Abstract
Microorganisms can be engineered to produce useful products, including chemicals and fuels from sugars derived from renewable feedstocks, such as plant biomass. An alternative method is to use low potential reducing power from nonbiomass sources, such as hydrogen gas or electricity, to reduce carbon dioxide directly into products. This approach circumvents the overall low efficiency of photosynthesis and the production of sugar intermediates. Although significant advances have been made in manipulating microorganisms to produce useful products from organic substrates, engineering them to use carbon dioxide and hydrogen gas has not been reported. Herein, we describe a unique temperature-dependent approach that confers on a microorganism (the archaeon Pyrococcus furiosus, which grows optimally on carbohydrates at 100°C) the capacity to use carbon dioxide, a reaction that it does not accomplish naturally. This was achieved by the heterologous expression of five genes of the carbon fixation cycle of the archaeon Metallosphaera sedula, which grows autotrophically at 73°C. The engineered P. furiosus strain is able to use hydrogen gas and incorporate carbon dioxide into 3-hydroxypropionic acid, one of the top 12 industrial chemical building blocks. The reaction can be accomplished by cell-free extracts and by whole cells of the recombinant P. furiosus strain. Moreover, it is carried out some 30°C below the optimal growth temperature of the organism in conditions that support only minimal growth but maintain sufficient metabolic activity to sustain the production of 3-hydroxypropionate. The approach described here can be expanded to produce important organic chemicals, all through biological activation of carbon dioxide.
Keywords: anaerobe, archaea, biotechnology, metabolic engineering, thermophile
Metabolically engineered microorganisms can be used to produce a variety of products, ranging from bulk chemicals and fuels to complex pharmaceutical molecules. The largest effort in biofuel production is currently based on renewable plant biomass (1–3). First-generation biofuels include ethanol from corn fermentation and fatty acid methyl esters from oils and fats; second-generation biofuels use cellulosic biomass as feedstocks and can generate higher alcohols (4). An alternative method for the microbial production of both fuels and chemicals is to use low potential reducing power from sources such as hydrogen gas, reduced metals, or electricity to reduce carbon dioxide directly to useful products. This circumvents the overall low efficiency experienced in generating both plant and algal photosynthetic products (5). Moreover, such electron sources can potentially be used to reduce carbon dioxide directly to produce liquid fuels or “electrofuels” (6) or to produce industrial chemicals without a sugar intermediate. However, although significant advances have been made in manipulating microorganisms to produce various fuels from organic substrates (4, 7, 8), the engineering of microorganisms to use carbon dioxide and hydrogen gas has not been reported.
We have developed a unique temperature-dependent approach (9) to confer on a microorganism that cannot naturally use carbon dioxide but that grows on sugars optimally at 100°C the capacity to use carbon dioxide near 70°C. Hydrogen gas is used as the reductant to incorporate the carbon of carbon dioxide to produce 3-hydroxypropionic acid (3-HP), which is one of the top 12 industrial chemical building blocks used in the production of acrylic acid, acrylamide, and 1,3-propanediol (10, 11). Furthermore, the metabolic burden of the engineered microorganism during chemical production from hydrogen and carbon dioxide is minimized by its strategic operation at temperatures that are suboptimal for its growth.
The hyperthermophilic archaeon Pyrococcus furiosus is an obligate heterotroph that grows optimally (Topt) at 100°C by fermenting sugars to hydrogen, carbon dioxide, and acetate (12). It cannot use carbon dioxide as its sole carbon source. A genetic system is available for P. furiosus based on a competent strain with a known sequence (13) that has allowed both homologous (14, 15) and heterologous (9) overexpression of genes. A novel means of metabolic control was recently reported in P. furiosus that exploited the difference in the temperature dependence of the host’s metabolism and the inserted foreign synthetic pathway (9). For example, expression in P. furiosus of the gene encoding lactate dehydrogenase from a moderately thermophilic bacterium (Caldicellulosiruptor bescii, Topt of 78°C) resulted in temperature-dependent lactate formation (9). Moreover, the engineered pathway was active near 70°C, conditions under which the host metabolism of P. furiosus is minimal, as it is nearly 30°C below its optimal temperature. Hence, the host will require minimal maintenance energy and, as a result, incur minimal metabolic burden, whereas the engineered pathway that it contains is optimally active. We have used this temperature-dependent strategy for optimal bioproduct generation by expressing in P. furiosus genes encoding carbon dioxide fixation and 3-HP synthesis from the thermoacidophilic archaeon Metallosphaera sedula (Topt 73°C) (16).
Results and Discussion
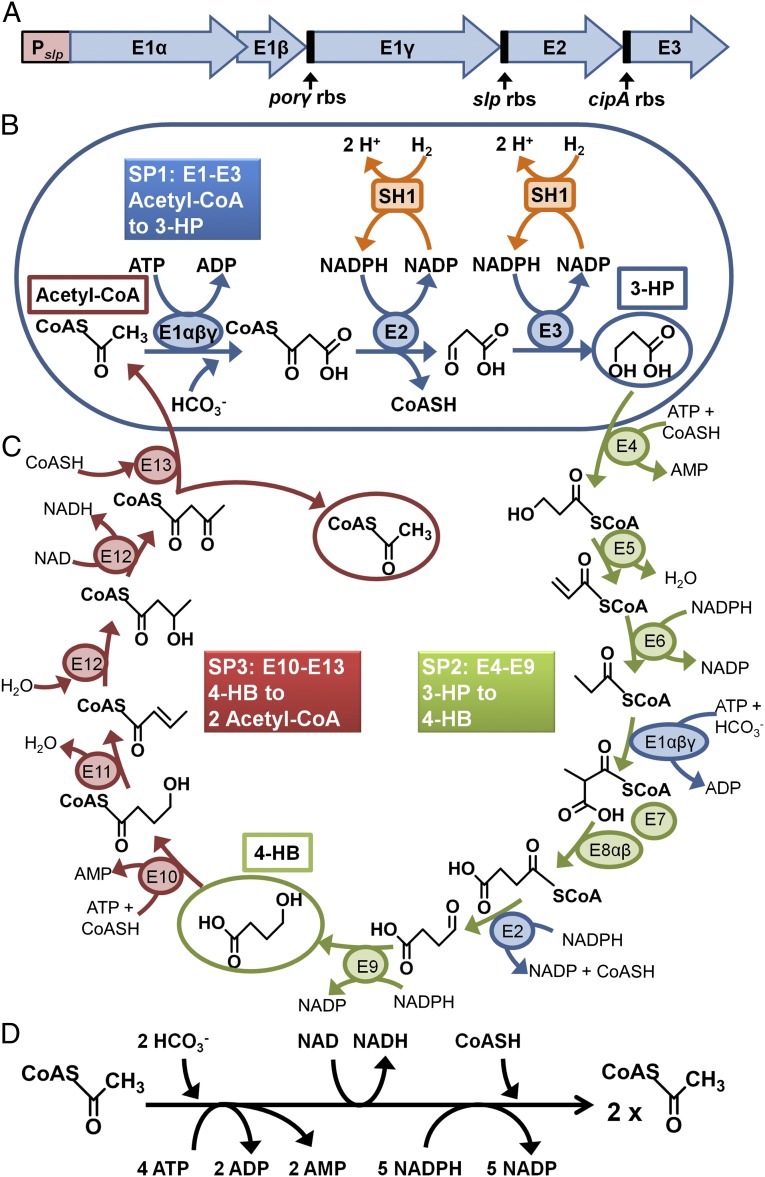
The genes that were incorporated into P. furiosus to enable it to use carbon dioxide are the first part of the 3-HP/4-hydroxybutyrate (4-HB) pathway of M. sedula, which consists of 13 enzymes in total (17). In one turn of the cycle, two molecules of carbon dioxide are added to one molecule of acetyl coenzyme A (acetyl-CoA) to generate a second molecule of acetyl-CoA (Fig. 1C). The cycle can be divided into three subpathways (SP1 to SP3) in which SP1 generates 3-HP from acetyl-CoA and carbon dioxide, SP2 generates 4-HB from 3-HP and carbon dioxide, and SP3 converts 4-HB to two molecules of acetyl-CoA. The reducing equivalents and energy for the pathway are supplied by NADPH and ATP, respectively (Fig. 1D). Notably, the 3-HP/4-HB pathway is purportedly more energetically efficient than carbon dioxide fixation by the ubiquitous Calvin cycle (18).
Fig. 1.
(A) The synthetic operon constructed to express the M. sedula genes encoding E1 (αβγ), E2, and E3 in P. furiosus under the control of Pslp. This includes P. furiosus RBSs from highly expressed genes encoding pyruvate ferredoxin oxidoreductase subunit γ (porγ, PF0971), the S-layer protein (slp, PF1399), and cold-induced protein A (cipA, PF0190). (B) The first three enzymes of the M. sedula 3-HP/4-HB cycle produce the key intermediate 3-HP. E1 is acetyl/propionyl-CoA carboxylase (αβγ, encoded by Msed_0147, Msed_0148, Msed_1375), E2 is malonyl/succinyl-CoA reductase (Msed_0709), and E3 is malonate semialdehyde reductase (Msed_1993). NADPH is generated by P. furiosus soluble hydrogenase 1 (SH1), which reduces NADP with hydrogen gas. (C) The first three enzymes (E1 to E3) are shown in context of the complete 3-HP/4-HP cycle for carbon dioxide fixation by M. sedula showing the three subpathways, SP1 (blue), SP2 (green), and SP3 (red). (D) The horizontal scheme shows the amount of energy (ATP), reductant (NADPH), oxidant (NAD), and coenzyme A (CoASH) required to generate 1 mol acetyl-CoA from 2 mol carbon dioxide.
The first three enzymes of the M. sedula 3-HP/4-HB cycle make up the SP1 pathway, and together they produce 3-HP from carbon dioxide and acetyl-CoA (Fig. 1B). The three enzymes are referred to here as E1 (acetyl/propionyl-CoA carboxylase, encoded by Msed_0147, Msed_0148, and Msed_1375), E2 (malonyl/succinyl-CoA reductase, Msed_0709), and E3 (malonate semialdehyde reductase, Msed_1993) (18–20). E1 carboxylates acetyl-CoA using bicarbonate and requires ATP. E2 breaks the CoA–thioester bond and, with E3, reduces the carboxylate to an alcohol with NADPH as the electron donor. E1 and E2 are bifunctional and are also involved in the SP2 part of the cycle (Fig. B and C). To demonstrate the concept, we expressed the M. sedula SP1 pathway in P. furiosus so that the organism could use carbon dioxide for the production of 3-HP, using hydrogen as the electron donor. Hydrogen is used in P. furiosus by the native soluble hydrogenase I (SHI) that reduces NADP to NADPH (21). SHI is extremely active, even at 70°C, and a P. furiosus strain engineered to overexpress the enzyme has been previously developed (14).
The five genes encoding the three enzymes (E1αβγ, E2, E3) of M. sedula SP1 were combined into a single synthetic operon with transcription driven by a P. furiosus S-layer promoter (Pslp), a native, constitutive promoter of the highly expressed S-layer protein (encoded by locus PF1399) of P. furiosus (14). The M. sedula ribosomal binding sites (RBS) for E1(γ), E2, and E3 were replaced with RBSs for known highly expressed P. furiosus proteins (Fig. 1A). The M. sedula RBS for E1β was retained, as the two genes (E1α and E1β) appear to be translationally coupled. The SP1 operon was inserted into P. furiosus (strain COM1) at two genome locations. In P. furiosus strain PF506, the SP1 operon was inserted at the site of the arginine decarboxylase gene, pdaD (encoded by locus PF1623; Fig. S1). The MW56 strain contained the SP1 operon between convergently transcribed genes (encoded by loci PF0574 and PF0575; Figs. S2 and S3) within a ∼100-bp region having little to no transcriptional activity, according to a previous tiling array study of P. furiosus (22). The P. furiosus strains used here are summarized in Table S1.
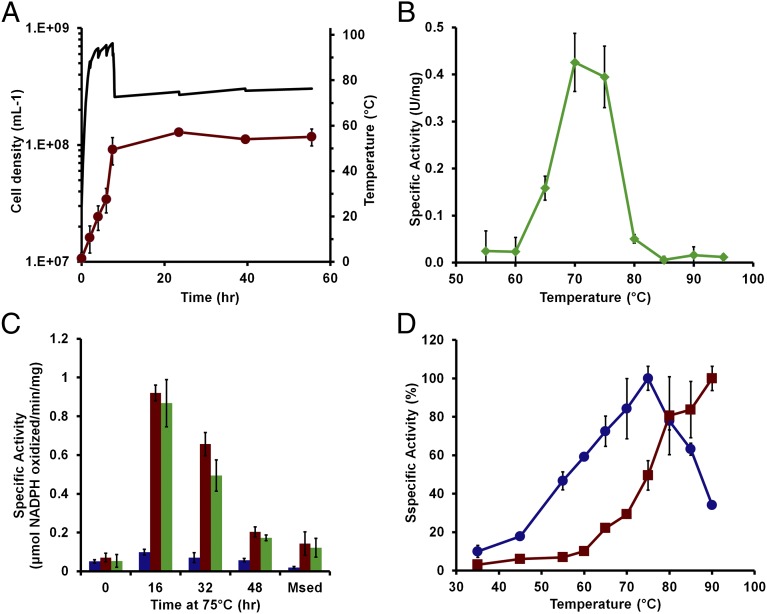
The premise for the temperature-dependent strategy is that P. furiosus (Topt 100°C) shows little growth and has very low metabolic activity (23) near the temperature at which the enzymes from M. sedula (Topt 73°C) are expected to be optimally active. In the recombinant P. furiosus strains (PF506 and MW56), the SP1 operon was under the control of a temperature-independent, constitutive promoter (Pslp); hence, the operon will be transcribed at both 100°C and 75°C. However, the resulting enzymes E1 to E3 should be stable and active only near 75°C. P. furiosus strains PF506 and MW56 were, therefore, grown at 98°C (to ∼1 × 108 cells/mL) in closed static cultures and then transferred to 75°C (Fig. 2A). There was no measurable activity of E1, E2, or E3 in cell-free extracts before the temperature change, but all three activities were present in cells after 16 h at 75°C. Moreover, specific activities were comparable to those measured in extracts of M. sedula cells grown autotrophically on hydrogen and carbon dioxide and to values reported by others (Fig. 2C) (16, 17). Indeed, when grown in a stirred, pH-controlled culture, the activity of the linked E2 + E3 enzymes in strain MW56 continued to increase over a 50-h period, growing to more than eightfold greater than that measured in M. sedula (Fig. 3C). When strain PF506 was grown at 95°C and then incubated for 16 h at temperatures between 55°C and 95°C, the maximum specific activity of the linked E2 + E3 enzymes was measured in cultures incubated at 70°C and 75°C, with dramatically lower values at 65°C and 80°C (Fig. 2B). This clearly indicates that the M. sedula enzymes functioned optimally in P. furiosus at 70°–75°C, especially as significant E2 + E3 activity could be measured at assay temperatures above 75°C, using cell-free extracts prepared from cultures incubated at 70°–75°C (Fig. 2D). Moreover, the enzymes are very thermostable, with a half-life of ∼60 min at 90°C (Fig. S5). This suggests that the lack of enzyme activity of the M. sedula enzymes (and of 3-HP production) in cultures that were incubated at 80°C or higher is not a result of the thermal instability of the M. sedula enzymes per se but, rather, to the temperature sensitivity of the protein-folding process during the synthesis of these enzymes, which is optimal in the 70°–75°C range.
Fig. 2.
Temperature-dependent production of the SP1 pathway enzymes in P. furiosus strain PF506. (A) Growth of triplicate cultures at 98°C (red circles) and temperature (black line) for the temperature shift from 98°C to 75°C are shown. (B) Specific activity (μmol NADPH oxidized⋅min–1⋅mg–1) of the coupled activity of E2+E3 in cell-free extracts from cultures grown at 95°C to a high cell density of 1 × 108 cells/mL and then incubated for 18 h at the indicated temperature. (C) Activities of E1, E2+E3, and E1+E2+E3 after the temperature shift to 75°C for the indicated period (Fig. S4). The activities of a cell-free extract of autotrophically grown M. sedula cells is also shown (labeled Msed). The specific activities are E1+E2+E3-coupled assay with acetyl-CoA and bicarbonate (blue), E2+E3-coupled assay with malonyl-CoA (red), and E2 with succinyl-CoA (green) as substrates. (D) Temperature dependence of the coupled activity of E2+E3 (blue circles) in the cell-free extracts after induction at 72°C for 16 h. The activity of P. furiosus glutamate dehydrogenase in the same cell-free extracts is also shown (red squares).
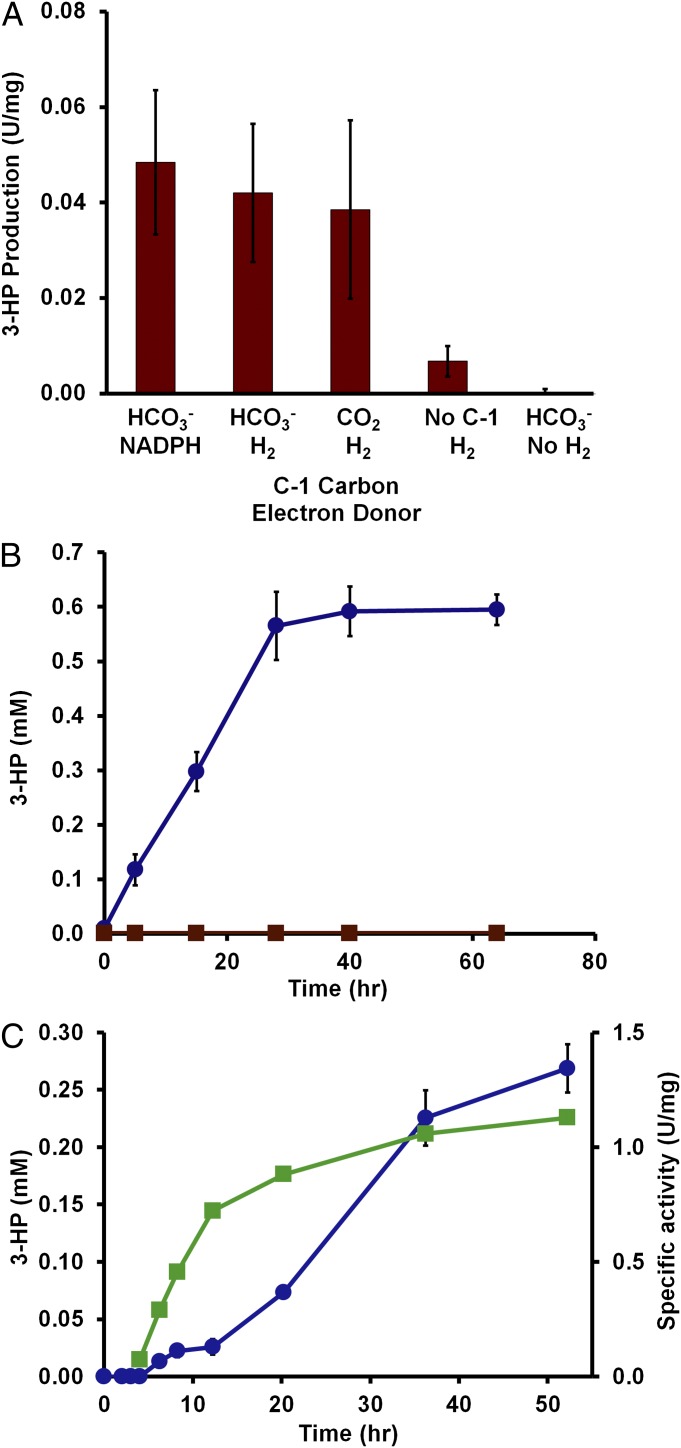
Fig. 3.
3-HP production by P. furiosus. Cells were grown at 95°C and then incubated at 72°C for 16 h to produce the SP1 enzymes. (A) In vitro 3-HP production from acetyl-CoA performed in triplicate. The sources of the C1 carbon (CO2 or HCO3−) and reducing equivalents (NADPH or NADP/H2) are indicated. Rates are expressed as micromoles 3-HP produced⋅min–1⋅mg–1. (B) In vivo 3-HP production by whole cells (static) using maltose as the source of acetyl-CoA in the presence of hydrogen gas and bicarbonate using cells grown in a 100-mL sealed bottle without pH control. The P. furiosus strains are MW56 (circles, blue) and COM1 (squares, red). (C) In vivo 3-HP production by whole cells (stirred) of MW56 using maltose as the source of acetyl-CoA (circles, blue) and E2+E3 specific activity of the cell-free extracts (squares, green) using cells grown in a 20 L fermenter with pH control (pH 6.8).
To determine the nature of the products of the SP1 pathway, recombinant P. furious strains PF506 and MW56 were grown at 95°C (to ∼1 × 108 cells/mL) and then transferred to 70°C for 16 h (Fig. S7). In extracts of these cells, the specific activities of the E1, E2, and E3 enzymes were comparable to those measured in extracts of autotrophically grown M. sedula cells (Fig. S7). Two methods were used to detect 3-HP and to confirm its production by the SP1 pathway in the recombinant P. furiosus strains. In the presence of acetyl-CoA, NaHCO3, and either NADPH or hydrogen gas as the electron donor, the 2-nitrophenylhydrazide-derivative (3-HP/HZ; m/z 224) was identified by electrospray ionization mass spectrometry (ESI-MS) in cell-free extracts of PF506 but was not detected in extracts of the parent P. furiosus strain (Fig. S8). This was confirmed by gas chromatography–mass spectrometry (GC-MS) of the O-trimethylsilylate derivative of 3-HP (3-HP/TMS), using malonyl-CoA and either NADPH or hydrogen gas as the electron donor (Table S2). The GC-MS also allowed quantitation of 3-HP/TMS and showed that ∼150 µM 3-HP was produced from malonyl-CoA after a 2 h incubation at 72°C with extracts of PF506 containing NADP under hydrogen gas (Table S2).
For routine analysis of 3-HP, a method was developed to extract 3-HP/HZ and separate and quantitate it by HPLC. As shown in Fig. 3A, this method was used to confirm 3-HP production from acetyl-CoA and carbon dioxide by the combined action of the enzymes E1, E2, and E3 in cell-free extracts. As expected, P. furiosus did not appear to further metabolize 3-HP, as the compound was stable when added to P. furiosus cultures. Moreover, the production of 3-HP from acetyl-CoA was dependent on either bicarbonate or carbon dioxide as the C-1 carbon source and either NADPH or hydrogen gas (and NADP) as the electron donor (Fig. 3A). The incorporation of electrons from hydrogen gas and the carbon from carbon dioxide into a single desired product is essentially the paradigm for electrofuels (6).
P. furiosus grows by fermenting sugars (such as the disaccharide maltose) to acetate, carbon dioxide, and hydrogen and can also use pyruvate as a carbon source (12). Acetyl-CoA and carbon dioxide are generated as the product of the pyruvate ferredoxin oxidoreductase reaction (Fig. S9). The reduced ferredoxin is oxidized by a membrane-bound hydrogenase to generate hydrogen gas (24).
Although growth is limited at 75°C (23), it was expected that when whole cells were incubated at 75°C with maltose or pyruvate, sufficient acetyl-CoA would be produced by the low metabolic activity of P. furiosus for the SP1 enzymes to produce 3-HP. This was confirmed by HPLC detection and quantitation of 3-HP as the 2-nitrophenylhydrazide derivative. For example, high–cell density suspensions (≥ 1010 cells/mL) of P. furiosus strains PF506 and MW56 produced up to 0.2 mM 3-HP after 1 h incubation at 75°C in the presence of maltose, hydrogen gas, and bicarbonate (Fig. S10), and 3-HP production was dependent on the presence of maltose or pyruvate (Table S3). Moreover, recombinant P. furiosus strains PF506 and MW56, grown in static cultures to late-log phase (∼1 × 108 cells/mL) at 98°C on maltose, produced up to 0.6 mM 3-HP (60 mg/L) when subsequently incubated at 72°C for up to 40 h (Fig. 3B). Furthermore, in a stirred, pH-controlled culture, strain MW56 produced 3-HP continuously during a 50-h period at 72°C (Fig. 3C). Overall, there appeared to be no significant difference between the two recombinant P. furiosus strains in terms of 3-HP production. This indicates that the genome location of the synthetic operon derived from M. sedula was not a determining factor, which bodes well for the insertion of additional synthetic operons in P. furiosus to extend the results reported here to other industrial chemicals.
In summary, this work demonstrates the use of hydrogen as the electron donor for carbon dioxide fixation into a product of great utility in the chemical industry; namely, 3-HP. Moreover, it is carried out by an engineered heterotrophic hyperthermophile some 30°C below the optimal growth temperature of the organism (conditions that support minimal growth), but sufficient metabolic activity is retained to sustain the production of 3-HP (6). The reaction can be accomplished by cell-free extracts and by whole cells in culture using sugar (maltose) as the source of the acetyl-CoA and ATP in a hydrogen- and carbon dioxide–dependent manner. The feasibility of using hydrogen gas as the source of reducing power (NADPH) for chemical synthesis, in this case 3-HP, is also of high significance, given the availability of relatively inexpensive natural gas as a hydrogen source (25). It is important to note that the low metabolic activity of P. furiosus at 72°C was sufficient to provide the ATP needed for carbon dioxide fixation. These results are a significant step forward toward the overall goal of incorporating into P. furiosus the complete M. sedula 3-HP/4-HB pathway, in which two molecules of carbon dioxide are reduced to acetyl-CoA that can then be converted into a variety of valuable products including biofuels (6). Clearly, there will be a balance between using a fixed carbon source (sugar) via the low metabolic activity of the host to produce ATP and the high catalytic activity of the heterologous enzymes to generate the desired product. The hydrogen-dependent fixation of carbon dioxide has enormous potential for the production of a variety of chemicals and fuels through strategic use of established biosynthetic pathways and exploiting the hyperthermophilicity of metabolically engineered microbial hosts (3, 4, 8, 25).
Materials and Methods
Construction of a Synthetic SP1 Operon.
PCR was performed using P. furiosus or M. sedula genomic DNA to generate the individual PCR products of the Pslp and the five M. sedula SP1 genes, consisting of coupled E1αβ (Msed_0147-Msed_0148), E1γ (Msed_1375), E2 (Msed_0709), and E3 (Msed_1993). P. furiosus RBSs, consisting of 11–14 bp of sequence upstream of highly expressed proteins, were added in front of E1γ (5′-ggaggtttgaag, sequence upstream from pyruvate ferredoxin oxidoreductase subunit γ (PF0791), E2 (5′-gggaggtggagcat, sequence upstream from slp, PF1399), and E3 (5′-ggtgatatgca, sequence upstream from cold-induced protein A, PF0190). The primer sequences are given in Table S4. Splicing by overlap extension and PCR (SOE-PCR) (26) was performed to combine the individual PCR products and generate the expression cassette for SP1 (Fig. 1A).
Construction of Vectors for Insertion of the SP1 Operon into P. furiosus.
The SP1 expression cassette (Fig. 1B) was cloned into plasmid pSPF300 (15), generating the plasmid pALM506-1, to be used for targeted insertion of the synthetic SP1 operon into the P. furiosus ΔpdaD strain (Fig. S1). SOE-PCR (26) was used to combine ∼0.5-kb flanking regions targeting homologous recombination in the intergenic space between convergent genes PF0574-PF0575, with a marker cassette, including restriction sites for cloning. The marker cassette for uracil prototrophic selection consisted of the pyrF gene driven by the promoter region of the glutamate dehydrogenase gene (consisting of a 157 base sequence immediately upstream of locus PF1602) and terminated with 12 bases of the 3′ UTR of the hpyA1 gene (5′-aatcttttttag, locus PF1722). A 65-b sequence of the 3′ end of the marker cassette (5′-ctaaaaaagattttatcttgagctccattctttcacctcctcgaaaatcttcttagcggcttccc) was repeated at the beginning of the cassette to serve as a homologous recombination region for selection of marker removal (27). Plasmid pGL007 targeting homologous recombination at the PF0574-PF0575 intergenic space was constructed by cloning the SOE-PCR product into plasmid pJHW006 (28) (Fig. S2). The SP1 expression cassette was PCR-amplified from plasmid pALM506-1. A terminator sequence was added to the 3′ end of the operon (5′-aatcttttttag, from the 3′ UTR of PF1722), and the construct was cloned into the AscI and NotI restriction enzyme sites of plasmid pGL007 to make plasmid pGL010 (Fig. S3), for targeted insertion of the SP1 operon at the PF0574-PF0575 intergenic space. Transformation of P. furiosus ΔpdaD strain was performed as previously described for COM1 (28), except that the defined medium contained maltose instead of cellobiose as the carbon source and was supplemented with 0.1% wt/vol casein hydrolysate. Transformation of P. furiosus strain COM1 was performed as previously described (28), except that linear plasmid DNA was used for transformation.
Growth of P. furiosus.
Strains were cultured as previously described in a seawater-based medium containing 5 g/L maltose, 5 g/L yeast extract, 0.5 μg/L riboflavin, and 20 μM uracil or 4 mM agmatine, as needed (28). Cultures were grown at 95°C until ∼1 × 108 cells/mL and then cooled at 23°C until the temperature reached 70–75°C and was maintained there for up to 48 h. For growth in a 20-L fermenter, the culture was sparged with 10% CO2/90% (vol/vol) N2 and stirred, and the pH was maintained at 6.8 by addition of 10% (wt/vol) NaHCO3. Cell extracts prepared anaerobically as described previously (28) in 100 mM 3-morpholinopropane-1-sulfonic acid (MOPS) at pH 7.5, reconcentrated three times with a 3-kDa centrifugation filter, and stored at −80°C.
Growth of M. sedula for Biochemical Assays and Product Analysis.
M. sedula (DSM 5348) was grown autotrophically at 70°C with microbubblers feeding 1 mL/min 80/20 H2/CO2 and 100 mL/min air in defined medium at pH 2.0, as previously described (29). To obtain cell-free extracts, frozen cell pellets were anaerobically suspended in 50 mM Tris HCl at pH 8.0 containing 0.5 μg/mL DNase 1 and stirred for 1 h in an anaerobic chamber. The cell extract was centrifuged at 100,000 × g for 1 h, and the supernatant was stored at −80°C.
E1, E2, and E3 Assays.
All reactions were carried out in sealed anaerobic cuvettes at 75°C containing 100 mM Mops at pH 7.5, 5 mM MgCl2, and 5 mM DTT. After the addition of NADPH (to an absorbance at 340 nm ∼1.0) and the relevant substrate, NADPH oxidation was measured at 340 nm. The substrates for the E2, E2+E3, and E1+E2+E3 assays were succinyl-CoA, malonyl-CoA, and acetyl CoA (each 1 mM), respectively. The latter assay also contained 1 mM ATP and 10 mM NaHCO3. E1 activity was measured by phosphate release. The assay contained 10 mM NaHCO3, 1 mM ATP, and 1 mM acetyl-CoA. Samples (20 μL) were removed at 2–4 min and diluted with water (180 µL), and the BioVision phosphate assay reagent (20 µL) was added. The phosphate produced was calculated using a molar extinction coefficient of 90,000 M−1cm−1 at 650 nm.
Measurement of 3-HP.
The 3-HP [H0297, 30% (wt/vol), in water] was obtained from TCI America (Portland, OR). By HPLC and 1H NMR, it was 75% pure, with the remaining 25% as 3,3′-oxydipropanoic acid. For GC-MS analysis, inositol was the internal standard. Samples were freeze-dried, incubated in 2 M trifluoroacetic acid at 80°C for 1 h, dried under nitrogen, and per-O-trimethylsilylated by treatment with Tri-Sil (Pierce) at 80°C for 30 min. GC-MS analysis was performed on an AT 7890n GC interfaced to a 5975C MSD, using a Grace EC-1 column (30 m × 0.25 mm). The exact mass of 3-HP/TMS is 162. Derivatization of 3-HP with 2-nitrophenyl hydrazine was carried out as described previously (30). The 3HP-hydrazide was extracted by adding 1.0 mL of 1 M KPO4 buffer at pH 7.0 and 1.5 mL of ether to 800 μL of the sample, centrifuging for 10 min at 6,000 × g to separate the phases, removing the top ether layer, and evaporating. The dried sample was resuspended in 200 μL ethanol, and 10–50 μL aliquots were analyzed by HPLC using a Supelco LiChrosorb RP-8 (5 μm) with solvent system [A: 0.05% (wt/vol) TFA, B: 100% acetonitrile] and run conditions as follows: 0–100% B for 20 min followed by 2 min at 100% B, using a flow rate of 1 mL/min and temperature at 30°C. For ESI-MS analysis, the dried derivative was dissolved in methanol and directly injected on a Perkin-Elmer API 1 plus in negative mode. The mass of the anionic 3-HP-hydrazide derivative is 224.
Production of 3-HP in Vitro from Malonyl-CoA by E2+E3 and from Acetyl-CoA by E1+E2+E3.
To the P. furiosus extract (1–2 mg/mL) in 100 mM Mops at pH 7.5, 5 mM MgCl2, and 5 mM DTT, we added 1–2 mM malonyl-CoA (for E2+E3) or 10 mM NaHCO3 (or 100% CO2 in the gas phase), 2 mM ATP, and 2 mM acetyl-CoA (for E1+E2+E3). The electron source was 2 mM NADPH or 0.5 mM NADP with 20% H2 in the headspace. Sealed anaerobic vials containing the reaction mixture were incubated at 75°C for up to 2 h. Samples were derivatized with 2-nitrophenyl hydrazine and analyzed for 3-HP by HPLC as described earlier.
Product Analysis of E1+E2+E3 Activities in Whole Cells.
P. furiosus strains PF506 and MW56 were grown in 2-L cultures at 95°C for 10 h until cell densities of 1 × 108 cells/mL and then cooled and incubated at 75°C for 16 h. Harvested cells were suspended to 5 × 1010 cells/mL in 100 mM Mops at pH 7.5 and base salts (28 g/L NaCl, 3.5 g/L MgSO4⋅7 H2O, 2.7 g/L MgCl2⋅6 H2O, 0.33 g/L KCl, 0.25 g/L NH4Cl, 0.14 g/L CaCl2⋅2H2O). The cell suspension was sealed in a serum vial and degassed with argon, and cysteine HCl (0.5 g/L), NaHCO3 (10 mM), and either maltose (10 mM) or pyruvate (40 mM) were added. The vials were degassed and flushed with H2 and incubated at 75°C for 60 min. Samples for 3-HP analysis were derivatized with 2-nitrophenyl hydrazine, using 1 mM p-hydroxyphenyl acetic acid as an internal standard; ether-extracted; and analyzed by HPLC, as described earlier.
Analysis of the P. furiosus Culture Medium for 3-HP.
P. furiosus strains PF506, MW56, and COM1 were grown at 98°C in 50-mL cultures with maltose (10 mM) as the carbon source until a cell density of 8 × 107 cells/mL was reached. The incubation temperature was then shifted to 72°C for up to 4 d. Samples (1 mL) were periodically removed and centrifuged (10,000 × g, 10 min), and to a 100-μL aliquot of the supernatant (the spent medium), 1 mM p-hydroxyphenyl acetic acid was added as an internal standard. The sample was derivatized with 2-nitrophenyl hydrazine, ether-extracted, and analyzed by HPLC, as described earlier.
Supplementary Material
Acknowledgments
We thank Brian Vaccaro, Dennis Phillips, and Zhirui Wang (University of Georgia) for running the NMR, ESI-MS, and GC-MS analyses of 3-HA, respectively. The GC-MS facility is supported in part by the Center for Plant and Microbial Complex Carbohydrates, funded by the US Department of Energy (DE-FG02-93ER-20097). This work was supported by the US Department of Energy as part of the Electrofuels Program of the Advanced Research Projects Agency - Energy (Grant DE-AR0000081).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222607110/-/DCSupplemental.
References
- 1.Somerville C, Youngs H, Taylor C, Davis SC, Long SP. Feedstocks for lignocellulosic biofuels. Science. 2010;329(5993):790–792. doi: 10.1126/science.1189268. [DOI] [PubMed] [Google Scholar]
- 2.Olson DG, McBride JE, Shaw AJ, Lynd LR. Recent progress in consolidated bioprocessing. Curr Opin Biotechnol. 2012;23(3):396–405. doi: 10.1016/j.copbio.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 3.Steen EJ, et al. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature. 2010;463(7280):559–562. doi: 10.1038/nature08721. [DOI] [PubMed] [Google Scholar]
- 4.Peralta-Yahya PP, Keasling JD. Advanced biofuel production in microbes. Biotechnol J. 2010;5(2):147–162. doi: 10.1002/biot.200900220. [DOI] [PubMed] [Google Scholar]
- 5.Wackett LP. Engineering microbes to produce biofuels. Curr Opin Biotechnol. 2011;22(3):388–393. doi: 10.1016/j.copbio.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 6.Hawkins AS, et al. Extremely thermophilic routes to microbial electrofuels. ACS Catalysis. 2011;1:1043–1050. [Google Scholar]
- 7.Shen CR, et al. Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli. Appl Environ Microbiol. 2011;77(9):2905–2915. doi: 10.1128/AEM.03034-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connor MR, Liao JC. Microbial production of advanced transportation fuels in non-natural hosts. Curr Opin Biotechnol. 2009;20(3):307–315. doi: 10.1016/j.copbio.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Basen M, Sun J, Adams MW. Engineering a hyperthermophilic archaeon for temperature-dependent product formation. MBio. 2012;3(2):e00053–e12. doi: 10.1128/mBio.00053-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paster M, Pellegrino JL, Carole TM. Industrial bioproducts: Today and tomorrow. Columbia, MD: US Department of Energy and Energetics Inc.; 2004. [Google Scholar]
- 11.Werpy T, Petersen G. 2004. Top value added chemicals from biomass: Volume 1—Results of screening for potential candidates from sugars and synthesis gas. Dept of Energy, 102004-1992, 10.2172/15008859.
- 12.Fiala G, Stetter KO. Pyrococcus furiosus sp. nov., represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch Microbiol. 1986;145:56–61. [Google Scholar]
- 13.Bridger SL, Lancaster WA, Poole FL, 2nd, Schut GJ, Adams MW. Genome sequencing of a genetically tractable Pyrococcus furiosus strain reveals a highly dynamic genome. J Bacteriol. 2012;194(15):4097–4106. doi: 10.1128/JB.00439-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandrayan SK, et al. Engineering hyperthermophilic archaeon Pyrococcus furiosus to overproduce its cytoplasmic [NiFe]-hydrogenase. J Biol Chem. 2012;287(5):3257–3264. doi: 10.1074/jbc.M111.290916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hopkins RC, et al. Homologous expression of a subcomplex of Pyrococcus furiosus hydrogenase that interacts with pyruvate ferredoxin oxidoreductase. PLoS ONE. 2011;6(10):e26569. doi: 10.1371/journal.pone.0026569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berg IA, et al. Autotrophic carbon fixation in archaea. Nat Rev Microbiol. 2010;8(6):447–460. doi: 10.1038/nrmicro2365. [DOI] [PubMed] [Google Scholar]
- 17.Ramos-Vera WH, Weiss M, Strittmatter E, Kockelkorn D, Fuchs G. Identification of missing genes and enzymes for autotrophic carbon fixation in crenarchaeota. J Bacteriol. 2011;193(5):1201–1211. doi: 10.1128/JB.01156-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berg IA, Kockelkorn D, Buckel W, Fuchs G. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science. 2007;318(5857):1782–1786. doi: 10.1126/science.1149976. [DOI] [PubMed] [Google Scholar]
- 19.Hügler M, Krieger RS, Jahn M, Fuchs G. Characterization of acetyl-CoA/propionyl-CoA carboxylase in Metallosphaera sedula. Carboxylating enzyme in the 3-hydroxypropionate cycle for autotrophic carbon fixation. Eur J Biochem. 2003;270(4):736–744. doi: 10.1046/j.1432-1033.2003.03434.x. [DOI] [PubMed] [Google Scholar]
- 20.Alber B, et al. Malonyl-coenzyme A reductase in the modified 3-hydroxypropionate cycle for autotrophic carbon fixation in archaeal Metallosphaera and Sulfolobus spp. J Bacteriol. 2006;188(24):8551–8559. doi: 10.1128/JB.00987-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma K, Adams MWW. Hydrogenases I and II from Pyrococcus furiosus. Methods Enzymol. 2001;331:208–216. doi: 10.1016/s0076-6879(01)31059-5. [DOI] [PubMed] [Google Scholar]
- 22.Yoon SH, et al. Parallel evolution of transcriptome architecture during genome reorganization. Genome Res. 2011;21(11):1892–1904. doi: 10.1101/gr.122218.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weinberg MV, Schut GJ, Brehm S, Datta S, Adams MW. Cold shock of a hyperthermophilic archaeon: Pyrococcus furiosus exhibits multiple responses to a suboptimal growth temperature with a key role for membrane-bound glycoproteins. J Bacteriol. 2005;187(1):336–348. doi: 10.1128/JB.187.1.336-348.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sapra R, Bagramyan K, Adams MW. A simple energy-conserving system: Proton reduction coupled to proton translocation. Proc Natl Acad Sci USA. 2003;100(13):7545–7550. doi: 10.1073/pnas.1331436100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kreysa G. Climate protection by an alternative use of methane—the carbon moratorium. ChemSusChem. 2009;2(1):49–55. doi: 10.1002/cssc.200800232. [DOI] [PubMed] [Google Scholar]
- 26.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: Gene splicing by overlap extension. Gene. 1989;77(1):61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 27.Farkas J, et al. Recombinogenic properties of Pyrococcus furiosus strain COM1 enable rapid selection of targeted mutants. Appl Environ Microbiol. 2012;78(13):4669–4676. doi: 10.1128/AEM.00936-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lipscomb GL, et al. Natural competence in the hyperthermophilic archaeon Pyrococcus furiosus facilitates genetic manipulation: Construction of markerless deletions of genes encoding the two cytoplasmic hydrogenases. Appl Environ Microbiol. 2011;77(7):2232–2238. doi: 10.1128/AEM.02624-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han Y, Hawkins AS, Adams MW, Kelly RM. Epimerase (Msed_0639) and mutase (Msed_0638 and Msed_2055) convert (S)-methylmalonyl-coenzyme A (CoA) to succinyl-CoA in the Metallosphaera sedula 3-hydroxypropionate/4-hydroxybutyrate cycle. Appl Environ Microbiol. 2012;78(17):6194–6202. doi: 10.1128/AEM.01312-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miwa H. High-performance liquid chromatographic determination of mono-, poly- and hydroxycarboxylic acids in foods and beverages as their 2-nitrophenylhydrazides. J Chromatogr A. 2000;881(1-2):365–385. doi: 10.1016/s0021-9673(00)00284-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.