribute to HIF-1α degradation.
Activators of CMA Inhibit HIF-1 Activity
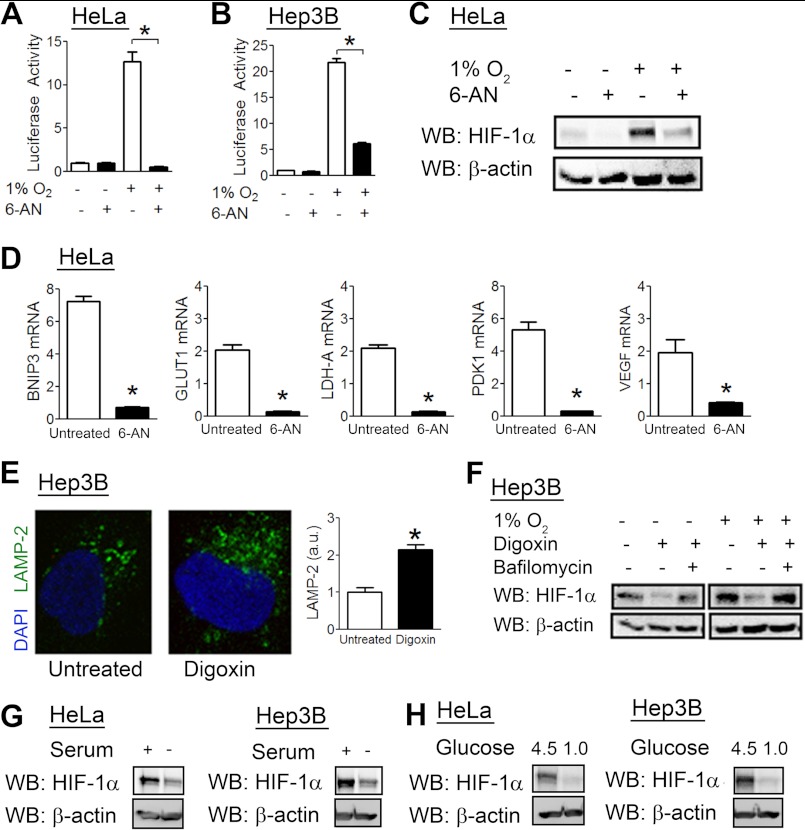
We analyzed the effect of 6-aminonicotinamide (6-AN), a commonly used activator of CMA (40). In both HeLa (Fig. 3A) and Hep3B (Fig. 3B) cells, 6-AN treatment blocked the induction of HIF-1 reporter activity by hypoxia. 6-AN treatment also decreased endogenous HIF-1α levels in hypoxic HeLa cells (Fig. 3C). Finally, cells treated with 6-AN had lower levels of GLUT1, PDK1, VEGF, BNIP3, and LDHA mRNAs compared with untreated cells (Fig. 3D).
FIGURE 3.
CMA activators decrease HIF-1α protein levels and HIF-1 activity. HeLa (A) and Hep3B (B) cells were transfected with the HIF reporter plasmid p2.1 and the control Renilla luciferase reporter pSV-RL, exposed to 1% O2, and left untreated or treated with 6-AN (50 mm) for 24 h. Afterward, cells were lysed, and reporter activity was measured. C, HeLa cells were left untreated or treated with 6-AN (50 mm) at either 20% or 1% O2. 8 h later, cells were lysed, and lysates were probed with the indicated antibodies. WB, Western blot. D, HeLa cells were left untreated or treated with 6-AN (50 mm) at 20% or 1% O2 for 24 h. Afterward, RNA was isolated, and RT-qPCR was performed. Fold induction of expression under hypoxic versus non-hypoxic conditions is shown for both treated and untreated cells. E, Hep3B cells were left untreated or treated with digoxin (200 nm) for 8 h, after which cells were fixed, stained with anti-LAMP2A antibody, and imaged by confocal microscopy, and staining per cell was quantified and normalized to the untreated condition. a.u., arbitrary units. F, Hep3B cells were treated with digoxin (200 nm) or bafilomycin (10 nm) as indicated and exposed to 20% or 1% O2 for 8 h. Afterward, cells were lysed, and lysates were subjected to immunoblot assays with the indicated antibodies. G, HeLa and Hep3B cells were either serum-starved or cultured in regular medium for 24 h in the presence of MG132 (10 μm). Afterward, cells were lysed, and lysates were probed with the indicated antibodies. H, HeLa and Hep3B cells were exposed to regular medium (containing 4.5 g/liter glucose) or low glucose medium (1.0 g/liter glucose) for 24 h in the presence of MG132 (10 μm). Afterward, cells were lysed, and lysates were probed with the indicated antibodies. Results are shown as means ± S.E. *, p < 0.01.
Cardiac glycosides, such as digoxin, inhibit HIF-1 activity by decreasing HIF-1α protein levels (41). Recently, cardiac glycosides were also identified as activators of protein flux through the lysosomal degradation pathway (42). The mechanisms by which this occurs are unclear, but consistent with this report, treatment of Hep3B cells with digoxin led to an increase in LAMP2-positive lysosomes compared with untreated cells (Fig. 3E). Concurrent treatment with bafilomycin abolished the effect of digoxin on HIF-1α protein levels in Hep3B cells (Fig. 3F), suggesting that digoxin enhances lysosomal degradation of HIF-1α.
CMA is increased under conditions of glucose or serum deprivation. To determine whether CMA promotes HIF-1α degradation in response to a physiological stimulus, we analyzed HIF-1α levels after serum starvation. Cells were pretreated with MG132 to exclude effects mediated through the proteasomal pathway. Both HeLa and Hep3B cells had reduced HIF-1α levels when serum-starved (Fig. 3G). Similarly, both cell types had decreased HIF-1α levels when exposed to low glucose culture medium (Fig. 3H).
TFEB Negatively Regulates HIF-1α Levels and HIF-1 Activity
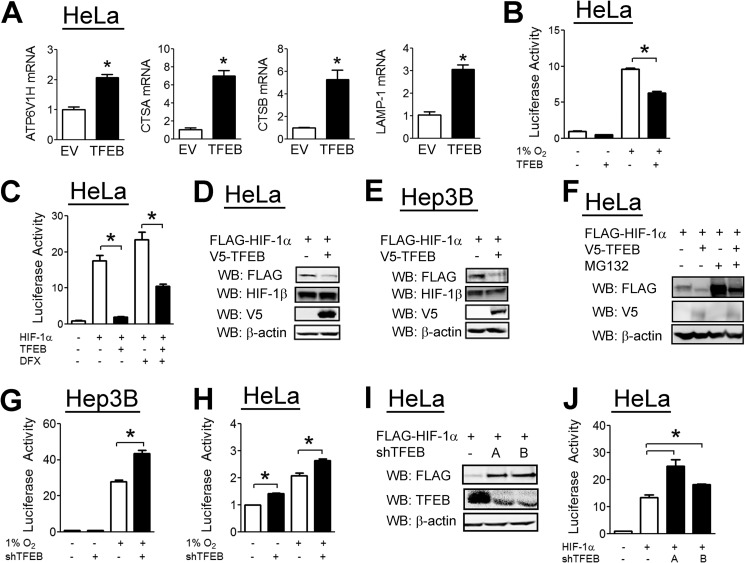
TFEB was recently identified as a master regulator of lysosomal biogenesis (28) that is required for autophagy (29). Overexpression of TFEB led to increased expression of several genes that are involved in lysosomal biogenesis (Fig. 4A). TFEB overexpression inhibited HIF-1 reporter activity in hypoxic HeLa cells (Fig. 4B). TFEB had a similar effect on HIF-1 reporter activity induced by HIF-1α overexpression, even in the presence of the hydroxylase inhibitor DFX (Fig. 4C). TFEB overexpression led to decreased HIF-1α protein levels in both HeLa and Hep3B cells while having no effect on HIF-1β levels (Fig. 4, D and E). This effect was maintained in the presence of MG132 (Fig. 4F), indicating that the effect of TFEB is independent of the proteasomal degradation pathway. In both Hep3B (Fig. 4G) and HeLa (Fig. 4H) cells, TFEB knockdown led to an increase in HIF-1 activity. To exclude an off-target effect of the TFEB shRNA, we constructed a second shRNA vector targeting a different sequence within TFEB. TFEB knockdown with either shRNA led to an increase in HIF-1α protein levels (Fig. 4I) and HIF-1 reporter activity (Fig. 4J).
FIGURE 4.
TFEB decreases HIF-1α levels and HIF-1 activity. A, HeLa cells were transfected with empty vector (EV) or TFEB expression vector. 24 h post-transfection, RNA was isolated, and RT-qPCR was performed. B, HeLa cells were cotransfected with the HIF reporter p2.1, pSV-RL, and empty vector or vector encoding TFEB. 24 h post-transfection, cells were exposed to 20% or 1% O2 for 24 h, after which cells were lysed, and reporter activity was measured. C, HeLa cells were cotransfected with p2.1, pSV-RL, FLAG-HIF-1α expression vector, and either empty vector or TFEB vector. Cells were left untreated or treated with DFX (100 μm) for 24 h, after which cells were lysed, and reporter activity was measured. HeLa (D) and Hep3B (E) cells were cotransfected with FLAG-HIF-1α vector and either empty vector or V5-TFEB vector. 48 h post-transfection, cells were lysed, and lysates were probed with the indicated antibodies. WB, Western blot. F, HeLa cells were cotransfected with FLAG-HIF-1α vector and either empty vector or V5-TFEB vector. 40 h post-transfection, cells were left untreated or treated with MG132 (10 μm) for an additional 8 h. Cells were then lysed, and lysates were probed with the indicated antibodies. Hep3B (G) and HeLa (H) cells were cotransfected with p2.1, pSV-RL, and either empty shRNA vector or vector encoding shRNA against TFEB (shTFEB). 24 h post-transfection, cells were incubated for an additional 24 h under 20% or 1% O2. Cells were then lysed, and reporter activity were measured. I, HeLa cells were transfected with FLAG-HIF-1α vector and either empty shRNA vector or one of two vectors encoding an shRNA against TFEB (TFEB-A and TFEB-B). 48 h post-transfection, cells were lysed, and lysates were probed with the indicated antibodies. J, HeLa cells were transfected with p2.1, pSV-RL, FLAG-HIF-1α vector, and either empty shRNA vector or one of two vectors encoding an shRNA against TFEB. 48 h post-transfection, cells were lysed, and lysates were probed with the indicated antibodies. Results are shown as means ± S.E. *, p < 0.01.
HIF-1α Interacts with Key CMA Effectors
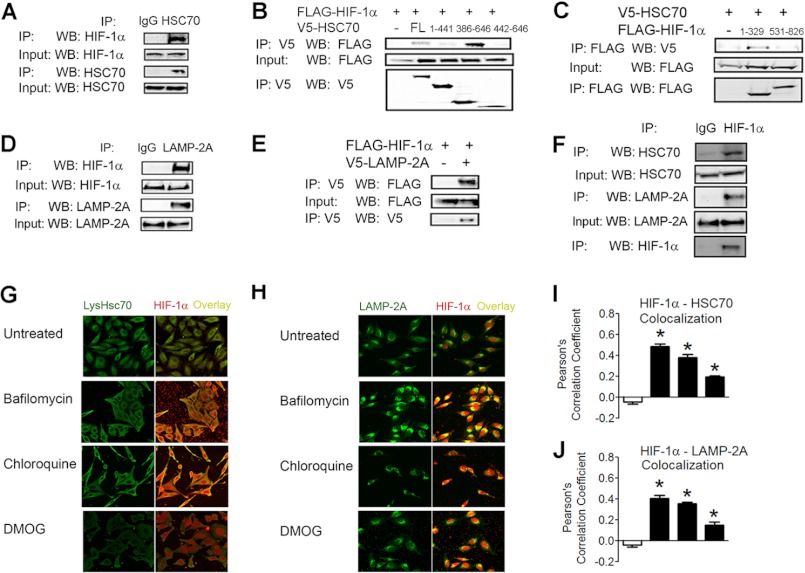
CMA requires the activity of the cytosolic chaperone HSC70, which binds to target proteins and mediates their interaction with LAMP2A, which then translocates target proteins to the lysosome for degradation. Co-immunoprecipitation assays in 293T cells treated with bafilomycin revealed interaction between endogenous HIF-1α and HSC70 (Fig. 5A). Using a series of HSC70 deletion mutants, we found that HIF-1α bound to amino acid residues 386–646 of HSC70 (Fig. 5B). To localize the binding site on HIF-1α, we used a series of HIF-1α deletion constructs and found that HSC70 bound to the amino acid residues 1–329 of HIF-1α (Fig. 5C). This result is consistent with other validated CMA substrates, which usually contain an HSC70-binding site within their N terminus (25).
FIGURE 5.
HIF-1α interacts with key CMA effectors. A, co-immunoprecipitation assay of 293T cells treated with bafilomycin (10 nm) for 8 h, after which cells were lysed, and lysates were immunoprecipitated (IP) with either IgG or anti-HSC70 antibody and probed for HIF-1α or HSC70. WB, Western blot. B, co-immunoprecipitation assay of 293T cells transfected with FLAG-HIF-1α vector and either empty vector or vector encoding the indicated V5-HSC70 deletion construct. FL, full-length. C, co-immunoprecipitation assay of 293T cells transfected with V5-HSC70 vector and empty vector or vector encoding FLAG-HIF-1α(1–329) or FLAG-HIF-1α(531–826). D, co-immunoprecipitation assay of 293T cells treated with bafilomycin (10 nm) for 8 h, after which cells were lysed, and lysates were immunoprecipitated with either IgG or anti-LAMP2A antibody and probed for HIF-1α or LAMP2A. E, co-immunoprecipitation assay of 293T cells transfected with FLAG-HIF-1α vector and either empty vector or V5-LAMP2A vector. F, co-immunoprecipitation assay of 293T cells exposed to 1% O2 for 8 h. Afterward, cells were lysed, and lysates were immunoprecipitated with either IgG or anti-HIF-1α antibody and probed for HIF-1α, LAMP2A, and HSC70. G, HeLa cells were left untreated or treated with bafilomycin (10 nm), chloroquine (50 μm), or dimethyloxalylglycine (DMOG; 1 mm) for 8 h, after which they were fixed, stained with anti-lysosomal HSC70 and anti-HIF-1α antibodies, and analyzed by two-photon microscopy. H, cells (treated as described for G) were stained with anti-LAMP2A and anti-HIF-1α antibodies and analyzed by two-photon microscopy. I and J, Pearson's correlation coefficient was calculated (for the conditions shown in G and H). Results are shown as means ± S.E. *, p < 0.01.
Co-immunoprecipitation assays also demonstrated an interaction between endogenous LAMP2A and HIF-1α in bafilomycin-treated cells (Fig. 5D), as well as between exogenous V5-tagged LAMP2A and FLAG-tagged HIF-1α in untreated cells (Fig. 5E). Finally, immunoprecipitation of endogenous HIF-1α in hypoxic 293T cells demonstrated an interaction between endogenous HSC70 and LAMP2A (Fig. 5F).
Because HIF-1α has not previously been shown to localize inside the lysosome or on the lysosomal membrane, we analyzed whether HIF-1α translocates inside lysosomes in whole cells. Two-photon microscopy demonstrated that HIF-1α and lysosomal HSC70, a marker of CMA-competent lysosomes, co-localized in HeLa cells when lysosomal degradation was blocked by bafilomycin or chloroquine treatment (Fig. 5G). In contrast, lysosomal localization of HIF-1α was not observed when expression was induced by treatment with dimethyloxalylglycine, which is a hydroxylase inhibitor that blocks proteasomal degradation of HIF-1α. Inhibitors of lysosomal degradation also increased the co-localization of HIF-1α with LAMP2A. Taken together, the data in Fig. 5 indicate that HIF-1α and key CMA effectors interact and co-localize in lysosomes.
HSC70 and LAMP2A Negatively Regulate HIF-1α Levels and HIF-1 Activity
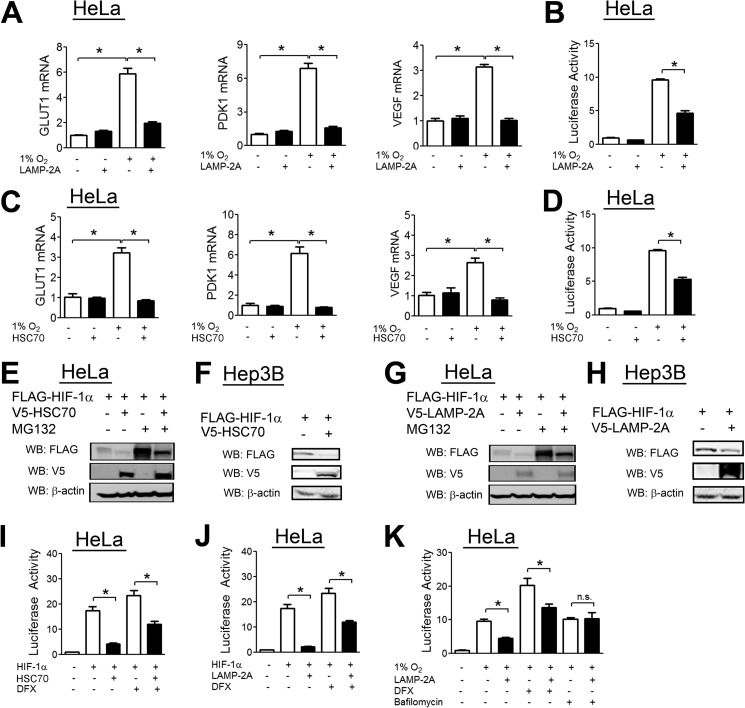
To determine the effect of HSC70 on HIF-1 transcriptional activity, we analyzed hypoxic induction of the HIF target genes GLUT1, PDK1, and VEGF upon HSC70 overexpression. RT-qPCR results demonstrated increased expression of HIF target genes upon exposure to hypoxia, which was abolished by LAMP2A overexpression (Fig. 6A), whereas there was no effect under non-hypoxic conditions. Hypoxia-induced HIF reporter activity was significantly decreased by LAMP2A overexpression (Fig. 6B). HSC70 overexpression also decreased HIF target gene expression (Fig. 6C) and HIF reporter activity (Fig. 6D).
FIGURE 6.
HSC70 and LAMP2A decrease HIF-1α levels and HIF-1 activity. A, HeLa cells were transfected with either empty vector or V5-LAMP2A expression vector. 24 h post-transfection, cells were exposed to 20% or 1% O2 for an additional 24 h, after which RNA was isolated, and RT-qPCR was performed. B, HeLa cells were transfected with the HIF reporter p2.1, the control reporter pSV-RL, and either empty vector or V5-LAMP2A expression vector. 24 h post-transfection, cells were exposed to either 20% or 1% O2 for an additional 24 h, after which cells were lysed, and reporter activity was measured. C, HeLa cells were transfected with empty vector or V5-HSC70 vector. 24 h post-transfection, cells were exposed to 20% or 1% O2 for an additional 24 h, after which RNA was isolated, and RT-qPCR was performed. D, HeLa cells were transfected with p2.1, pSV-RL, and empty vector or V5-HSC70 vector. 24 h post-transfection, cells were exposed to 20% or 1% O2 for 24 h, after which cells were lysed, and reporter activity was measured. E, HeLa cells were cotransfected with FLAG-HIF-1α expression vector and either empty vector or V5-HSC70 vector. 24 h post-transfection, cells were left untreated or treated with the proteasomal inhibitor MG132 (10 μm) for 8 h. Cells were then lysed, and lysates were probed with the indicated antibodies. WB, Western blot. F, Hep3B cells were cotransfected with FLAG-HIF-1α vector and either empty vector or V5-HSC70 vector. 24 h post-transfection, cells were then lysed, and lysates were probed with the indicated antibodies. G, HeLa cells were cotransfected with FLAG-HIF-1α vector and either empty vector or V5-LAMP2A vector. 24 h post-transfection, cells were left untreated or treated with MG132 (10 μm) for 8 h. Cells were then lysed, and lysates were probed with the indicated antibodies. H, Hep3B cells were cotransfected with FLAG-HIF-1α vector and either empty vector or V5-LAMP2A vector. 24 h post-transfection, cells were lysed, and lysates were probed with the indicated antibodies. I, HeLa cells were cotransfected with p2.1, pSV-RL, FLAG-HIF-1α vector, and either empty vector or V5-HSC70 vector. 24 h post-transfection, cells were left untreated or treated with DFX (100 μm) for 24 h. Afterward, cells were lysed, and reporter activity was measured. J, HeLa cells were cotransfected with p2.1, pSV-RL, FLAG-HIF-1α vector, and either empty vector or V5-LAMP2A vector. 24 h post-transfection, cells were left untreated or treated with DFX (100 μm) for 24 h. Afterward, cells were lysed, and reporter activity was measured. K, HeLa cells were cotransfected with p2.1, pSV-RL, FLAG-HIF-1α vector, and either empty vector or V5-LAMP2A vector. 24 h post-transfection, cells were exposed to 1% O2 for 24 h and either left untreated or treated with DFX (100 μm) or bafilomycin (5 nm). Afterward, cells were lysed, and reporter activity was measured. Results are shown as means ± S.E. *, p < 0.01; n.s., not significant.
Overexpression of HSC70 in HeLa (Fig. 6E) or Hep3B (Fig. 6F) cells led to a decrease in HIF-1α protein levels that was not blocked by the proteasomal inhibitor MG132 (Fig. 6E). Similar results were obtained upon overexpression of LAMP2A (Fig. 6, G and H). Similarly, overexpression of either HSC70 or LAMP2A led to a decrease in HIF reporter activity induced by HIF-1α overexpression, which was resistant to the effects of the hydroxylase inhibitor DFX (Fig. 6, I and J). Importantly, however, bafilomycin treatment completely abolished the inhibitory effect of LAMP2A overexpression on HIF activity (Fig. 6K).
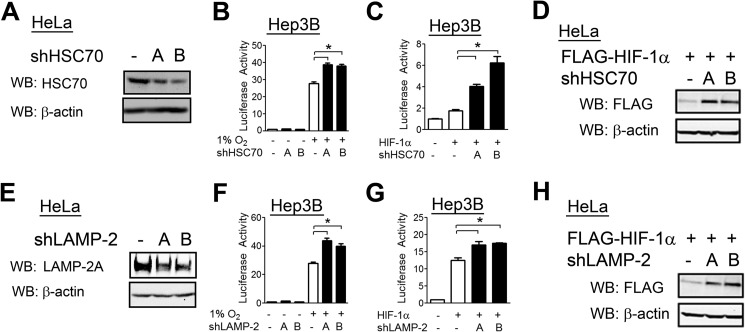
To confirm that HSC70 inhibits HIF-1 activity at endogenous levels, we constructed two shRNA vectors targeting distinct sequences in HSC70 (Fig. 7A). In Hep3B cells, knockdown of HSC70 with either vector led to an increase in HIF-1 reporter activity induced by exposure to hypoxia (Fig. 7B) or by overexpression of HIF-1α under non-hypoxic conditions (Fig. 7C). HSC70 knockdown also increased FLAG-HIF-1α protein levels (Fig. 7D). These results confirm that HSC70 regulates HIF-1α protein abundance because the FLAG-HIF-1α expression vector lacked the transcriptional and translational regulatory signals that are present in the endogenous gene.
FIGURE 7.
Knockdown of HSC70 or LAMP2 expression increases HIF-1α levels and HIF-1 activity. A, HeLa cells were transfected with either empty shRNA vector or one of two vectors expressing shRNA targeting HSC70 (shHSC70-A and shHSC70-B). 48 h post-transfection, cells were lysed, and lysates were probed with the indicated antibodies. WB, Western blot. B, Hep3B cells were transfected with p2.1, pSV-RL, and empty shRNA vector or HSC70-A or HSC70-B shRNA. 24 h post-transfection, cells were exposed to 20% or 1% O2 for 24 h, after which cells were lysed, and reporter activity was measured. C, Hep3B cells were cotransfected with p2.1, pSV-RL, empty vector or FLAG-HIF-1α vector, and empty shRNA vector or HSC70-A or HSC70-B shRNA. WB, Western blot. D, HeLa cells were cotransfected with FLAG-HIF-1α vector and empty shRNA vector or HSC70-A or HSC70-B shRNA. 48 h post-transfection, cells were lysed, and lysates were probed with the indicated antibodies. E, HeLa cells were transfected with either empty shRNA vector or one of two vectors expressing an shRNA targeting LAMP2 (shLAMP2-A and shLAMP2-B). 48 h post-transfection, cells were lysed, and lysates were probed with the indicated antibodies. F, Hep3B cells were transfected with p2.1, pSV-RL, and either empty shRNA vector or LAMP2-A or LAMP2-B shRNA. 24 h post-transfection, cells were exposed to either 20% or 1% O2 for 24 h, after which cells were lysed, and reporter activity was measured. G, Hep3B cells were transfected with p2.1, pSV-RL, empty vector or FLAG-HIF-1α vector, and empty shRNA vector or LAMP2-A or LAMP2-B shRNA. H, HeLa cells were cotransfected with FLAG-HIF-1α vector and empty shRNA vector or LAMP2-A or LAMP2-B shRNA. 48 h post-transfection, cells were lysed, and lysates were probed with the indicated antibodies. Results are shown as means ± S.E. *, p < 0.01.
We also constructed two shRNA vectors targeting different sequences in LAMP2 (Fig. 7E). Knockdown of LAMP2 with either shRNA vector led to an increase in HIF-1 reporter activity induced by hypoxia (Fig. 7F) or FLAG-HIF-1α overexpression (Fig. 7G) and increased FLAG-HIF-1α protein levels (Fig. 7H). We conclude that, at endogenous levels, the CMA effectors HSC70 and LAMP2A regulate HIF-1 transcriptional activity by promoting HIF-1α degradation.
Hypoxia Increases the Expression of Genes Involved in CMA and Lysosomal Biogenesis
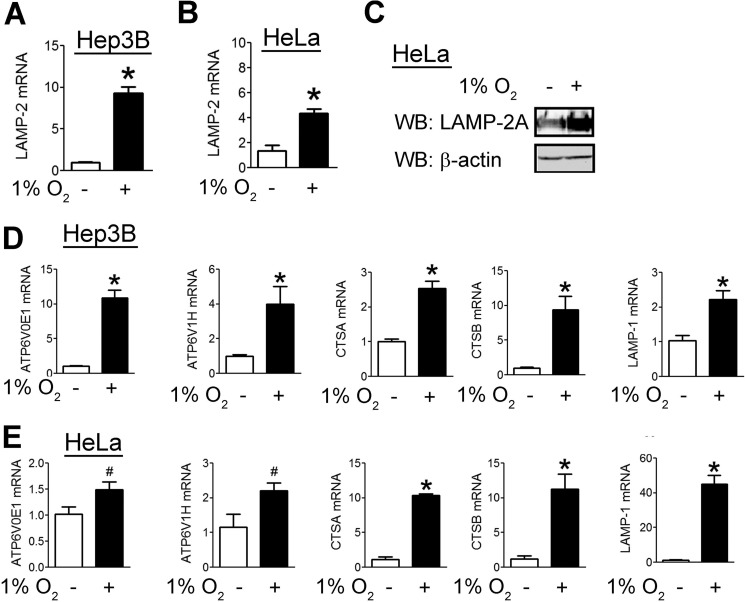
To determine whether CMA was induced by hypoxia as part of a negative feedback loop, we analyzed LAMP2 mRNA levels. Hypoxia increased LAMP2 mRNA levels in HeLa and Hep3B cells (Fig. 8, A and B), and we detected a corresponding increase in LAMP2A protein levels (Fig. 8C). Hypoxia also increased the expression of five other genes involved in lysosomal biogenesis in both Hep3B (Fig. 8D) and HeLa (Fig. 8E) cells.
FIGURE 8.
Hypoxia induces expression of mRNAs encoding CMA components. Hep3B (A) and HeLa (B) cells were exposed to 20% or 1% O2 for 24 h. Afterward, RNA was isolated, and RT-qPCR was performed. C, HeLa cells were exposed to 20% or 1% O2 for 48 h, after which cells were lysed, and lysates were probed with the indicated antibodies. WB, Western blot. Hep3B (D) and HeLa (E) cells were exposed to 20% or 1% O2 for 24 h. Afterward, RNA was isolated, and RT-qPCR was performed. Results are shown as means ± S.E. *, p < 0.01.
DISCUSSION
In this study, we have delineated a novel pathway for HIF-1α degradation that is independent of the proteasome. We demonstrated that HIF-1α interacted at endogenous levels with HSC70 and LAMP2A, which are key CMA effectors, and co-localized with lysosomal HSC70 and LAMP2A using two-photon microscopy. Overexpression of HSC70 or LAMP2A led to a marked decrease in HIF-1α levels, whereas knockdown of HSC70 or LAMP2A had the opposite effect. Inhibition of lysosomal degradation with bafilomycin or chloroquine increased HIF-1α levels and HIF target gene expression. Activation of CMA by pharmacological treatment with 6-AN or digoxin or by a physiological stimulus, such as serum starvation or glucose deprivation, decreased HIF-1α levels. Overexpression of TFEB, a transcription factor that promotes lysosomal biogenesis and autophagy, led to decreased HIF-1α levels. Thus, a variety of pharmacological and genetic approaches demonstrate that HIF-1α is regulated by lysosomal degradation mediated by CMA. We obtained evidence supporting a role for lysosomal degradation of HIF-1α in HeLa cervical carcinoma cells, Hep3B hepatocellular carcinoma cells, human foreskin fibroblasts, and MEFs, suggesting that this is a widespread mechanism by which HIF-1α is regulated.
We also demonstrated that hypoxia increased LAMP2A expression in two cancer cell lines, which is consistent with a previous report that hypoxia increases LAMP2A expression in neural tissue (43). Another recent report demonstrated increased expression of several genes encoding lysosomal components in hypoxic mouse fibroblasts (44). Although the mechanism is unclear, the effects of hypoxia may partially explain recent findings that CMA is increased in several cancers (45) because hypoxia is a common feature of the tumor microenvironment.
Previous work from our group identified HSP70, a protein with a high degree of sequence similarity to HSC70, as a regulator of HIF-1α degradation (24). However, HSP70 is involved in proteasomal degradation of HIF-1α rather than lysosomal degradation. Binding assays demonstrated that whereas HSC70 bound the N-terminal portion of HIF-1α (Fig. 5C), HSP70 bound the C-terminal region. In addition, HIF-1α bound the C terminus of HSC70 but the N-terminal end of HSP70 (Fig. 5B). Taken together, these two studies indicate that despite the high degree of sequence similarity, HSC70 and HSP70 enhance the degradation of HIF-1α by distinct molecular mechanisms. This duality has also been observed for several other protein families. LIMD1 enhances proteasomal degradation of HIF-1α (46), whereas the related FHL (four-and-a-half LIM domain) family members utilize distinct mechanisms to inhibit HIF-1α transactivation domain function (47, 48). Members of the MCM (minichromosome maintenance) family modulate O2-dependent regulation of HIF-1α stability or transactivation domain function (49). Finally, the homologs SSAT1 (spermidine/spermine N1-acetyltransferase) (50) and SSAT2 (51) stimulate O2-independent and O2-dependent HIF-1α degradation, respectively.
Recent work has shown that PKM2 (pyruvate kinase M2) undergoes degradation via CMA in cells cultured under high glucose concentrations (52). PKM2 acts as a coactivator for HIF-1 transcriptional activity by increasing HIF-1 binding and p300 recruitment to HIF-1 target genes (53). Degradation of PKM2 may act as an indirect mechanism by which CMA reduces HIF-1 activity, in combination with the direct degradation of HIF-1α by CMA.
The regulation of HIF-1α levels by lysosomal degradation may help explain previous results regarding the role of TFEB in cancer. TFEB mutations have been identified in renal carcinomas (54), which often have activation of HIF-1 due to loss of function of VHL, which is required for proteasomal degradation of HIF-1α. Because increased TFEB levels lead to decreased HIF-1α levels and HIF-1 activity, TFEB loss of function may contribute to the increased HIF-1α levels that are observed in many human cancers (55).
We have previously reported that digoxin decreases HIF-1α levels and inhibits tumor growth and metastasis in mouse models (41, 56–59), and epidemiological studies suggest an anticancer effect of digoxin in humans as well (60). Although the mechanism is unclear, a recent screen identified cardiac glycosides as drugs that enhance flux through the lysosomal pathway (36). Our results suggest that this effect of digoxin may contribute to its inhibition of HIF-1α protein levels and raise the possibility that other agents that modulate lysosomal degradation of HIF-1α may have therapeutic potential.
Acknowledgment
We thank K. Padgett (Novus Biologicals) for providing anti-HSC70 and anti-TFEB antibodies.
This work was supported in part by National Institutes of Health Contracts N01-HV28180 and HHS-N268201000032c from the United States Public Health Service. This work was also supported by the Johns Hopkins Institute for Cell Engineering and Dean's Research Funding and American Heart Association Predoctoral Fellowship 10PRE4160120.
- HIF
- hypoxia-inducible factor
- DFX
- desferrioxamine
- CMA
- chaperone-mediated autophagy
- TFEB
- transcription factor EB
- MEF
- mouse embryonic fibroblast
- RT-qPCR
- real-time quantitative RT-PCR
- 3-MA
- 3-methyladenine
- 6-AN
- 6-aminonicotinamide.
REFERENCES
- 1. Semenza G. L., Wang G. L. (1992) A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol. Cell. Biol. 12, 5447–5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang G. L., Jiang B. H., Rue E. A., Semenza G. L. (1995) Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. U.S.A. 92, 5510–5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Iyer N. V., Kotch L. E., Agani F., Leung S. W., Laughner E., Wenger R. H., Gassmann M., Gearhart J. D., Lawler A. M., Yu A. Y., Semenza G. L. (1998) Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1α. Genes Dev. 12, 149–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yu A. Y., Shimoda L. A., Iyer N. V., Huso D. L., Sun X., McWilliams R., Beaty T., Sham J. S., Wiener C. M., Sylvester J. T., Semenza G. L. (1999) Impaired physiological responses to chronic hypoxia in mice partially deficient for hypoxia-inducible factor 1α. J. Clin. Invest. 103, 691–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Semenza G. L. (2009) Regulation of oxygen homeostasis by hypoxia-inducible factor 1. Physiology 24, 97–106 [DOI] [PubMed] [Google Scholar]
- 6. Ebert B. L., Firth J. D., Ratcliffe P. J. (1995) Hypoxia and mitochondrial inhibitors regulate expression of glucose transporter-1 via distinct cis-acting sequences. J. Biol. Chem. 270, 29083–29089 [DOI] [PubMed] [Google Scholar]
- 7. Kim J. W., Tchernyshyov I., Semenza G. L., Dang C. V. (2006) HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 3, 177–185 [DOI] [PubMed] [Google Scholar]
- 8. Papandreou I., Cairns R. A., Fontana L., Lim A. L., Denko N. C. (2006) HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 3, 187–197 [DOI] [PubMed] [Google Scholar]
- 9. Forsythe J. A., Jiang B. H., Iyer N. V., Agani F., Leung S. W., Koos R. D., Semenza G. L. (1996) Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol. Cell. Biol. 16, 4604–4613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Semenza G. L., Jiang B. H., Leung S. W., Passantino R., Concordet J. P., Maire P., Giallongo A. (1996) Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 271, 32529–32537 [DOI] [PubMed] [Google Scholar]
- 11. Bruick R. K. (2000) Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc. Natl. Acad. Sci. U.S.A. 97, 9082–9087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hubbi M. E., Kshitiz, Gilkes D. M., Rey S., Wong C. C., Luo W., Kim D. H., Dang C. V., Levchenko A., Semenza G. L. (2013) A nontranscriptional role for HIF-1α as a direct inhibitor of DNA replication. Sci. Signal. 6, ra10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Patel S. A., Simon M. C. (2008) Biology of hypoxia-inducible factor-2α in development and disease. Cell Death Differ. 15, 628–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Epstein A. C., Gleadle J. M., McNeill L. A., Hewitson K. S., O'Rourke J., Mole D. R., Mukherji M., Metzen E., Wilson M. I., Dhanda A., Tian Y. M., Masson N., Hamilton D. L., Jaakkola P., Barstead R., Hodgkin J., Maxwell P. H., Pugh C. W., Schofield C. J., Ratcliffe P. J. (2001) C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107, 43–54 [DOI] [PubMed] [Google Scholar]
- 15. Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., Kaelin W. G., Jr. (2001) HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464–468 [DOI] [PubMed] [Google Scholar]
- 16. Jaakkola P., Mole D. R., Tian Y. M., Wilson M. I., Gielbert J., Gaskell S. J., von Kriegsheim A., Hebestreit H. F., Mukherji M., Schofield C. J., Maxwell P. H., Pugh C. W., Ratcliffe P. J. (2001) Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472 [DOI] [PubMed] [Google Scholar]
- 17. Yu F., White S. B., Zhao Q., Lee F. S. (2001) HIF-1α binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc. Natl. Acad. Sci. U.S.A. 98, 9630–9635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lando D., Peet D. J., Whelan D. A., Gorman J. J., Whitelaw M. L. (2002) Asparagine hydroxylation of the HIF transactivation domain: a hypoxic switch. Science 295, 858–861 [DOI] [PubMed] [Google Scholar]
- 19. Lando D., Peet D. J., Gorman J. J., Whelan D. A., Whitelaw M. L., Bruick R. K. (2002) FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 16, 1466–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jiang B. H., Zheng J. Z., Leung S. W., Roe R., Semenza G. L. (1997) Transactivation and inhibitory domains of hypoxia-inducible factor 1α. Modulation of transcriptional activity by oxygen tension. J. Biol. Chem. 272, 19253–19260 [DOI] [PubMed] [Google Scholar]
- 21. Liu Y. V., Baek J. H., Zhang H., Diez R., Cole R. N., Semenza G. L. (2007) RACK1 competes with HSP90 for binding to HIF-1α and is required for O2-independent and HSP90 inhibitor-induced degradation of HIF-1α. Mol. Cell 25, 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu Y. V., Hubbi M. E., Pan F., McDonald K. R., Mansharamani M., Cole R. N., Liu J. O., Semenza G. L. (2007) Calcineurin promotes hypoxia-inducible factor 1α expression by dephosphorylating RACK1 and blocking RACK1 dimerization. J. Biol. Chem. 282, 37064–37073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Koh M. Y., Darnay B. G., Powis G. (2008) Hypoxia-associated factor, a novel E3-ubiquitin ligase, binds and ubiquitinates hypoxia-inducible factor 1α, leading to its oxygen-independent degradation. Mol. Cell. Biol. 28, 7081–7095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Luo W., Zhong J., Chang R., Hu H., Pandey A., Semenza G. L. (2010) Hsp70 and CHIP selectively mediate ubiquitination and degradation of hypoxia-inducible factor (HIF)-1α but not HIF-2α. J. Biol. Chem. 285, 3651–3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Montagner M., Enzo E., Forcato M., Zanconato F., Parenti A., Rampazzo E., Basso G., Leo G., Rosato A., Bicciato S., Cordenonsi M., Piccolo S. (2012) SHARP1 suppresses breast cancer metastasis by promoting degradation of hypoxia-inducible factors. Nature 487, 380–384 [DOI] [PubMed] [Google Scholar]
- 26. Kaushik S., Bandyopadhyay U., Sridhar S., Kiffin R., Martinez-Vicente M., Kon M., Orenstein S. J., Wong E., Cuervo A. M. (2011) Chaperone-mediated autophagy at a glance. J. Cell Sci. 124, 495–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chiang H. L., Terlecky S. R., Plant C. P., Dice J. F. (1989) A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science 246, 382–385 [DOI] [PubMed] [Google Scholar]
- 28. Cuervo A. M., Dice J. F. (1996) A receptor for the selective uptake and degradation of proteins by lysosomes. Science 273, 501–503 [DOI] [PubMed] [Google Scholar]
- 29. Konecki D. S., Foetisch K., Zimmer K. P., Schlotter M., Lichter-Konecki U. (1995) An alternatively spliced form of the human lysosome-associated membrane protein-2 gene is expressed in a tissue-specific manner. Biochem. Biophys. Res. Commun. 215, 757–767 [DOI] [PubMed] [Google Scholar]
- 30. Fujiwara Y., Furuta A., Kikuchi H., Aizawa S., Hatanaka Y., Konya C., Uchida K., Yoshimura A., Tamai Y., Wada K., Kabuta T. (2013) Discovery of a novel type of autophagy targeting RNA. Autophagy 9, 403–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sardiello M., Palmieri M., di Ronza A., Medina D. L., Valenza M., Gennarino V. A., Di Malta C., Donaudy F., Embrione V., Polishchuk R. S., Banfi S., Parenti G., Cattaneo E., Ballabio A. (2009) A gene network regulating lysosomal biogenesis and function. Science 325, 473–477 [DOI] [PubMed] [Google Scholar]
- 32. Settembre C., Di Malta C., Polito V. A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S. U., Huynh T., Medina D., Colella P., Sardiello M., Rubinsztein D. C., Ballabio A. (2011) TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dice J. (2000) Molecular Biology Intelligence Unit: Lysosomal Pathways of Protein Degradation, Landes Bioscience, Austin, TX [Google Scholar]
- 34. Rubinsztein D. C., Gestwicki J. E., Murphy L. O., Klionsky D. J. (2007) Potential therapeutic applications of autophagy. Nat. Rev. Drug Discov. 6, 304–312 [DOI] [PubMed] [Google Scholar]
- 35. Kshitiz, Hubbi M. E., Ahn E. H., Downey J., Afzal J., Kim D. H., Rey S., Chang C., Kundu A., Semenza G. L., Abraham R. M., Levchenko A. (2012) Matrix rigidity controls endothelial differentiation and morphogenesis of cardiac precursors. Sci. Signal. 5, ra41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee K., Zhang H., Qian D. Z., Rey S., Liu J. O., Semenza G. L. (2009) Acriflavine inhibits HIF-1 dimerization, tumor growth, and vascularization. Proc. Natl. Acad. Sci. U.S.A. 106, 17910–17915 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37. Seglen P. O., Gordon P. B. (1982) 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc. Natl. Acad. Sci. U.S.A. 79, 1889–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ekström P., Kanje M. (1984) Inhibition of fast axonal transport by erythro-9-[3-(2-hydroxynonyl)]adenine. J. Neurochem. 43, 1342–1345 [DOI] [PubMed] [Google Scholar]
- 39. Zhang H., Bosch-Marce M., Shimoda L. A., Tan Y. S., Baek J. H., Wesley J. B., Gonzalez F. J., Semenza G. L. (2008) Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J. Biol. Chem. 283, 10892–10903 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40. Finn P. F., Mesires N. T., Vine M., Dice J. F. (2005) Effects of small molecules on chaperone-mediated autophagy. Autophagy 1, 141–145 [DOI] [PubMed] [Google Scholar]
- 41. Zhang H., Qian D. Z., Tan Y. S., Lee K., Gao P., Ren Y. R., Rey S., Hammers H., Chang D., Pili R., Dang C. V., Liu J. O., Semenza G. L. (2008) Digoxin and other cardiac glycosides inhibit HIF-1α synthesis and block tumor growth. Proc. Natl. Acad. Sci. U.S.A. 105, 19579–19586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hundeshagen P., Hamacher-Brady A., Eils R., Brady N. R. (2011) Concurrent detection of autolysosome formation and lysosomal degradation by flow cytometry in a high-content screen for inducers of autophagy. BMC Biol. 9, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dohi E., Tanaka S., Seki T., Miyagi T., Hide I., Takahashi T., Matsumoto M., Sakai N. (2012) Hypoxic stress activates chaperone-mediated autophagy and modulates neuronal cell survival. Neurochem. Int. 60, 431–442 [DOI] [PubMed] [Google Scholar]
- 44. Peña-Llopis S., Vega-Rubin-de-Celis S., Schwartz J. C., Wolff N. C., Tran T. A. T., Zou L., Xie X. J., Corey D. R., Brugarolas J. (2011) Regulation of TFEB and V-ATPases by mTORC1. EMBO J. 30, 3242–3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kon M., Kiffin R., Koga H., Chapochnick J., Macian F., Varticovski L., Cuervo A. M. (2011) Chaperone-mediated autophagy is required for tumor growth. Sci. Transl. Med. 3, 109ra117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Foxler D. E., Bridge K. S., James V., Webb T. M., Mee M., Wong S. C., Feng Y., Constantin-Teodosiu D., Petursdottir T. E., Bjornsson J., Ingvarsson S., Ratcliffe P. J., Longmore G. D., Sharp T. V. (2012) The LIMD1 protein bridges an association between the prolyl hydroxylases and VHL to repress HIF-1 activity. Nat. Cell Biol. 14, 201–208 [DOI] [PubMed] [Google Scholar]
- 47. Hubbi M. E., Gilkes D. M., Baek J. H., Semenza G. L. (2012) Four-and-a-half LIM domain proteins inhibit transactivation by hypoxia-inducible factor 1. J. Biol. Chem. 287, 6139–6149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lin J., Qin X., Zhu Z., Mu J., Zhu L., Wu K., Jiao H., Xu X., Ye Q. (2012) FHL family members suppress vascular endothelial growth factor expression through blockade of dimerization of HIF1α and HIF1β. IUBMB Life 64, 921–930 [DOI] [PubMed] [Google Scholar]
- 49. Hubbi M. E., Luo W., Baek J. H., Semenza G. L. (2011) MCM proteins are negative regulators of hypoxia-inducible factor 1. Mol. Cell 42, 700–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Baek J. H., Liu Y. V., McDonald K. R., Wesley J. B., Zhang H., Semenza G. L. (2007) Spermidine/spermine-N1-acetyltransferase-1 binds to hypoxia-inducible factor-1α (HIF-1α) and RACK1 and promotes ubiquitination and degradation of HIF-1α. J. Biol. Chem. 282, 33358–33366 [DOI] [PubMed] [Google Scholar]
- 51. Baek J. H., Liu Y. V., McDonald K. R., Wesley J. B., Hubbi M. E., Byun H., Semenza G. L. (2007) Spermidine/spermine-N1-acetyltransferase 2 is an essential component of the ubiquitin ligase complex that regulates hypoxia-inducible factor 1α. J. Biol. Chem. 282, 23572–23580 [DOI] [PubMed] [Google Scholar]
- 52. Lv L., Li D., Zhao D., Lin R., Chu Y., Zhang H., Zha Z., Liu Y., Li Z., Xu Y., Wang G., Huang Y., Xiong Y., Guan K. L., Lei Q. Y. (2011) Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol. Cell 42, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Luo W., Hu H., Chang R., Zhong J., Knabel M., O'Meally R., Cole R. N., Pandey A., Semenza G. L. (2011) Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell 145, 732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Argani P., Ladanyi M. (2005) Translocation carcinomas of the kidney. Clin. Lab. Med. 25, 363–378 [DOI] [PubMed] [Google Scholar]
- 55. Zhong H., De Marzo A. M., Laughner E., Lim M., Hilton D. A., Zagzag D., Buechler P., Isaacs W. B., Semenza G. L., Simons J. W. (1999) Overexpression of hypoxia-inducible factor 1α in common human cancers and their metastases. Cancer Res. 59, 5830–5835 [PubMed] [Google Scholar]
- 56. Zhang H., Wong C. C., Wei H., Gilkes D. M., Korangath P., Chaturvedi P., Schito L., Chen J., Krishnamachary B., Winnard P. T., Jr., Raman V., Zhen L., Mitzner W. A., Sukumar S., Semenza G. L. (2012) HIF-1-dependent expression of angiopoietin-like 4 and L1CAM mediates vascular metastasis of hypoxic breast cancer cells to the lungs. Oncogene 31, 1757–1770 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57. Wong C. C., Zhang H., Gilkes D. M., Chen J., Wei H., Chaturvedi P., Hubbi M. E., Semenza G. L. (2012) Inhibitors of hypoxia-inducible factor 1 block breast cancer metastatic niche formation and lung metastasis. J. Mol. Med. 90, 803–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schito L., Rey S., Tafani M., Zhang H., Wong C. C., Russo A., Russo M. A., Semenza G. L. (2012) Hypoxia-inducible factor 1-dependent expression of platelet-derived growth factor B promotes lymphatic metastasis of hypoxic breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 109, E2707–E2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chaturvedi P., Gilkes D. M., Wong C. C., Kshitiz, Luo W., Zhang H., Wei H., Takano N., Schito L., Levchenko A., Semenza G. L. (2013) Hypoxia-inducible factor-dependent breast cancer-mesenchymal stem cell bidirectional signaling promotes metastasis. J. Clin. Invest. 123, 189–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Platz E. A., Yegnasubramanian S., Liu J. O., Chong C. R., Shim J. S., Kenfield S. A., Stampfer M. J., Willett W. C., Giovannucci E., Nelson W. G. (2011) A novel two-stage, transdisciplinary study identifies digoxin as a possible drug for prostate cancer treatment. Cancer Discov. 1, 68–77 [DOI] [PMC free article] [PubMed] [Google Scholar]