Abstract
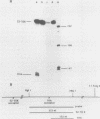
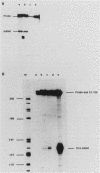
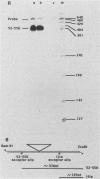
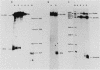
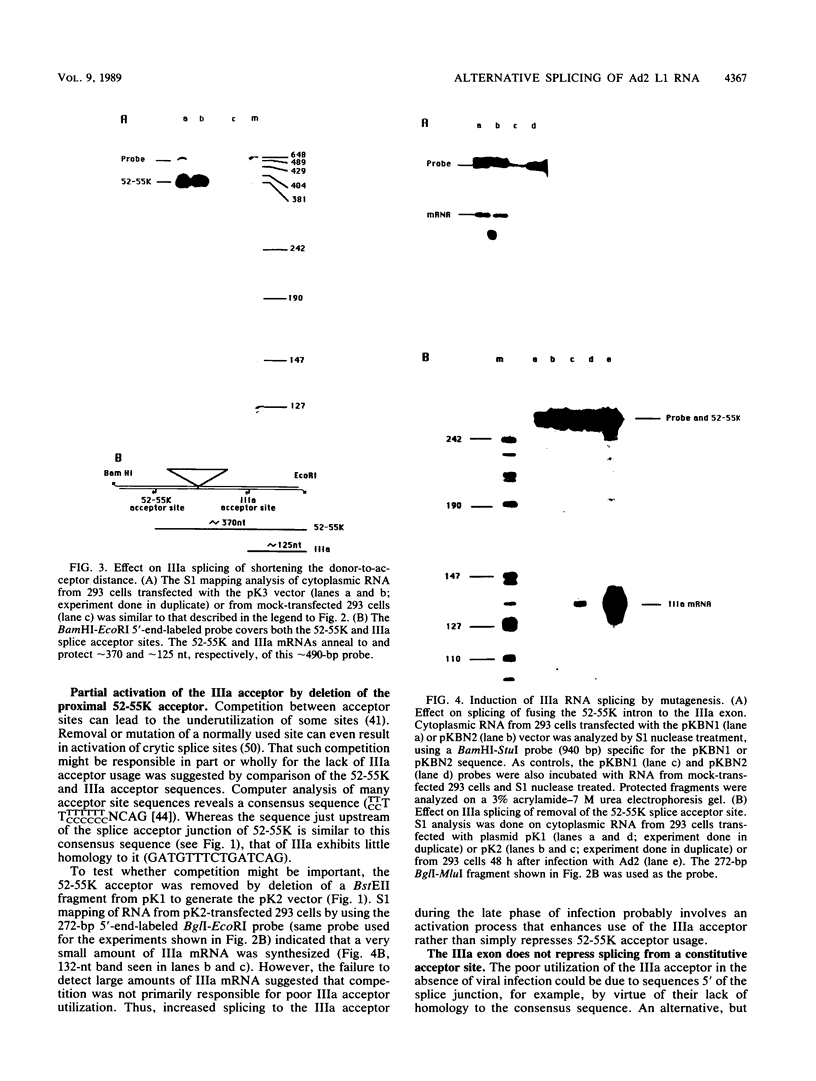
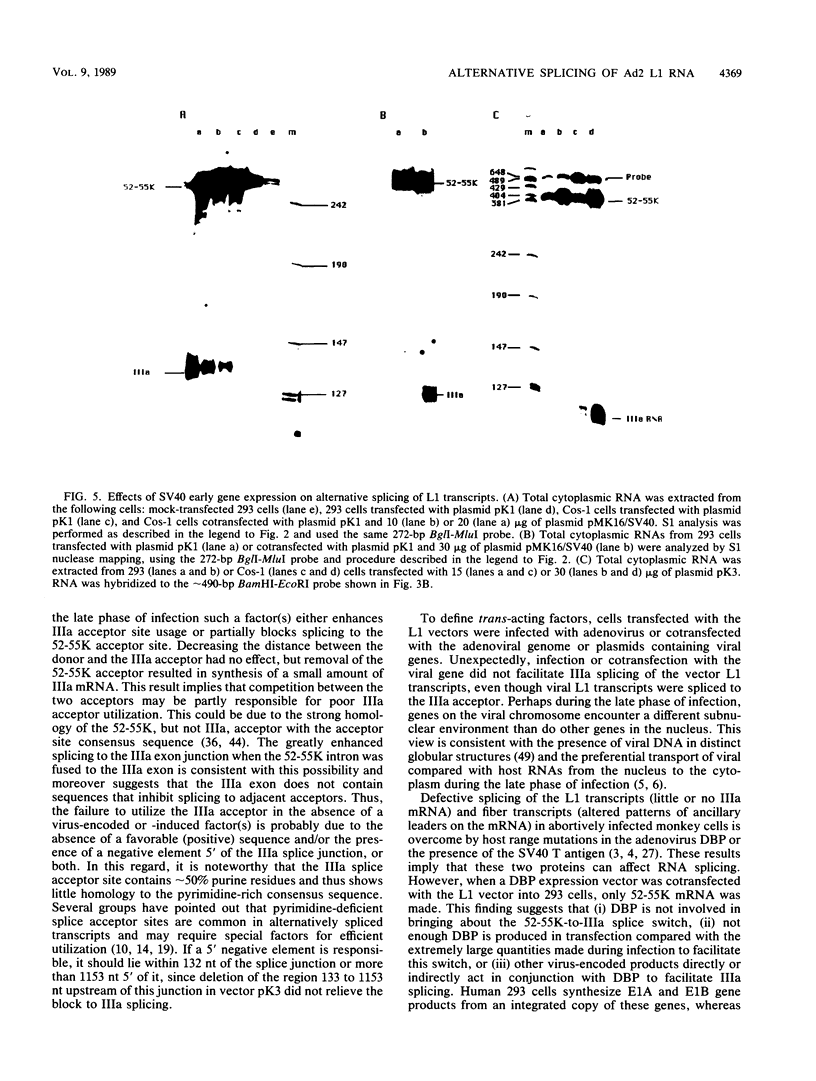
Expression of the L1 region of adenovirus is temporally regulated by alternative splicing to yield two major RNAs encoding the 52- to 55-kilodalton (52-55K) and IIIa polypeptides. The distal acceptor site (IIIa) is utilized only during the late phase of infection, whereas the proximal site (52-55K) is used at both early and late times. Several parameters that might affect this alternative splicing were tested by using expression vectors carrying the L1 region or mutated versions of it. In the absence of a virus-encoded or -induced factor(s), only the 52-55K acceptor was used. Decreasing the distance between the donor and the IIIa acceptor had no effect. Removal of the 52-55K acceptor induced IIIa splicing slightly, implying competition between the two acceptors. Fusion of the IIIa exon to the 52-55K intron greatly enhanced splicing of the IIIa junction, suggesting that the IIIa exon does not contain sequences that inhibit splicing. Thus, the lack of splicing to the IIIa acceptor in the absence of a virus-encoded or -induced factor(s) is probably due to the absence of a favorable sequence and/or the presence of a negative element 5' of the IIIa splice junction, or both. The presence of several adenovirus gene products, including VA RNAs, the E2A DNA-binding protein, and the products of E1A and E1B genes, did not facilitate use of the IIIa acceptor. In contrast, the simian virus 40 early proteins, probably large T antigen, induced IIIa splicing. This result, together with those of earlier studies, suggest that T antigen plays a role in modulation of alternative RNA splicing.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akusjärvi G. Anatomy of region L1 from adenovirus type 2. J Virol. 1985 Dec;56(3):879–886. doi: 10.1128/jvi.56.3.879-886.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akusjärvi G., Persson H. Controls of RNA splicing and termination in the major late adenovirus transcription unit. Nature. 1981 Jul 30;292(5822):420–426. doi: 10.1038/292420a0. [DOI] [PubMed] [Google Scholar]
- Anderson C. W., Hardy M. M., Lewis J. B. Abnormal expression of a late gene family L1 protein in monkey cells abortively infected with adenovirus type 2. Virus Genes. 1988 Mar;1(2):149–164. doi: 10.1007/BF00555934. [DOI] [PubMed] [Google Scholar]
- Anderson K. P., Klessig D. F. Altered mRNA splicing in monkey cells abortively infected with human adenovirus may be responsible for inefficient synthesis of the virion fiber polypeptide. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4023–4027. doi: 10.1073/pnas.81.13.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babich A., Feldman L. T., Nevins J. R., Darnell J. E., Jr, Weinberger C. Effect of adenovirus on metabolism of specific host mRNAs: transport control and specific translational discrimination. Mol Cell Biol. 1983 Jul;3(7):1212–1221. doi: 10.1128/mcb.3.7.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltz G. A., Flint S. J. Inhibition of HeLa cell protein synthesis during adenovirus infection. Restriction of cellular messenger RNA sequences to the nucleus. J Mol Biol. 1979 Jun 25;131(2):353–373. doi: 10.1016/0022-2836(79)90081-0. [DOI] [PubMed] [Google Scholar]
- Benyajati C., Spoerel N., Haymerle H., Ashburner M. The messenger RNA for alcohol dehydrogenase in Drosophila melanogaster differs in its 5' end in different developmental stages. Cell. 1983 May;33(1):125–133. doi: 10.1016/0092-8674(83)90341-0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Structure of the adenovirus 2 early mRNAs. Cell. 1978 Jul;14(3):695–711. doi: 10.1016/0092-8674(78)90252-0. [DOI] [PubMed] [Google Scholar]
- Bermingham J. R., Jr, Scott M. P. Developmentally regulated alternative splicing of transcripts from the Drosophila homeotic gene Antennapedia can produce four different proteins. EMBO J. 1988 Oct;7(10):3211–3222. doi: 10.1002/j.1460-2075.1988.tb03188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein S. I., Hansen C. J., Becker K. D., Wassenberg D. R., 2nd, Roche E. S., Donady J. J., Emerson C. P., Jr Alternative RNA splicing generates transcripts encoding a thorax-specific isoform of Drosophila melanogaster myosin heavy chain. Mol Cell Biol. 1986 Jul;6(7):2511–2519. doi: 10.1128/mcb.6.7.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart R. E., Andreadis A., Nadal-Ginard B. Alternative splicing: a ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu Rev Biochem. 1987;56:467–495. doi: 10.1146/annurev.bi.56.070187.002343. [DOI] [PubMed] [Google Scholar]
- Brough D. E., Rice S. A., Sell S., Klessig D. F. Restricted changes in the adenovirus DNA-binding protein that lead to extended host range or temperature-sensitive phenotypes. J Virol. 1985 Jul;55(1):206–212. doi: 10.1128/jvi.55.1.206-212.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtis K. C., Baker B. S. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell. 1989 Mar 24;56(6):997–1010. doi: 10.1016/0092-8674(89)90633-8. [DOI] [PubMed] [Google Scholar]
- Chow L. T., Broker T. R., Lewis J. B. Complex splicing patterns of RNAs from the early regions of adenovirus-2. J Mol Biol. 1979 Oct 25;134(2):265–303. doi: 10.1016/0022-2836(79)90036-6. [DOI] [PubMed] [Google Scholar]
- Forbes D. J., Kirschner M. W., Caput D., Dahlberg J. E., Lund E. Differential expression of multiple U1 small nuclear RNAs in oocytes and embryos of Xenopus laevis. Cell. 1984 Oct;38(3):681–689. doi: 10.1016/0092-8674(84)90263-0. [DOI] [PubMed] [Google Scholar]
- Fu X. Y., Colgan J. D., Manley J. L. Multiple cis-acting sequence elements are required for efficient splicing of simian virus 40 small-t antigen pre-mRNA. Mol Cell Biol. 1988 Sep;8(9):3582–3590. doi: 10.1128/mcb.8.9.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X. Y., Ge H., Manley J. L. The role of the polypyrimidine stretch at the SV40 early pre-mRNA 3' splice site in alternative splicing. EMBO J. 1988 Mar;7(3):809–817. doi: 10.1002/j.1460-2075.1988.tb02879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X. Y., Manley J. L. Factors influencing alternative splice site utilization in vivo. Mol Cell Biol. 1987 Feb;7(2):738–748. doi: 10.1128/mcb.7.2.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Gluzman Y., Sambrook J. F., Frisque R. J. Expression of early genes of origin-defective mutants of simian virus 40. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3898–3902. doi: 10.1073/pnas.77.7.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., Smiley J., Russell W. C., Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977 Jul;36(1):59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- Green M. R. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- Helfman D. M., Cheley S., Kuismanen E., Finn L. A., Yamawaki-Kataoka Y. Nonmuscle and muscle tropomyosin isoforms are expressed from a single gene by alternative RNA splicing and polyadenylation. Mol Cell Biol. 1986 Nov;6(11):3582–3595. doi: 10.1128/mcb.6.11.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman D. M., Ricci W. M., Finn L. A. Alternative splicing of tropomyosin pre-mRNAs in vitro and in vivo. Genes Dev. 1988 Dec;2(12A):1627–1638. doi: 10.1101/gad.2.12a.1627. [DOI] [PubMed] [Google Scholar]
- Johnston J. M., Anderson K. P., Klessig D. F. Partial block to transcription of human adenovirus type 2 late genes in abortively infected monkey cells. J Virol. 1985 Nov;56(2):378–385. doi: 10.1128/jvi.56.2.378-385.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman R. J. Identification of the components necessary for adenovirus translational control and their utilization in cDNA expression vectors. Proc Natl Acad Sci U S A. 1985 Feb;82(3):689–693. doi: 10.1073/pnas.82.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klessig D. F., Chow L. T. Incomplete splicing and deficient accumulation of the fiber messenger RNA in monkey cells infected by human adenovirus type 2. J Mol Biol. 1980 May 15;139(2):221–242. doi: 10.1016/0022-2836(80)90306-x. [DOI] [PubMed] [Google Scholar]
- Klessig D. F., Grodzicker T. Mutations that allow human Ad2 and Ad5 to express late genes in monkey cells map in the viral gene encoding the 72K DNA binding protein. Cell. 1979 Aug;17(4):957–966. doi: 10.1016/0092-8674(79)90335-0. [DOI] [PubMed] [Google Scholar]
- Leff S. E., Evans R. M., Rosenfeld M. G. Splice commitment dictates neuron-specific alternative RNA processing in calcitonin/CGRP gene expression. Cell. 1987 Feb 13;48(3):517–524. doi: 10.1016/0092-8674(87)90202-9. [DOI] [PubMed] [Google Scholar]
- Leff S. E., Rosenfeld M. G., Evans R. M. Complex transcriptional units: diversity in gene expression by alternative RNA processing. Annu Rev Biochem. 1986;55:1091–1117. doi: 10.1146/annurev.bi.55.070186.005303. [DOI] [PubMed] [Google Scholar]
- Marsh J. L., Erfle M., Wykes E. J. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene. 1984 Dec;32(3):481–485. doi: 10.1016/0378-1119(84)90022-2. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins J. R., Wilson M. C. Regulation of adenovirus-2 gene expression at the level of transcriptional termination and RNA processing. Nature. 1981 Mar 12;290(5802):113–118. doi: 10.1038/290113a0. [DOI] [PubMed] [Google Scholar]
- Noble J. C., Prives C., Manley J. L. Alternative splicing of SV40 early pre-mRNA is determined by branch site selection. Genes Dev. 1988 Nov;2(11):1460–1475. doi: 10.1101/gad.2.11.1460. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986 Aug 29;46(5):681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro M. B., Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987 Sep 11;15(17):7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw A. R., Ziff E. B. Transcripts from the adenovirus-2 major late promoter yield a single early family of 3' coterminal mRNAs and five late families. Cell. 1980 Dec;22(3):905–916. doi: 10.1016/0092-8674(80)90568-1. [DOI] [PubMed] [Google Scholar]
- Somasekhar M. B., Mertz J. E. Exon mutations that affect the choice of splice sites used in processing the SV40 late transcripts. Nucleic Acids Res. 1985 Aug 12;13(15):5591–5609. doi: 10.1093/nar/13.15.5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Voelkerding K., Klessig D. F. Identification of two nuclear subclasses of the adenovirus type 5-encoded DNA-binding protein. J Virol. 1986 Nov;60(2):353–362. doi: 10.1128/jvi.60.2.353-362.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieringa B., Hofer E., Weissmann C. A minimal intron length but no specific internal sequence is required for splicing the large rabbit beta-globin intron. Cell. 1984 Jul;37(3):915–925. doi: 10.1016/0092-8674(84)90426-4. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yen T. J., Gay D. A., Pachter J. S., Cleveland D. W. Autoregulated changes in stability of polyribosome-bound beta-tubulin mRNAs are specified by the first 13 translated nucleotides. Mol Cell Biol. 1988 Mar;8(3):1224–1235. doi: 10.1128/mcb.8.3.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziff E. B. Transcription and RNA processing by the DNA tumour viruses. Nature. 1980 Oct 9;287(5782):491–499. doi: 10.1038/287491a0. [DOI] [PubMed] [Google Scholar]