Abstract
All fully sequenced strains of Streptococcus pneumoniae (pneumococcus) contain a version of the blp locus which is responsible for the regulation and secretion of a variable repertoire of pneumococcal bacteriocins called pneumocins and their associated immunity proteins. Pneumocins mediate intra- and interspecies competition in vitro and have been shown to provide a competitive advantage in vivo. Pneumocin production is stimulated by extracellular accumulation of the peptide pheromone, BlpC. Both BlpC and the functional pneumocins are secreted out of the cell via the Blp transporter, BlpAB. The conserved surface-expressed serine protease, HtrA, has been shown to limit activation of the locus and secretion of functional pneumocins. In this work, we demonstrate that htrA mutants stimulate the blp locus at lower cell density and to a greater extent than strains expressing wild-type HtrA. This effect is not due to direct proteolytic degradation of secreted pheromone by the protease, but instead is a result of HtrA-mediated disruption of peptide processing and secretion. Because pneumocins are secreted through the same transporter as the pheromone, this finding explains why pheromone supplementation cannot completely restore pneumocin inhibition to strains expressing high levels of HtrA despite restoration of blp transcriptional activity. HtrA restricts pneumocin production to high cell density by limiting the rate of accumulation of BlpC in the environment. Importantly, HtrA does not interfere with the ability of a strain to sense environmental pheromones, which is necessary for the induction of protective immunity in the face of pneumocin-secreting competitors.
INTRODUCTION
The species Streptococcus pneumoniae is characterized by significant genomic diversity which allows for adaptation to changing pressures in the host environment, including pressure from the resident flora (1–3). The blp locus, which encodes the blp bacteriocins (pneumocins), associated immunity proteins, and the regulatory and secretion proteins required for pneumocin production, is an example of how selective pressure exerted by other members of the nasopharyngeal flora influences genomic content. Although all strains examined to date have some version of the blp locus, the locus is characterized by significant genetic diversity in both the peptide pheromone and pneumocin content (4–7). The blp locus includes genes encoding a typical two-component regulatory system (BlpR and BlpH), a peptide pheromone involved in quorum sensing (BlpC), various bacteriocins/immunity proteins located in the bacteriocin-immunity region (BIR), and an ABC transporter complex (BlpAB) required for the processing and secretion of BlpC and the bacteriocin peptides (Fig. 1A and B) (4, 5, 7). Pneumocin production is stimulated by the binding of secreted BlpC to the histadine kinase, BlpH (Fig. 1A). Pneumocins, such as the two-peptide bacteriocin pneumocin MN, have been shown to play an important role in intraspecies competition in a murine model (4). We had previously demonstrated that variations in the integrity of BlpA, the BlpC specificity of BlpH, and the specific content of bacteriocin and immunity peptides all play an important role in dictating the outcome of competition between different pneumococcal strains (8). As an example, a highly conserved 4-bp insertion in blpA can be found in nearly half of strains studied. This insertion results in a frameshift mutation that renders the secretion apparatus nonfunctional. Strains with a nonfunctional blpA gene are unable to secrete pheromone or pneumocins but are still able to respond to exogenous pheromone due to a preserved two-component regulatory system allowing for the induction of functional immunity genes (8). These “cheater” strains were presumably selected for because they avoid the energetic cost of pheromone and pneumocin secretion.
Fig 1.
HtrA alters the density of activation of the blp locus in the three most common pherotypes. (A) Diagramatic representation of the sequence of BlpC-induced stimulation of the blp locus. 1, BlpC is produced as a preprotein inside the cell (small circles); 2, pre-BlpC is cleaved at a double glycine motif (GG) and transported out of the cell by the BlpAB complex; 3, active BlpC binds to the histidine kinase, BlpH, resulting in (4) phosphor transfer to the response regulator, BlpR; 5, phospho-BlpR upregulates all genes in the blp locus by binding to specific inverted repeats in the blp locus. (B) Diagramatic representation of the blp locus and the reporter plasmid integration used for subsequent studies. Letter designations refer to blp alleles; BIR refers to a variable region containing bacteriocins and immunity proteins; Afr refers to the 5′ fragment of blpA cloned into the reporter plasmid; large arrows denote ORFs and direction of transcription; and an X denotes the region of plasmid integration. Small black arrowheads signify the locations and directions of transcription of promoters and the site of phosphor-BlpR binding. After plasmid integration, lacZ is driven by the proximal BIR promoter. (C to F) R6-derived reporter strains expressing pherotypes BlpC164 (triangles), BlpCR6 (squares), or BlpC6A (circles) were assessed for activation of the blp locus throughout the growth phase. Dotted lines in gray represent the respective growth curves. Reporter strains with wild-type htrA are represented by closed symbols; strains containing an in-frame unmarked deletion of htrA are represented by open symbols. The active site HtrA mutation, HtrAS234A, was tested in the R6-responsive background as indicated in panel C (closed diamonds). (F) Pherotype BlpCR6-expressing strains with htrA (closed squares) and without htrA (open squares) were assessed for blp activation at 32°C. (G) Western blot analysis of lysates from strain PSD100 at different OD620 values for HtrA expression levels. Equivalent amounts of total protein were loaded in each lane; membranes were probed with polyclonal HtrA antiserum.
Previous studies have demonstrated that the expression of pneumocins in the prototypic pneumocin-producing strain, 6A, is repressed by the outer-surface serine protease, HtrA (9). HtrA is a member of the DegP family of proteases and has been shown to also play a role in regulating pneumococcal competence and survival during high-temperature growth (10–12). In the 6A background, HtrA had two distinct roles in reducing pneumocin expression: it both repressed pneumocin production by altering the dose-response for the peptide pheromone BlpC and decreased inhibition by reducing the amount of functional pneumocin (9). The most straightforward hypothesis for this observation is that HtrA activity either directly or indirectly results in the inactivation of either the secretion apparatus or the secreted peptides themselves, including both pheromone and structural bacteriocins. Because the ability to sense exogenous peptide from competitor strains is vital for the survival of the significant number of cheater strains in the population, we were interested in determining if HtrA activity would affect pheromone detection and, therefore, the induction of protective immunity. In this work, we demonstrate that HtrA production reduces secretion of the three most common BlpC types (pherotypes) in the same manner, suggesting that the effect of HtrA is independent of the amino acid sequence of the peptides. Despite the fact that the protease is primarily associated with the outer surface of the bacterium (11, 13), HtrA affects only the amount of pheromone that is secreted but does not appreciably lower the concentration of exogenous environmental pheromones. The repressive effect of HtrA can be overcome at high cell density through accumulation of a sufficient concentration of BlpC. In this way, HtrA protease activity effectively controls the density-dependent regulation of the locus without interfering with pheromone sensing, preserving the ability of bacteria to sense pneumocin-producing competitors in the nasopharynx.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The Streptococcus pneumoniae strains used are described in Table 1. S. pneumoniae was grown at 37°C in Todd-Hewitt broth supplemented with 0.5% yeast extract (THY) or at 37°C in 5% CO2 on tryptic soy agar plates supplemented with Catalase (Worthington, Lakewood, NJ) (4,741 U) or 5% sheep blood (SBA). Escherichia coli strains were grown in Luria-Bertani (LB) broth or LB agar supplemented with the appropriate antibiotics at 37°C. Antibiotic concentrations used were as follows: for S. pneumoniae, 500 μg/ml kanamycin, 100 μg/ml streptomycin, 2 μg/ml chloramphenicol, 1 μg/ml erythromycin, and 200 μg/ml spectinomycin; and for E. coli, 50 μg/ml kanamycin, 20 μg/ml chloramphenicol, 100 μg/ml erythromycin, and 100 μg/ml spectinomycin.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| R6x | Unencapsulated laboratory strain with rpsLK56T; Str | 29 |
| R6 | Unencapsulated laboratory strain; St sensitive | 30 |
| P376 | Serotype 6A isolate; exhibits BlpC6A secretion | 31 |
| P764 | Serotype 6B isolate; exhibits BlpC164 secretion; Emr | 32 |
| D39 | Serotype 2 | 30 |
| P76 | D39 (rspL1-htrA::kan-rpsL+) (Janus); Knr | M. E. Sebert personal collection |
| P81 | P76 (rspL1ΔhtrA298–1152); Str | M. E. Sebert personal collection |
| P639 | P764 × pE57, resulting in a BlpC164 reporter; Cmr Emr | 8 |
| P1822 | P1720 (htrAS234A) active site mutation; Str | 9 |
| PSD100 | R6x × P639, resulting in a BlpCR6 reporter; Str Cmr | This study |
| PSD101 | PSD100 (blpC::aad9); Cmr Spr | This study |
| PSD102 | PSD101 (htrA::kan-rpsL+); Cmr Knr Spr | This study |
| PSD103 | PSD101 but ΔhtrA298–1152; Cmr Str | This study |
| PSD104 | PSD100 (htrA::kan-rpsL+); Cmr Knr Spr | This study |
| PSD105 | PSD100 but ΔhtrA298–1152; Cmr Str | This study |
| PSD106 | PSD100 (blpC::kan-rpsL+); Cmr Knr | This study |
| PSD107 | PSD100 but blpHC6A; Cmr Str | This study |
| PSD108 | PSD107 (htrA::kan-rpsL+); Cmr Knr | This study |
| PSD109 | PSD107 but ΔhtrA298–1152; Cmr Str | This study |
| PSD110 | PSD107 but blpC::aad9; Cmr Spr Str | This study |
| PSD111 | PSD109 but blpC::aad9; Cmr Spr Str | This study |
| PSD114 | PSD100 but blpHC164; Cmr Str | This study |
| PSD115 | PSD107 (blpC::kan-rpsL+); Cmr Knr | This study |
| PSD116 | PSD114 (htrA::kan-rpsL+); Cmr Knr | This study |
| PSD117 | PSD114 but ΔhtrA298–1152; Cmr Str | This study |
| PSD118 | PSD107 but blpH6A-blpCR6; Cmr Str | This study |
| PSD119 | PSD118 (htrA::kan-rpsL+); Cmr Knr | This study |
| PSD120 | PSD118 but ΔhtrA298–1152; Cmr Str | This study |
| PSD121 | PSD114 (blpC::aad9); Cmr Spr | This study |
| PSD122 | PSD121(htrA::kan-rpsL+); Cmr Knr | This study |
| PSD123 | PSD121 but ΔhtrA298–1152; Cmr Str | This study |
| PSD124 | PSD100 but htrAS234A; Cmr Str | This study |
| PSD125 | PSD100 but blpCFLAG; Cmr Str | This study |
| PSD126 | PSD125 (htrA::kan-rpsL+); Cmr Knr | This study |
| PSD127 | PSD125 but ΔhtrA298–1152; Cmr Str | This study |
| PSD128 | PSD125 (blpA::kan-rspL+); Cmr Knr | This study |
| PSD129 | PSD125 but ΔblpA5–707; Cmr Str | This study |
| PSD130 | PSD127 (blpA::kan-rpsL+); Cmr Knr | This study |
| PSD131 | PSD127 but ΔblpA5–707; Cmr Str | This study |
| PSD132 | R6 × PSD100, resulting in a BlpCR6 reporter; Cmr | This study |
| PSD133 | PSD132 (htrA::erm); Cmr Emr | This study |
| PSD134 | PSD132 (rpsLK56T); Cmr Str | This study |
| PSD135 | PSD133 (rpsLK56T); Cmr Str Emr | This study |
| Plasmids | ||
| pEVP3 | Reporter plasmid for integration in S. pneumoniae; Cmr | 33 |
| pE57 | pEVP3 derivative with promoter of blpQ from strain P764 cloned upstream of lacZ; Cmr | 8 |
| pE104 | blpC region from R6 cloned into TOPO vector; Amr Knr | This study |
| pE133 | Derivative of pE104 with an in-frame deletion of blpC generated by inverse PCR | This study |
| pE132 | pE133 derivative with blpC::aad9 deletion cloned in place of blpC; Spr Amr Knr | This study |
Abbreviations used: Sp, spectinomycin; Am, ampicillin; Cm, chloramphenicol; Kn, kanamycin; Em, erythromycin; St, streptomycin; r, resistance. ΔhtrA298–1152 is htrA with nucleotides 298 to 1152 deleted; ΔblpA5–707 is blpA with nucleotides 5 to 707 deleted.
Construction and analysis of reporter constructs, chimeras, and HtrA mutants in R6.
In order to detect the transcriptional activity of the blp locus, we utilized an existing R6 derivative (PSD100) that contained an active blp locus from a serotype 6B strain (P639). The construction of this strain has been previously described (8). In addition to a portion of the type 6B blp locus that includes blpA downstream of the BIR, this strain contains a reporter plasmid integration at the proximal BIR promoter, resulting in a promoterless lacZ gene fused to the promoter driving pneumocin gene expression (Fig. 1B). Sequence analysis verified that this strain contains R6-derived sequence beginning upstream of the blpB gene, including the R6-derived blpC allele and corresponding blpRH genes but an intact blpA gene and BIR region derived from the 6B strain. We then created a series of reporter strains with different blpC and blpH alleles by cloning the region flanking the blpC gene from R6x into pCR2.1 using primers 4 and 5, creating plasmid pE104 (primers are listed in Table 2). blpC was then deleted from this plasmid using inverse PCR with primers 1 and 2, introducing a SmaI site in place of the blpC gene, creating plamid pE133. The Janus cassette (14), which contains the gene for kanamycin resistance and the rpsL gene for streptomycin sensitivity, was amplified and ligated into the SmaI site. The ligation product was used to transform PSD100, and kanamycin-resistant, streptomycin-sensitive transformants were obtained. Allelic exchange of the blpC gene for the Janus cassette was verified by PCR. The Janus cassette was replaced with the blpCH region from P376, resulting in BlpC6A secretion and responsiveness, or with the corresponding region from strain P764 (resulting in BlpC164 secretion and responsiveness), using PCR products created using primers 3 and 4, creating strains PSD107 and PSD114, respectively. The resulting kanamycin-sensitive streptomycin-resistant strains were sequenced to verify correct replacement and streaked on media containing 500 ng/ml of each synthetic active peptide (sBlpC) (Genscript, Piscataway, NJ) and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) to verify correct pherotype responsiveness. BlpC amino acid sequences are listed in Table 3; the sBlpC peptides used consisted of only the active peptide region. BlpC deletion strains were created in each background using plasmid pE132 containing a spectinomycin cassette in place of the blpC gene created by ligating a blunt-ended spectinomycin cassette into the SmaI site of plasmid pE133. Strains were screened for expected responsiveness by plating on media with sBlpC and X-Gal as described above. Chimeric strains were created by introducing the Janus cassette into the blpC gene of the PSD107 strain. Strains with the Janus cassette in the blpC gene but maintaining responsiveness to sBlpC6A were chosen for allelic replacement with the blpC region from R6. The resulting streptomycin-resistant transformants were screened by PCR and sequencing for replacement of only the blpC gene and retention of the 6A-derived blpH allele. An epitope-tagged version of BlpCR6 was created by amplifying DNA from strain R6 with primer pair 5 and 6 and primer pair 4 and 7 that engineers two in-frame FLAG tags to the C-terminal end of BlpCR6 followed by a BamHI site, digesting with BamHI, ligating products, and transformation into strains with the exchangeable Janus cassette in blpC, resulting in strain PSD125. Streptomycin-resistant strains were analyzed for Janus replacement as described above, and the exchanged region was sequenced using primers 3 and 4. The amino acid sequence of the resultant BlpCFLAG is included in Table 3.
Table 2.
Primers used in this study
| Primer | Sequence | Description |
|---|---|---|
| 1 | CAG AGG CCC GGG CAT GTT ATG ATT CTC CTT TTT | Forward primer for inverse PCR to make deletion in blpC |
| 2 | GAG GCG CCC GGG TAA AAA CAA GAC CGA GAA AC | Reverse primer for inverse PCR to make deletion in blpC |
| 3 | TCC AAC TAA AGC CCA TAC CG | Forward primer in blpS |
| 4 | CAA CAA ATT GGA GAA TAT CAA GAG | Reverse primer in blpB |
| 5 | TTG GAT ACC TAT TTG AAG GA | Forward primer in blpH |
| 6 | GAC CGG ATC CCT ATA AAA ACA AGA CCG AGA AAC | Reverse primer in blpC to introduce BamHI site |
| 7 | GAC GGA TCC TTA TTT ATC ATC ATC ATC TTT ATA ATC TTT ATC ATC ATC ATC TTT ATA ATC TAG CTG AAT AGG TAG TTC AAG TGC | Forward primer in blpC introducing two FLAG tags and a BamHI site |
| 8 | GAC TTG CCC GGG TCA TTA GCT TTT TTA GTG GA | Forward primer used to amplify Janus cassette and introduce SmaI site |
| 9 | GAC TTC CCC GGG GAG CAC TTT GTA AGT CTG TTG | Reverse primer used to amplify Janus cassette and introduce SmaI site |
| 10 | GAT AAA AAT GGC TCC TCT GC | Forward primer in blpB |
| 11 | CTG GAT GTT TCT ATT CTG ATT CT | Reverse primer in blpA |
| 12 | GAG CGG ATC CAT AAG AAG TCA TTG GCA ATT CC | Forward primer in blpA introducing a BamHI site |
| 13 | GAG CGG ATC CCA AGG TTT CTA CCA TCA TCT | Reverse primer in blpA introducing a BamHI site |
| 14 | TCA AAA ATA ATT CGC GTC TG | Reverse primer in blp lacZ |
| 15 | TTA AGA AAA AAG CCG GGA AAA TTC C | Forward primer in htrA |
| 16 | CGC GTG CTT CTT CTA TTG GGT TTA A | Reverse primer in htrA |
| 17 | CTA TTT TGT GCT TCC ATT TTT G | Forward primer in blpH 312 bp from ATG |
Table 3.
BlpC type and amino acid sequence
| BlpC type | Signal sequence | sBlpC sequence | MWa |
|---|---|---|---|
| BlpC164 | MDKKQNLTSFQELTTTELNQIIGG | GWWEDFLYRFNIIEQKNTKGFYQPIQL | 6.1/3.4 |
| BlpCR6 | MDKKQNLTSFQELTTTELNQITGG | GWWEELLHETILSKFKITKALELPIQL | 5.9/3.2 |
| BlpC6A | MDKKQNLTSFQELTTTELNQITGG | GLWEDILYSLNIIKHNNTKGLHHPIQL | 5.8/3.2 |
| BlpCFLAG | MDKKQNLTSFQELTTTELNQITGG | GWWEELLHETILSKFKITKALELPIQLDYKDDDDKDYKDDDDK | 7.9/5.2 |
MW, predicted molecular weight of preprotein/mature protein.
blpA deletions were created by cloning a fragment of the R6 region of blpA into pCR2.1 Topo vector (Invitrogen), digesting the plasmid with NdeI, blunt ending with T4 DNA polymerase, and ligating the Janus cassette amplified with primers 8 and 9 cut with SmaI. The ligation reaction was used to transform the BlpCFLAG-containing PSD125 strain, creating strain PSD128. Insertion was confirmed by PCR. In-frame, unmarked blpA deletions were made by amplifying R6 DNA with primer pair 10 and 13 and primer pair 12 and 14. Products were digested with BamHI, ligated, and transformed into the Janus PSD128 strain described above, creating strain PSD129. Deletions were confirmed by PCR and sequencing.
In-frame, unmarked htrA mutations were introduced into each background using genomic DNA from a homologous D39 strain containing a Janus cassette in place of the internal portion of the htrA gene. In-frame deletions were constructed using genomic DNA derived from P81, a D39 strain with an in-frame, unmarked deletion of the htrA gene. Transformations were confirmed by PCR and complete loss of expression of HtrA by Western blot analysis. To create the PSD100 strain containing the htrA allele with an active site mutation, a PCR product from strain P1822 with primers 15 and 16 giving 500 bp flanking either side of the htrA open reading frame (ORF) was used to replace the Janus cassette in htrA, resulting in strain PSD124.
To determine the effect of streptomycin resistance on the role of HtrA in the density-dependent control of blp transcription, DNA from strain PSD100 was used to transform the streptomycin-sensitive strain, R6, creating strain PSD132. Transformants were confirmed to be streptomycin sensitive after they were selected for chloramphenicol resistance and self-induction on plates containing X-Gal. Because Janus exchange cannot be used in streptomycin-sensitive strains, a deletion in htrA was introduced into this strain background by moving a PCR product generated using primers 15 and 16 from an existing htrA::erm deletion mutant into the R6 reporter background, creating strain PSD133. To directly analyze the role of streptomycin resistance in HtrA activity, the streptomycin resistance gene was moved into both the HtrA-sufficient and -deficient backgrounds using genomic DNA from the R6x strain and selecting for streptomycin resistance, creating strains P134 and P135, respectively.
Pneumococcal transformations were performed as previously described (9, 15) except that strains were grown in THY medium to an optical density at 620 nm (OD620) of 0.2 to 0.4 and then diluted into C+Y (pH 8.0) medium (15) prior to addition of CSP1 (EMRLSKFFRDFILQRKK) and DNA.
Determination of transcriptional activity of reporter strains.
Transcriptional activity of the reporter strains was determined by assessing β-galactosidase activity using the substrate o-nitrophenyl-β-d-galactopyranoside (ONPG). For strains with intact loci, pneumococcal strains were streaked onto SBA and inoculated into THY medium with chloramphenicol and grown to an OD620 of 0.5. Cultures were frozen in 20% glycerol. For growth curve assays, 100 μl of thawed culture was added to 15 ml of THY. To avoid the potential effect of antibiotic exposure on HtrA activity, chloramphenicol was not added to the media during growth for any natural or stimulated assay. Plasmid integration stability was assessed by comparing colony counts of select reporter strains on media with and without chloramphenicol. No evidence of plasmid excision was noted even after multiple passages without selection (not shown). Growth was followed by assessing OD620 every 30 min, and samples were taken every 30 min when the OD620 reached 0.050. Samples (100 μl) were lysed by the addition of 1 μl 10% Triton X-100 and stored on ice until completion of the assay. Plates were incubated at 37°C for 10 min and examined for complete lysis of cells. A 25-μl volume of ONPG in 5× Z-buffer (5 mM MgCl, 50 mM KCl, 0.3 M Na2HPO4, 0.2 M NaH2PO4, 250 mM β-mercaptoethanol, 4 mg/ml ONPG) was added, and the reaction was allowed to continue until color change was appreciated or for 45 min if no color change was appreciated. A 50-μl volume of 1 M NaCO3 was added to stop the reaction, and plates were read at OD415 and OD550. Miller units were determined as previously described (16). Sample procedures were performed in triplicate.
For strains with blpC disruptions, 100-μl glycerol stocks prepared as described above were diluted into 10 ml of fresh THY medium and allowed to grow to an OD620 of 0.10. Aliquots (100 μl) of cells were added to a 96-well plate containing 2-fold dilutions of sBlpC in 10 μl of sterile water. Blank wells with medium alone and cells exposed to water only were used as controls. Plates were incubated for 1 h at 37°C before lysis with 1 μl of 10% Triton X-100, and Miller units were determined as described above. To calculate the value at which 50% of maximal activation is reached for each strain (50% effective concentration [EC50]), concentrations of sBlpC were converted to log values and Miller units were normalized by setting the top value of each sample at 100% and the bottom value at 0%. EC50s and 95% confidence intervals were calculated using Prism 5.0.
For quantification of BlpC in supernatants of stimulated chimeric strains, glycerol stocks of the PSD118 strain with and without htrA deletions were grown to an OD620 of 0.3 in 5 ml THY medium. sBlpC6A at the indicated concentrations was then added to the culture and allowed to incubate for 1 h at 37°C. Cells were removed by centrifugation at 5,000 × g for 5 min, and culture supernatant was sterile filtered using a 0.22-μm-pore-size syringe filter. Overall secretion of the pheromone was detected by the use of a blpC deletion reporter strain, PSD101. PSD101 was allowed to grow to an OD620 of 0.4, and 50 μl of filtered supernatant was added to 50 μl of the reporter strain in a 96-well plate. Unstimulated supernatant and blank medium were used as controls. Reporter strains plus filtered supernatant were allowed to incubate for 1 h at 37°C, and a β-galactosidase assay was performed as described above. To create a dose-response curve corresponding to sBlpCR6 data, 2-fold dilutions of sBlpCR6 were used in place of blank medium for PSD101 cells and assayed in parallel with the supernatants described above. A dose-response curve was generated after converting concentrations to log values and after transforming data by setting low and high values to 0% and 100%, respectively. BlpCR6 concentrations in supernatants were derived using this equation.
Western blot analysis of pneumococcal proteins.
Pneumococcal strains were grown in THY medium to the indicated optical density. Cells were then prepared as pellets and resuspended in CelLytic B lysis buffer (Sigma-Aldrich). Protein concentrations were determined following the manufacturer's specifications by the use of Micro BCA (Pierce). Equal amounts of protein were boiled in sample buffer for 5 min and then separated on Tris-HCl (4% to 15%) or 16% Tris-Tricine polyacrylamide gels. To evaluate stimulated FLAG constructs, cultures were grown to an OD620 of 0.2 and stimulated with 100 ng/ml of sBlpCR6. Samples were taken at the indicated time points, prepared as pellets, and lysed. To evaluate unstimulated FLAG-expressing constructs, strains were grown to an OD620 of 0.5 and 2 ml of cells was prepared as pellets and resuspended directly in 50 μl of loading buffer to obtain maximally concentrated samples. Gels were transferred to a 0.45 μM polyvinylidene difluoride (PVDF) membrane (Immobilon; Millipore). The membrane was blocked with Tris-buffered saline (TBS) and 5% nonfat dry milk for 1 h. The membrane was first probed with primary antibody and anti-FLAG M2 antibody (Stratagene) at a 1:2,000 dilution or HtrA polyclonal antiserum (11) at a 1:2,000 dilution overnight and then washed 3 times with TBS with 0.5% Tween 20. The membrane was then incubated with a horseradish peroxidase-conjugated anti-mouse secondary antibody (FLAG) or an anti-rabbit secondary antibody (HtrA). After washing, the membranes were incubated with SuperSignal West Pico chemiluminescent reagents (Pierce). For quantification of processed and unprocessed forms of epitope-tagged BlpC, images were obtained with a FluorChem M image processor (Protein Simple) and band quantification was performed using Alphaview software. Local background was subtracted from each band. Top and bottom bands were added and percentage values determined for each band as a fraction of the total signal.
Proteinase K treatment of FLAG-expressing contructs.
Strains were grown to an OD620 of 0.2 and induced with 100 ng/ml of BlpCR6 for 60 min at 37°C. Three separate 1-ml aliquots of each culture were prepared as pellets by centrifugation at 2,500 × g for 5 min. Pellets were resuspended in 100 μl of phosphate-buffered saline (PBS) alone or PBS with 0.2 mg/ml of proteinase K and incubated at 37°C for 15 min. One pellet of each strain was resuspended in CelLytic B lysis buffer (Sigma-Aldrich) prior to addition of proteinase K. After 15 min, phenylmethylsulfonyl fluoride (PMSF) was added to reach a final concentration of 5 mM to inhibit further digestion, unlysed samples were prepared as pellets, and pellets were resuspended in Tricine loading dye. Loading dye was added directly to lysed samples.
RESULTS
HtrA restricts activation of the blp locus at low cell densities in all three pherotypes.
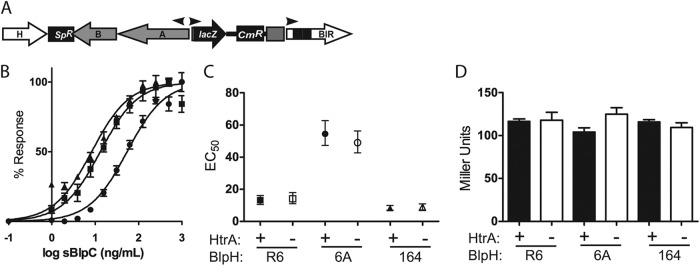
In order to assess the role of HtrA in strains with different pherotypes, we created a series of blp reporter strains based on the R6 laboratory strain that differed only by their blpHC sequences. These constructs were created in a derivative of the R6 laboratory strain in which the existing, nonfunctional blp locus was replaced with a functional locus derived from strain P764 (8). These strains contain a reporter plasmid insertion at the 5′ end of the BIR region that results in a lacZ gene fused to the promoter driving pneumocin and immunity protein production (Fig. 1B). Using allelic exchange, three reporter strains were created that contain the three most common BlpC types found in the pneumococcal population (BlpCR6, BlpC6A, and BlpC164) along with their corresponding BlpH alleles. Cultures were inoculated at a low OD620 of <0.005, and samples were taken every 30 min beginning at an OD620 of 0.05 to assess natural activation of the blp locus. Strains expressing the three different BlpC pherotypes had similar patterns of induction, with activation of BIR transcription occurring during the mid-exponential phase at an OD620 of between 0.4 and 0.5 (Fig. 1C to E). Each pherotype-specific strain had a distinct peak level of activation; the BlpC6A-expressing strain had a significantly lower maximal level of activation than the R6 and P164 pherotype strains (Fig. 1D). Unmarked deletions of htrA were introduced into each reporter strain by allelic exchange, and growth and activation curve analyses were repeated. Activation of the blp locus occurred at lower cell densities in strains expressing all three pherotypes, and peak levels of transcription were higher in the htrA deletion strains than in the matched wild-type (WT) strains (Fig. 1C to E). The BlpC6A pherotype strain with an htrA deletion showed significantly higher and faster transcription than the BlpC6A strain with wild-type htrA, but peak levels of activation were lower than those appreciated with the htrA mutants in the R6 and P164 pherotype strains.
HtrA is required for normal growth of S. pneumoniae under conditions of high incubation temperatures (10). In E. coli, proteolytic activity of the HtrA homologue, DegP, is enhanced at elevated temperatures whereas chaperone-like functions predominate at lower temperatures (17). To address the potential role of incubation temperature in the early induction seen with htrA mutants, we assessed the induction patterns of wild-type and htrA deletion strains in the BlpCR6 background at a low incubation temperature of 32°C. HtrA mutants had similar early and elevated levels of blp transcription compared with the wild-type strain at 37°C (Fig. 1F). To determine if HtrA protease activity was required for the delayed activation in strains expressing wild-type HtrA, the htrAS234A allele, containing a serine-to-alanine mutation of the active site serine residue, was introduced into the BlpCR6 reporter strain via allelic exchange. The reporter strain with the active site mutation had a phenotype of blp activation that was indistinguishable from that of the htrA deletion strain (Fig. 1C), demonstrating that HtrA proteolytic activity is required for the density-dependent control of blp locus activation.
We have shown previously that certain naturally occurring phenotypic variants of S. pneumoniae demonstrate growth-phase-dependent expression of HtrA (9). Opaque variants have a decrease in HtrA levels at the late exponential phase which is thought to explain their high levels of pneumocin activity compared with those of transparent variants in which HtrA levels are relatively constant. To determine if a decrease in HtrA expression at higher cell density was responsible for the activation of the locus in wild-type strains, HtrA levels were assessed throughout the growth phase by Western blot analysis using anti-HtrA antiserum (Fig. 1G). HtrA levels did not appreciably decrease during the exponential phase, suggesting that variations in the amount of HtrA produced do not contribute to the density-dependent activation of the locus in this strain background.
HtrA does not affect the response to exogenously added BlpC.
Evaluation of wild-type reporter strains demonstrated that, in the absence of htrA, initiation of BIR transcription occurs at a lower cell density. To address whether the HtrA-mediated control of blp activation could affect the response to environmental pheromone, we introduced a deletion in the blpC gene of each of the reporter strains and the corresponding htrA mutants (Fig. 2A). These constructs allowed us to perform accurate dose-response curve analyses with defined concentrations of sBlpC by eliminating the positive-feedback amplification of transcriptional activation due to BlpC secretion. Cultures at an OD620 of 0.1 were induced with increasing concentrations of the appropriate sBlpC, and transcriptional activity was assessed 1 h after stimulation. Dose-response curves were evaluated in each background and EC50s calculated (Fig. 2B and C). In each case, HtrA mutants had EC50s identical to those of their WT counterparts (Fig. 2C). The BlpC6A pherotype strains had a higher EC50 than the BlpCR6 and BlpC164 pherotype strain pairs, suggesting that the lower maximal activation levels that were seen in strains expressing the 6A pherotype under natural induction conditions were due to a dampened transcriptional response to pheromone that is independent of HtrA (Fig. 1E and 2B and C). When saturating concentrations of BlpC were added, however, all pherotypes reached similar levels of maximal transcriptional activity (Fig. 2D).
Fig 2.
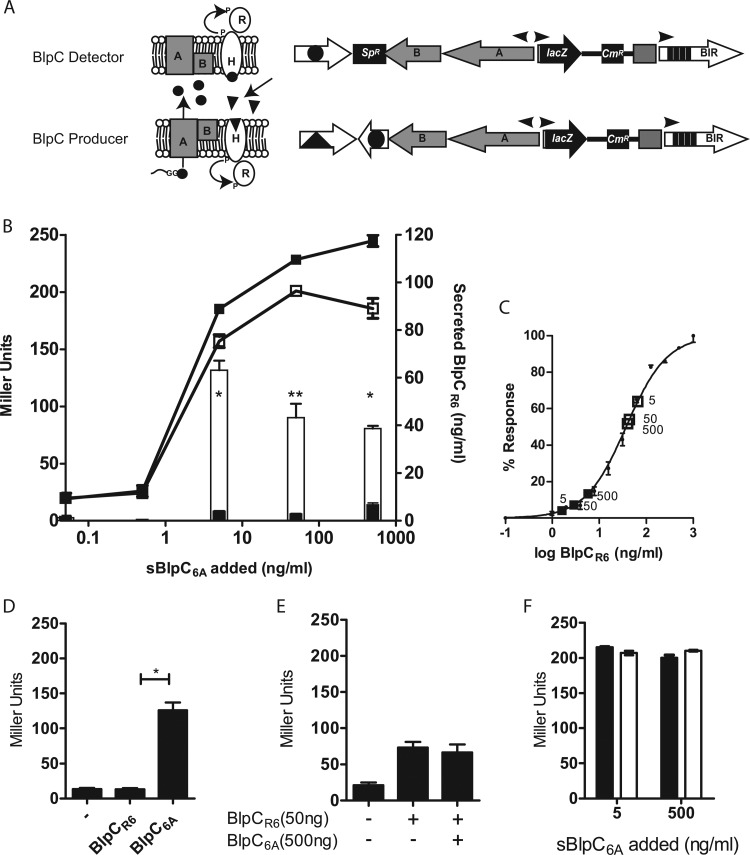
HtrA does not affect the dose-response curve corresponding to blp activation with respect to exogenous BlpC of the three most common pherotypes or the maximal level of induction. (A) Diagrammatic representation of the blp reporter strains with a deletion in the blpC gene. All three pherotype-responsive strains were tested separately. (B) BlpC knockout strains were stimulated with increasing concentrations of their cognate BlpC types. Triangles designate BlpC164 responsive, squares BlpCR6 responsive, and circles BlpC6A-responsive strains. (C) EC50s were computed from dose-response curves in all three pherotype-responsive strains by comparing strains with (closed symbols) and without (open symbols) HtrA expression. Bars denote 95% confidence intervals. (D) Miller units following induction with saturating concentrations (500 ng/ml) of sBlpC. The WT strains of all three pherotypes are indicated by black bars; the respective htrA mutants are indicated by white bars. Differences between wild-type and htrA-deficient strains were not significant by a Student t test.
HtrA affects the amount of BlpC that is secreted into the environment.
Because natural induction experiments suggested that HtrA suppresses activation of the blp locus at low cell densities, but pheromone knockout strains did not support a role for HtrA in degradation of exogenous peptide, we hypothesized that HtrA may impact the amount of secreted pheromone. To address this, we created a series of chimeric strains that would allow us to examine the role of HtrA in endogenous BlpC production alone. Strains were created that contain a mismatched blpH and blpC pair. This approach allowed us to activate the locus with sBlpC of one type and assess the amount of BlpC secreted into the culture supernatant of another type (Fig. 3A). This strategy eliminated the contribution of BlpC amplification of its own signal to the overall activation of the secreting strain. Using allelic exchange, we created the PSD118 strain. This strain was confirmed to respond to exogenous sBlpC6A by activation of the BIR promoter in a dose-dependent manner similar to that seen with our blpC deletion reporters (Fig. 3B). Endogenous BlpCR6 secretion by the chimeric strain was detected by assaying filtered supernatants of stimulated cultures for the presence of BlpCR6 using PSD101, a BlpCR6-responsive reporter strain containing a deletion in blpC. Concentrations of secreted BlpCR6 in the supernatant were determined by comparing levels of activation seen with cell-free supernatant to those seen with known concentrations of synthetic peptide (Fig. 3C). We confirmed that the chimeric strain does not respond to its own BlpCR6 peptide (Fig. 3D). We also confirmed that the presence of high levels of sBlpC6A in the supernatant does not interfere with the sensitivity of the BlpCR6 reporter strain to secreted BlpCR6 (Fig. 3E). In-frame unmarked htrA deletion mutations were introduced in the chimeric reporter strain to assess the role of HtrA in secreted pheromone levels. The chimeric blpH6A-blpCR6-containing strains with and without htrA were stimulated with 10-fold dilutions of sBlpC6A ranging from 0.5 to 500 ng/ml. BlpCR6 secretion could be detected in the media with concentrations of sBlpC6A that resulted in activation of the BIR promoter in the chimeric strain (5 to 500 ng/ml). Although levels of BIR transcription in the chimeric strains with and without htrA were induced by exogenous sBlpC6A to similar degrees, secreted BlpCR6 levels differed markedly between the two strains (Fig. 3B). Significantly more BlpCR6 was detected in the medium of the htrA mutant. Extrapolated BlpCR6 concentrations in the media demonstrated that over 90% of active BlpCR6 is eliminated by the presence of HtrA. Similar results were noted for the reverse chimeric strain pair that was engineered to respond to sBlpCR6 and secrete BlpC6A (data not shown).
Fig 3.
HtrA affects secretion of endogenous BlpC. (A) Diagrammatic representation of the proposed mechanisms and genetic compositions of the BlpC producer and detector strains used in this assay. PSD118, a reporter strain with a BlpC6A-responsive BlpH (designated by a triangle) but containing a blpCR6 allele (designated by a circle), was stimulated with exogenous sBlpC6A. Secretion of BlpCR6 into the medium by this strain was detected by the BlpCR6-specific, blpC knockout PSD101 strain. (B) Chimeric strain PSD118 (filled squares and bars) or the chimeric strain with a deletion in htrA, PSD120 (open squares and bars), was induced with increasing concentrations of sBlpC6A for 1 h. Stimulation of the chimeric strains with synthetic peptide was determined by a Miller assay (line graph). Secretion of endogenous BlpCR6 was assessed by exposing the BlpCR6-responsive reporter strain to filtered supernatant, and corresponding BlpCR6 concentrations were determined by interpolation based on the dose-response curve with respect to sBlpCR6 (bar graph). Asterisks denote differences between wild-type and ΔhtrA strains determined by a Student t test. *, P = 0.0001; **, P = 0.002. (C) Secreted BlpCR6 amounts for each of the three stimulatory concentrations of BlpC6A. Numbers placed next to boxes denote the concentrations of stimulatory sBlpC6A used. Secreted values after conversion to log values are shown as closed (wild-type) or open (ΔhtrA) squares. Small circles denote the dose-response curve with respect to the BlpCR6-responsive reporter for synthetic BlpCR6. (D) The specificity of the chimeric strain for BlpC6A was confirmed by performing a Miller assay on the chimeric strain after exposure to 500 ng/ml of BlpC6A or BlpCR6 for 1 h. *, P = 0.0006. (E) sBlpC6A does not interfere with detection of BlpCR6 by the BlpCR6-specific reporter strain. Response to the noted concentrations of BlpCR6 with and without the addition of a 10-fold excess of BlpC6A was measured by determining Miller units 1 h after stimulation. (F) Comparison of the responses of BlpCR6-specific reporter strains with (black bars) and without (open bars) a functional htrA gene to supernatant-derived BlpCR6 secreted by strain PSD120 following induction with 5 or 500 ng/ml of sBlpC6A. Values were not statistically different by a Student t test.
HtrA does not affect the response to extracellular, naturally produced BlpC.
Because the dose-response experiments on reporter strains with blpC disruptions were performed with synthetic BlpC, we could not address whether uncharacterized posttranslational modifications of the peptide pheromone that could be present in endogenously produced BlpC were responsible for the difference in wild-type and htrA deletion strains. To address this possibility, supernatants containing BlpCR6 derived from the stimulation of the chimeric strain with a mutation in htrA were used to stimulate the BlpCR6-responsive reporter strains with and without a deletion in htrA. No difference in the levels of responsiveness to nonsynthetic BlpCR6 was noted in comparisons of the two strains, demonstrating that HtrA does not affect the dose-response curve of naturally secreted BlpCR6 when it is exogenously applied (Fig. 3F).
HtrA decreases BlpC secretion by affecting ABC transporter processing following pheromone-mediated stimulation of the blp locus.
Our observation that HtrA decreases the amount of secreted peptide pheromone but does not act upon environmental peptides suggests that HtrA either was degrading newly synthesized peptides immediately upon transport out of the cell or was interfering with transport via disruption of ABC transporter function. To better address these possibilities, we created a pair of active reporter strains with and without htrA in which the blpCR6 gene had been replaced with a blpC gene encoding two FLAG tags at the C terminus of the peptide, resulting in production of BlpCFLAG. To demonstrate the role of BlpA in processing and secretion of the BlpCFLAG peptide, a BlpCFLAG-expressing strain was also made in the presence of an in-frame, unmarked deletion of the blpA gene. Miller assays performed on broth-grown cells without synthetic peptide addition did not show evidence of BlpCFLAG-mediated activation, suggesting that the epitope tag interferes with normal function of the pheromone (data not shown). To examine the role of HtrA during pheromone-mediated induction of the locus, cells were induced with sBlpCR6 and samples were taken at 0, 15, 30, and 60 min postinduction. BlpCFLAG populations in whole-cell lysates were assessed by Western blot analysis and BIR transcript levels determined by β-galactosidase assays. Strains induced with sBlpCR6 showed rapid accumulation of two forms of FLAG-tagged peptide with molecular weights (MW) consistent with the presence of a full-length prepeptide (preBlpCFLAG; predicted MW, 7.9) and a processed form of the peptide (BlpCFLAG; predicted MW, 5.2) (Fig. 4A). As expected, induced ΔblpA strains produce only the larger, preBlpCFLAG form, following induction due to the loss of BlpA-derived peptidase function. Despite identical levels of BIR transcription following induction (Fig. 4B), the strain lacking htrA produced more BlpCFLAG than the wild-type strain and less preBlpCFLAG. This difference was noted at 30 min and was particularly evident at 60 min postinduction, when the htrA mutant strain primarily contained the processed form of BlpCFLAG. Of note, although levels of BlpCFLAG increased over time in the wild-type and htrA mutant strains, there was no increase in signal noted in the preBlpCFLAG band produced by the ΔblpA strain, suggesting that the preBlpCFLAG peptide is relatively unstable compared with the processed form. To address the role of HtrA in the stability of preBlpCFLAG production, a strain was created with both blpA and htrA deletions. After 1 h of induction, this strain had an identical amount of preBlpC produced, demonstrating that HtrA is not involved in the apparent instability of preBlpCFLAG (Fig. 4C).
Fig 4.

Epitope-tagged BlpC processing is enhanced in htrA mutants under basal and stimulated conditions. Whole-cell lysates of R6 derivatives containing a BlpCR6 with two FLAG tags encoded on the C-terminal end of the peptide were compared for the presence of unprocessed and processed forms of BlpCFLAG. Strains used in these assays were BlpCFLAG-expressing strains with wild-type htrA (1 or closed circle), ΔhtrA (2 or open circle), ΔblpA (3 or open triangle), or ΔhtrA blpA (4) or a ΔblpC strain lacking the FLAG epitope (5). (A) Strains were stimulated for the indicated times with sBlpCR6. Cell lysates were separated on 16% Tricine gels, transferred, and probed with anti-FLAG antibody. Top (%preC) and bottom (%C) band relative densities were compared by determining the ratio of each band to the total density for the two bands combined. (B) Miller assay of the samples used as described for panel A following stimulation with sBlpCR6. (C) Western blot analysis of strains by the use of anti-FLAG antibody after 1 h of induction with sBlpCR6. (D) Western blot analysis of concentrated whole-cell lysates of unstimulated strains 1 to 3 and strain 5 grown to the late exponential phase. (E) Western blot analysis of induced FLAG-expressing strains following mock or proteinase K (Prot K) treatment on either whole cells or SDS-exposed lysates as indicated. Blots were probed with either anti-FLAG antibody or anti-HtrA antibody as noted to the right of each blot.
HtrA decreases BlpC secretion by affecting basal levels of ABC transporter processing in unstimulated cells.
To demonstrate the pattern of unstimulated accumulation of BlpC, reporter strains with the BlpCFLAG construct were grown to the late exponential phase. There was no difference in BIR transcript levels in uninduced strains with and without htrA deletions (data not shown). Uninduced wild-type cells contained primarily preBlpCFLAG, while htrA mutants had both processed and unprocessed forms present (Fig. 4D), suggesting that HtrA also interferes with peptide processing under basal expression conditions.
Processed epitope-tagged pheromones are not secreted.
Unlike the chimeric peptides, we were unable to detect BlpCFLAG in the media under any condition, including in the supernatant of induced HtrA mutants (data not shown), suggesting either that secretion is blocked by the presence of the epitope tags or that the peptides are secreted and either tightly adherent to the outer surface of the bacterium or rapidly degraded by a protease other than HtrA. To demonstrate that the processed form of the BlpCFLAG peptide seen in Western blots of whole-cell lysates remains largely in the intracellular compartment and would therefore not be accessible to extracellular HtrA, BlpCFLAG-expressing whole cells with either wild-type htrA or an htrA deletion were induced for 60 min with sBlpCR6 and then subjected to proteinase K digestion. Blots were first probed with anti-FLAG antibody and then stripped and reprobed with anti-HtrA antibody (Fig. 4E). Incubation with proteinase K substantially reduced the HtrA signal but did not significantly affect the amount of the processed form of BlpCFLAG, consistent with the majority of detected peptide remaining in an intracellular location.
HtrA activity with respect to BlpC accumulation is affected by the rate of miscoding errors.
Stevens et al. have recently shown that stimulation of pneumococcal competence is affected by the rate of accumulation of misfolded proteins that arise due to decoding errors (18). Stimulation of competence through secretion of the competence pheromone, CSP, is both blunted and delayed when the rate of misfolded proteins is low compared with stimulation in cells with a high rate of protein misfolding. In assessing the role of HtrA in control of competence in these different backgrounds, the authors found that HtrA blunts and delays the stimulation of competence only when levels of protein misfolding are low. The effect of HtrA on competence induction is significantly less dramatic when protein misfolding rates are high. The rpsLK56T streptomycin resistance allele (encoding a lysine-to-threonine change at position 56) that is present in our reporter strains has been shown to reduce the rate of ribosomal miscoding errors (18). To determine if low rates of miscoding in this background would impact the effect of htrA mutations on density-dependent stimulation of blp transcription, we assessed the activity of the BlpCR6-responsive reporters during growth in broth in matched low-error-rate rpsLK56T and medium-error-rate wild-type rpsL backgrounds (Fig. 5). Compared with the pair of rpsLK56T reporter strains, both the htrA-sufficient and -deficient strains with a wild-type rpsL allele had higher peak levels of blp transcription. Despite a higher peak in the medium-error-rate background in htrA-sufficient cells, the htrA deletion mutant in this background still demonstrated an earlier and higher peak in blp activation than the matched wild-type htrA control.
Fig 5.
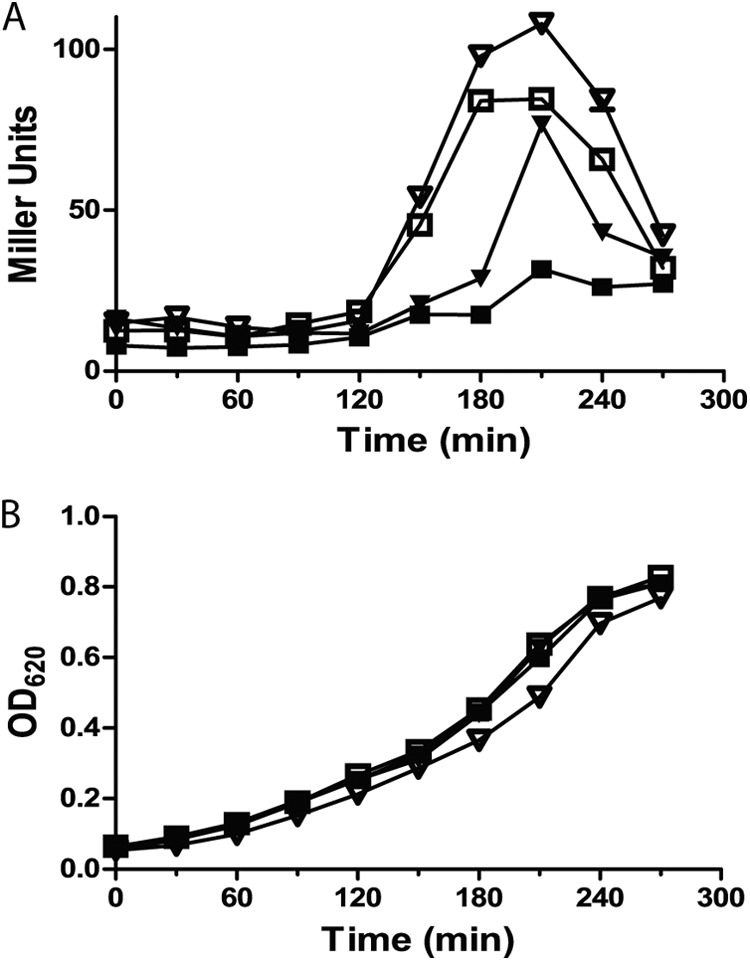
The rate of ribosomal miscoding affects the role of HtrA in the control of blp locus stimulation. Paired reporter strains were created with either the medium-error-rate wild-type rpsL streptomycin sensitivity (inverted triangles) or the low-error-rate rpsLK56T streptomycin resistance (squares) allele. Strains were tested for natural stimulation of the locus with (closed symbols) or without (open symbols) an intact htrA gene. Strains were grown in broth at 37°C and assayed every 30 min for β-galactosidase activity as quantified by Miller units (A) and OD620 (B).
DISCUSSION
HtrA is a surface-expressed serine protease that is highly conserved in S. pneumoniae. Levels of the protease are controlled by the CiaRH two-component regulatory system (19, 20). Although the specific trigger for Cia activation is unknown, ciaRH and htrA transcripts are induced during competence and thought to play a role in repression of the competence state (21). Similar to their role in competence, we had previously shown that both CiaRH and HtrA are involved in the repression of pneumocin expression (9). Naturally occurring opacity variants that differ only moderately in relative HtrA expression levels demonstrated clear phenotypic differences in pneumocin-mediated inhibition that were dependent on HtrA activity (9). In transparent variants with higher htrA expression, addition of sBlpC restored the defect in transcriptional activation of the locus but did not restore pneumocin-mediated inhibition. Only a deletion in htrA resulted in transparent strains that produced appreciable inhibitory activity (9).
Because this observation suggested that HtrA affects pheromone and pneumocin levels separately, we initially hypothesized that HtrA functioned by degrading both BlpC pheromones and pneumocins on the surface of the cell, decreasing their effective concentration. A similar mechanism of CSP degradation has been proposed for HtrA-mediated control of competence (13, 18). Our data do not support this hypothesis. If HtrA activity on the surface of the cell were to result in pheromone digestion, we would expect that strains expressing HtrA would require increased concentrations of BlpC to stimulate the blp locus. We have shown that the presence of HtrA does not affect the dose-response curve with respect to either synthetic or cell-derived pheromones when added exogenously to reporter strains that cannot secrete their own pheromone.
HtrA does, however, affect the amount of BlpC secreted into the supernatant. While the presence of an intact htrA gene in chimeric strains with mismatched blpH and blpC alleles did not affect stimulation of the reporter gene compared with htrA mutant strain results, strains expressing wild-type HtrA secreted less than 7% of the amount of BlpC secreted by htrA mutants following stimulation. Despite the significant reduction in secreted BlpC, accumulation of the pheromone at higher cell densities allows for sufficient concentrations of secreted BlpC to trigger activation of the locus. In this way, HtrA controls the density at which pneumocin expression can be stimulated and effectively delays this stimulation until a higher cell density is reached.
Based on the pattern of BlpC production in strains expressing the epitope-tagged pheromone, the mechanism of HtrA-mediated density control of blp activation appears to involve a functional disruption of the ABC transporter that participates in pheromone and pneumocin processing and secretion rather than direct degradation of the secreted peptides themselves. Under noninducing conditions, htrA mutants have evidence of BlpA-mediated processing of the BlpC signal sequence whereas wild-type strains primarily accumulate the unprocessed form of BlpC. Under induced conditions, htrA mutants produce more processed BlpC than wild-type strains, with a corresponding reduction in the unprocessed form. Because the epitope-tagged peptide was not secreted, it would not be accessible to HtrA. Hence, the decreased amount of processed BlpC detected in HtrA-sufficient cells compared with htrA mutants cannot be due to direct degradation of the peptide by HtrA. Although a small amount of HtrA protein remains detectable following proteinase K treatment, all known bacterial HtrA homologues function in the periplasm, on the surface of the cell, or in the secreted form (13, 22–25), making it unlikely that a small amount of intracellular HtrA could account for the specific decrease in processed BlpC noted only in wild-type strains.
Our data support the hypothesis that, prior to stimulation, HtrA affects the amount of BlpC signal sequence cleavage. Based on homology to other peptide pheromone-controlled systems, proteolytic processing of the BlpC signal sequence is thought to occur during transport by the BlpAB complex (26). The effect of HtrA on pheromone processing is presumably due to an impact on either the quantity or function of BlpAB. After pheromone-initiated stimulation of the locus, htrA mutants process pre-BlpC more effectively than wild-type strains. Because blpA, blpB, and blpC are all located on the same operon (5) and, thus, production of both the transporter and pheromone would be expected to be upregulated following stimulation to the same degree, BlpC-mediated stimulation of this transcript can at least partially overwhelm the effect of HtrA on BlpC transport. We hypothesize that HtrA-disrupted ABC transporters that accumulate in the preinduction period are replaced following stimulation, allowing partial restoration of peptide and pneumocin processing and secretion.
To address whether HtrA interferes with the integrity of BlpAB and to determine if HtrA and BlpAB are colocalized on the cell surface, we have attempted to raise antibodies specific for BlpA or BlpB using both purified fragments of the proteins (BlpA and BlpB) and epitope tags (BlpA). Unfortunately, none of the antibodies were able to detect BlpA or BlpB in either the wild-type or htrA mutant strain background. Although data from the active site mutant of HtrA demonstrate that protease activity of HtrA is required for control of BlpC secretion, because we are not able to detect BlpAB protein levels, we cannot address whether the effect of HtrA is direct (via degradation of either BlpA or BlpB) or indirect (via disruption of proteins that stabilize BlpAB expression or function). The BlpA transporter is a transmembrane protein in which both N- and C-terminal domains are predicted to be intracellular, leaving very little of this protein accessible to HtrA (27). In contrast, a large portion of the single transmembrane domain protein of BlpB is predicted to be outside the cell based on its homology to other accessory proteins, possibly implicating this protein as a target for HtrA protease activity (28).
Recently, Cassone et al. demonstrated that HtrA degrades CSP in vitro, using purified recombinant HtrA and synthetic CSP, and in vivo by demonstrating that cleavage of exogenously applied fluorescently labeled peptide occurred only in HtrA-sufficient cells (13). We cannot exclude the possibility that HtrA also degrades BlpC to some extent. In fact, degradation of synthetic forms of BlpC by purified recombinant HtrA can be demonstrated in vitro (data not shown); however, our in vivo data do not support the idea that this mechanism is the primary means by which HtrA decreases pneumocin expression. Similarly, although Cassone et al. showed that HtrA can degrade CSP, they did not demonstrate that extracellular degradation of CSP was responsible for the effects of HtrA on competence induction. To our knowledge, the amount of endogenous CSP secretion in HtrA-sufficient and -deficient cells has not been specifically assessed. It is interesting that HtrA plays such strikingly similar roles in the control of competence and pneumocin production in S. pneumoniae. Given the homology between the Com and Blp transporter proteins at the amino acid level, it is possible that HtrA controls competence, at least in part, via a similar mechanism of disruption of ComAB function.
As we have shown previously, small alterations in HtrA expression such as those found in opacity variants can dramatically change the culture density required for stimulation of the blp locus, allowing tight control of this locus under a variety of environmental conditions (9). Even when expression levels of HtrA are stable, HtrA activity for specific target proteins can be altered according to the availability of alternative substrates. Stevens et al. have recently demonstrated that stimulation of competence in S. pneumoniae is affected by the accumulation of misfolded proteins that arise under antibiotic stress conditions due to decoding errors (18). This effect is mediated by HtrA activity. The authors hypothesize that the presence of alternative substrates for HtrA in the form of misfolded proteins results in increased competence because the relative effect of HtrA on the com system is diminished. There is a similar correlation between the rate of ribosomal miscoding and the effect of HtrA on BlpC pheromone stimulation. High-stress conditions that result in accumulation of misfolded proteins decrease the effect of HtrA on both pneumocin and competence induction, allowing both pathways to induce at lower cell densities.
In addition to controlling density-dependent stimulation of the blp locus by HtrA, it appears that some strains regulate the activity of the blp locus by altering the responsiveness of the BlpR-BlpH two-component system to BlpC. The type 6A BlpC-expressing strains had a markedly dampened response to endogenous and exogenous BlpC compared with other pherotype-expressing strains. This effect was independent of HtrA activity because htrA mutants grown under natural inducing conditions did not reach the same peak of transcriptional activation and pheromone knockout strains had a significantly higher EC50 than in the R6- and 164-responsive strains. The 6A-responsive strains could achieve levels of transcriptional activation similar to those seen with other pherotypes with saturating concentrations of BlpC, suggesting that the difference was not due to an alteration downstream of BlpC/BlpH binding but was a result of decreased affinity of BlpH for BlpC. In this way, increased concentrations of BlpC are required to initiate transcription of the locus. This represents an alternative strategy for control of the locus and may explain why, in our previous work, we were unable to detect activation of a pherotype 6A locus in an encapsulated isolate under conditions of broth growth (9).
In this work, we demonstrated that HtrA affects BlpC secretion but does not alter the dose-response curve with respect to exogenous peptide pheromone when strains lack the capacity to secrete their own pheromone. This is an important distinction, because the density-dependent regulation of pheromone secretion does not interfere with the ability of the organism to detect environmental signals from either other members of the population or competitors, allowing an appropriate response to pheromone signaling. Pneumocin expression is presumably energetically costly enough to restrict expression solely to high-density culture conditions, when the number of organisms would be sufficient to produce inhibitory quantities of pneumocins; however, sensing of competitors must occur at low cell density so that the production of immunity proteins can be stimulated. This is particularly true in strains that lack the ability to stimulate their own pneumocin locus due to a frameshift mutation in blpA. HtrA activity restricts only self-signaling but does not affect pheromone sensing, effectively controlling the density at which activation can occur without interfering with the induction of protective immunity.
ACKNOWLEDGMENTS
This work was supported by the Elizabeth E. Kennedy Research Award and grant AI078538 funded by the National Institute of Allergy and Infectious Diseases.
We thank Jeffrey Weiser and Michael Sebert for their advice and critical reading of an early version of the manuscript.
Footnotes
Published ahead of print 25 January 2013
REFERENCES
- 1. Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, Pichon B, Baker S, Parry CM, Lambertsen LM, Shahinas D, Pillai DR, Mitchell TJ, Dougan G, Tomasz A, Klugman KP, Parkhill J, Hanage WP, Bentley SD. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331:430–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hiller NL, Ahmed A, Powell E, Martin DP, Eutsey R, Earl J, Janto B, Boissy RJ, Hogg J, Barbadora K, Sampath R, Lonergan S, Post JC, Hu FZ, Ehrlich GD. Generation of genic diversity among Streptococcus pneumoniae strains via horizontal gene transfer during a chronic polyclonal pediatric infection. PLoS Pathog. 6:e1001108 doi:10.1371/journal.ppat.1001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hiller NL, Janto B, Hogg JS, Boissy R, Yu S, Powell E, Keefe R, Ehrlich NE, Shen K, Hayes J, Barbadora K, Klimke W, Dernovoy D, Tatusova T, Parkhill J, Bentley SD, Post JC, Ehrlich GD, Hu FZ. 2007. Comparative genomic analyses of seventeen Streptococcus pneumoniae strains: insights into the pneumococcal supragenome. J. Bacteriol. 189:8186–8195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dawid S, Roche AM, Weiser JN. 2007. The blp bacteriocins of Streptococcus pneumoniae mediate intraspecies competition both in vitro and in vivo. Infect. Immun. 75:443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. de Saizieu A, Gardes C, Flint N, Wagner C, Kamber M, Mitchell TJ, Keck W, Amrein KE, Lange R. 2000. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J. Bacteriol. 182:4696–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lux T, Nuhn M, Hakenbeck R, Reichmann P. 2007. Diversity of bacteriocins and activity spectrum in Streptococcus pneumoniae. J. Bacteriol. 189:7741–7751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reichmann P, Hakenbeck R. 2000. Allelic variation in a peptide-inducible two-component system of Streptococcus pneumoniae. FEMS Microbiol. Lett. 190:231–236 [DOI] [PubMed] [Google Scholar]
- 8. Son MR, Shchepetov M, Adrian PV, Madhi SA, de Gouveia L, von Gottberg A, Klugman KP, Weiser JN, Dawid S. 2011. Conserved mutations in the pneumococcal bacteriocin transporter gene, blpA, result in a complex population consisting of producers and cheaters. mBio 2:e00179–11 doi:10.1128/mBio.00179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dawid S, Sebert ME, Weiser JN. 2009. Bacteriocin activity of Streptococcus pneumoniae is controlled by the serine protease HtrA via posttranscriptional regulation. J. Bacteriol. 191:1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ibrahim YM, Kerr AR, McCluskey J, Mitchell TJ. 2004. Role of HtrA in the virulence and competence of Streptococcus pneumoniae. Infect. Immun. 72:3584–3591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sebert ME, Patel KP, Plotnick M, Weiser JN. 2005. Pneumococcal HtrA protease mediates inhibition of competence by the CiaRH two-component signaling system. J. Bacteriol. 187:3969–3979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ibrahim YM, Kerr AR, McCluskey J, Mitchell TJ. 2004. Control of virulence by the two-component system CiaR/H is mediated via HtrA, a major virulence factor of Streptococcus pneumoniae. J. Bacteriol. 186:5258–5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cassone M, Gagne AL, Spruce LA, Seeholzer SH, Sebert ME. 2012. The HtrA protease from Streptococcus pneumoniae digests both denatured proteins and the competence-stimulating peptide. J. Biol. Chem. 287:38449–38459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sung CK, Li H, Claverys JP, Morrison DA. 2001. An rpsL cassette, Janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 67:5190–5196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lacks S, Hotchkiss RD. 1960. A study of the genetic material determining an enzyme in Pneumococcus. Biochim. Biophys. Acta 39:508–518 [DOI] [PubMed] [Google Scholar]
- 16. Miller J. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 17. Spiess C, Beil A, Ehrmann M. 1999. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97:339–347 [DOI] [PubMed] [Google Scholar]
- 18. Stevens KE, Chang D, Zwack EE, Sebert ME. 2011. Competence in Streptococcus pneumoniae is regulated by the rate of ribosomal decoding errors. mBio 2:e00071–11 doi:10.1128/mBio.00071-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sebert ME, Palmer LM, Rosenberg M, Weiser JN. 2002. Microarray-based identification of htrA, a Streptococcus pneumoniae gene that is regulated by the CiaRH two-component system and contributes to nasopharyngeal colonization. Infect. Immun. 70:4059–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zähner D, Kaminski K, van der Linden M, Mascher T, Meral M, Hakenbeck R. 2002. The ciaR/ciaH regulatory network of Streptococcus pneumoniae. J. Mol. Microbiol. Biotechnol. 4:211–216 [PubMed] [Google Scholar]
- 21. Peterson SN, Sung CK, Cline R, Desai BV, Snesrud EC, Luo P, Walling J, Li H, Mintz M, Tsegaye G, Burr PC, Do Y, Ahn S, Gilbert J, Fleischmann RD, Morrison DA. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 51:1051–1070 [DOI] [PubMed] [Google Scholar]
- 22. Poquet I, Saint V, Seznec E, Simoes N, Bolotin A, Gruss A. 2000. HtrA is the unique surface housekeeping protease in Lactococcus lactis and is required for natural protein processing. Mol. Microbiol. 35:1042–1051 [DOI] [PubMed] [Google Scholar]
- 23. Kim KI, Park SC, Kang SH, Cheong GW, Chung CH. 1999. Selective degradation of unfolded proteins by the self-compartmentalizing HtrA protease, a periplasmic heat shock protein in Escherichia coli. J. Mol. Biol. 294:1363–1374 [DOI] [PubMed] [Google Scholar]
- 24. Rosch JW, Caparon MG. 2005. The ExPortal: an organelle dedicated to the biogenesis of secreted proteins in Streptococcus pyogenes. Mol. Microbiol. 58:959–968 [DOI] [PubMed] [Google Scholar]
- 25. Hoy B, Geppert T, Boehm M, Reisen F, Plattner P, Gadermaier G, Sewald N, Ferreira F, Briza P, Schneider G, Backert S, Wessler S. 2012. Distinct roles of secreted HtrA proteases from gram-negative pathogens in cleaving the junctional protein and tumor suppressor E-cadherin. J. Biol. Chem. 287:10115–10120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nes IF, Diep DB, Havarstein LS, Brurberg MB, Eijsink V, Holo H. 1996. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Van Leeuwenhoek 70:113–128 [DOI] [PubMed] [Google Scholar]
- 27. Franke CM, Tiemersma J, Venema G, Kok J. 1999. Membrane topology of the lactococcal bacteriocin ATP-binding cassette transporter protein LcnC. Involvement of LcnC in lactococcin A maturation. J. Biol. Chem. 274:8484–8490 [DOI] [PubMed] [Google Scholar]
- 28. Franke CM, Leenhouts KJ, Haandrikman AJ, Kok J, Venema G, Venema K. 1996. Topology of LcnD, a protein implicated in the transport of bacteriocins from Lactococcus lactis. J. Bacteriol. 178:1766–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tiraby JG, Fox MS. 1973. Marker discrimination in transformation and mutation of pneumococcus. Proc. Natl. Acad. Sci. U. S. A. 70:3541–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Avery OT, MacLeod CM, McCarty M. 1979. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Inductions of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J. Exp. Med. 149:297–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim JO, Weiser JN. 1998. Association of intrastrain phase variation in quantity of capsular polysaccharide and teichoic acid with the virulence of Streptococcus pneumoniae. J. Infect. Dis. 177:368–377 [DOI] [PubMed] [Google Scholar]
- 32. King SJ, Hippe KR, Gould JM, Bae D, Peterson S, Cline RT, Fasching C, Janoff EN, Weiser JN. 2004. Phase variable desialylation of host proteins that bind to Streptococcus pneumoniae in vivo and protect the airway. Mol. Microbiol. 54:159–171 [DOI] [PubMed] [Google Scholar]
- 33. Claverys JP, Dintilhac A, Pestova EV, Martin B, Morrison DA. 1995. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene 164:123–128 [DOI] [PubMed] [Google Scholar]